Abstract
Uracil-DNA glycosylase (UDG) is responsible for the removal of uracil from DNA. It has previously been demonstrated that UDG exhibits some sequence dependence in its activity, although this has not been well characterised. This study has investigated the sequence-dependent activity of UDG from herpes simplex virus type-1 (HSV-1). A more detailed analysis has been possible by using both kinetic and binding assays with a variety of different oligonucleotide substrates. The target uracil has been placed in substrates with either A-T-rich or G-C-rich flanking sequences and analyses have been performed on both the single- and double-stranded forms of each substrate. In the latter the uracil has been placed in both a U·A base pair and a U·G mismatch. It is observed that the sequences flanking the target uracil have a greater effect on UDG activity than the partner base of the uracil. Furthermore, the sequence context effects extend to single-stranded DNA. Systematic examination of the kinetics and binding of UDG with these different substrates has enabled us to examine the origin of the sequence preferences. We conclude that the damage recognition step in the HSV-1 UDG reaction pathway is modulated by local DNA sequence.
INTRODUCTION
Uracil can arise in DNA through the spontaneous deamination of cytosine. This gives rise to a G·U mismatch which, if not repaired, will lead to an A·T transition mutation in one of the daughter duplexes following DNA replication. Cytosine deamination is thought to occur at least 100 times a day in human cells (1) and is thus a significant threat to the integrity of the genome. Indeed, analysis of 179 nonsense mutations in human genetic disease has revealed that 65% were caused by C→T transition mutations (2).
Uracil-DNA glycosylase (UDG) is responsible for the removal of uracil from DNA by hydrolysis of the N-glycosidic bond that links the base to the deoxyribose backbone, leaving an apyrimidinic (AP) site (3). AP sites are themselves potentially mutagenic and are repaired by the base excision repair pathway (4). UDG is a highly conserved enzyme found in many species. Detailed studies have been performed on the human, Escherichia coli and herpes simplex virus type 1 (HSV-1) enzymes, which are highly homologous at both the amino acid and structural levels (5–7). While some minor discrepancies exist, these different enzymes appear to have very similar properties: they are able to cleave uracil from both single- and double-stranded DNA, whether it is in a U·A base pair or a U·G mismatch (8).
Structural studies have revealed that UDG has an active site pocket that is highly specific for uracil (7,9). This prevents UDG from cleaving any normal bases from DNA or, indeed, uracil from RNA. The enzyme binds uracil by flipping the target nucleotide out of the double helix and into the active site pocket via the major groove (5,9). A well-conserved leucine loop on the enzyme surface interacts with the minor groove of DNA. The highly conserved leucine from which this loop takes its name intercalates into the vacant space in the helix once the uracil has been flipped out. However, the precise mechanism of nucleotide flipping remains unclear.
Nucleotide flipping has been qualitatively described as either a destabilisation of the stacked conformation, a ‘push’, or a stabilisation of the unstacked conformation, a ‘pull’, by the enzyme (5,10). However, without quantitative data it is difficult to make precise conclusions as to what drives the process of nucleotide flipping. Other studies with E.coli UDG have demonstrated an active role for the enzyme in flipping out the target uracil (11). These studies have also demonstrated that the rate of base flipping is independent of the stability of the base pair, suggesting that the enzyme pays the energetic cost of unstacking the uracil in a DNA deformation step that precedes the actual flipping event. This conclusion is supported by crystallographic studies of human UDG–DNA complexes, which reveal that the backbone of the DNA is significantly compressed at the site of nucleotide flipping (12). It has been suggested that this compression, which is mediated by conserved enzyme loops, is responsible for flipping out the uracil-containing nucleotide.
Biochemical studies have shown that both human and E.coli UDG exhibit sequence preference for the removal of uracil. Although this behaviour has not been examined in detail, it can be seen that in general uracil is removed more efficiently from sites where it is flanked by A-T-rich sequences, while sites with G-C-rich flanking sequences make poorer substrates (13–15). The relative rates of uracil cleavage from within the lacI gene have also been correlated with known mutational hotspots, indicating that the sequence preference of UDG may have biologically significant implications (14).
Here we examine the sequence preference of UDG in greater detail with the aim of understanding the basis of this sequence discrimination. A combination of steady-state kinetics and equilibrium binding has been used to investigate the interaction of HSV-1 UDG with oligonucleotide substrates, where the target uracil has been placed in both A-T- and G-C-rich sequence contexts. These assays were applied to both single- and double-stranded substrates: in the latter we have placed the uracil in both a U·A base pair and a U·G mismatch. We have analysed both the kinetic and binding properties of UDG with these different substrates. This detailed examination of sequence context significantly extends our understanding of UDG–substrate interactions.
MATERIALS AND METHODS
Protein overexpression and purification
HSV-1 UDG was overexpressed from a recombinant plasmid (supplied by Dr R. Savva, Birkbeck College, London, UK) in E.coli BL834, as previously described (16). The harvested cells were lysed by sonication in buffer A [20 mM Tris–HCl pH 8.25, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol (DTT)]. Streptomycin sulphate (0.1 vol, 10% w/v) was added to remove nucleic acids and the lysate was clarified by centrifugation (40 000 r.p.m., 40 min). The clarified lysate was loaded onto a DEAE–cellulose column equilibrated in buffer A and the flow-through was run directly onto an SP-Sepharose column, also equilibrated in buffer A. After washing, the HSV-1 UDG was eluted from the SP-Sepharose column with a gradient of 0–1.6 M NaCl. The fractions were assayed by SDS–PAGE and the fractions containing UDG were pooled and concentrated by ultrafiltration. The concentrated fractions were diluted with buffer A, so that the NaCl concentration was <0.02 M. The partially pure protein was then applied to a poly(U)–Sepharose affinity column equilibrated in buffer A and, after washing, the UDG was eluted with a gradient of 0–0.5 M NaCl. The fractions were assayed and those containing pure UDG were concentrated as above. The concentration of the protein was calculated from the OD280, based on an extinction coefficient of 46.6 × 103 M–1 cm–1. Glycerol was added to either 10 or 50% and the pure enzyme stored at –20°C. The enzyme was removed from the frozen stocks in small aliquots and diluted in buffer (20 mM Tris pH 8.25, 1 mM EDTA, 1 mM DTT, 0.1 mg ml–1 BSA, 150 mM NaCl, 1 mM spermine) prior to use. The final glycerol concentration in reactions was always <5%. The D88N mutant enzyme was expressed and purified from a construct (R. Savva) in exactly the same manner.
DNA synthesis and purification
Oligodeoxynucleotides were synthesised by either Cruachem (UK) or Len Hall (University of Bristol). All modifications were incorporated into oligonucleotides at the point of synthesis using conventional phosphoramidite chemistry. The phosphoramidites of 2′-deoxyuridine and hexachlorofluorescein (HEX) were obtained from Cruachem, while the fluorescent base analogue 2-aminopurine was obtained from Glenn Research (USA). Oligonucleotides were purified by HPLC as previously described (17). Double-stranded substrates were made by mixing equimolar amounts of complementary strands, heating to 90°C and cooling slowly to room temperature.
The apyrimidinic oligonucleotides 1HAP and 2HAP were prepared by incubating a 100 µM solution of the equivalent uracil-containing substrate (1HU and 2HU, respectively) in reaction buffer with 1 µM UDG overnight at 25°C. This procedure did not lead to the accumulation of any detectable amounts of cleaved abasic DNA and the abasic reaction products were used without further purification.
Kinetic assays
All steady-state reactions were performed on a SPEX Fluoromax spectrofluorometer. The 2-aminopurine fluorescence was measured with an excitation wavelength of 310 nm and an emission wavelength of 368 nm using a 5 mm cuvette. Assays were performed at 25°C in reaction buffer (50 mM Tris pH 8.0, 1 mM EDTA, 150 mM NaCl, 100 µg ml–1 BSA, 2 mM MgCl2) with substrate concentrations ranging from 0.2 to 40 µM. Reactions were initiated by adding UDG to give a final concentration of 0.5, 1.0 or 2.0 nM, with the AT-rich substrate, or 10 or 20 nM, for the GC-rich substrate. The end-point of the reaction was determined by addition of a second aliquot of high concentration UDG. The steady-state velocity (µM s–1) was calculated from the initial linear slope of the fluorescence increase and the total change in fluorescence. The rate (s–1) was then calculated by normalising to the total enzyme concentration. Steady-state kinetic parameters were determined by fitting the change in rate with substrate concentration to the Michaelis–Menten equation using Grafit 4 (Erithacus Software). Steady-state assays were performed with the substrates shown in Table 1.
Table 1. Substrates used in steady-state assays.
| Abbreviation |
Sequence |
| 1U | 5′-GAC TAP UAA TGA CTG CG-3′ |
| 1U·A (X = A) | 5′-GAC TAP UAA TGA CTG CG-3′ |
| 1U·G (X = G) | 3′-CTG ATT XTT ACT GAC GC-5′ |
| 2U | 5′-GAG GCP UCC ACG CTG CG-3′ |
| 2U·A (X = A) | 5′-GAG GCP UCC ACG CTG CG-3′ |
| 2U·G (X=G) | 3′-CTC CGC XGG TGC GAC GC-5′ |
Oligonucleotides were synthesised containing the fluorescent base 2-aminopurine (P), as indicated, and the double-stranded substrates were made by annealing the appropriate strands. U, 2′-deoxyuridine.
Equilibrium binding assays
Equilibrium binding measurements were made on a SPEX Fluoromax spectrofluorometer fitted with polarising filters using a 5 mm square cuvette. Fluorescence anisotropy was used to monitor the binding of D88N UDG to the 5′-HEX-labeled oligonucleotides shown in Table 2. Absolute fluorescence intensity was used to monitor the binding of D88N UDG to oligonucleotide 1U. Data were recorded at an excitation wavelength of 530 nm and an emission wavelength of 550 nm to monitor HEX fluorescence or 310 and 368 nm to monitor 2-aminopurine fluorescence. Binding assays were performed at 25°C in reaction buffer by titrating fixed concentrations of the fluorescent DNA with increasing amounts of the D88N mutant enzyme. The observed anisotropy, or 2-aminopurine fluorescence intensity, was plotted against the concentration of enzyme and the data fitted to binding equation 1 using Grafit 4. The value of [DNA]T was set as a constant equal to the concentration of DNA used in each experiment and the values of Kd, AD and ADE determined by curve fitting.
Table 2. Substrates used in equilibrium binding assays.
| Abbreviation |
Sequence |
| 1HU | 5′-(HEX) GAC TAT UAA TGA CTG CG-3′ |
| 1HU·A (X = A) | 5′-(HEX) GAC TAT UAA TGA CTG CG-3′ |
| 1HU·G (X = G) | 3′-CTG ATA XTT ACT GAC GC-5′ |
| 1HAP (X = abasic) | 5′-(HEX) GAC TAT XAA TGA CTG CG-3′ |
| 2HU | 5′-(HEX) GAG GCG UCC ACG CTG CG-3′ |
| 2HU·A (X = A) | 5′-(HEX) GAG GCG UCC ACG CTG CG-3′ |
| 2HU·G (X = G) | 3′-CTC CGC XGG TGC GAC GC-5′ |
| 2HAP (X = abasic) | 5′-(HEX) GAG GCG XCC ACG CTG CG-3′ |
The chromophore hexachlorofluorescein (HEX) was incorporated at the 5′-end during synthesis, as indicated, and the double-stranded substrates were made by annealing the appropriate strands. U, 2′-deoxyuridine.
A = AD + (ADE – AD) × ({(Kd + [E]0 + [DNA]T) – √(– Kd – [E]0 – [DNA]T2 – 4[E]0[DNA]T)}/(2[DNA]T)) 1
where A is the observed anisotropy, AD is the anisotropy of free DNA, ADE is the anisotropy of enzyme-bound DNA, Kd is the equilibrium dissociation constant, [DNA]T is the total concentration of DNA and [E]0 is the enzyme concentration.
RESULTS
Steady-state analysis
A fluorescence assay was used to monitor the cleavage of uracil from oligonucleotide substrates by UDG. A fluorescent base analogue, 2-aminopurine, was incorporated adjacent to the target uracil. Cleavage of the uracil thus leads to a change in the environment and hence fluorescence of the 2-aminopurine, which can be used to monitor the turnover of UDG under steady-state conditions (18). The increase in fluorescence was monitored over time and the steady-state rate was determined from the initial linear increase in fluorescence.
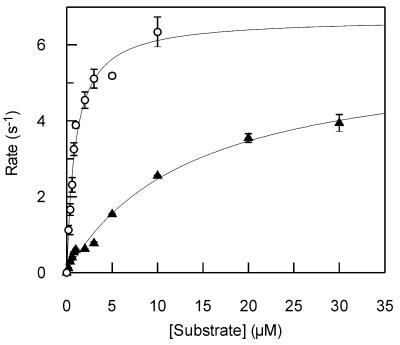
A variety of different substrates were used in which the target uracil was placed in either an A-T-rich or G-C-rich sequence context (Table 1). In each case, reactions were performed on both the single-stranded (1U and 2U) and double-stranded forms of the substrate: in the latter the uracil was placed in both a U·A base pair (1U·A and 2U·A) and a U·G mismatch (1U·G and 2U·G). Reactions were performed over a range of concentrations for each substrate and the rate constants showed a hyperbolic dependence on the concentration of substrate. The parameters kcat and Km were determined by fitting the data to the Michaelis–Menten equation (Fig. 1). This analysis was performed for each of the above substrates (Table 3).
Figure 1.
Steady-state reactions. Steady-state reactions were performed by monitoring the fluorescence of 2-aminopurine-containing substrates (Table 1) after addition of UDG (Materials and Methods). Rates were determined from the initial increase in fluorescence at different concentrations of substrates. The observed rates are shown plotted against concentration for substrates 1U (open circles) and 2U (closed triangles). The solid lines are the best fit of the data to the Michaelis–Menten equation, with the values shown in Table 3.
Table 3. Steady-state data for UDG.
| Substrate |
kcat (s–1) |
Km (µM) |
kcat/Km (µM–1 s–1) |
| 1U | 6.7 ± 0.3 | 0.9 ± 0.1 | 7.1 ± 1.1 |
| 1U·A | 2.1 ± 0.1 | 1.6 ± 0.2 | 1.3 ± 0.2 |
| 1U·G | 2.4 ± 0.2 | 0.9 ± 0.2 | 2.7 ± 0.8 |
| 2U | 5.8 ± 0.4 | 13.7 ± 2.2 | 0.42 ± 0.10 |
| 2U·A | 1.7 ± 0.1 | 5.4 ± 0.8 | 0.31 ± 0.06 |
| 2U·G | 3.6 ± 0.3 | 5.8 ± 1.0 | 0.62 ± 0.16 |
Steady-state reactions were performed with UDG and the substrates above using the fluorescence assay (Materials and Methods). The kinetic parameters were determined by fitting the observed rates as a function of substrate concentration (Fig. 1).
Equilibrium binding
The binding of UDG to DNA substrates was monitored under equilibrium conditions. To prevent turnover of the substrate, an inactive mutant of UDG was used, where the active site Asp was replaced with Asn (D88N). We were unable to detect any activity with D88N using the fluorescence assay described above, even when reactions were left for several hours with high concentrations of enzyme (data not shown).
Fluorescence anisotropy was used to monitor the binding of D88N to substrates containing a 5′ HEX label. Binding of enzyme to the labeled oligonucleotide leads to an increase in the rotational correlation time of the chromophore due to the increased mass of the enzyme–DNA complex relative to free DNA, and hence a change in anisotropy (19). This provides a very sensitive assay that can be used at low concentrations of substrate and, as a solution-based technique, it is not prone to the deviations due to mass transport effects experienced with surface plasmon resonance techniques. Moreover, the fluorescence group is remote from the target uracil and so does not interfere with the interactions of enzyme and DNA.
A series of substrates analogous to those used in the steady-state assays were synthesised with 5′ HEX labels (Table 2). The target uracil was placed in single-stranded substrates in either an A-T-rich (1HU) or G-C-rich (2HU) sequence context. The double-stranded forms of these substrates were also used and the effects of placing the uracil in either a U·A base pair (1HU·A and 2HU·A) or a U·G mismatch (1HU·G and 2HU·G) were investigated.
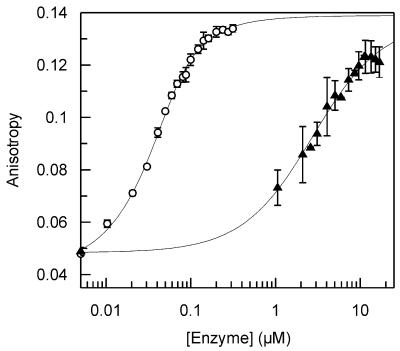
Binding isotherms were obtained for each substrate by adding increasing concentrations of the D88N enzyme to a solution of oligonucleotide and measuring the change in anisotropy (Fig. 2). The concentration of oligonucleotide used was set at approximately the same value as the Kd and so varied from 50 nM to 5 µM, depending on the substrate: the appropriate range was established from pilot experiments. The Kd for each substrate was determined by fitting the data to equation 1 (Table 4).
Figure 2.
Equilibrium binding. Equilibrium binding experiments were performed by fluorescence anisotropy using oligonucleotide substrates that contain a 5′ HEX chromophore (Materials and Methods and Table 2). The increase in anisotropy for substrates 1HU (open circles) and 2HU (closed triangles) is shown plotted against the enzyme concentration on a logarithmic scale. The solid lines show the best fit of the data to equation 1 with the following values: 1HU, Kd = 0.012 ± 0.001 µM, AD = 0.042 ± 0.001, ADE = 0.140 ± 0.001; 2HU, Kd = 2.5 ± 0.3 µM, AD = 0.048 ± 0.002, ADE = 0.137 ± 0.002.
Table 4. Equilibrium binding.
| Oligonucleotide |
Kd (µM) |
| 1HU | 0.012 ± 0.002 |
| 1HU·A | 0.52 ± 0.11 |
| 1HU·G | 0.83 ± 0.15 |
| 2HU | 2.5 ± 0.3 |
| 2HU·A | 3.0 ± 0.4 |
| 2HU·G | 4.2 ± 0.9 |
| 1HAP | 1.5 ± 0.2 |
| 2HAP | 4.0 ± 0.7 |
The equilibrium dissociation constant (Kd) for D88N UDG was determined with different substrates by fluorescence anisotropy. The oligonucleotides 1HAP and 2HAP are the abasic forms of oligonucleotides 1HU and 2HU, where the uracil was removed by incubation with wild-type UDG prior to the determination of Kd (Materials and Methods).
The Kd values show significant variations between the different substrates examined in these experiments (Table 4). The single-stranded forms of the two substrates (1HU and 2HU) have tighter binding than either of their double-stranded equivalents. In all cases each form of the A-T-rich oligonucleotide 1 has tighter binding than the equivalent G-C-rich oligonucleotide 2: with the single-stranded substrates there is a two orders of magnitude difference (Fig. 2), while with the double-stranded forms there is an ∼5-fold difference. These results clearly show that the ability of D88N UDG to bind the target uracil is dependent upon the sequence context of the uracil.
The binding of D88N to the single-stranded abasic reaction products 1HAP and 2HAP was also investigated (Table 4). This revealed that D88N binds the reaction product of the A-T-rich oligonucleotide 100-fold less tightly than the substrate DNA. However, with the G-C-rich oligonucleotide there is virtually no difference in binding affinity between the substrate and product.
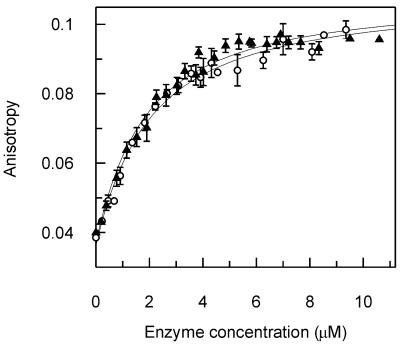
The use of the abasic reaction product also allowed us to compare the binding of the wild-type and D88N mutant forms of UDG. The dissociation constant for 1HAP was determined with both enzymes. This revealed them to have an identical affinity for the product DNA (Fig. 3). Whilst this does not exclude possible differences between D88N and wild-type UDG in their ability to bind substrate DNA, it does demonstrate that D88N is properly folded and that the interactions with the DNA backbone are unaffected.
Figure 3.
Binding of wild-type and D88N mutant UDG to abasic DNA. Equilibrium binding experiments were performed with the HEX-labeled oligonucleotide 1HAP (Materials and Methods). Fluorescence anisotropy was used to follow the binding of wild-type (open circles) and D88N mutant (closed triangles) UDG to this abasic DNA. Both data sets are shown with the best fit to equation 1, with the following values: D88N, Kd = 1.5 ± 0.20 µM, AD = 0.037 ± 0.002, ADE = 0.11 ± 0.002; wild-type, Kd = 1.7 ± 0.24 µM, AD = 0.036 ± 0.002, ADE = 0.11 ± 0.003
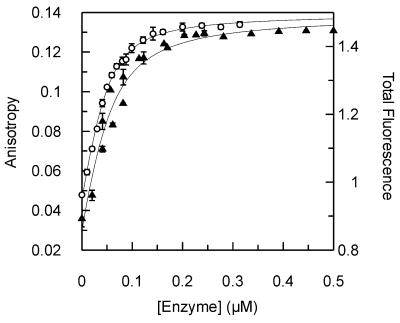
Since the binding of UDG to DNA is sequence dependent, we were concerned that the incorporation of 2-aminopurine might affect local sequence structure and hence UDG binding. To test this, 2-aminopurine fluorescence was used as a measure of UDG binding. The observed increase in fluorescence emission when a uracil that is adjacent to a 2-aminopurine is removed by UDG has been described above. A similar change in fluorescence was also observed when oligonucleotides containing 2-aminopurine adjacent to the target uracil were bound by D88N. This is due to the change in the environment of the 2-aminopurine and is most likely associated with flipping of the target uracil out of the DNA double helix and into the enzyme active site (11). The fluorescence intensity for oligonucleotide 1U showed a hyperbolic increase with increasing concentrations of D88N that closely resembled the increase in anisotropy of oligonucleotide 1HU with D88N (Fig. 4). The Kd values were within the range of experimental error for each oligonucleotide. It was therefore concluded that the incorporation of 2-aminopurine into the substrates does not have a discernable effect on the ability of UDG to interact with this substrate. It was not possible to measure the binding of oligonucleotide 2U by this method since, under weak binding conditions, the concentration of enzyme required was so large that the 2-aminopurine fluorescence signal was swamped by the inherent tryptophan fluorescence of UDG.
Figure 4.
Binding of D88N mutant UDG to 2-aminopurine and HEX-labeled oligonucleotides. Equilibrium binding experiments were performed with the HEX-labeled oligonucleotide 1HU and the 2-aminopurine-containing oligonucleotide 1U (Materials and Methods). Fluorescence anisotropy (left scale) was used to follow the binding of D88N to 1HU (open circles; Figure 2), while total 2-aminopurine fluorescence intensity (right scale) was used to measure binding to 1U (closed triangles). Both data sets are shown with the best fit to equation 1, with the following values: 1HU, Kd = 0.012 ± 0.001 µM, AD = 0.042 ± 0.001, ADE = 0.140 ± 0.001; 1U, Kd = 0.024 ± 0.006 µM, FD = 0.86 ± 0.03, FDE = 1.49 ± 0.02 (where FD and FDE are the total fluorescence of free and enzyme-bound DNA, respectively)
DISCUSSION
In this study sequence context effects on HSV-1 UDG were examined using a combination of steady-state kinetics and equilibrium binding. The single-stranded substrate 1U contains a uracil with A-T-rich flanking sequences and UDG is able to cleave the uracil from this substrate very efficiently. In contrast, oligonucleotide 2U has the target uracil located within a G-C-rich sequence context and UDG exhibits very different characteristics with this substrate. While the values of kcat for the two substrates are very similar, Km for the G-C-rich oligonucleotide 2 is significantly higher. Overall, 2U is a much poorer substrate than 1U, with a kcat/Km that is an order of magnitude lower (Table 3). Clearly, sequence context has a significant impact on the ability of UDG to excise uracil from DNA.
Equilibrium binding assays also revealed a large difference in the binding affinity of UDG for single-stranded substrates of different sequence. The A-T-rich oligonucleotide 1HU is bound two orders of magnitude tighter than the G-C-rich oligonucleotide 2HU (Fig. 2). The ability of UDG to bind uracil is thus modulated by the sequences flanking the target base.
It is also of note that Km > Kd with the single-stranded substrates tested here. In considering the scheme for a steady-state reaction:
k1 k2
E + S ↔ E·S → E + P
k–1
Km = (k–1 + k2)/k1
Kd = k–1/k1
it follows that Km > Kd when k2 ≥ k–1. Also, when Km >> Kd, as with substrate 1U, then k2 > k–1. The effect of this is that once the enzyme–substrate complex has been formed, the forward rate, k2, is faster than the reverse rate, k–1. Hence, the enzyme rapidly proceeds through catalysis and product dissociation much faster than the rate at which the enzyme–substrate complex dissociates without reacting. The UDG–1U enzyme–substrate complex will thus not accumulate during the reaction, as it will rapidly proceed further through the reaction pathway.
The increase in Km with 2U compared to 1U (Table 3) can be accounted for by the corresponding increase in Kd (Table 4). The change in binding equilibrium demonstrates that the ability of UDG to locate uracil lesions is modulated by the sequence context of the uracil. Target location by UDG will be more complicated than the single step shown here, as it will involve both non-specific binding and base flipping of the target nucleotide to form the catalytically competent enzyme–substrate complex. However, the observed changes in Km and Kd reveal that damage recognition by UDG is modulated by DNA sequence.
UDG exhibited a weaker binding affinity for the double-stranded G-C-rich substrate compared to the double-stranded A-T-rich substrate. But, once again, no significant differences were observed between a U·G mismatch and a U·A base pair with either substrate, in agreement with other studies (13–15). With the double-stranded substrates Km increased so that it was approximately equal to Kd, unlike the single-stranded substrates, where Km > Kd. This difference may be accounted for by a change in the substrate binding affinity, such that k–1 > k2. This explanation is consistent with the overall weaker equilibrium binding that we observe with the double-stranded substrates compared to the single-stranded substrates (Table 4).
The differences in sequence-dependent activity that we have observed are greatest on single-stranded DNA, although with the double-stranded substrates a 5-fold difference in the kcat/Km values was observed. While Eftedal et al. (13) reported that E.coli UDG has sequence preference, they only observed this with double-stranded DNA. Whilst this may be true of their particular sequence, it should be noted that their conclusions were based on relative rates determined at a single substrate concentration. However, this can lead to significant errors in interpretation: it can be seen from Figure 1 that relative rates may vary significantly with substrate concentration. Without determining kinetic constants, it is thus difficult to draw firm conclusions regarding the behaviour of enzymes with different substrates. Our findings clearly demonstrate that the sequence preference of UDG extends to single-stranded DNA. Therefore, the increased rate of uracil cleavage from within A-T-rich sequences cannot be due to increased local strand melting, as previously suggested (13).
The sequence-dependent differences in catalytic activity of HSV-1 UDG that we have observed were largely due to changes in Km, while kcat remained fairly constant; a similar trend has been noted in another recent report (20). These differences produce changes in kcat/Km of up to 17-fold (Table 3). While this is not large when compared to the overall catalytic power of the enzyme (20), it should be noted that kcat/Km represents the ability of the enzyme to discriminate between different substrates (21). The differences in sequence specificity that we have observed may thus be sufficient to produce mutational hotspots, as it only requires a single unrepaired error to create a point mutation.
Examination of the binding affinity of UDG to the abasic reaction products shows that there is little difference between the two DNA products that we have tested (Table 4). However, there is a significant difference in the relative binding affinity of UDG for the substrate and product of each sequence. With the A-T-rich oligonucleotide 1HU the substrate is bound two orders of magnitude tighter than the reaction product. This is anticipated, as we would expect the uracil to contribute to binding interactions. However, with the weaker binding G-C-rich oligonucleotide 2HU no difference in binding affinity is observed between the substrate and product forms. This suggests that with the weaker binding substrate, interactions with the target uracil contribute little to the binding affinity. This may be explained if the enzyme–substrate complex with 2HU is relatively strained, such that it is not able to maximise all of the possible interactions with both the DNA backbone and the target uracil simultaneously. This is consistent with observations that backbone interactions have an important role in the ability of UDG to flip and bind a deoxyuridine nucleotide (20,22).
The results presented here differ from those observed with human UDG, where the abasic reaction product is bound more tightly than the substrate (12). These observations led to the hypothesis that the binding energy develops stereoelectronic strain and that this contributes to the catalytic potential of the enzyme (23). The active sites of the human and HSV-1 UDG enzymes are essentially identical and it would seem very unlikely that they operate via different chemical mechanisms. However, the same argument does not apply to the HSV-1 UDG as our results show that interactions with the uracil contribute positively to binding energy. We believe that the different binding affinities of human and HSV-1 UDG for abasic DNA are due to primary structure variations at positions remote from the active site, which are responsible for stabilising the DNA-bound enzyme conformation.
Implications for damage recognition
All of the results reported here are consistent with the proposition that DNA damage recognition by UDG is dependent upon the sequence context of the target uracil. The molecular basis for this behaviour is likely to be a result of two possibilities. The stability of the target uracil within the DNA structure will change with sequence, so the energy required to flip the uracil into the active site of the enzyme will also change and this may affect the rate of excision. Alternatively, the ability of UDG to interact with the target uracil may be modulated by local DNA structure.
There is now considerable evidence that DNA helix stability is affected by sequence. However, a comparison of our results with recent studies detailing the relative stability of all possible tetranucleotide permutations (24) did not reveal any correlation. Also, the increased stability of a U·A base pair in comparison to a U·G mismatch did not result in a significant difference in DNA binding or catalytic activity (Tables 3 and 4). Work by Stivers and colleagues has also failed to reveal a correlation between base stability and the observed rate of nucleotide flipping (11). Hence, it seems unlikely that the stability of the target base within the DNA structure is the determining factor in the observed sequence preferences.
It is now well established that DNA structure is dependent upon local sequence (25). It is thus possible that the ability of UDG to bind its target uracil is dependent upon the local DNA structure surrounding the uracil residue. The structure of human UDG bound to DNA shows a distortion in the DNA backbone, such that the distance between two of the phosphates flanking the uracil is reduced from 12 to 8 Å (12). It has been suggested that this backbone ‘pinch’ is the mechanism by which UDG destabilises the stacked conformation of the nucleotide, thus inducing it to flip into the active site. It may thus be expected that any changes in the inter-phosphate distance of the DNA backbone will affect the interactions between these phosphates and UDG. It is quite possible that the sequence-specific effects that we have observed in this study arise from structural differences in the substrates. Further investigation of this would require structural analysis of the substrates that we have used.
The data presented provide a further insight into the origin of the sequence dependence of UDG activity. Our results are consistent with the proposition that DNA damage recognition by UDG is modulated by DNA sequence and that this is most likely due to local perturbations in DNA structure. There is no biological reason for UDG to possess sequence discrimination. But, as with other ‘non-specific’ enzymes that act on DNA, such as DNase I, some sequence preference is observed (26). It appears impossible to find enzymes that act on DNA which are truly non-specific. This general observation may be due to the heterogeneous nature of DNA. However, with a DNA repair enzyme this lack of specificity may have significant consequences. Poor sites of repair by UDG have been linked to mutational hotspots in the lacI gene (14) and may be a concern in other genetic diseases caused by C→T transition mutations.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Dr Renos Savva for supplying the overexpressing constructs of HSV-1 UDG and for his freely given advice and discussions, Prof. Stephen Halford for his ever-insightful help, advice and discussions and Dr Daniel Barsky, whose ideas stimulated the remarks on non-specific enzymes that act on DNA. We would particularly like to thank the BBSRC who have funded this work.
References
- 1.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson J. and Martin,R. (1994) Mutations to nonsense codons in human genetic-disease—implications for gene-therapy by nonsense suppressor transfer-RNAs. Nucleic Acids Res., 22, 1327–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindahl T. (1974) An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl Acad. Sci. USA, 9, 3649–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeberg E., Eide,L. and Bjoras,M. (1995) The base excision-repair pathway. Trends Biochem. Sci., 20, 391–397. [DOI] [PubMed] [Google Scholar]
- 5.Slupphaug G., Mol,C.D., Kavli,B., Arvai,A.S., Krokan,H.E. and Tainer,J.A. (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature, 384, 87–92. [DOI] [PubMed] [Google Scholar]
- 6.Xiao G.Y., Tordova,M., Jagadeesh,J., Drohat,A.C., Stivers,J.T. and Gilliland,G.L. (1999) Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited. Proteins, 35, 13–24. [PubMed]
- 7.Savva R., Mcauleyhecht,K., Brown,T. and Pearl,L. (1995) The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature, 373, 487–493. [DOI] [PubMed] [Google Scholar]
- 8.Verri A., Mazzarello,P., Spadari,S. and Focher,F. (1992) Uracil-DNA glycosylases preferentially excise mispaired uracil. Biochem J., 287, 1007–1010. [DOI] [PMC free article] [PubMed]
- 9.Mol C.D., Arvai,A.S., Slupphaug,G., Kavli,B., Alseth,I., Krokan,H.E. and Tainer,J.A. (1995) Crystal-structure and mutational analysis of human uracil-DNA glycosylase—structural basis for specificity and catalysis. Cell, 80, 869–878. [DOI] [PubMed] [Google Scholar]
- 10.Barrett T.E., Savva,R., Panayotou,G., Barlow,T., Brown,T. and Jiricny,J. (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. [DOI] [PubMed] [Google Scholar]
- 11.Stivers J.T., Pankiewicz,K.W. and Watanabe,K.A. (1999) Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry, 38, 952–963. [DOI] [PubMed] [Google Scholar]
- 12.Parikh S.S., Mol,C.D., Slupphaug,G., Bharati,S., Krokan,H.E. and Tainer,J.A. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eftedal I., Guddal,P.H., Slupphaug,G., Volden,G. and Krokan,H.E. (1993) Consensus sequences for good and poor removal of uracil from double-stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res., 21, 2095–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsen H., Yazdankhah,S.P., Eftedal,I. and Krokan,H.E. (1995) Sequence specificity for removal of uracil from U·A pairs and U·G mismatches by uracil-DNA glycosylase from Escherichia coli and correlation with mutational hotspots. FEBS Lett., 362, 205–209. [DOI] [PubMed] [Google Scholar]
- 15.Slupphaug G., Eftedal,I., Kavli,B., Bharati,S., Helle,N.M., Haug,T., Levine,D.W. and Krokan,H.E. (1995) Properties of a recombinant human uracil-DNA glycosylase from the ung gene and evidence that ung encodes the major uracil-DNA glycosylase. Biochemistry, 34, 128–138. [DOI] [PubMed] [Google Scholar]
- 16.Savva R. and Pearl,L.H. (1993) Crystallization and preliminary-x-ray analysis of the uracil-DNA glycosylase DNA-repair enzyme from herpes-simplex virus type-1. J. Mol. Biol., 234, 910–912. [DOI] [PubMed] [Google Scholar]
- 17.Baldwin G.S., Vipond,I.B. and Halford,S.E. (1995) Rapid reaction analysis of the catalytic cycle of the EcoRV restriction endonuclease. Biochemistry, 34, 705–714. [DOI] [PubMed] [Google Scholar]
- 18.Stivers J.T. (1998) 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res., 26, 3837–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell L.M., Connolly,B.A. and Dryden,D.T.F. (1998) The DNA binding characteristics of the trimeric EcoKI methyltransferase and its partially assembled dimeric form determined by fluorescence polarisation and DNA footprinting. J. Mol. Biol., 283, 947–961. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y.L. and Stivers,J.T. (2001) Reconstructing the substrate for uracil DNA glycosylase: tracking the transmission of binding energy in catalysis. Biochemistry, 40, 7710–7719. [DOI] [PubMed] [Google Scholar]
- 21.Cornish-Bowden A. (1995) Fundamentals of Enzyme Kinetics. Portland Press, London.
- 22.Werner R.M., Jiang,Y.L., Gordley,R.G., Jagadeesh,G.J., Ladner,J.E., Xiao,G., Tordova,M., Gilliland,G.L. and Stivers,J.T. (2000) Stressing-out DNA? The contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase. Biochemistry, 39, 12585–12594. [DOI] [PubMed] [Google Scholar]
- 23.Parikh S.S., Walcher,G., Jones,G.D., Slupphaug,G., Krokan,H.E., Blackburn,G.M. and Tainer,J.A. (2000) Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl Acad. Sci. USA, 97, 5083–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Packer M.J., Dauncey,M.P. and Hunter,C.A. (2000) Sequence-dependent DNA structure: tetranucleotide conformational maps. J. Mol. Biol., 295, 85–103. [DOI] [PubMed] [Google Scholar]
- 25.Neidle S. (1998) New insights into sequence-dependent DNA structure. Nature Struct. Biol., 5, 754–756. [DOI] [PubMed] [Google Scholar]
- 26.Evans S., Shipstone,E., Maughan,W. and Connolly,B. (1999) Site-directed mutagenesis of phosphate-contacting amino acids of bovine pancreatic deoxyribonuclease I. Biochemistry, 38, 3902–3909. [DOI] [PubMed] [Google Scholar]