Abstract
Pontocerebellar hypoplasia (PCH) is a heterogeneous group of rare recessive disorders with prenatal onset, characterized by hypoplasia of pons and cerebellum. Mutations in a small number of genes have been reported to cause PCH, and the vast majority of PCH cases are explained by mutations in TSEN54, which encodes a subunit of the tRNA splicing endonuclease complex. Here we report three families with homozygous truncating mutations in TBC1D23 who display moderate to severe intellectual disability and microcephaly. MRI data from available affected subjects revealed PCH, small normally proportioned cerebellum, and corpus callosum anomalies. Furthermore, through in utero electroporation, we show that downregulation of TBC1D23 affects cortical neuron positioning. TBC1D23 is a member of the Tre2-Bub2-Cdc16 (TBC) domain-containing RAB-specific GTPase-activating proteins (TBC/RABGAPs). Members of this protein family negatively regulate RAB proteins and modulate the signaling between RABs and other small GTPases, some of which have a crucial role in the trafficking of intracellular vesicles and are involved in neurological disorders. Here, we demonstrate that dense core vesicles and lysosomal trafficking dynamics are affected in fibroblasts harboring TBC1D23 mutation. We propose that mutations in TBC1D23 are responsible for a form of PCH with small, normally proportioned cerebellum and should be screened in individuals with syndromic pontocereballar hypoplasia.
Keywords: pontocerebellar hypoplasia, microcephaly, TBC1D23, small normally proportioned cerebellum
Introduction
Based on clinical and molecular features, there are currently ten described subtypes of PCH. These subtypes are classified based on the nature of the brain anomalies and the presence of associated clinical features,1 such as microcephaly, seizures, and extra-neurological features, and are further delineated by the underlying gene defect. Despite the identification of mutations in 13 genes to date, the principal one being TSEN54 (MIM: 608755), more than 25% of PCH cases remain uncharacterized at the molecular level.1
PCH1 was first characterized by degeneration of cells from the anterior horn of the spinal cord. Mutations in VRK1 (MIM: 602168) (PCH1A [MIM: 607596]) and EXOSC3 (MIM: 606489) (PCH1B [MIM: 614678]) have been reported in affected individuals.2, 3 In recent publications, EXOSC3 accounted for between 40% and 75% of PCH1-affected subjects.1, 2 PCH2, the most common form of PCH, is characterized by dyskinesia and absence of spinal muscular atrophy. Approximately 88% of subjects harbor the recurrent homozygous missense mutations in exon 8 of TSEN54 (PCH2A [MIM: 277470]), a subunit of the transfer RNA splicing endonuclease complex. The remaining PCH2-affected individuals harbor mutations in TSEN2 (MIM: 608753) (PCH2B [MIM: 612389]), TSEN34 (MIM: 608754) (PCH2C [MIM: 612390]),4 and SEPSECS (MIM: 613009) (PCH2D [MIM: 613811]).5 PCH3 (MIM: 608027)-affected individuals are characterized by PCH associated with seizures, facial dysmorphism, and optic atrophy. Recently, homozygous nonsense mutations of PCLO (MIM: 604918) have been reported in subjects with PCH3.6 PCH4 (MIM: 225753) is similar to PCH2 but with a more severe presentation and C-shaped inferior olives on brain MRI.7 Subjects with PCH4 have been reported with association of two deleterious mutations in TSEN54, one common missense mutation and one nonsense mutation. PCH5 (MIM: 610204) is characterized by prenatal seizure-like activity. Compound heterozygous mutations in TSEN54 have been reported in these individuals.4 PCH6 (MIM: 611523)-affected subjects are distinct from other types of PCH by the occurrence of intractable epilepsy with elevated CSF lactate. This PCH phenotype is caused by mutations in RARS2 (MIM: 611524), encoding a mitochondrial arginyl transfer synthetase.8 PCH7 (MIM: 614969) is a recent subtype of PCH associated with hypogonadism.9 To date, no gene has been found to be involved in this form of PCH. PCH8 (MIM: 614961) subtype is distinguished by joint contractures and growth retardation. Mutations in CHMP1A (MIM: 164010) have been reported in two families.10 Recently, two additional PCH subtypes were described that are not yet embedded in the official classification: PCH9 (MIM: 615809) and PCH10 (MIM: 615803). PCH9-affected subjects present with microcephaly and progressive contractures and mutations in AMPD2 (MIM: 102771) that encodes an adenosine monophosphate deaminase.11 The last subtype, PCH10, is characterized by microcephaly, hypertonia, and increased tendon reflexes. Exome sequencing revealed mutations in CLP1 (MIM: 608757), a kinase involved in tRNA splicing.12 Recently mutations in RELN (MIM: 600514) and VLDLR (MIM: 192977), which both belong to the same pathophysiological Reelin pathway, were reported in subjects with severe cerebellar hypoplasia, intellectual disability (ID), and a small and dysplastic cerebellar vermis. Individuals with RELN mutations often exhibit cortical abnormalities and neonatal seizures whereas persons with VLDLR mutations presented with non-progressive congenital cerebellar ataxia.13
Here, we report three consanguineous families with an autosomal-recessive form of ID and global developmental delay associated with homozygous truncating mutations in TBC1D23. Interestingly, all subjects for whom brain MRI was performed have PCH in common. Moreover, re-examination of MRI data revealed the presence of a small, normally proportioned cerebellum associated with corpus callosum abnormalities.
We further show that downregulation of TBC1D23 leads to anomalies in neuronal positioning in the mouse cortex during embryonic and early post-natal stages. Additionally, we demonstrate intracellular trafficking defects associated with TBC1D23 mutations.
Subjects and Methods
Subjects
Families for this study were identified through sharing of exome data of individuals with brain developmental anomalies between members involved in Gencodys European research consortium. For exome sequencing (ES) and validation of variants, blood or DNA samples from affected individuals, healthy parents, and other family members and informed consent were obtained from all participants in accordance with the site-specific Institutional Review Boards (IRB). Written parents’ consents were obtained for conducting the study and showing MRIs and photographs of their affected children.
Exome Sequencing and Variant Validation
Exome sequencing (ES) of selected family members included in this study was performed using in solution exome capture kits (Sure Select Human All Exome 50MB kit, Agilent Technology, or Illumina TruSeq Exome Enrichment, Illumina) and Illumina HiSeq sequencing platforms (Illumina) to generate paired end reads sequences. Sequence analysis pipeline included alignment with BWA-mem against the hg19 genome reference, GATK for base recalibration, indel realignment, and the GATK Unified Genotyper for variant calling. Variants called were generated in the standard VCF version and filtered for homozygous exonic and splice site variants, and excluding known SNPs in the dbSNP131 database, 1000 Genomes, Exome Variant Server, the Exome Aggregation Consortium (ExAC), and a local ES databases. Sequence validation of potential relevant variants and segregation analysis were performed by Sanger sequencing using standard procedures.
Plasmid Constructs
Three different 29-mer shRNA sequences targeting mouse Tbc1d23 (GenBank: NM_026254) commercially designed and provided by Origene were cloned in the psiStrike vector under the control of a U6 promoter: shRNA-1, targeting the sequence 5′-AGAATCCATCTGAGTTTGCACAGTCAGT-3′, specific for both mouse and human TBC1D23; shRNA-2, targeting the sequence 5′-TTACATCCAGTCTCGGCAAGCACTAAATT-3′ and specific for mouse Tbc1d23; and shRNA-3, targeting the sequence 5′-TGTTAGCATTGCCAGTGGAGGATTTATGG-3′, also specific for mouse Tbc1d23. A 29-mer non-effective oligonucleotide (Origene) cloned in the same vector was used as control. Mouse Tbc1d23 cDNA fused to GFP was cloned in the psiStrike vector under the control of a CAG (chicken beta-actin promoter with CMV enhancer) promoter. For in utero electroporation (IUE) experiment, a pCAGGS-IRES-Tomato (RFP) vector was used as a reporter. For time-lapse experiments, we used neuropeptide Y (NPY) fused to GFP and expressed under the β-actin promoter. This construct was a kind gift from M. Silverman.
Cell Culture
Mouse neuroblastoma N2a cells, control, and subject’s human fibroblasts carrying the TBC1D23 p.Val492Glyfs∗8 variant were cultured in DMEM (GIBCO) supplemented with 1 g/L of glucose and 5% fetal calf serum. Transfection was performed using Lipofectamine2000 (Invitrogen) according to the manufacturer’s instructions and cells were then cultured for 48 hr.
Primary cortical neurons were prepared from E17 mouse embryos. Cortices were dissected in cold PBS with 2.56 mg/mL D-glucose, 3 mg/mL BSA, and 1.16 mM MgSO4. Tissue digestion was performed enzymatically in a solution containing 0.25 mg/mL trypsin which was then inhibited with 0.5 mg/mL trypsin soybean inhibitor, followed by mechanical dissociation. 5 million neurons in suspension were electroporated using Amaxa Nucleofector kit for mouse neurons (Lonza) following manufacturer’s instructions.
Western Blot
Cells were lysed in RIPA buffer (50 mM Tris-HCl [pH 7.7], 0.15 M NaCl, 1 mM EDTA, and 1% Triton X-100) supplemented with proteases inhibitors (Roche). Protein concentrations were determined with a Bradford assay. Equal amounts of lysates were then loaded on polyacrylamide gels and then transferred onto nitrocellulose membranes. Membranes were blocked in 5% milk solution in TBS-0.1% Tween and then were incubated with the following primary antibodies overnight at 4°C: rabbit TBC1D23 antibody (ab183478, Abcam) used at 1:500 and mouse actin antibody (IGBMC) used at 1:1,000. HRP (horseradish)-conjugated secondary antibodies (Thermo Fischer) were used at 1:10,000.
In Utero Electroporation
In utero electroporation was performed as described previously14 using timed pregnant Swiss mice (Janvier). Animal experimentations were performed at the IGBMC animal facilities. The study was conducted according to national and international guidelines (authorization number 2017062811273521, French MESR) and the procedures followed were in accordance with the ethical standards of the responsible committee on mouse experimentation (Comité d’éthique pour l’expérimentation animale [Strasbourg, France]). At E14.5, the pregnant mice were anaesthetized with isoflurane (2 L/min of oxygen, 3% isoflurane during induction and 2.5% during surgery). The uterine horns were exposed, and a lateral ventricle of each embryo was injected using pulled glass capillaries (Harvard Apparatus) with Fast Green (2 μg/mL; Sigma) combined with the DNA constructs prepared with EndoFree plasmid purification kit (Macherey Nagel) and the pCAGGS-IRES-Tomato reporter vector at 1 μg/μL. For knockdown experiments, different shRNAs and control shRNA were electroporated at 1 μg/μL. For rescue experiments, the concentrations used were either 1 μg/μL for shRNA and cDNA construct or 1 μg/μL shRNA with 3 μg/μL cDNA. Mouse cDNA construct alone was electroporated at 1 μg/μL. Plasmids were further electroporated into the neuronal progenitors adjacent to the ventricle by delivering five electric pulses at 50 V for 50 ms at 950-ms intervals using a CUY21EDIT electroporator (Sonidel). After electroporation, embryos were placed back in the abdominal cavity and development was allowed to continue until E16, E18, or P3. Embryo or pup brains were dissected and fixed in 4% paraformaldehyde in PBS (phosphate-buffered saline) overnight and then sectioned at 80 μm slices using a VT1000S vibratome (Leica biosystems).
Immunohistochemistry and Immunofluorescence
Free floating sections were blocked in 2% donkey serum in PBS-0.2% Triton-X and then incubated overnight at 4°C with the following primary antibodies: rabbit CDP (Cux1) (M-222) (sc-13024, Santa Cruz) at 1:100; rabbit Ki67 (IHC-00375, Bethyl laboratories) at 1:200; rabbit prosphohistone 3 (06-570, Millipore) at 1:500. Alexa-coupled secondary antibodies (Thermo Fischer) were used at 1:500. Images were acquired using a confocal microscope TCS SP8 X (Leica microsystems).
Human fibroblasts were fixed in 4% PFA, blocked in 5% donkey serum in PBS-0.3% Triton-X, and then incubated for 1 hr with rabbit TBC1D23 (17002-1-AP, Proteintech) primary antibody at 1:200. Alexa-coupled secondary antibody (Thermo Fischer) was used at 1:500. Images were acquired using a confocal microscope TCS SP5 (Leica microsystems).
Live-Cell Imaging and Analysis
Human fibroblasts were cultured in 35 mm bottom glass dishes (Fluorodish, WPI) and either tranfected with NPY-GFP or treated with lysotracker Green (Molecular Probes) following manufacturer’s instructions.
Primary cortical neurons electroporated with BDNF-mCherry and either control shRNA or shRNA-2 were resuspended in neurobasal medium supplemented with 1xB-27, 0.5 mM L-glutamine, and 100 IU/mL penicillin/streptomycin at 37°C in 5% CO2 and were plated in AXIS microfluidic chambers (Millipore).
Trafficking was monitored using a Confocal Spinning disk (Nikon-Ti) with a 60× PLAN APO VC objective (Nikon Instruments) and driven by Metamorph 7.0 (Molecular Devices). Cells were kept at 37°C and 5% CO2 during acquisition. Images were taken every 0.2 s for 1 min. Kymographs were generated and analyzed using KymoToolBox plugin for ImageJ (NIH, USA). Kymographs generated from maximal projections of the acquired movies were calibrated in space and time. Vesicles’ trajectories were manually analyzed in order to obtain the dynamic parameters of the tracked particles. Static vesicles were not taken into account and velocities below 0.2 μm/s were considered as not moving segments.
Neurite Outgrowth Assay
Neuro2a cells were electroporated using the Amaxa V Nucleofector kit (Lonza) according to the manufacturer’s protocol with either control shRNA or shRNA-2 together with the GFP-derived reporter protein Venus. Cells were then mixed at a ratio of 1:3 with non-nucleofected Neuro2a cells, plated on 13 mm glass coverslips, and cultured overnight. The next day, to induce differentiation, the medium was replaced by DMED without serum and supplemented with 1 μM Retinoic acid. After 48 hr cells were fixed in 4% PFA and immunostained for GFP (Thermo Fisher, A10262, 1:1,000), tubulin (Abcam, ab6160, 1:1,000), and DAPI. Measurements of neurites were made using the Simple Neurite Tracer plugin in ImageJ.15 Each coverslip was evaluated using at least 10 different fields representing in total between 50 and 100 cells. The means of three independent experiments were used.
Statistical Analysis
All experiments were done in at least three independent replicates. All statistics were calculated with GraphPad Prism 6. Data are presented as mean ± SEM. For in utero electroporation data, a two-way ANOVA was performed followed by a Tukey’s multiple comparison test. For western blot quantification, a one-way ANOVA was performed, followed by a Dunett’s multiple comparison test. For trafficking analysis and neurite outgrowth assays, unpaired two-tailed Student’s t test was used. Data distribution was assumed to be normal.
Results
Truncating Mutations in TBC1D23 Associated with Pontocerebellar Hypoplasia and Microcephaly
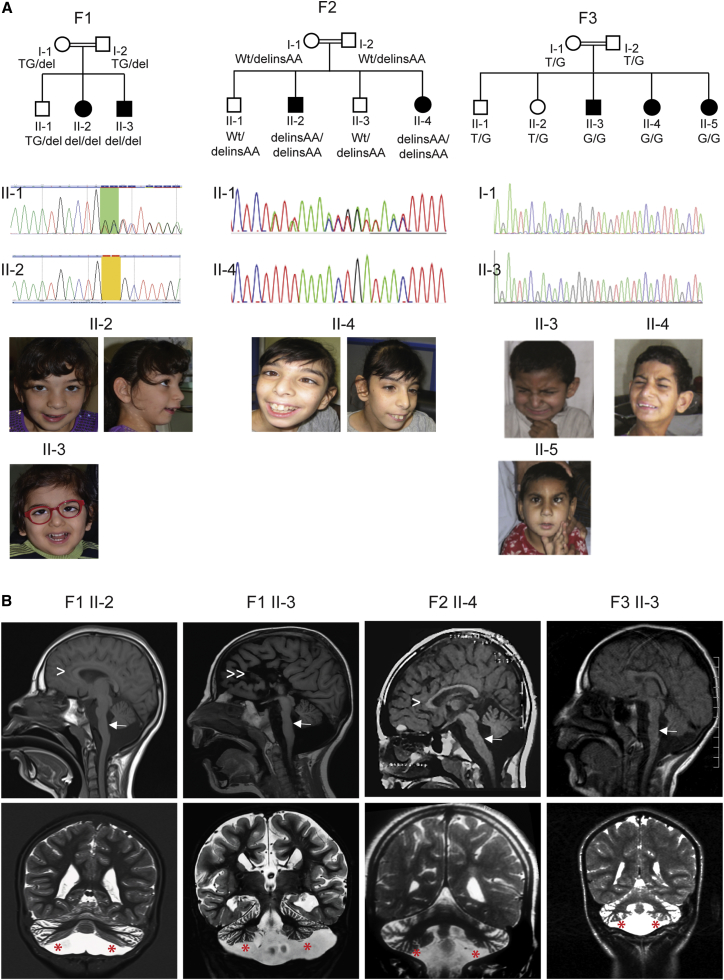
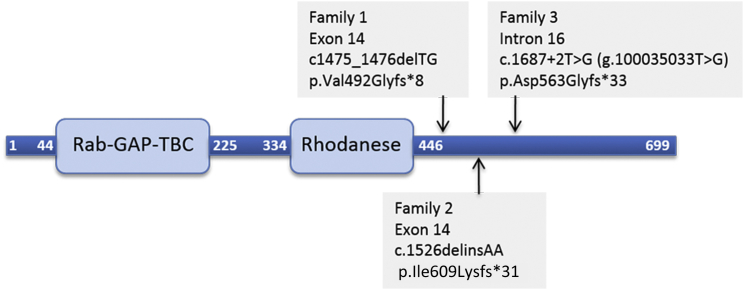
Given the consanguineous relationships in the investigated families, we hypothesized that the phenotype was caused by homozygous mutations in each of the families. For family 1, analysis of exome-sequencing data of parents-affected children trio (Figure 1A; individual II-2 and her parents) revealed a single potential pathogenic variant: an intragenic 2-bp deletion on chromosome 3 (100029306_100029307delTG), corresponding to a homozygous frameshift mutation in exon 14 of TBC1D23 (GenBank: NM_001199198.2): c.1475_1476delTG (p.Val492Glyfs∗8) (Figure 2). The mutation was confirmed by PCR and Sanger sequencing using exon 14-specific primers of TBC1D23 (Figure 1A). Segregation analysis by PCR and direct sequencing of this frameshift mutation using parental and sibships blood DNA showed that both affected individuals were homozygous and the healthy parents and son were heterozygous.
Figure 1.
Mutations in TBC1D23 Cause PCH
(A) Each of the three families is represented by a pedigree showing the segregation of the identified mutation, and representative photographs of affected individuals illustrating the happy appearance and dysmorphic features. For each family, the mutation is also illustrated by Sanger sequencing chromatograms of heterozygous healthy parents and homozygous affected individuals. Each individual or his or her parents provided informed consent for the publication if these photographs.
(B) Representative sections of brain MRIs for affected individuals with TBC1D23 mutations. First row, sagittal images in four different individuals showing pontocerebellar hypoplasia (PCH) (white arrows); hypoplasia of the corpus callosum (F1 II-2 and F2 II-4, arrowheads), and agenesis of the corpus callosum (F1-II-3, double arrowheads). Second row, axial sections showing a small normally proportioned cerebellum (red asterisks).
Figure 2.
Representation of the TBC1D23 Polypeptide
Linear representation of TBC1D23 polypeptide showing positions of Rab-GAP-TBC and rhodanese domains, as well as positions of premature stop codon resulting from each of the mutation identified in the families reported in this study.
In family 2, analysis of ES data of individual II-4 (Figure 1A) and subsequent Sanger sequencing revealed a potentially pathogenic SNV in all affected individuals: a homozygous frameshift mutation c.1526delinsAA (p.Ile509Lysfs∗31) in exon 14 of TBC1D23 (Figure 2).
Finally, in family 3, ES of affected individuals II-3 and II-4 (Figure 1A) revealed a potential deleterious homozygous mutation, c.1687+2T>G (GenBank: NC_000003.11; g.100035033T>G on hg19) in the canonical donor splice site of intron 16 of TBC1D23, predicted to lead to a frameshift (p.Asp563Glyfs∗33) secondary to skipping of exon 16 (Figure 2). In accordance with this prediction, the disruptive impact of the mutation on the wild-type donor splice site is estimated at 100% by the prediction programs MaxEntScan, HSF, and NNsplice. Segregation analysis of the mutation in the family showed that all affected individuals are homozygous and healthy parents and sibships are heterozygous or wild-type at both alleles.
Overall, TBC1D23 variants identified in the three families reported in this study were unique and not represented in dbSNP, 1000 Genomes, or ExAC datasets and co-segregated with the phenotype in each of the three families. Moreover, all of the mutations were predicted to lead to a disruption of the reading frame and premature termination codons upstream of the 50–55 nucleotides threshold of the final exon-exon junction.
MRI scans were available for evaluation from four individuals (Figure 1B). All four presented PCH and small normally proportioned cerebellum. In three subjects, anomalies of the corpus callosum were revealed. Phenotypically, all individuals present severe cognitive delay associated in some with behavior abnormalities and autistic features and happy facial expression with dysmorphic features. Detailed clinical descriptions and radiological features of the reported individuals are summarized in Table 1 and the Supplementary Note.
Table 1.
Clinical and Radiological Features of Individuals with TBC1D23 Mutations
|
Family |
F1 |
F2 |
F3 |
||||
|---|---|---|---|---|---|---|---|
|
Consanguinity |
yes |
yes |
yes |
||||
| Individual | II-2 | II-3 | II-2 | II-4 | II-3 | II-4 | II-5 |
| Gender | F | M | M | F | M | F | F |
| Age at onset | postnatal | postnatal | postnatal | postnatal | postnatal | postnatal | postnatal |
| Development and growth milestones delay | + | + | + | + | + | + | + |
| Seizures | − | − | − | − | − | − | − |
| Age at last examination | 11 years | 3 years | 16 years | 4 years | 14 years | 16 years | 10 years |
| Weight (kg) | 18.7 (< P3) | 11.4 (< P3) | 21 (< P3) | 13 (< P3) | 24 (< P3) | 23 (< P3) | 8 (< P3) |
| Height (cm) | 122 (< P3) | 90 (P15) | 125 (< P3) | 93 (P15) | NA | 147.3 (< P3) | 104.1 (< P3) |
| HC (cm) | 45.5 (< P3) | 45 (< P3) | 42 (< P3) | 42 (< P3) | 45.7 (< P3) | 48.2 (< P3) | 45.7 (< P3) |
| Spastic hypertonia | + | + | + | + | + | + | + |
| Cerebellar syndrome | + | + | + | + | NR | NR | NR |
| Stereotypic movements | + | + | + | + | NR | NR | NR |
| Cognitive delay (ID) | severe | severe | severe | severe | severe | severe | severe |
| Motor weakness | NR | NR | + | + | + | + | + |
| Behavior abnormalities/autistic features | stereotypic movements, autistic behavior | stereotypic movements, autistic behavior, self-injurious behavior | NA | NA | NA | ADHD, poor communication and interaction, persistent repetitive motor behavior | poor communication and interaction, persistent repetitive motor behavior |
| Happy expression | + | + | + | + | + | + | + |
| Ocular abnormalities | NR | hyperopia, strabismus | − | − | coloboma | coloboma | coloboma, strabismus |
| Ears | large | large | large | large | large | large | large |
| Nose | bulbous tip | bulbous tip | bulbous tip | bulbous tip | bulbous tip | prominent nasal bridge | |
| Brain MRI | PCH, HCC | PCH, ACC | NA | PCH, HCCa | PCHb | NA | NA |
| Other features | − | feet and fingers abnormalities,c imperforate anus | flat feet | flat feet | dental anomalies | − | − |
Abbreviations are as follows: HC, head circumference; PCH, pontocerebellar hypoplasia; ACC, agenesis of the corpus callosum; HCC, hypoplasia of the corpus callosum; MRI, magnetic resonance imaging; NA, not available.
Features of Dandy-Walker variant
Features of mega cisterna magna
Feet and finger abnormalities consist of bilateral talipes equinovarus and clinodactyly of the fifth finger
In Vivo Effects of TBC1D23 Knockdown on Cortical Development
The identified mutations in TBC1D23 are predicted to result in a loss-of-function allele due to nonsense-mediated RNA decay and truncation of the protein (Figure 2). We therefore downregulated endogenous Tbc1d23 using short hairpin RNA (shRNA) constructions and we analyzed the effects on neurodevelopmental processes in the cortex using the in utero electroporation (IUE) method in mice.
Three different shRNAs targeting Tbc1d23 mRNA were used. Western blot experiments on Neuro2a neuroblastoma cells transfected with the shRNAs showed that all three of them lead to reduced levels of TBC1D23 (−23% for shRNA1, −47% for shRNA2, and −57% for shRNA3, compared to a scrambled control shRNA), confirming the efficacy of the shRNAs (Figures S1A–S1B).
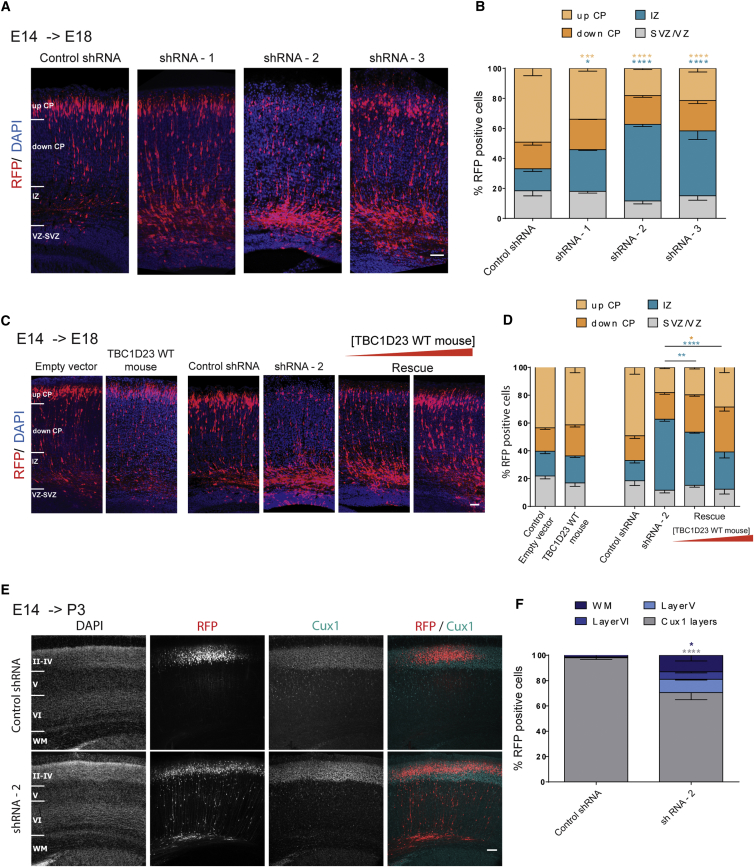
These shRNA constructs were individually electroporated, together with a pCAGGS-IRES-Tomato (RFP) reporter construct in progenitor cells adjacent to the ventricle in E14.5 embryonic mouse cortices. We then studied the effects on neuronal positioning at E18.5. In brain sections electroporated with the control shRNA, we observed a normal pattern of distribution for projection neurons with the majority of them reaching the cortical plate. By contrast, IUE of each of the three shRNAs directed against Tbc1d23 induced a faulty neuronal positioning at this stage (Figures 3A and 3B). An important percentage of the electroporated cells accumulate in the intermediate zone and fewer cells reach the upper cortical plate compared to the control shRNA, suggesting an important role of Tbc1d23 in neuronal positioning.
Figure 3.
Effects of TBC1D23 Downregulation on Neuronal Migration and Positioning
Short hairpin RNA and cDNA constructions were co-electroporated with a RFP-encoding reporter construction (Tomato) in progenitor cells adjacent to the ventricle in E14.5 embryonic mouse cortices. Analyses were done at E18.5 or P3.
(A) Coronal sections of E18.5 brains electroporated at E14.5 with either a control shRNA or one of the three different anti-TBC1D23 shRNAs. Sections were stained with DAPI. CP, cortical plate; IZ, intermediate zone; VZ/SVZ, ventricular zone/subventricular zone. Scale bar, 50 μm.
(B) Quantification of fluorescent neurons positioning in four different regions: up CP, down CP, IZ, and VZ/SVZ. Data are represented as mean ± SEM. shRNA-1 versus control, p = 0.0165 (IZ), p = 0.041 (up CP); shRNA-2 versus control, p < 0.0001 (IZ), p < 0.0001 (up CP); shRNA-3 versus control, p < 0.0001 (IZ), p < 0.0001 (up CP).
(C) Coronal sections of E18.5 brains electroporated at E14.5 with empty vector, mouse Tbc1d23-WT cDNA, control shRNA, shRNA-2, or shRNA-2 together with Tbc1d23-WT (Rescue). For rescue experiments, two different concentration ratios were tested: 1 μg/μL shRNA-2 + 1 μg/μL Tbc1d23-WT and 1 μg/μL shRNA-2 + 3 μg/μL Tbc1d23-WT. Sections were stained with DAPI. Scale bar, 50 μm.
(D) Quantification of fluorescent neurons positioning in four different regions: up CP, down CP, IZ, and VZ/SVZ. Data are represented as mean ± SEM. shRNA-2 versus shRNA-2 + Tbc1d23-WT p = 0.0078 (IZ); shRNA-2 versus shRNA-2+3∗Tbc1d23-WT p < 0.0001 (IZ), p = 0.0113 (down CP).
(E) Coronal sections of P3 brains electroporated at E14.5 with either a control shRNA or shRNA-2. Sections were stained for the upper layer marker Cux1 (cyan) and with DAPI in order to define upper and deep cortical layers and the white matter (WM). Scale bar, 100 μm.
(F) Fluorescent neurons were counted in four different regions: Cux1 layers (II–IV), layer V, layer VI, white matter. Data are represented as mean ± SEM. Control versus shRNA-2: p < 0.0001 (Cux1 layers), p = 0.025 (WM).
In order to validate the specificity of the shRNA-induced phenotype, we performed a rescue experiment by co-electroporating the shRNA inducing the most severe phenotype (shRNA-2) together with the wild-type mouse Tbc1d23 cDNA fused to GFP. All three designed shRNAs target the coding cDNA sequence of Tbc1d23, and therefore it is inevitable that they also target the cDNA used for the rescue experiment. In fact, western blot analysis in transfected Neuro2a cells showed a significant decrease in the GFP-fused TBC1D23 levels when either of the shRNAs is added (−89% for shRNA1, −87% for shRNA2, and −86% for shRNA3) (Figures S1C and S1D). In order to overcome this limitation, we tested two different ratios of cDNA/shRNA—either the same concentration of cDNA and shRNA or three times more cDNA than shRNA. It should be noted that overexpression of this mouse WT form alone does not induce a particular phenotype (Figures 3C and 3D). We saw a significant restoration in the phenotype in a dose-dependent fashion contingent upon cDNA/shRNA ratio (Figures 3C and 3D). Fewer cells accumulated in the IZ and more cells reached the cortical plate under these rescue conditions. These results are in favor of the specificity of the shRNA-induced phenotype.
We further assessed the effect on neuron positioning at post-natal day 3 (P3) by electroporation with the shRNA-2. We immunolabeled the brain sections for Cux1, a marker for the upper cortical layers II, III, and IV, and did a Hoechst staining in order to distinguish between these upper cortical layers, the deeper ones (V and VI), and the white matter (WM) (Figures 3E and 3F). In the control condition, electroporated neurons were almost exclusively found in the upper Cux1 layers of the cortex (98%). In the knockdown condition, a smaller proportion of these neurons was found in the Cux1 layers (70%) and there were also electroporated cells in the layers V (11%) and VI (6%) and in the white matter (13%). These results suggest that the phenotype caused by Tbc1d23 downregulation persists at this post-natal stage.
Further on, considering the presence of microcephaly in subjects, we sought to explore any Tbc1d23 knockdown-related alterations in the proliferation of neuronal progenitors that might be at the origin of this phenotype. We electroporated the shRNA-2 at E14.5 and studied progenitors proliferation 2 days later at E16.5. We saw no apparent anomalies in the percentage of mitotic cells (PH3 staining) nor in the percentage of cells actively engaged in the cell cycle (Ki67 staining) (Figures S1E–S1H). We also assessed cell death and saw no increase in the cleaved Caspase-3-positive cells electroporated with the shRNA (data not shown).
TBC1D23 Mutation Is Associated with Trafficking Defects
TBC1D23 is a Rab-specific GTPase-activating-protein (Rab-GAP). As a member of the TBC domain protein family, it contains the conserved Tbc domain known to inactivate RABs by accelerating their GTPase activity and thus promoting their inactive GDP-bound state.16, 17 As for RABs, they are membrane trafficking proteins highly conserved in eukaryotic cells that function in a variety of vesicle trafficking processes. Hence by controlling the activation state of Rabs, Rab-GAPs play an important role in vesicular trafficking.
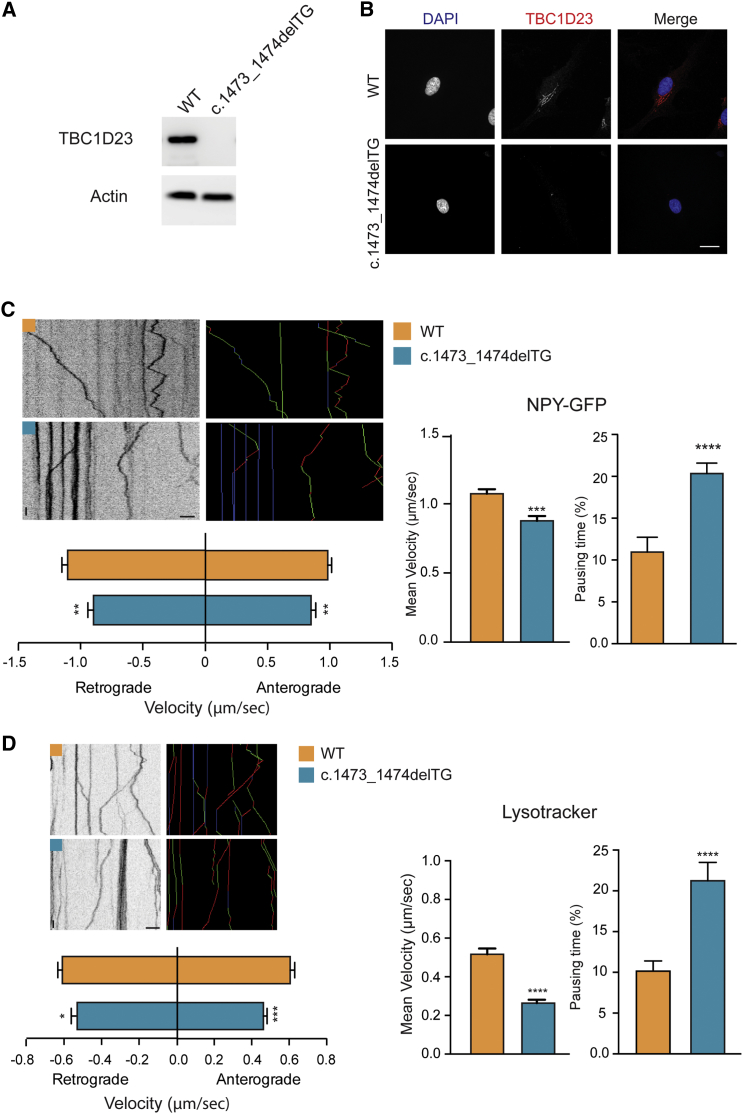
Thus, we wondered about the consequences of the TBC1D23 mutations on intracellular trafficking. To this end, we utilized available human fibroblasts from the individual harboring the TBC1D23 p.Val492Glyfs∗8 variant. We first characterized the protein levels of TBC1D23 in mutant fibroblasts and compared them to control fibroblasts. The truncating mutation leads to a lack of TBC1D23 as shown by western blot analysis (Figure 4A) and immunostaining in human fibroblasts (Figure 4B). We then transfected these cells with the dense core vesicle-resident neuropeptide Y tagged to GFP (NPY-GFP) and used live-cell imaging in order to study the dynamics of these vesicles. In control fibroblasts, NPY-GFP-containing vesicles processively moved both in the anterograde as in the retrograde directions, with vesicles changing directions and pausing, and with the presence of static vesicles as shown by the kymographs. In mutant fibroblasts, we found that the average velocity of NPY-containing vesicles was significantly decreased. The presence of the mutation leads to an increase in the time vesicles spend at pause and a reduction in the instantaneous velocity of particles moving in both directions (Figure 4C). We further validated these results by extending these observations to BDNF vesicles, also known to be present in dense core vesicles (data not shown). Rab-GTPases control the activation state of Rabs which in turn are known to be associated specifically to different vesicular compartments. However, whether TBC1D23 regulates one particular Rab is still unknown. Therefore, to establish whether TBC1D23 mutations alter specifically dense core vesicle dynamics, or whether the effect is general, we imaged and analyzed lysosomal movement. In line with the observations in dense core vesicles, the presence of TBC1D23 mutation also leads to less dynamic lysosomes with altered anterograde and retrograde transport (Figure 4D).
Figure 4.
TBC1D23 Truncating Mutation Alters Intracellular Transport
(A) Immunoblot from human fibroblasts showing the absence of TBC1D23 in the presence of the mutation.
(B) Immunofluoresence staining against endogenous TBC1D23 of control and mutant fibroblasts showing the lack of the protein in mutants. Scale bar, 20 μm.
(C) Kymographs and colored kymographs (green, anterograde; blue, static; red, retrograde) showing the trajectories of single NPY-GFP vesicles. Histograms represent mean ± SEM of average velocity (t = 3.864; p = 0.0028; n control = 220, n mutant = 362), %pausing time (t = 4.448; p < 0.0001; n control = 59, n mutant = 98), and instantaneous velocity (anterograde: t = 2.668; p = 0.0049; n control = 271, n mutant = 152; retrograde: t = 3.086; p = 0.0041; n control = 267, n mutant = 173) of NPY-GFP vesicles. Scale bars: 5 μm and 5 s.
(D) Kymographs and colored kymographs (green, anterograde; blue, static; red, retrograde) showing the trajectories of single lysosomes. Histograms represent mean ± SEM of average velocity (t = 6.119; p < 0.0001; n control = 224, n mutant = 137), %pausing time (t = 4.600; p < 0.0001; n control = 190, n mutant = 137), and instantaneous velocity (anterograde: t = 4.545; p < 0.0001; n control = 318, n mutant = 218; retrograde: t = 2.223; p = 0.0264; n control = 394, n mutant = 245) of lysosomes. Scale bars: 5 μm and 5 s.
In order to confirm the presence of such trafficking defect in neurons, we conducted similar experiments in mouse primary cortical neurons. Neurons were transfected with the shRNA-2 against Tbc1d23 and BDNF vesicles were tracked. Results were similar to the ones obtained in fibroblasts showing reduced instant and average velocity in both retrograde and anterograde transport and increased pausing time for neurons transfected with the shRNA-2 (Figure S2).
TBC1D23 Downregulation Affects Neurite Outgrowth
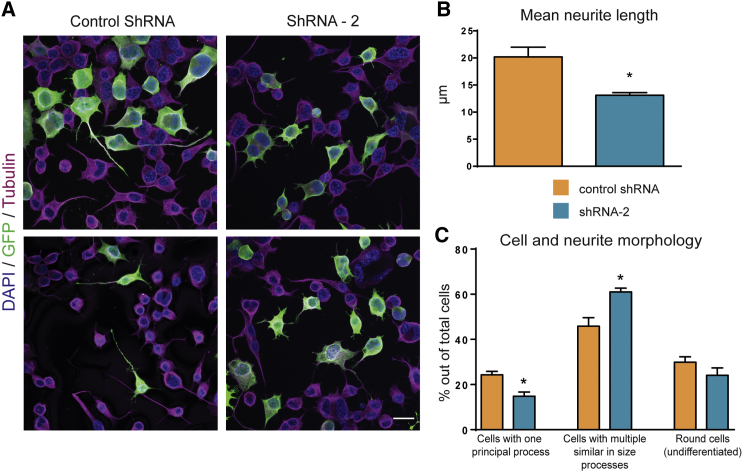
Given the effect of Tbc1d23 downregulation on neuronal positioning in vivo and the phenotype of altered vesicular trafficking in both fibroblasts and primary neurons, we assessed the implication of TBC1D23 in neurite outgrowth, since membrane trafficking and cargo delivery are essential for axonal and dendritic growth. To this end we used Neuro2a cells that we nucleofected with either the control shRNA or shRNA-2 targeting TBC1D23 and then cultured on serum-deprived medium in order to induce differentiation. Transfected cells were visualized with co-transfection of the GFP-derived Venus protein and processes were visualized with immunolabelling for tubulin. We measured the length of individual neurites and the number of neurites per cell. We also focused on the presence or the absence of a principal axon-like process. In the control condition, around 25% of the transfected GFP-positive cells had one principal axon-like process and one or several additional shorter processes (Figure 5). When N2a cells were nucleofected with the shRNA-2, fewer cells with this morphology could be observed (14.8%) (Figure 5C). The majority of the cells presented with multiple neurites similar in size (61%) with no principal process. Thus, when we measured the mean length of all neurites, we found a significantly higher value for the control condition (20.2 μm) than for the shRNA condition (13.1 μm) (Figure 5B). Additionally, cells transfected with the shRNA-2 tended to have more neurites per cell.
Figure 5.
TBC1D23 Downregulation Alters Neurite Outgrowth
Cells were electroporated with either control shRNA or shRNA-2 along with GFP, and then cultured in DMEM without serum to induce differentiation and neurites formation. After 48 hr cells were fixed in 4% PFA and immunostained for GFP, tubulin, and DAPI.
(A) Representative images of Neuro2a cells transfected with either control shRNA (left) or shRNA-2 targeting TBC1D23 (right); scale bar, 20 μm.
(B) Histograms representing the average neurite length (control shRNA versus shRNA-2, p = 0.0186). Data are represented as mean ± SEM.
(C) Cells were sorted in one of three categories: cells with one principal axon-like process; cells with multiple similar in size processes and round (most likely undifferentiated) cells. Histograms represent mean ± SEM percentage of each category out of the total cells counted. shRNA-2 versus control, p = 0.0167 (cells with one principal process), p = 0.0219 (cells with multiple similar in size processes).
Discussion
We report on three consanguineous families (seven affected individuals) with recessive forms of ID associated with PCH. In the three families, we identified homozygous truncating mutations (one splice and two frameshift mutations) predicted to lead to TBC1D23 loss of function. The common core phenotype of these individuals includes developmental milestone delay, microcephaly, and severe ID, but no history of seizure disorder. Additional features such as cerebellar syndrome, stereotypic movements, and coloboma were noticed in some subjects (Table 1). The postnatal course was also marked by a harmonious growth retardation and a common phenotype that included happy facial expression and dysmorphic features such as large ears, bulbous nasal tip, and eye abnormalities (Figure 1A). Moreover, brain MRI showed a small, normally proportioned cerebellum and that the corpus callosum was hypoplastic (Figure 1B). Interestingly, another distinctive cerebellum shape has been observed in individuals with TSEN54 mutations, namely a cerebellum with flattened cerebral hemispheres and a relatively preserved vermis.4, 7 Altogether these findings along with those reported by Marin-Valencia et al.18 suggest that TBC1D23-related PCH and associated clinical symptoms could not fit any of the reported PCH subtypes highlighted in the current classification of PCH.1 In addition, no mutation in the previously described PCH-related genes was found through analysis of exome data from each individual subjected to exome sequencing. It is worth mentioning the recently reported homozygous truncating mutation in TBC1D23 in individuals of a consanguineous family with severe non-syndromic ID.19 Though the type of the described mutation (truncated) is similar to those reported in this study and one can therefore predict similar resulting phenotypes, no MRI data were reported.
TBC1D23 belongs to the Tre2-Bub2-Cdc16 (TBC) family, a recently described family of proteins that act as Rab inhibitors.17, 20 Two TBC1D23 isoforms have been described, which differ by adjunction of an exon 15 of 45 bp in the longer form. The longer form contains 19 exons and encodes a 699 amino acid protein that contains two known functional domains (Figure 2). Expression of TBC1D23 transcripts seems to be relatively ubiquitous in human tissues, including the brain.
Although further investigations are required to assess consequences of the mutations described in this study on mRNA stability, it is worth mentioning that they are all truncating mutations including the splice mutation predicted to lead to exon16 skipping. The loss of the reading frame and resulting gained stop codons is predicted to trigger nonsense-mediated mRNA decay (NMD) as the premature termination codon occurred before the 50–55 nucleotides of the last exon-exon junction.21 This is in line with our observations of the lack of TBC1D23 in individuals’ fibroblasts.
The association between neurodevelopmental defects and TBC1D23 truncating mutations contrasts with the screening assay using RNAi that pinpointed a potential implication of TBC1D23 in the regulation of innate immune system.22 This potential finding was further investigated by the same group using one mutant mouse model developed by the International Gene Trap Consortium with a gene trap cassette inserted in the first intron of Tbc1d23. Mutant mice presented with increased response to many Toll-like receptors as demonstrated by higher IL-6 level compared to control. Interestingly, this response was time dependent as reported by Victorino et al.23 Cellular assay using mutant Tbc1d23 altered at the highly conserved arginine residue (p.Arg50Ala; GenBank: NM_026254) located in the RAB-GAP domain showed that Tbc1d23 strongly affects innate immunity signaling.22 The immune function of TBC1D23 is likely to be through the Rab-GAP signaling pathway. Though it is reasonable to hypothesize that nonsense mutations reported here lead to loss of function as reported in the transgenic mice, no symptom of innate immunity disorder has been pointed out in the herein reported individuals with PCH. Neither were any neurological features described for the Tbc1d23 transgenic mice.22, 23 Possibly, such neurological defects may have escaped attention, as phenotypic analyses were targeted at the immune system. Alternatively, these gene-trap mutants do not manifest any neurological phenotype, because of the residual Tbc1d23 expression.22, 23
Our in utero electroporation results show that downregulation of Tbc1d23 causes a defect in the positioning of cortical projection neurons at pre- and early post-natal stages, suggesting that functional TBC1D23 is required for correct cortical development. In affected subjects, we observed no gyration or lamination anomalies, nor heterotopia, which are usually but not always present in cases of altered neuronal positioning during development.24 However, for three of the four individuals for whom MRI images were available, a hypoplasia of the corpus callosum was present. The corpus callosum is made by myelinated axons of cortical projection neurons whose bodies reside in the upper cortical layers, principally in layers II–III but also V and VI.25 Aberrations in the development of these projection neurons such as delay in migration could affect their specification as callosally projecting neurons and could potentially alter the correct formation of their projections which could be at the origin of a partial or complete agenesis of the corpus callosum.26 Additionally, our findings in Neuro2a cells suggest an implication of TBC1D23 in neurite extension and in particular in the formation of a principal axon-like process. Downregulation or absence of TBC1D23 in neurons could disrupt axonogenesis and thus the proper formation of corpus callosum. The exact mechanisms leading to the corpus callosum abnormalities in affected individuals, though, remains to be further investigated.
We saw no effect of Tbc1d23 knockdown on progenitor proliferation or cell death that could provide insights to understand the microcephaly phenotype observed in affected persons. However, in our shRNA experiment, silencing of Tbc1d23 was not complete as shown by the presence of protein in the immunoblotting assays (Figure S1A), while no TBC1D23 was detected in fibroblasts from affected individuals (Figure 4A). This residual TBC1D23 could explain why no proliferation effect was observed.
The precise function of TBC1D23 and its role during brain development is poorly understood. However, like many other members of Tre2-Bub2-Cdc16 (TBC) family, TBC1D23 has a GTPase-activating protein (GAP) domain that catalyzes GTP hydrolysis. The proposed mechanism of how TBC works is through a unique feature of the Tre2-Bub2-Cdc16 domain, which uses two catalytic residues, Arg and Gln, to stabilize the transition state and catalyze the GTP hydrolysis of RABs.27 Strikingly, TBC1D23 is classified as an unconventional TBC due to the lack of these two indispensable residues.20 However, some TBC proteins that lack the Arg or Gln residues are conserved between subgroups, suggesting evolution constraints.28 Nevertheless, our results showing altered vesicular trafficking associated with TBC1D23 downregulation and pathogenic mutations suggest a possible role for TBC1D23 in Rab regulation. The exact mechanism by which this occurs and the impact on the proper development of the brain is still to be uncovered, even though neurite extension and outgrowth anomalies could be one underlying mechanism. It would be relevant to further investigate the in vivo consequences of the altered intracellular transport on membrane reorganization, trophic factors release, and signaling in the context of TBC1D23 deficiency. TBC1D23 is also the only member of TBC family to possess a rhodanese domain. The function of this domain is still elusive, but thiosulfate sulfutransferase activity with cyanide detoxification has been reported.29, 30 Until recently no substrates were known for TBC1D23.20 Interestingly, TBC1D23 is a central partner of an interacting network of proteins implicating WDR62, STK11, UBC, PRKAR2B, TBC1D8, KIAA1033, and VPS26A. Some of these proteins are also involved in neurodevelopmental disorders.31 Additionally, recessive mutations in the related TBC1D24 (MIM: 613577) were shown to be involved in a large spectrum of neurodevelopmental disorders, which include focal seizures with mild to moderate ID and brain abnormalities. MRI of individuals reported with mutations in TBC1D24 showed subtle cortical thickening with cerebellar atrophy.32 Moreover, implication of Tre2-Bub2-Cdc16 (TBC) proteins in developmental processes is also illustrated by association between mutations of TBC1D1 (MIM: 609850) and congenital anomalies of the kidneys and urinary tract (CAKUT [MIM: 610805]).33 It is well established that most of the previously reported genes responsible for PCH are involved in essential processes in protein synthesis in general and tRNA processing in particular.1, 7 Though these latter functions were not described for Tre2-Bub2-Cdc16 (TBC) proteins, in view of the findings reported in this study, one can question whether TBC1D23 as well could be implicated in one of these two pathways.
In this paper, we describe seven individuals in three families presenting with PCH and harboring homozygous nonsense or splice mutations in TBC1D23, likely to cause a loss of function. This work widened the role of TBC1D23 to a more severe and global phenotype in humans. We strongly believe that TBC1D23 is an additional gene responsible for PCH of a yet to be discovered mechanism. Based on these data, it appears legitimate to propose TBC1D23 as a PCH-associated disorder gene and to suggest molecular screening in individuals presenting syndromic PCH and negatives for known genes responsible for this disorder.
Acknowledgments
We are grateful to the individuals and their families for their participation. This work was supported by funding from Strasbourg University and the grant ANR-10-LABX-0030-INRT, a French State Fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02, the Fondation pour la Recherche Médicale (Equipe FRM; J.C–DEQ20130326477), the Fondation Maladies Rares, Agence Nationale de Recherche (ANR-ERare-012-01 - project E10107KP; ANR-13-BSV-0009-01), the EU-FP7 project GENECODYS (grant number 241995), and DESIRE (grant agreement 602531). Part of this work was also supported by a grant from the Iranian National Science Foundation. We thank the Imaging Center of IGBMC (ici.igbmc.fr), in particular Basile Gurchenkov and Elvire Guiot for their assistance in the microscope imaging experiments.
Published: August 17, 2017
Footnotes
Supplemental Data include Supplemental Note and two figures and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.07.010.
Accession Numbers
The accession numbers for the TBC1D23 c.1473_1474delTG variant reported in this work is ClinVar: CSV000537309, for the TBC1D23 c.1526delinsAA variant is ClinVar: CSV000537310, and for the TBC1D23 c.1687+2T>G variant is ClinVar: CSV000537311.
Web Resources
Allen Brain Atlas, http://www.brain-map.org/
BioGRID, http://thebiogrid.org/
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
Gencodys, http://www.gencodys.eu/
Human Splicing Finder, http://www.umd.be/HSF/
International Gene Trap Consortium, http://www.genetrap.org/
MaxENt Scan, http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html
NNsplice, https://omictools.com/nnsplice-tool
OMIM, http://www.omim.org/
Primer-BLAST, https://www.ncbi.nlm.nih.gov/tools/primer-blast/
STRING 9.0, http://www.string-db.org/
The Human Protein Atlas, http://www.proteinatlas.org/
Supplemental Data
References
- 1.Rudnik-Schöneborn S., Barth P.G., Zerres K. Pontocerebellar hypoplasia. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:173–183. doi: 10.1002/ajmg.c.31403. [DOI] [PubMed] [Google Scholar]
- 2.Eggens V.R., Barth P.G., Niermeijer J.M., Berg J.N., Darin N., Dixit A., Fluss J., Foulds N., Fowler D., Hortobágyi T. EXOSC3 mutations in pontocerebellar hypoplasia type 1: novel mutations and genotype-phenotype correlations. Orphanet J. Rare Dis. 2014;9:23. doi: 10.1186/1750-1172-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinograd-Byk H., Sapir T., Cantarero L., Lazo P.A., Zeligson S., Lev D., Lerman-Sagie T., Renbaum P., Reiner O., Levy-Lahad E. The spinal muscular atrophy with pontocerebellar hypoplasia gene VRK1 regulates neuronal migration through an amyloid-β precursor protein-dependent mechanism. J. Neurosci. 2015;35:936–942. doi: 10.1523/JNEUROSCI.1998-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Namavar Y., Chitayat D., Barth P.G., van Ruissen F., de Wissel M.B., Poll-The B.T., Silver R., Baas F. TSEN54 mutations cause pontocerebellar hypoplasia type 5. Eur. J. Hum. Genet. 2011;19:724–726. doi: 10.1038/ejhg.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anttonen A.K., Hilander T., Linnankivi T., Isohanni P., French R.L., Liu Y., Simonović M., Söll D., Somer M., Muth-Pawlak D. Selenoprotein biosynthesis defect causes progressive encephalopathy with elevated lactate. Neurology. 2015;85:306–315. doi: 10.1212/WNL.0000000000001787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed M.Y., Chioza B.A., Rajab A., Schmitz-Abe K., Al-Khayat A., Al-Turki S., Baple E.L., Patton M.A., Al-Memar A.Y., Hurles M.E. Loss of PCLO function underlies pontocerebellar hypoplasia type III. Neurology. 2015;84:1745–1750. doi: 10.1212/WNL.0000000000001523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Namavar Y., Barth P.G., Kasher P.R., van Ruissen F., Brockmann K., Bernert G., Writzl K., Ventura K., Cheng E.Y., Ferriero D.M., PCH Consortium Clinical, neuroradiological and genetic findings in pontocerebellar hypoplasia. Brain. 2011;134:143–156. doi: 10.1093/brain/awq287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lax N.Z., Alston C.L., Schon K., Park S.M., Krishnakumar D., He L., Falkous G., Ogilvy-Stuart A., Lees C., King R.H. Neuropathologic Characterization of Pontocerebellar Hypoplasia Type 6 Associated With Cardiomyopathy and Hydrops Fetalis and Severe Multisystem Respiratory Chain Deficiency due to Novel RARS2 Mutations. J. Neuropathol. Exp. Neurol. 2015;74:688–703. doi: 10.1097/NEN.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson C., Davies J.H., Lamont L., Foulds N. Early pontocerebellar hypoplasia with vanishing testes: A new syndrome? Am. J. Med. Genet. A. 2011;155A:667–672. doi: 10.1002/ajmg.a.33897. [DOI] [PubMed] [Google Scholar]
- 10.Mochida G.H., Ganesh V.S., de Michelena M.I., Dias H., Atabay K.D., Kathrein K.L., Huang H.T., Hill R.S., Felie J.M., Rakiec D. CHMP1A encodes an essential regulator of BMI1-INK4A in cerebellar development. Nat. Genet. 2012;44:1260–1264. doi: 10.1038/ng.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh A.P., Lukic V., Pope K., Bromhead C., Tankard R., Ryan M.M., Yiu E.M., Sim J.C., Delatycki M.B., Amor D.J. Complete callosal agenesis, pontocerebellar hypoplasia, and axonal neuropathy due to AMPD2 loss. Neurol Genet. 2015;1:e16. doi: 10.1212/NXG.0000000000000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karaca E., Weitzer S., Pehlivan D., Shiraishi H., Gogakos T., Hanada T., Jhangiani S.N., Wiszniewski W., Withers M., Campbell I.M., Baylor Hopkins Center for Mendelian Genomics Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell. 2014;157:636–650. doi: 10.1016/j.cell.2014.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valence S., Garel C., Barth M., Toutain A., Paris C., Amsallem D., Barthez M.A., Mayer M., Rodriguez D., Burglen L. RELN and VLDLR mutations underlie two distinguishable clinico-radiological phenotypes. Clin. Genet. 2016;90:545–549. doi: 10.1111/cge.12779. [DOI] [PubMed] [Google Scholar]
- 14.Tabata H., Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- 15.Longair M.H., Baker D.A., Armstrong J.D. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- 16.Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta - Rev. Cancer. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci. Rep. 2011;31:159–168. doi: 10.1042/BSR20100112. [DOI] [PubMed] [Google Scholar]
- 18.Marin-Valencia I., Gerondopoulos A., Zaki M.S., Ben-Omran T., Almureikhi M., Demir E., Guemez-Gamboa A., Gregor A., Issa M.Y., Appelhof B. Homozygous mutations in TBC1D23 lead to a non-degenerative form of pontocerebellar hypoplasia. AJHG. 2017;101:441–450. doi: 10.1016/j.ajhg.2017.07.015. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harripaul R., Vasli N., Mikhailov A., Rafiq M.A., Mittal K., Windpassinger C., Sheikh T.I., Noor A., Mahmood H., Downey S. Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol. Psychiatry. 2017 doi: 10.1038/mp.2017.60. Published online April 11, 2017. [DOI] [PubMed] [Google Scholar]
- 20.Frasa M.A., Koessmeier K.T., Ahmadian M.R., Braga V.M. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat. Rev. Mol. Cell Biol. 2012;13:67–73. doi: 10.1038/nrm3267. [DOI] [PubMed] [Google Scholar]
- 21.Lykke-Andersen S., Jensen T.H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 22.De Arras L., Yang I.V., Lackford B., Riches D.W., Prekeris R., Freedman J.H., Schwartz D.A., Alper S. Spatiotemporal inhibition of innate immunity signaling by the Tbc1d23 RAB-GAP. J. Immunol. 2012;188:2905–2913. doi: 10.4049/jimmunol.1102595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Victorino F., Alper S. Identifying novel spatiotemporal regulators of innate immunity. Immunol. Res. 2013;55:3–9. doi: 10.1007/s12026-012-8344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barkovich A.J., Guerrini R., Kuzniecky R.I., Jackson G.D., Dobyns W.B. A developmental and genetic classification for malformations of cortical development: update 2012. Brain. 2012;135:1348–1369. doi: 10.1093/brain/aws019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fame R.M., MacDonald J.L., Macklis J.D. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011;34:41–50. doi: 10.1016/j.tins.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards T.J., Sherr E.H., Barkovich A.J., Richards L.J. Clinical, genetic and imaging findings identify new causes for corpus callosum development syndromes. Brain. 2014;137:1579–1613. doi: 10.1093/brain/awt358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan X., Eathiraj S., Munson M., Lambright D.G. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- 28.Gabernet-Castello C., O’Reilly A.J., Dacks J.B., Field M.C. Evolution of Tre-2/Bub2/Cdc16 (TBC) Rab GTPase-activating proteins. Mol. Biol. Cell. 2013;24:1574–1583. doi: 10.1091/mbc.E12-07-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krepinsky K., Leimkühler S. Site-directed mutagenesis of the active site loop of the rhodanese-like domain of the human molybdopterin synthase sulfurase MOCS3. Major differences in substrate specificity between eukaryotic and bacterial homologs. FEBS J. 2007;274:2778–2787. doi: 10.1111/j.1742-4658.2007.05811.x. [DOI] [PubMed] [Google Scholar]
- 30.Remelli W., Guerrieri N., Klodmann J., Papenbrock J., Pagani S., Forlani F. Involvement of the Azotobacter vinelandii rhodanese-like protein RhdA in the glutathione regeneration pathway. PLoS ONE. 2012;7:e45193. doi: 10.1371/journal.pone.0045193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riazuddin S., Hussain M., Razzaq A., Iqbal Z., Shahzad M., Polla D.L., Song Y., van Beusekom E., Khan A.A., Tomas-Roca L., UK10K Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol. Psychiatry. 2016 doi: 10.1038/mp.2016.109. Published online July 26, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balestrini S., Milh M., Castiglioni C., Lüthy K., Finelli M.J., Verstreken P., Cardon A., Stražišar B.G., Holder J.L., Jr., Lesca G. TBC1D24 genotype-phenotype correlation: Epilepsies and other neurologic features. Neurology. 2016;87:77–85. doi: 10.1212/WNL.0000000000002807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosfeld A., Kreuzer M., Daniel C., Brand F., Schäfer A.K., Chadt A., Weiss A.C., Riehmer V., Jeanpierre C., Klintschar M. Whole-exome sequencing identifies mutations of TBC1D1 encoding a Rab-GTPase-activating protein in patients with congenital anomalies of the kidneys and urinary tract (CAKUT) Hum. Genet. 2016;135:69–87. doi: 10.1007/s00439-015-1610-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.