Abstract
Pontocerebellar hypoplasia (PCH) represents a group of recessive developmental disorders characterized by impaired growth of the pons and cerebellum, which frequently follows a degenerative course. Currently, there are 10 partially overlapping clinical subtypes and 13 genes known mutated in PCH. Here, we report biallelic TBC1D23 mutations in six individuals from four unrelated families manifesting a non-degenerative form of PCH. In addition to reduced volume of pons and cerebellum, affected individuals had microcephaly, psychomotor delay, and ataxia. In zebrafish, tbc1d23 morphants replicated the human phenotype showing hindbrain volume loss. TBC1D23 localized at the trans-Golgi and was regulated by the small GTPases Arl1 and Arl8, suggesting a role in trans-Golgi membrane trafficking. Altogether, this study provides a causative link between TBC1D23 mutations and PCH and suggests a less severe clinical course than other PCH subtypes.
Keywords: TBC1D23, pontocerebellar hypoplasia, intellectual disability, microcephaly, ataxia
Main Text
Originally named by Brun in 1917,1 pontocerebellar hypoplasia (PCH) is a devastating neurological disorder characterized by impaired growth and/or degeneration of cerebral structures, primarily the pons and cerebellum. To date, ten different clinical subtypes of PCH have been described, the majority leading to a neurodegenerative course, manifesting with progressive intellectual and motor decline.2, 3 Treatments are only palliative and the prognosis is poor, as most affected individuals die during infancy or childhood. Despite the expansion of known genes associated with PCH, most individuals remain without genetic diagnosis, suggesting that additional causes remain to be identified.
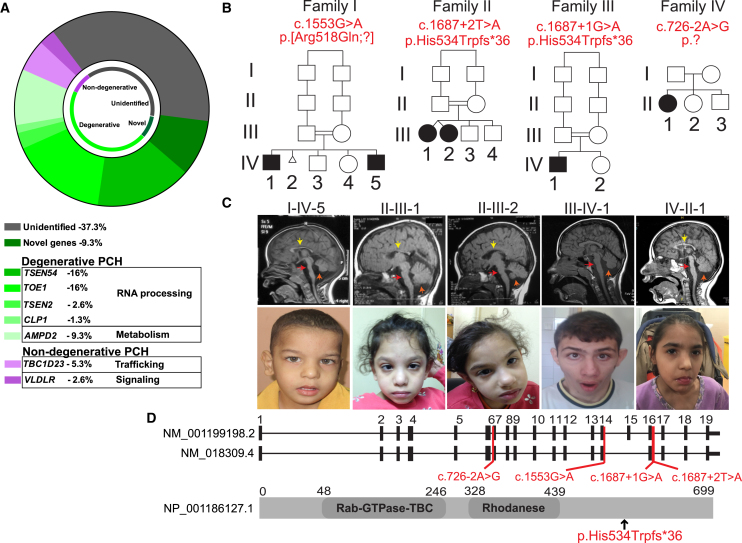
We recruited a cohort of 75 families with likely autosomal-recessive PCH, of which 53 (70.6%) documented parental consanguinity and 19 (25.3%) had two or more affected individuals. All affected members were clinically evaluated by a pediatric neurologist and geneticist, blood and/or saliva samples and skin biopsies were collected from participating individuals after obtaining proper informed consent, and DNA for whole-exome sequencing (WES) was extracted from at least one affected member of each family as described.4 The study followed the IRB guidelines and was approved by the ethical committees of UC San Diego, The Rockefeller University, and other participating institutions. In consanguineous families, we emphasized homozygous, rare (<0.1% allele frequency in our exome database of 5,000 individuals), and potentially damaging variants (Genomic Evolutionary Rate Profile [GERP] score > 4 or phastCons > 0.9).5 Likely causative mutations were identified in 47 families (62.6% of the total) (Figure 1A). Seven families (9.3%) had a likely causative variant in a gene not previously implicated in PCH. A total of 34 families (45.3%) carried mutations in genes previously associated with degenerative forms of PCH, encoding proteins involved in tRNA splicing, mRNA processing, and protein synthesis.5, 6, 7, 8 Six families (8%) demonstrated a non-degenerative course of PCH, and among these, TBC1D23 mutations were identified as a cause for this condition. TBC1D23 has been recently linked also with autosomal-recessive intellectual disability.9 We found six affected individuals with mutations in TBC1D23 from four unrelated families from Egypt (families I and II), Turkey (family III), and Lebanon (family IV) (Figures 1B and S1).
Figure 1.
Homozygous Mutations in TBC1D23 Lead to Pontocerebellar Hypoplasia
(A) Summary of exome sequencing results from a cohort of 75 families with pontocerebellar hypoplasia. Most PCH subtypes showed neurodegenerative features and the genes found in this group were primarily associated with RNA processing (TSEN54, TSEN2, TOE1, and CLP1) and with metabolism (AMPD2). In the non-degenerative group, TBC1D23 was the most frequent gene, accounting for 5.3% of families, and VLDLR was found in 2.6% of the families.
(B) Pedigrees showing consanguinity in three families. A total of six affected subjects were identified with different deleterious mutations in TBC1D23. Mutation in brackets has a presumed effect on splicing.
(C) Available midline sagittal T1-weighed MRIs and facies from affected individuals are shown. All individuals manifested severe hypoplasia of pons (red arrow) and moderate to severe hypoplasia of the cerebellum (orange arrow) and all but III-IV-1 had hypoplasia of corpus callosum (yellow arrow). All were microcephalic and none of them showed obvious signs of facial dysmorphism. Subject II-III-2 had head deviation due to neck hypotonia.
(D) TBC1D23 exons as ticks and location of mutations are indicated in both transcripts. A schematic of the protein structure is illustrated along with the protein mutation of families II and III.
Family I presented with two affected boys of 16 years (I-IV-1) and 2 years (I-IV-5) of age, family II presented with two non-identical girl twins of 4 years of age (II-III-1 and II-III-2), family III presented with one boy of 14 months of age (III-IV-1), and family IV presented with a girl of 6 months of age (IV-II-1) (Figure 1B). Some of these individuals manifested reduced head circumference at birth (≥−2 SD standard deviations [SD] below the mean) and all showed signs of global psychomotor deficits since early infancy, involving gross and fine motor skills, language (expressive > receptive), and social interaction due to communication impairment (Table 1). In the most recent clinical evaluation, all subjects were microcephalic (≥−3 SD), and height and weight ranged from normal to −5 SD. Neurological exam was remarkable for generalized weakness (6/6 affected subjects), global hypotonia (5/6), and cerebellar deficits such as uncoordinated limb movements (4/6), hyporeflexia (3/6), and impaired or no ambulation (6/6). Brainstem symptoms including dysphagia and dysarthria were present in subjects from family I. None of the six individuals manifested clinical signs of neurological deterioration and, hitherto, they are all alive. Brain MRI showed pontocerebellar hypoplasia in all subjects, along with thin corpus callosum (I-IV-5, II-III-1, II-III-2, IV-II-1) and cortical hypoplasia (I-IV-5, II-III-1, II-III-2) (Figure 1C). The radiological findings of subject III-IV-1 did not change appreciably over a 3-year interval (Figure S2), which is consistent with the non-progressive course of this form of PCH.
Table 1.
Description of Clinical Findings of Individuals with TBC1D23 Mutations
| I-IV-1 | I-IV-5 | II-III-1 | II-III-2 | III-VI-1 | IV-II-1 | |
|---|---|---|---|---|---|---|
| Mutation (genomic hg19) | chr3:g.100029386G>A | chr3:g.100029386G>A | chr3:g.100035033T>A | chr3:g.100035033T>A | chr3:g.100035032G>A | chr3:g.100014144A>G |
| Mutation (cDNA) (NM_001199198.2) | c.1553G>A | c.1553G>A | c.1687+2T>A | c.1687+2T>A | c.1687+1G>A | c.726−2A>G |
| Mutation (protein) | p.[Arg518Gln;?] | p.[Arg518Gln;?] | p.His534Trpfs∗36 | p.His534Trpfs∗36 | p.His534Trpfs∗36 | p.? |
| Gestational age | 40 | 38 | 37 | 37 | N/A | N/A |
| Weight at birth (kg) | 3 (−0.90 SD) | 2.8 (−1.22 SD) | 1.6 (−3.47 SD) | 1.5 (−3.66 SD) | 3.8 (+0.49 SD) | 8.8 at age 2 years (−3 SD) |
| Length at birth (cm) | 50 (−0.06 SD) | 48 (−0.81 SD) | 48 (−0.65 SD) | 47 (−1.10 SD) | N/A | N/A |
| HC at birth (cm) | 32.2 (−1.62 SD) | 32 (−1.71 SD) | 32 (−1.90 SD) | 31.5 (−2.24 SD) | 42 at 14 months (−3.84 SD) | 41.2 at 2 years (−4.4 SD) |
| Age at diagnosis | delayed since infancy, but first seen at 16 years of age | 2 years | delayed since infancy, but first seen at 4 years of age | delayed since infancy, but first seen at 4 years of age | 14 months | 6 months |
| Weight (kg), age at last examination | 38, 16 years (−2.65 SD) | 14, 6 years (−2.96 SD) | 14.5, 4 years (−0.79 SD) | 11.5, 4 years (−2.62 SD) | N/A | 11.9, 7.5 years (−4.11 SD) |
| Height (cm), age at last examination | 143, 16 years (−3.56 SD) | 103, 6 years (−2.44 SD) | 103, 4 years (+0.50 SD) | 97, 4 years (−0.89 SD) | N/A | 98, 7.5 years (−4.89 SD) |
| HC (cm), age at last examination | 48, 16 years (−4.77 SD) | 44.5, 6 years (−5.27 SD) | 43.5, 4 years (−3.96 SD) | 41.5, 4 years (−5.25 SD) | 50, 16 years (−3.4 SD) | 42, 7.5 years (−7.77 SD) |
| Psychomotor Development | ||||||
| Gross motor | delayed; can walk alone | delayed; sits only | delayed; can walk alone | delayed; walks supported | delayed | delayed |
| Fine motor | delayed | delayed | delayed | delayed | absent | delayed |
| Language | delayed | delayed | delayed | delayed | absent | delayed (babbling at 7 years of age) |
| Social | delayed | delayed | delayed | delayed | absent | delayed |
| Regression of acquired milestones | – | – | – | – | – | – |
| Neurological Findings | ||||||
| Brainstem findings | dysarthria. no history of apnea, hearing deficit, dizziness, or dysphagia | minimal dysphagia. no history of apnea, hearing deficit, or dizziness. | – | – | N/A | – |
| Cerebellar deficits | truncal and appendicular ataxia | truncal and appendicular ataxia | truncal and appendicular ataxia | truncal and appendicular ataxia | – | – |
| Muscle strength (scale 0->5 in upper and lower extremities) | grade 4/5 | grade 4/5 | grade 4/5 | grade 4/5 | grade 5/5 | grade 3/5 |
| Muscle tone | hypotonia | hypotonia | hypotonia | hypotonia | normal muscle tone | hypotonia; reduced muscle tone at 2 years, but developed spasticity especially in lower limbs at 7 years. |
| Deep tendon reflexes | normal | normal | hyporeflexia | hyporeflexia | normal | hyporeflexia; last examined at 2 years of age |
| Gait | wide base unsteady gait, ataxia | can sit and only stand supported with wide base gait | wide base gait, ataxia | non-ambulatory | wide base gait | crawling, unsupported sitting, and cannot bear weight in left limb. |
| Seizures | ||||||
| Onset | – | – | – | – | 11 years | 2 years |
| Type | – | – | – | – | focal seizures with secondary generalization | myoclonic seizures |
| Other Systemic Findings | recurrent respiratory infections, sepsis, muscle atrophy | recurrent respiratory infections, sepsis | strabismus, recurrent respiratory infections, hypoplasia of labia minora | strabismus, recurrent respiratory infections, muscle atrophy, hypoplasia of labia minora | esotropia of left eye, proximal interphalangeal joint contractures | – |
Abbreviations are as follows: plus sign (+), present; minus sign (-) absent, (N/A) not assessed. SD, standard deviation; HC, head circumference. Mutations in brackets have a presumed effect on splicing.
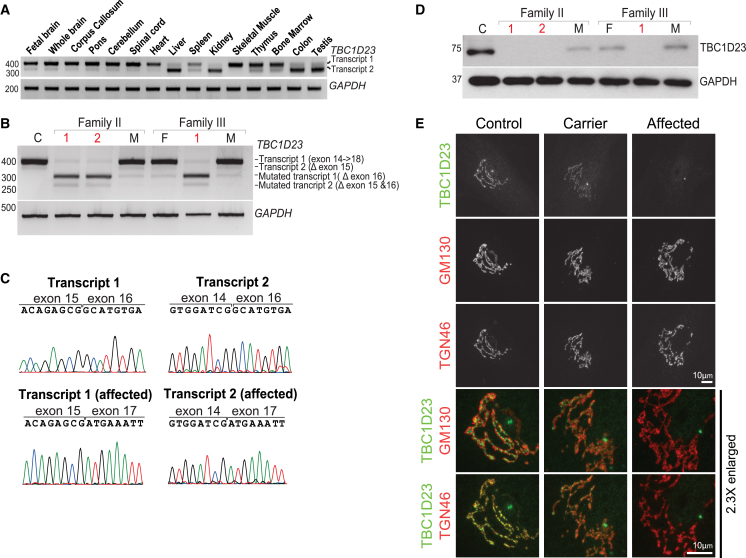
All six affected individuals carried mutations predicted to result in altered splicing, occurring at or near canonical splice sites (Figure 1D; Table 1), so we used genome build hg19 and transcript GenBank: NM_001199198.2 (transcript 1) to annotate splicing effects. We found that both TBC1D23 transcripts were differently expressed in human tissues, with transcript 1 primarily expressed in the fetal and adult brain and spinal cord (Figure 2A). To study potential effects of the variants on splicing, we used RT-PCR to amplify annotated transcripts from fibroblasts of families II and III, who carried mutations c.1687+2T>A and c.1687+1G>A, respectively (primers are available upon request). Affected individuals had shorter transcripts relative to control individuals (Figure 2B), and sequencing of the amplified PCR products confirmed that shorter transcripts had skipped exon 16 (Figure 2C), leading to a shift in the reading frame and truncated protein (p.His534Trpfs∗36) (Figures 1D and 2D). Fibroblasts from families I and IV were not available to assess splicing. Family IV carried a variant in a canonical splice site (c.726−2A>G) and family I carried a missense mutation at the last base of exon 14 (c.1553G>A), both of which were also expected to compromise splicing. Cellular localization of endogenous TBC1D23 was examined in control, carrier, and affected individuals’ fibroblasts using specific antibodies. In control fibroblasts, TBC1D23 overlapped with the trans-Golgi marker TGN46 and showed signal adjacent to the cis-Golgi marker GM130 (Figure 2E). This trans-Golgi staining pattern of TBC1D23 was absent in cells from affected individuals, whereas cells from carriers showed reduced staining intensity. Despite the loss of detectable TBC1D23 in affected subjects (Figures 2D and 2E), there was no obvious alteration in the relative positions of the cis/trans-Golgi markers or the ribbon-like structure of the Golgi (Figure 2E).
Figure 2.
Splicing Mutations in TBC1D23 Cause Splicing Defects and Prevent Protein Synthesis
(A) RT-PCR of both TBC1D23 transcripts in different human tissues reveals that transcript 1 is predominantly expressed in the central nervous system.
(B) RT-PCR from control (C), unaffected parents (F, father; M, mother), and affected subjects (1 and 2 from family II, 1 from family III). Parents (carriers) showed splicing defect in the mutated allele, represented by the lack of exon 16 in both transcripts. Homozygous individuals manifested splicing defect in both alleles.
(C) Sequencing of PCR bands confirms the absence of exon 16 in both mutant transcripts.
(D) Western blot shows that TBC1D23 is absent in affected individuals and decreased in carriers compared to control.
(E) Representative immunofluorescence images of fibroblasts from control, carrier, and affected. In control and carrier fibroblasts, TBC1D23 (rabbit 17002; Proteintech, 1:1,000) is located toward the trans-Golgi network as it overlaps with TGN46 (sheep; AbD Serotec; 1:1,000) and not with GM130 (mouse clone 35; BD; 1:1,000). In agreement with western blot results, cells from the affected individual did not show signal except non-specific staining of the centrosome. The enlarged region shows details of TBC1D23 with the different Golgi markers. Scale bars represent 10 μm.
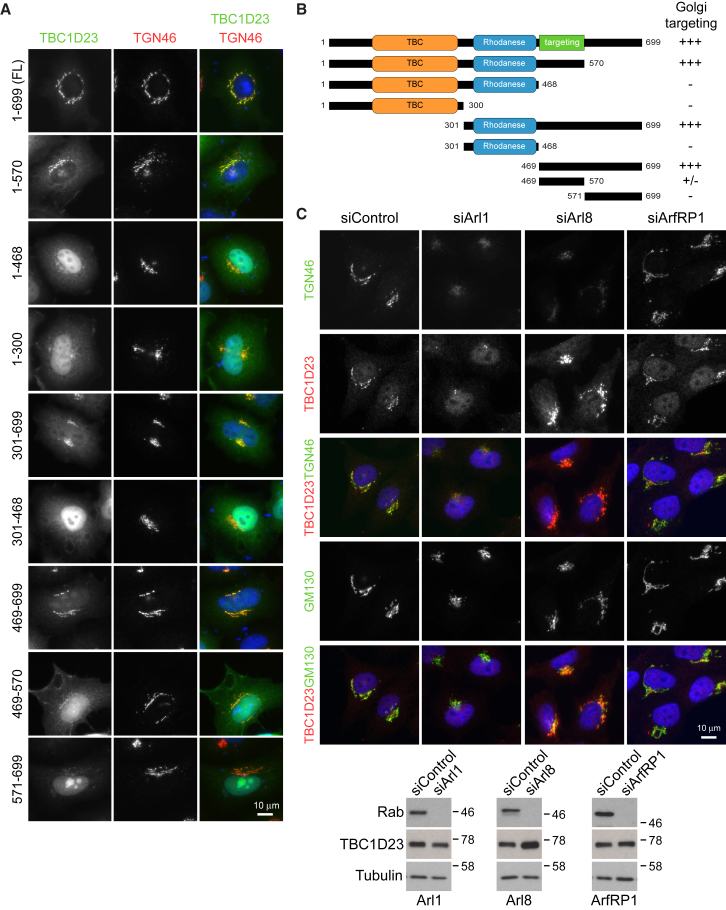
TBC1D23 belongs to a family of Tre2-Bub2-Cdc16 (TBC) domain-containing Rab-specific GTPase-activating proteins (TBC/RabGAPs) that regulate membrane trafficking by inactivating Rabs.10, 11, 12 Most TBC/RabGAPs contain two catalytic residues, Arg and Gln, to stimulate the hydrolysis of GTP in Rab proteins.13, 14 TBC1D23 falls in the category of unconventional TBC/RabGAPs since it lacks the catalytic Arg-Gln residues and it might, a priori, work through a different mechanism to induce GTP hydrolysis14 or it might have a Rab-independent function. When compared to TBC1D20, which acts on Rab1, none of the 55 Rabs tested showed robust activation of GTP hydrolysis in the presence of purified TBC1D2315 (Figure S3). This raised the possibility that TBC1D23 is a Rab-binding protein, or effector, rather than a Rab regulator and it may target to the Golgi via this means. However, two lines of evidence argue against this. First, a region in TBC1D23 (469–570 aa), C-terminal to the TBC1 and Rhodanese domains, is responsible for its targeting to the trans-Golgi (Figures 3A and 3B). Second, TBC1D23 remains associated with trans-Golgi membranes when Rabs are depleted, with just a subset (Rab1a/b, Rab2a/b, Rab6a/b, Rab7a, Rab14a/b) giving rise to altered TBC1D23 localization due to their effects on Golgi structure or trafficking to and from the Golgi (Figure S3). This suggested a Rab-independent targeting mechanism.
Figure 3.
Trans-Golgi Location of TBC1D23 Depends on the C-Terminal Region and Is Regulated by Arl1 and Arl8
(A and B) HeLa cells were transfected using Mirus LT1 (Mirus LCC) with full-length GFP-tagged TBC1D23 (A) or with deletion constructs outlined in the schematic (B). After 20 hr of the transfection, cells were fixed with 3% PFA (wt/vol) for 15 min, permeabilized with 0.1% Triton X-100 (vol/vol) for 7 min, and then stained for TGN46 using a standard protocol. TBC1D23 was visualized using GFP. As illustrated, the region between amino acids 469 and 570 is responsible for the targeting of TBC1D23 to the trans-Golgi.
(C) HeLa cells were transfected using Oligofectamine (Life Technologies) with a specific library of siRNA duplexes targeting Arfs and Arls (Dharmacon) for 72 hr (a subset of this library is shown). Cells were then fixed and stained as mentioned previously with antibodies against TBC1D23, TGN46, and GM130. The depletion of Arl1 caused complete loss of TBC1D23 from the trans-Golgi, whereas knocking down Arl8 increased TBC1D23 staining at the trans-Golgi. Depletion of ArfRP1, which controls Arl1 targeting to the Golgi, did not alter TBC1D23 expression or localization. Western blot demonstrates efficient knockdown of targeted Rabs by siRNA duplexes and shows the resulting expression of TBC1D23. In this case, HeLa cells were transfected with siRNA duplexes to Arl1, ArfRP1, and Arl8 or non-specific control for 72 hr and transfected with expressing EGFP-tagged Arl1, ArfRP1, and Arl8 20 hr before collection for western blot. Antibodies against EGFP (raised against full-length GFP in sheep) and Tubulin (mouse DM1A; Sigma-Aldrich) were used.
Scale bars represent 10 μm.
Like Rabs, Ras superfamily GTPases of the Arl and Arf group are known to be involved in recruitment of cytosolic proteins to membrane surfaces. Strikingly, depletion of Arl1, but not ArfRP1 or other Arfs or Arls, resulted in the complete loss of TBC1D23 from the trans-Golgi (Figure 3C). Conversely, knocking down Arl8 resulted in elevated staining for TBC1D23 at the trans-Golgi (Figure 3C). These findings connect TBC1D23 to an Arl1-dependent trafficking process at the trans-Golgi16, 17, 18, 19 and to Arl8 function in the endosome-lysosome system.20, 21, 22 Thus far, these two Arls have not been associated with human disease and the impact of their interaction with TBC1D23 on brain development requires additional studies.
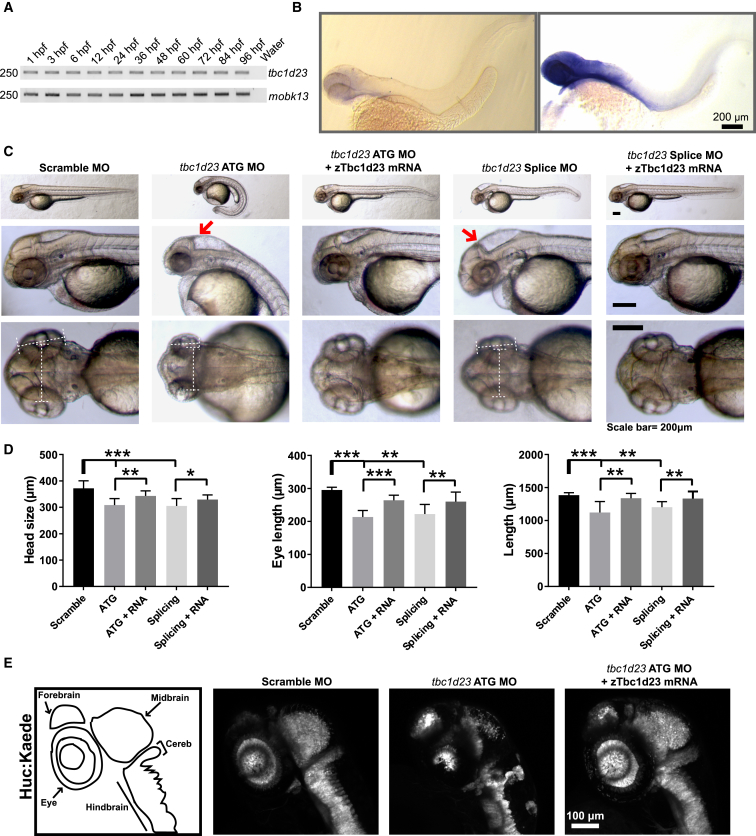
To investigate the role of TBC1D23 in brain development, we designed a zebrafish model of disease. A single tbc1d23 ortholog (GenBank: NM_200487) encodes a protein with 77% identity with the human TBC1D23 amino acid sequence. tbc1d23 transcript was detected as early as the first hours post-fertilization (hpf) by RT-PCR, and by in situ RNA hybridization tbc1d23 was primarily localized in the head at 48 hpf (Figures 4A and 4B), suggesting a role in brain development. To test whether knockdown tbc1d23 in zebrafish replicates the human phenotype, we knocked down tbc1d23 using a translation blocking morpholino targeting the ATG start codon (tbc1d23-ATG MO; 5′-CTTCCCCTACAGCATCCGCCATTGC-3′) and a splice blocking morpholino targeting intron 4 to exon 5 (tbc1d23-splice MO; 5′-GCAGTCTCTGCAAAAGGCAATATGC-3′). In contrast to scramble MO, both ATG and splice MO-injected embryos (3 ng each) had reduced brain and eye size and manifested curved tails at 48 hpf (more severe in ATG MO embryos), and this phenotype was partially rescued with injection of zebrafish tbc1d23 mRNA (Figures 4C and 4D). These findings were corroborated in Tg(HuC:Kaede) transgenic zebrafish line, which expresses the fluorescent protein Kaede in neurons. The ATG MO injected Tg(HuC:Kaede) zebrafish showed reduced signal in the neural axis and manifested altered morphology of forebrain, brainstem, and cerebellum relative to scramble MO embryos (Figure 4E). Altogether, tbc1d23 disruption in zebrafish replicates the human phenotype by impairing brain growth and development.
Figure 4.
A Zebrafish Model Reproduces the Human Phenotype
(A) RT-PCR shows expression of zebrafish tbc1d23 at different developmental stages relative to control mobk13.23
(B) In situ RNA hybridization in AB embryos at 48 hr post-fertilization (hpf) with sense (negative control) and antisense (signal) probes for tbc1d23. The expression of tbc1d23 occurs primarily in the head, pointing out its potential role in the development of cerebral structures.
(C) Representative images of fishes injected with scramble, ATG, and splicing morpholinos (MO, all 3 ng) and rescued with zTbc1d23 mRNA (150–200 ng). Red arrows show reduced size of cerebellum and brainstem along with enlarged IV ventricle in ATG and splicing MO fishes relative to scramble MO and rescued fishes.
(D) Quantification of morphometric parameters (9–11/condition). Reduced head and eye size and length of both ATG and splicing MO fish was partially rescued with zTbc1d23 mRNA (n = 10/condition).
(E) Transgenic line Huc:Kaede (CNS label) showed reduced signal and size of the neural axis of ATG MO compared to scramble MO, which improved in the rescued fish. Drawings on the left illustrate the brain regions labeled in each transgenic fish.
All numerical values were expressed as mean ± SEM and statistical analysis was performed using two-tailed Student’s t test. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars in (B) and (C) represent 200 μm, and in (E) represents 100 μm.
This study enhances the genetic diagnosis and expands the phenotypic spectrum of PCH. In contrast to most subtypes, individuals with TBC1D23-associated PCH did not show clinical neurological deterioration and MRI findings did not worsen over time. None of the affected individuals have died so far and the oldest are currently 16 years old (I-IV-1 and III-IV-1). This distinctive clinical course is therefore highly valuable for family counseling and prognostication since most individuals with other PCH subtypes show progressive worsening and typically succumb during infancy or early childhood.2 TBC1D23 individuals shared some neurological manifestations with other forms of PCH, such as psychomotor impairment, microcephaly, brainstem deficits, and ataxia.2 In addition to severe volume loss of pons and cerebellum, TBC1D23 individuals manifested hypoplasia of cortex and of corpus callosum as seen in other forms of PCH3 (Figure 1C). At the systemic level, the most common findings in all affected subjects were recurrent respiratory infections and even sepsis (Table 1). This could relate to the essential role of the brainstem to swallowing function and handling respiratory secretions. However, it has been reported that TBC1D23 may have inhibitory effects on innate immunity and on LPS-induced cytokine release in mice,24 and we cannot exclude the possibility that loss of TBC1D23 could exacerbate the inflammatory response against bacterial infections and lead to more severe clinical manifestations. At baseline, affected individuals did not show significant increase of inflammatory markers and cytokine levels compared to control subjects (Table S1). Thus, more studies are necessary to evaluate the immune system of these individuals in order to determine their inflammatory response to infections.
Thus far, deleterious mutations in two TBC/RabGAPs genes have been implicated in disorders of brain development: TBC1D24 (MIM: 613577), which causes focal and familial infantile myoclonic epilepsy,25, 26, 27 and TBCK (MIM: 616899), which has been associated with severe infantile syndromic encephalopathy.28, 29, 30 Like TBC1D23, both TBC1D24 and TBCK are part of the unconventional group of TBC/RabGAPs since they lack the Arg and/or Gln fingers of the TBC domain. TBC1D24 regulates neuronal migration and maturation by inactivating ADP ribosylation factor (ARF) 6, a small GTPase involved in vesicle trafficking.26 Whether ARF6 inactivation occurs by direct or indirect induction of GTP hydrolysis is still unknown. On the other hand, the target GTPase of TBCK has not been yet identified. TBCK regulates cell growth and proliferation by modulating transcription of several constituents of the mTOR pathway.30 Structural and molecular modeling analyses of mutations in the TBC domain suggests that TBCK may also have Rab GAP activity and that the loss of GAP function is associated with disease.28 Our results point toward a Rab-independent trafficking role of TBC1D23 that may be critical during hindbrain formation, but not contributory to degeneration. Ivanova et al.31 showed that vesicle trafficking in fibroblasts from affected individuals lacking TBC1D23 is significantly slower than control subjects. How this finding relates to disruption of brain development needs to be elucidated.
Another PCH subtype that is not associated with neurodegeneration is PCH8.32 This condition is caused by mutations in CHMP1A (MIM: 164010), a gene that regulates proliferation of neural progenitor cells. CHMP1A has two potential functions—as a charged multivesicular body protein and as a chromatin-modifying protein33, 34—such that it links cytoplasmic signals with chromatin modifications to regulate proliferation of progenitor cells.32 Individuals with mutations in CHMP1A manifest severe hypoplasia of pons and cerebellum and cortical atrophy, and none of them have shown progression of the clinical and radiological findings, suggesting that this is a developmental and not a degenerative disorder.32 Another non-degenerative form of PCH not included in the current classification is VLDLR-associated PCH (MIM: 192977). Along with apoE receptor 2 (ApoER2), VLDLR serves as a Reelin receptor to regulate microtubule function in migrating neurons.35 Individuals with damaging mutations in VLDLR manifest non-progressive cerebellar hypoplasia with flattened pons and cortical dysplasia.36, 37 The majority of PCH subtypes, however, are associated with disruption of protein synthesis, for example by altering RNA processing (i.e., TSEN genes, RARS2, VRK1, TOE1, CLP1)6, 8, 38, 39, 40 or GTP-dependent protein synthesis (i.e., AMPD2),41 functions that are critical for brain development and considered causative of degeneration when disrupted.2, 7 In PCH, how mutations in genes that regulate protein synthesis cause neurodegeneration whereas genes involved in trafficking and signaling impairs primarily brain development, and why all these genes preferentially involve the hindbrain are questions that remain unsolved. Whether there are key molecular pathways involved in hindbrain formation where these genes converge and lead to a pontocerebellar phenotype is a matter that needs further investigation.
Acknowledgments
The authors thank all families for participation in this study. We thank Tessa van Dijk for coordinating data gathering from family 4572. Thanks to the Rockefeller and UCSD Microscopy Cores (P30 NS047101) for imaging support. I.M.-V. was sponsored by Pilot Grant awarded by the Center for Basic and Translational Research on Disorders of the Digestive System at The Rockefeller University through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust. We thank the Broad Institute (U54HG003067 to E. Lander and UM1HG008900 to D. MacArthur) and the Yale Center for Mendelian Disorders (U54HG006504 to R. Lifton and M. Gunel). This work was supported by NIH grants P01HD070494, 1R01NS098004, R01NS048453, R01NS052455, and UL1TR001866 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program, the Simons Foundation Autism Research Initiative (275275), Howard Hughes Medical Institute (to J.G.G.), Qatar National Research Foundation NPRP 6-1463-3-351 (to T.B.-O. and J.G.G.), NIH grant K99NS089943 (to A.G.-G.), American Academy of Neurology Clinical Research Training Scholarship 2017-205 (to I.M.-V.), Joshua Deeth Foundation (to F.B.), and a Wellcome Trust Senior Investigator Award 097769/Z/11/Z (to F.A.B.).
Published: August 17, 2017
Footnotes
Supplemental Data include three figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.07.015.
Accession Numbers
The accession number for the TBC1D23 sequence reported in this paper is Genbank: NM_001199198.2.
Web Resources
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
GME Variome, http://igm.ucsd.edu/gme
Mutation Assessor, http://mutationassessor.org/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PROVEAN, http://provean.jcvi.org
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Brun R. Zur Kenntnis der Bildungsfehler des Kleinhirns. Epikritische Bemerkungen zur Entwicklungspathologie, Morphologie und Klinik der umschriebenen Entwicklungshemmungen des Neozerebellums. Schweiz. Arch. Neurol. Psychiatr. 1917;1:48–105. [Google Scholar]
- 2.Rudnik-Schöneborn S., Barth P.G., Zerres K. Pontocerebellar hypoplasia. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:173–183. doi: 10.1002/ajmg.c.31403. [DOI] [PubMed] [Google Scholar]
- 3.Namavar Y., Barth P.G., Poll-The B.T., Baas F. Classification, diagnosis and potential mechanisms in pontocerebellar hypoplasia. Orphanet J. Rare Dis. 2011;6:50. doi: 10.1186/1750-1172-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon-Salazar T.J., Silhavy J.L., Udpa N., Schroth J., Bielas S., Schaffer A.E., Olvera J., Bafna V., Zaki M.S., Abdel-Salam G.H. Exome sequencing can improve diagnosis and alter patient management. Sci. Transl. Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akizu N., Cantagrel V., Zaki M.S., Al-Gazali L., Wang X., Rosti R.O., Dikoglu E., Gelot A.B., Rosti B., Vaux K.K. Biallelic mutations in SNX14 cause a syndromic form of cerebellar atrophy and lysosome-autophagosome dysfunction. Nat. Genet. 2015;47:528–534. doi: 10.1038/ng.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaffer A.E., Eggens V.R., Caglayan A.O., Reuter M.S., Scott E., Coufal N.G., Silhavy J.L., Xue Y., Kayserili H., Yasuno K. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell. 2014;157:651–663. doi: 10.1016/j.cell.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Namavar Y., Barth P.G., Kasher P.R., van Ruissen F., Brockmann K., Bernert G., Writzl K., Ventura K., Cheng E.Y., Ferriero D.M., PCH Consortium Clinical, neuroradiological and genetic findings in pontocerebellar hypoplasia. Brain. 2011;134:143–156. doi: 10.1093/brain/awq287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budde B.S., Namavar Y., Barth P.G., Poll-The B.T., Nürnberg G., Becker C., van Ruissen F., Weterman M.A., Fluiter K., te Beek E.T. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat. Genet. 2008;40:1113–1118. doi: 10.1038/ng.204. [DOI] [PubMed] [Google Scholar]
- 9.Harripaul R., Vasli N., Mikhailov A., Rafiq M.A., Mittal K., Windpassinger C., Sheikh T.I., Noor A., Mahmood H., Downey S. Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol. Psychiatry. 2017 doi: 10.1038/mp.2017.60. Published online April 11, 2017. [DOI] [PubMed] [Google Scholar]
- 10.Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci. Rep. 2011;31:159–168. doi: 10.1042/BSR20100112. [DOI] [PubMed] [Google Scholar]
- 12.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 13.Pan X., Eathiraj S., Munson M., Lambright D.G. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- 14.Frasa M.A., Koessmeier K.T., Ahmadian M.R., Braga V.M. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat. Rev. Mol. Cell Biol. 2012;13:67–73. doi: 10.1038/nrm3267. [DOI] [PubMed] [Google Scholar]
- 15.Haas A.K., Fuchs E., Kopajtich R., Barr F.A. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat. Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- 16.Panic B., Perisic O., Veprintsev D.B., Williams R.L., Munro S. Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol. Cell. 2003;12:863–874. doi: 10.1016/s1097-2765(03)00356-3. [DOI] [PubMed] [Google Scholar]
- 17.Wu M., Lu L., Hong W., Song H. Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat. Struct. Mol. Biol. 2004;11:86–94. doi: 10.1038/nsmb714. [DOI] [PubMed] [Google Scholar]
- 18.Lu L., Horstmann H., Ng C., Hong W. Regulation of Golgi structure and function by ARF-like protein 1 (Arl1) J. Cell Sci. 2001;114:4543–4555. doi: 10.1242/jcs.114.24.4543. [DOI] [PubMed] [Google Scholar]
- 19.Lu L., Tai G., Hong W. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-golgi network. Mol. Biol. Cell. 2004;15:4426–4443. doi: 10.1091/mbc.E03-12-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donaldson J.G., Jackson C.L. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakae I., Fujino T., Kobayashi T., Sasaki A., Kikko Y., Fukuyama M., Gengyo-Ando K., Mitani S., Kontani K., Katada T. The arf-like GTPase Arl8 mediates delivery of endocytosed macromolecules to lysosomes in Caenorhabditis elegans. Mol. Biol. Cell. 2010;21:2434–2442. doi: 10.1091/mbc.E09-12-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pu J., Schindler C., Jia R., Jarnik M., Backlund P., Bonifacino J.S. BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell. 2015;33:176–188. doi: 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y., Xie S., Yao J. Identification of novel reference genes suitable for qRT-PCR normalization with respect to the zebrafish developmental stage. PLoS ONE. 2016;11:e0149277. doi: 10.1371/journal.pone.0149277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Arras L., Yang I.V., Lackford B., Riches D.W., Prekeris R., Freedman J.H., Schwartz D.A., Alper S. Spatiotemporal inhibition of innate immunity signaling by the Tbc1d23 RAB-GAP. J. Immunol. 2012;188:2905–2913. doi: 10.4049/jimmunol.1102595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corbett M.A., Bahlo M., Jolly L., Afawi Z., Gardner A.E., Oliver K.L., Tan S., Coffey A., Mulley J.C., Dibbens L.M. A focal epilepsy and intellectual disability syndrome is due to a mutation in TBC1D24. Am. J. Hum. Genet. 2010;87:371–375. doi: 10.1016/j.ajhg.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falace A., Buhler E., Fadda M., Watrin F., Lippiello P., Pallesi-Pocachard E., Baldelli P., Benfenati F., Zara F., Represa A. TBC1D24 regulates neuronal migration and maturation through modulation of the ARF6-dependent pathway. Proc. Natl. Acad. Sci. USA. 2014;111:2337–2342. doi: 10.1073/pnas.1316294111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falace A., Filipello F., La Padula V., Vanni N., Madia F., De Pietri Tonelli D., de Falco F.A., Striano P., Dagna Bricarelli F., Minetti C. TBC1D24, an ARF6-interacting protein, is mutated in familial infantile myoclonic epilepsy. Am. J. Hum. Genet. 2010;87:365–370. doi: 10.1016/j.ajhg.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong J.X., Caputo V., Phelps I.G., Stella L., Worgan L., Dempsey J.C., Nguyen A., Leuzzi V., Webster R., Pizzuti A., University of Washington Center for Mendelian Genomics Recessive inactivating mutations in TBCK, encoding a Rab GTPase-activating protein, cause severe infantile syndromic encephalopathy. Am. J. Hum. Genet. 2016;98:772–781. doi: 10.1016/j.ajhg.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhoj E.J., Li D., Harr M., Edvardson S., Elpeleg O., Chisholm E., Juusola J., Douglas G., Guillen Sacoto M.J., Siquier-Pernet K. Mutations in TBCK, encoding TBC1-domain-containing kinase, lead to a recognizable syndrome of intellectual disability and hypotonia. Am. J. Hum. Genet. 2016;98:782–788. doi: 10.1016/j.ajhg.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Yan X., Zhou T. TBCK influences cell proliferation, cell size and mTOR signaling pathway. PLoS ONE. 2013;8:e71349. doi: 10.1371/journal.pone.0071349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanova E.L., Mau-Them F.T., Riazuddin S., Kahrizi K., Laugel V., Schaefer E., de Saint Martin A., Runge K., Iqbal Z., Marie-Aude S. Homozygous truncating variants in TBC1D23 cause pontocerebellar hypoplasia and alter cortical development. Am. J. Hum. Genet. 2017;101:428–440. doi: 10.1016/j.ajhg.2017.07.010. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mochida G.H., Ganesh V.S., de Michelena M.I., Dias H., Atabay K.D., Kathrein K.L., Huang H.T., Hill R.S., Felie J.M., Rakiec D. CHMP1A encodes an essential regulator of BMI1-INK4A in cerebellar development. Nat. Genet. 2012;44:1260–1264. doi: 10.1038/ng.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howard T.L., Stauffer D.R., Degnin C.R., Hollenberg S.M. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 2001;114:2395–2404. doi: 10.1242/jcs.114.13.2395. [DOI] [PubMed] [Google Scholar]
- 34.Stauffer D.R., Howard T.L., Nyun T., Hollenberg S.M. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J. Cell Sci. 2001;114:2383–2393. doi: 10.1242/jcs.114.13.2383. [DOI] [PubMed] [Google Scholar]
- 35.Hiesberger T., Trommsdorff M., Howell B.W., Goffinet A., Mumby M.C., Cooper J.A., Herz J. Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron. 1999;24:481–489. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 36.Sonmez F.M., Gleeson J.G., Celep F., Kul S. The very low density lipoprotein receptor-associated pontocerebellar hypoplasia and dysmorphic features in three Turkish patients. J. Child Neurol. 2013;28:379–383. doi: 10.1177/0883073812441065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozcelik T., Akarsu N., Uz E., Caglayan S., Gulsuner S., Onat O.E., Tan M., Tan U. Mutations in the very low-density lipoprotein receptor VLDLR cause cerebellar hypoplasia and quadrupedal locomotion in humans. Proc. Natl. Acad. Sci. USA. 2008;105:4232–4236. doi: 10.1073/pnas.0710010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lardelli R.M., Schaffer A.E., Eggens V.R., Zaki M.S., Grainger S., Sathe S., Van Nostrand E.L., Schlachetzki Z., Rosti B., Akizu N. Biallelic mutations in the 3′ exonuclease TOE1 cause pontocerebellar hypoplasia and uncover a role in snRNA processing. Nat. Genet. 2017;49:457–464. doi: 10.1038/ng.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edvardson S., Shaag A., Kolesnikova O., Gomori J.M., Tarassov I., Einbinder T., Saada A., Elpeleg O. Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am. J. Hum. Genet. 2007;81:857–862. doi: 10.1086/521227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renbaum P., Kellerman E., Jaron R., Geiger D., Segel R., Lee M., King M.C., Levy-Lahad E. Spinal muscular atrophy with pontocerebellar hypoplasia is caused by a mutation in the VRK1 gene. Am. J. Hum. Genet. 2009;85:281–289. doi: 10.1016/j.ajhg.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akizu N., Cantagrel V., Schroth J., Cai N., Vaux K., McCloskey D., Naviaux R.K., Van Vleet J., Fenstermaker A.G., Silhavy J.L. AMPD2 regulates GTP synthesis and is mutated in a potentially treatable neurodegenerative brainstem disorder. Cell. 2013;154:505–517. doi: 10.1016/j.cell.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.