Abstract
In five separate families, we identified nine individuals affected by a previously unidentified syndrome characterized by growth retardation, spine malformation, facial dysmorphisms, and developmental delays. Using homozygosity mapping, array CGH, and exome sequencing, we uncovered bi-allelic loss-of-function CDK10 mutations segregating with this disease. CDK10 is a protein kinase that partners with cyclin M to phosphorylate substrates such as ETS2 and PKN2 in order to modulate cellular growth. To validate and model the pathogenicity of these CDK10 germline mutations, we generated conditional-knockout mice. Homozygous Cdk10-knockout mice died postnatally with severe growth retardation, skeletal defects, and kidney and lung abnormalities, symptoms that partly resemble the disease’s effect in humans. Fibroblasts derived from affected individuals and Cdk10-knockout mouse embryonic fibroblasts (MEFs) proliferated normally; however, Cdk10-knockout MEFs developed longer cilia. Comparative transcriptomic analysis of mutant and wild-type mouse organs revealed lipid metabolic changes consistent with growth impairment and altered ciliogenesis in the absence of CDK10. Our results document the CDK10 loss-of-function phenotype and point to a function for CDK10 in transducing signals received at the primary cilia to sustain embryonic and postnatal development.
Keywords: CDK10, growth retardation, ETS2, cilia, metabolism, spine malformation, Al Kaissi syndrome knockout mice, congenital disorder
Introduction
Individuals with severe growth retardation arising from developmental defects often suffer from additional medical problems that make diagnosing these diseases difficult. Here, we studied nine individuals (from five families) who display a closely matched array of phenotypes characterized by severe growth retardation, spine malformation, facial dysmorphisms, developmental delay, and intellectual disability. Using next-generation sequencing, we discovered that all affected individuals harbor germline homozygous mutations in cyclin-dependent kinase 10 (CDK10 [MIM: 603464]).
Consisting of 20 members in mammals, the group of protein kinases known as CDKs play critical roles in cell-cycle control, transcription, and development.1, 2 CDKs (CDK1–CDK6) mostly promote the progression from one cell-cycle phase to the next, and CDK1 is essential for proliferation. CDKs have been characterized extensively, and some of their substrates and in vivo functions are known.1, 3 Another group of CDKs, e.g., CDK7–CDK9 and CDK11–CDK13, are mostly involved in transcription. Most of the remaining CDKs are “orphan” kinases, which have been poorly studied. CDK10 is an orphan kinase that was cloned on the basis of its homology to CDK1.4, 5 CDK10 binds the transcription factor ETS2 and modulates its transactivation.6 CDK10 kinase activity is regulated by the binding of CDK10 to cyclin M, which is the product of FAM58A (MIM: 300708).7 CDK10/cyclin M complexes also have been shown to phosphorylate ETS2, leading directly to degradation of ETS2. X-linked mutations in FAM58A have been reported to cause STAR syndrome (MIM: 300707).8, 9 Recently, it was revealed that CDK10/cyclin M complexes phosphorylate the protein kinase PKN2, which affects cilia growth.10 In zebrafish, morpholino-mediated silencing of cdk10 impaired neurogenesis by modulating raf1a expression.11 In addition, CDK10 was one locus implicated by a genome-wide association study (GWAS) for determining height in Han Chinese.12
Germline mutations in CDK genes are exceedingly rare with the exception of an activating germline mutation (encoding p.Arg24Cys) predisposing to malignant melanoma13, 14 in CDK4 (MIM: 123829) and CDK6 mutations that cause autosomal-recessive primary microcephaly 12 (MCPH12 [MIM: 616080]).15 To the best of our knowledge, no other loss-of-function mutations in any member of the CDK family have been linked to a Mendelian syndrome.
Material and Methods
Array CGH Analysis
The Human Genome CGH Microarray Kit 180k (Agilent Technologies) was used according to the manufacturer’s instructions. The data were analyzed by Agilent Cytogenomics software with the statistical algorithm ADM-2 and a 3-probe minimum aberration call and were compared with the Database of Genomic Variants. Coordinates of copy-number variations are based on the GRCh36/hg18 assembly.
Exome Sequencing
The exome library was prepared on an Ion OneTouch System and sequenced on an Ion Proton instrument (Life Technologies) with one Ion PI chip. Sequence reads were aligned to the human GRCh37/hg19 assembly (UCSC Genome Browser). Variants were filtered for common SNPs against the NCBI’s “common and no known medical impacts” database (ClinVar), the Exome Aggregation Consortium (ExAC) Browser, the NHLBI Exome Sequencing Project, and an in-house database of 518 sequenced samples. Exome sequencing of both affected individuals of the Tunisian family was carried out by Macrogen on the Illumina HiSeq 1000 platform with Illumina TruSeq Exome Enrichment.
Tomography Analysis of Skeleton
Micro-computed tomography (CT) scans were performed with a μCT 50 system (SCANCO Medical) operated at 70 kV with a 0.5 mm Al filter and 500 ms exposure time per projection. 1,000 projections were captured over 180°. Images with 7.4 μm resolution were obtained after 3D data reconstruction. The images were downsized with a 0.5 scaling factor, and a 3D mean filter was applied to reduce noise. The segmentation was performed with a global threshold. The 3D rendering and the measurement were performed with Amira software (v6.3, FEI Visualization Sciences Group). After globally observing the entire skeleton, we assessed the volume of five skeleton pieces, the length of the left femur, and the shape of the cervical vertebrae.
Histology
Different mouse tissues were isolated at postnatal day 0 (P0), fixed in 10% neutral buffered formalin (Sigma-Aldrich, HT501128), and transferred into 70% ethanol before being embedded in paraffin and sectioned. Histological sections were stained with hematoxylin and eosin.
RT-PCR and qPCR
Total RNA was extracted with the NucleoSpin RNA kit according to the manufacturer’s protocol (Macherey-Nagel, 740955). For each RT-PCR reaction, first-strand cDNA was generated from 1 μg of total RNA with the Maxima First Strand cDNA Synthesis Kit (Thermo Scientific, K1642). PCR amplification was carried out with the Maxima SYBR Green qPCR Master Mix (Fermentas, K0252) and the appropriate primer pair (Table S4). The reactions were monitored continuously in a Rotor-Gene thermal cycler (Corbett Research) according to the following program: 95°C for 10 min followed by 50 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. All data were normalized to the expression levels of housekeeping genes Eef2 (mouse) and GAPDH (human) according to the (2−ΔΔCt) method.
Exon Trapping Assay
Cloning and Plasmid Preparation
Exons 7–11 of CDK10 (GenBank: NM_052988) were cloned into the pET01 Exontrap vector from MoBiTec with primers flanked by restriction enzyme sites XhoI and BamHI (cdk10_pET01_XhoI_f and cdk10_pET01_BamHI_r, respectively). The vector and resulting PCR product were both digested with XhoI and BamHI (NEB), and the digested products were gel purified with the QIAquick Gel Extraction Kit (QIAGEN) and ligated with T4 DNA Ligase (Promega) overnight at 4°C. Resulting clones were verified by colony PCR with QIAGEN HotStarTaq Master Mix and primers pET01_cdk10_5r and pET01_cdk10_4f. Plasmids of two clones were purified with the EndoFree Plasmid Maxi Kit (QIAGEN), and the sequence was verified (see primers pENT01_cdk10_1/5-f/r in Table S4).
Cell Culture and Transfection
AD293 cells (Agilent) were transfected with 10 μg of plasmid DNA with the NEON Transfection System (Invitrogen) according to the following electroporation conditions: 1,100 V, 20 ms, and 2 pulse.
RNA Isolation and cDNA Screening
24 hr after transfection, cells were collected with 1 mL Trizol (Invitrogen), and RNA was isolated according to the manufacturer’s protocol. cDNA was generated with the QuantiTech Reverse Transcription Kit (QIAGEN). For verification of the mutational impact on the splice donor site of exon 9, target cDNA was amplified with primers ETPR06 and ETPR07 from the Exontrap kit and analyzed by Sanger sequencing with ChromasLite software (Technelysium) and the UCSC Genome Browser.16, 17
Homozygosity Mapping
Both affected individuals of the Tunisian family were genotyped on Affymetrix GeneChip Human Mapping 250K NspI arrays with dCHIP software18 at the Center for Medical Research at the Medical University of Graz.
Generation of Cdk10flox Mice and Other Transgenic Lines
The Cdk10 mouse strain was generated essentially as described previously for Cdk1flox.19 Mouse genomic DNA harboring the Cdk10 locus was isolated from the BAC clone pBACe3.6 RP23-297L8 (PKB983, Invitrogen). 5′ and 3′ retrieval homology arms were amplified by PCR (primers PKO1087 + PKO1091 for the 5′ arm and primers PKO1092 + PKO1093 for the 3′ arm) and subcloned into the pBlight-TK vector (PKB1009) according to the bacterial recombineering technique.20 For generation of the targeting vector, the obtained pBlight-TK plasmid containing the entire Cdk10 locus (PKB1015) was modified by the addition of a unique LoxP site in the intron downstream of exon 8 of Cdk10 and a FRT-LoxP-eng2SA-Neo-FRT-LoxP cassette in the intron upstream of exon 2 of Cdk10. The resulting targeting vector (PKB1037) was linearized by PvuI digestion and introduced into embryonic stem (ES) cells by electroporation. After positive and negative selection with Geneticin and ganciclovir, respectively, genomic DNA of surviving ES cell colonies was screened for homologous recombination by Southern hybridization using 5′ (290 bp) and 3′ (323 bp) external probes generated by PCR from BAC DNA with the use of primer pairs PKO1046 + PKO1047 and PKO1048 + PKO1049, respectively. Digest with restriction enzymes EcoRV and NotI and hybridization with the 5′ probe characterized clones that carried the wild-type (WT) band at 15.8 kb and the targeted band at 6.2 kb. Digest with NheI and hybridization with the 3′ probe resulted in clones that carried the WT band at 14.3 kb and the targeted band at 6.1 kb. Three correctly targeted ES cell clones (4021, 4023, and 4032) were selected for microinjection into blastocysts and derivation of chimeric mice for the generation of the Cdk10-conditional-knockout mouse strain.
Obtained mice carrying the eng2SA-neomycin cassette (Cdk10+/neo) were bred with β-actin-Flpe transgenic mice21 (strain B6.Cg-Tg(ACTFLPe) 9205Dym/J; stock no. 005703; the Jackson Laboratory) for the removal of the eng2SA-neomycin cassette and generation of the Cdk10flox allele. The Cdk10null allele, in which the deletion of exons 2–8 creates a frameshift, was the result of crossing Cdk10+/neo mice with β-actin-Cre transgenic mice22 (strain FVB/N-Tg(ACTB-cre)2Mrt/J; stock no. 003376; the Jackson Laboratory). Mice were housed under standard conditions with food and water available ad libitum and maintained on a 12 hr light-dark cycle. Mice were fed a standard chow diet containing 6% crude fat and were treated humanely in compliance with the guidelines of the institutional animal care and use committee at the Biological Resource Center (BRC) at Biopolis, Singapore.
Genotyping
For genotyping PCR of Cdk10 WT, flox, and null alleles, primers Pr1 (PKO1229), Pr2 (PKO2647), and Pr3 (PKO2648) (Table S4) were used at 1 μM final concentration. In brief, cells or tissue pieces to be genotyped were lysed by being boiled in lysis solution (25 mM NaOH [pH 12] and 0.2 mM EDTA) for 20–30 min for the extraction of genomic DNA.23 Alkaline pH was neutralized by the addition of an equal volume of neutralization buffer (40 mM Tris-HCl [pH 5]). 1 μL of the resultant genomic DNA solution was used as a template in 20 μL of PCR reaction with 0.5 units of DreamTaq polymerase (Bioline). 30 PCR cycles with 30 s denaturation at 94°C and 1 min annealing and extension at 62°C were performed for amplification of different alleles of Cdk10, resulting in a band of 283 bp (Cdk10WT), 398 bp (Cdk10flox), or 508 bp (Cdk10null).
Gene Expression Analysis
For microarray analysis, RNA from mouse tissue (kidney, lung, heart, brain, stomach, intestine, thymus, and liver) was isolated with the Invitrogen PureLink RNA Mini Kit. RNA was prepared with the Illumina TotalPrep RNA Amplification Kit according to the manufacturer’s protocol. The raw Illumina MouseRef-8 v2.0 Expression BeadChip array data were subjected to quality-control inspection, during which probes were removed for the following reasons: the probe quality was bad, the probe had no match, probe sequence was not from chromosomes 1–19, or the gene represented by the probe was not found in Entrez or Ensembl. The data intensity was not further normalized given that the gene expression levels were highly consistent within each organ (between the knockout and the heterozygous control). The expression data were converted into a ratio between the knockout and the control in each organ and log2 transformed for statistical analysis. A model-based method called EBprot24 was applied to ratio data from each organ and reported the posterior probability of differential expression and associated false-discovery rate (FDR). A gene was considered to be differentially expressed if it was statistically significantly different in at least two organs (FDR 1% and minimal fold change 50%). Enrichment of biological functions, represented by Gene Ontology and CPDB,25 was evaluated with an in-house implementation of a hypergeometric test.
Isolation and Culture of Primary Mouse Embryonic Fibroblasts
Primary mouse embryonic fibroblasts (MEFs) of WT (Cdk10+/+) and knockout (Cdk10−/−) genotypes were isolated from embryonic day 13.5 (E13.5) mouse embryos as described previously.19 In brief, the head and the visceral organs were removed, the embryonic tissue was chopped into fine pieces with a razor blade and trypsinized for 15 min at 37°C, and finally tissue and cell clumps were dissociated by pipetting. Cells were plated in a 10 cm culture dish (passage 0) and grown in HyClone high-glucose DMEM (GE Healthcare, SH30243) supplemented with 10% fetal bovine serum (GIBCO, 26140) and 1% penicillin-streptomycin (Invitrogen, 15140-122). Primary Cdk10+/+ and Cdk10−/− MEFs were immortalized by serial passaging 30 times according to a modified 3T3 protocol.26
Primary human dermal fibroblasts were isolated from fresh skin biopsies obtained from subject 6 and an ethnically matched healthy control individual. Fibroblasts were cultured in high-glucose DMEM supplemented with 1% L-glutamine, 10% fetal bovine serum, and 1% penicillin-streptomycin. Both primary and immortalized MEFs and human fibroblasts were cultured in a humidified incubator with 5% CO2 and 3% O2.
Proliferation Assay and FACS Analysis
For alamarBlue proliferation assays, 1,500 cells in a volume of 150 μL were plated per well of 96-well plates in 6 replicates for each clone. Starting 24 hr after seeding, cells were incubated for 4 hr in a final volume of 300 μL after the addition of 150 μL of assay medium (1:5 ratio of alamarBlue [AbD Serotec, BUF012B] to growth medium), and metabolic activity was quantified via measurement of the fluorescence at 585 nm.
For BrdU labeling and fluorescence-activated cell sorting (FACS) analysis, primary MEFs serum starved for 72 hr were trypsinized and replated in 10 cm dishes with DMEM supplemented with 10% fetal bovine serum for the induction of synchronized entry into the cell cycle. For monitoring S phase, cells were labeled with 100 μM BrdU (BD PharMingen, 550891) for 1 hr before being collected at different time points. At the end of each time point, cells were trypsinized and fixed in −20°C 70% ethanol and stained with anti-BrdU antibodies (BD PharMingen, 555627) followed by Alexa-Fluor-647-labeled goat-anti-mouse secondary antibodies (Invitrogen, A21235). Cells were counterstained for DNA content with propidium iodide (Sigma, 81845). Cell-cycle analysis was performed by FACS on a LSRII cytometer (BD Biosciences), and the resulting data were analyzed by FlowJo 8 software.
Cell Lysis, SDS-PAGE, and Immunodetection
Cells were lysed in Laemmli buffer (60 mM Tris-HCl [pH 6.8], 10% glycerol, 100 mM DTT, and 2% SDS) and supplemented with protease inhibitors (10 μg/mL each of leupeptin, chymostatin, and pepstatin [Chemicon EI8, EI6, and EI10, respectively]). 10 or 20 μg of protein extracts was separated on 9% or 8% polyacrylamide gels, transferred onto polyvinylidene difluoride membranes (Millipore, IPVH0010) with a semi-dry system, and blocked in Tris-buffered saline with 0.1% Tween 20 and 4% non-fat dry milk (Bio-Rad, 1706404). Blots were probed with the appropriate primary antibodies overnight at 4°C and subsequently with secondary goat-anti-mouse (Pierce, 0031432) or anti-rabbit antibodies (Pierce, 0031462) conjugated to horseradish peroxidase and were developed with an enhanced chemiluminescence reagent (PerkinElmer, NEL105001EA). The antibodies used were cyclin A2 (Santa Cruz, SC-596), total pRB (BD-PharMingen, 554136), HSP90 (BD-Transduction Laboratories, 610419), and Ets2 (Santa Cruz, SC351).
Generation of Antibodies against CDK10
Two peptide sequences corresponding to the C terminus (HHRNKRAAPAAAEGQSKRC) and T-loop (CIMLQVLRGLQYLHR) of CDK10 were synthesized by Keck Biotechnology Resource Laboratory and GL Biochem (Shanghai), respectively. The sequences were then coupled to mcKLH as previously described.27 1 mg of mcKLH-conjugated peptides was mixed with Freud’s complete adjuvant and injected subcutaneously into two rabbits on day 0. All subsequent immunizations on days 14, 35, 56, 77, 98, 119, 140, 161, 182, 203, 224, 245, and 266 were in the presence of Freud’s incomplete adjuvant. Rabbits were bled on days 0, 49, 70, 91, 112, 133, 154, 175, 196, 217, 238, 259, and 280, and sera were stored at –80°C. Serum from Blk319R was subsequently affinity purified through a column covalently coupled to both peptides, and it was then concentrated. All purified antibodies were reconstituted to a final concentration of 1 mg/mL in PBS and 50% glycerol and were stored at –20°C.
Immunofluorescence Microscopy
Cells were grown on coverslips (Lab Tek) before serum starvation. At different time points, cells were fixed with 4% PFA for 10 min. The fixed cells were permeabilized for 10 min with 0.2% Triton X-100 and exposed to a blocking solution over the course of 1 hr (PBS, 0.2% Triton X-100, and 2% BSA [Sigma]). Cells were stained with primary antibodies directed against Arl13B (ProteinTech Group, 17711-1-AP) and gamma-tubulin (Sigma-Aldrich, T6557) for 1 hr at room temperature and then incubated with Alexa-Fluor-488-labeled goat-anti-mouse (Invitrogen, A11029) and Alexa-Fluor-555-labeled goat-anti-rabbit (Invitrogen, A21428) secondary antibodies for 1 hr. Cells were counterstained with DAPI. Images were acquired at 100× magnification on an Olympus FluoView upright laser scanning confocal microscope. 15 confocal images of cilia were randomly captured for each experiment. Ciliary length of the longest cilium in each confocal image was analyzed with Olympus FluoView FV1000 v3.1 software. Statistical analysis was done with GraphPad Prism and a Student’s t test, and the average ciliary length was calculated.
Results
We report here the clinical, genetic, and molecular etiology of a syndrome that was independently uncovered by five research teams and investigated collaboratively with the use of GeneMatcher.28 The investigations were performed in accordance with the ethical standards of the responsible committee on human experimentation. Parents of all affected individuals provided written informed consent for genetic analyses of their affected children and themselves in accordance with the ethical standards of the institutional review boards.
CDK10 Mutations Cause a Growth-Retardation Syndrome
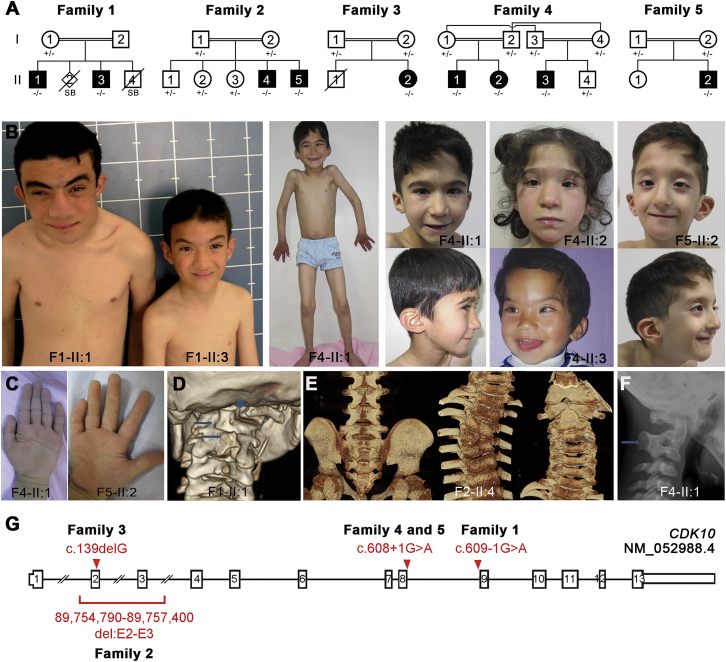
We have identified nine individuals with homozygous mutations in CDK10 from five unrelated families (Figure 1A). The detailed description of the clinical features can be found in the Supplemental Note, Table 1, and Table S1.
Figure 1.
Biallelic Germline Mutations in CDK10 in Nine Individuals from Five Consanguineous Families
(A) Pedigrees of families from Tunisia (1), Algeria (2), Saudi Arabia (3), and Turkey (4 and 5). The affected individuals carry homozygous germline mutations in CDK10. Open symbols represent unaffected individuals, and filled symbols represent affected individuals.
(B) Photographs of affected individuals show facial dysmorphisms.
(C) Hand anomalies with clinodactyly of the fifth finger and single transverse palmar crease.
(D) Severe mal-segmentation of the cervical vertebrae in proband F1-II:1.
(E) Lack of fusion of the posterior arches of S2–S5, right T4 hemivertebrae, and lack of fusion of the anterior arch of the atlas and partial fusion of C2 and C3 vertebrae in proband F2-II:4.
(F) Fusion of the cervical vertebrae in proband F4-II:1.
(G) Exon-intron structure of CDK10 on chromosome 16 shows the position of the four identified homozygous mutations.
Table 1.
Clinical Features of Nine Individuals with CDK10 Mutations
| Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | |||||
|---|---|---|---|---|---|---|---|---|---|
| CDK10 Mutation | |||||||||
| Genomic position (hg19) | chr16: 89,760,580 G>A | arr 16q24.3 89,754,790 −89,757,400 × 0 | chr16: 89,755,711delG | chr16: 89,759,876 G>A | chr16: 89,759,876 G>A | chr16: 89,759,876 G>A | chr16: 89,759,876 G>A | ||
| cDNA change | c.609−1G>A | ND | ND | c.139delG | c.608+1G>A | c.608+1G>A | c.608+1G>A | c.608+1G>A | |
| Demographic Data | |||||||||
| Individual (year of birth) | individual 1 (1997) | individual 2 (2002) | individual 3 (2009) | individual 4 (2011) | individual 5 (2013) | individual 6 (2008) | individual 7 (2010) | individual 8 (2013) | individual 9 (2008) |
| Sex | male | male | male | male | female | male | female | male | male |
| Individual ID | F1-II:1 | F1-II:3 | F2-II:4 | F2-II:5 | F3-II:2 | F4-II:1 | F4-II:2 | F4-II:3 | F5-II:2 |
| Ancestry | Tunisian | Tunisian | Algerian | Algerian | Saudi Arabian | Kurdish (from Turkey) | Kurdish (from Turkey) | Kurdish (from Turkey) | Turkish |
| Consanguinity | + | + | + | + | + | + | + | + | − |
| Growth Parameters | |||||||||
| Age at examination | 9 years | 4 years | 7 years 8 months | 4 years 11 months | 18 months | 8 years 11 months | 7 years 3 months | 3 years 9 months | 6 years 8 months |
| Height (SD) | 114 cm (−3) | 93 cm (−2.5) | 111.8 cm (−2.5) | 96 cm (−3) | 75 cm (−1.3) | 106 cm (−4.4) | 97.5 cm (−4.7) | 86 cm (−3.5) | 107 cm (−2.6) |
| Weight (SD) | 17 kg (−3) | 11.75 kg (−3) | 16.2 kg (−2.9) | 12.3 kg (−3.2) | 8.4 kg (−0.2) | 12.2 kg (−4.7) | 11.5 kg (−4.2) | 10.5 kg (−3.4) | 13.5 kg (−3) |
| Occipital frontal circumference (SD) | 50 cm (−2.3) | 47 cm (−3) | 51.5 cm (−0.8) | 49 cm (−2) | 50 cm (3) | 46.5 cm (−4.6) | 46 cm (−4.4) | 44 cm (−3.9) | 49.5 cm (−2) |
| Developmental Stages | |||||||||
| Age of sitting (months) | 17 | 15 | 18 | 18 | 30 | 18 | 20 | 24 | 11 |
| Age of walking (months) | 24 | 22 | 30 | 48 | cannot walk | 42 | 35 | 40 | 24 |
| Language skills | severely mentally impaired | severely mentally impaired | two-word combinations, no sentence | no words | two-word combinations, no sentence | simple two-word sentences, 40–50 words | compatible with 3 years of age | simple words, no word combinations | no sentences, two-word combinations |
| Cognitive Skills | |||||||||
| Learning disorders | + | + | + | + | + | + | + | + | + |
| Estimated degree of ID | moderate to severe | moderate to severe | moderate to severe | moderate to severe | severe | moderate to severe | moderate to severe | moderate to severe | moderate to severe |
| Dysmorphic Features | |||||||||
| Telecanthus | + | + | + | + | − | − | + | + | + |
| Bilateral epicanthal folds | + | + | + | + | + | + | + | + | + |
| Downslanting palpebral fissures | + | + | + | + | − | − | − | − | − |
| Depressed nasal bridge | + | + | + | + | + | wide | wide | + | wide |
| Broad nasal tip | + | + | + | + | + | + | + | + | + |
| Long philtrum | + | + | + | + | + | +, smooth | +, smooth | +, smooth | +, smooth |
| Small chin | + | + | + | + | + | − | − | − | − |
| Triangular face | + | + | + | + | − | − | − | − | − |
| Low-set ears | + | + | + | + | + | + | + | + | + |
| Posteriorly rotated ears | + | + | + | + | + | + | + | + | + |
| Other | macrodontia | macrodontia | nevus flammeus of the glabellar region | nevus flammeus of the glabellar region | high-arched palate, brachycephaly, synophrys | malar rash, high-arched palate, pointed chin | malar rash, high-arched palate, pointed chin | nevus flammeus of the glabella, malar rash | strabismus, malar hypoplasia, high-arched palate |
| Hand Examination | |||||||||
| Small hands | + | + | + | + | − | + | + | + | + |
| Deep palmar creases | + | + | + | + | − | −, bilateral single transverse palmar crease | − | − | − |
Seven boys and two girls originating from Turkey, Algeria, Tunisia, or Saudi Arabia were found to have a mean height of −3 SD (range: −1.3 to −4.5 SD), mean weight of −3.1 SD (range: −0.2 to −4.7 SD), and mean occipitofrontal circumference of −2.2 SD (range: −4.6 to +3 SD). They all displayed developmental delays, a mean age of sitting of 19 months (range: 11–30 months), a mean age of walking of 33 months (range: 22–48 months), and a pronounced language delay. The estimated degree of intellectual disability was moderate to severe, and all displayed a learning disorder. They shared dysmorphic facial features: a triangular face with a small pointed chin, low-set and posteriorly rotated ears, a depressed or wide nasal bridge with a broad nasal tip, telecanthus, and epicanthal folds. Orthopaedic anomalies included joint hyperlaxity and pes planus. A sacral dimple was observed in five of seven affected individuals. Brain MRI indicated hypoplasia of the corpus callosum in three individuals (Figure 1B, Table 1, and Table S1). A skeletal survey uncovered that all but one investigated individual had striking cervical spine anomalies, including clefting of the posterior arch of the atlas (C1) and partial fusion of C2 and C3 cervical vertebrae (Figures 1D–1F). The spine abnormalities are a hallmark of this syndrome.
A combination of array CGH, SNP-array-based whole-genome homozygosity mapping, and whole-exome sequencing in these five families revealed a common genetic etiology consisting of distinct germline CDK10 homozygous mutations. The intron 8 splice-acceptor-site mutation c.609−1G>A (GenBank: NM_052988.4) segregated in family 1, the single-nucleotide deletion c.139delG (p.Glu47ArgfsTer21) segregated in family 3, the splice-donor-site mutation c.608+1G>A segregated in families 4 and 5, and a small (2.6–28 kb) homozygous 16q24.3 microdeletion encompassing exons 2 and 3 of CDK10 segregated in family 2 (Figures 1A and 1G and Figure S1). We suggest naming this previously unrecognized congenital disease with CDK10 mutations as Al Kaissi syndrome.
CDK10 Splice-Site Mutations Behave as Loss-of-Function Alleles
Four differentially spliced transcripts expressed from the CDK10 locus were previously described29, 30 (Figure S1). These transcripts differ in their 5′ and 3′ UTRs and in exon 11. Most likely only two of these transcripts are functional: the full-length transcript (GenBank: NM_052988.4), which encodes the 360 amino acid CDK10 kinase; and a shorter transcript (GenBank: NM_052987.3), which encodes a truncated 272 amino acid CDK10 variant. The latter is missing the ATP-binding domain and therefore encodes most likely a catalytically inactive form of CDK10.
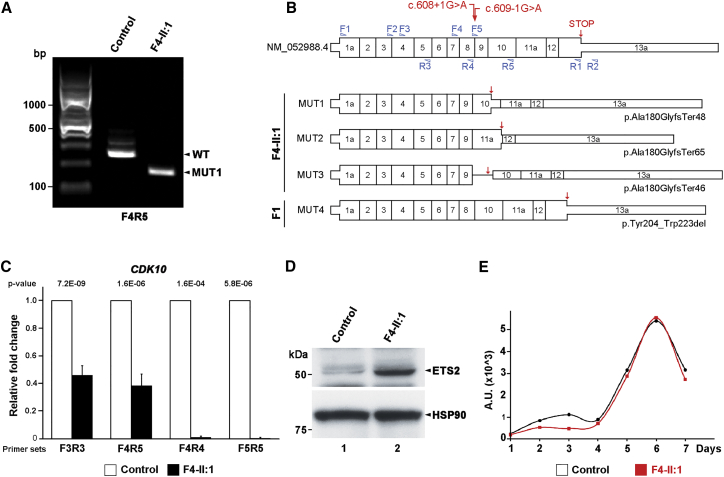
To test whether splicing of the endogenous CDK10 transcripts is affected in family 4, we grew and compared primary skin fibroblasts from proband II:1 with ethnically, age-, and sex-matched dermal fibroblasts from a healthy donor. Using primers spanning exons 7–10, we detected a shorter amplicon in the proband’s cells than in control cells (Figure 2A). WT and mutant amplicons were gel extracted for cloning, and multiple clones were sequenced. Several aberrant splice isoforms were identified: MUT1 skipped exon 8, MUT2 skipped exons 8 and 10, and MUT3 skipped exon 8 and retained intron 9 (Figure 2B). Compared with the longest CDK10 transcript (GenBank: NM_052988.4), each of these mis-spliced transcripts can result in truncated mRNAs (Figure 2B). Given that RNA samples were not available for family 1, we used an exon trap vector system (pET01, MoBiTec GmbH, Göttingen, Germany) to address the consequences of the mutation on the cDNA level. Sequencing of the resulting transcripts revealed exon 9 skipping, which presumably leads to the in-frame deletion p.Tyr204_Trp223del (Figure 2B). In conclusion, all mutant transcripts resulted in frameshifts or internal truncations, establishing the deleterious effect of the donor c.608+1G>A and acceptor c.609−1G>A splice-site mutations in intron 8.
Figure 2.
CDK10 Splice-Site Mutations Reduce Endogenous mRNA Levels and Increase ETS2 Levels
(A) RT-PCR performed on primary dermal fibroblasts derived from affected (F4-II:1) and control individuals indicates aberrant splicing of endogenous CDK10 mRNA.
(B) Schematic representations of the wild-type CDK10 isoform and four mutant transcripts cloned from primary fibroblasts with intron 8 donor and acceptor splice-site mutations.
(C) qRT-PCR analysis demonstrates lower endogenous CDK10 mRNA levels in cells derived from affected individuals than in control cells, suggestive of nonsense-mediated decay.
(D) Whole-cell lysates of human fibroblasts from control and affected individuals were separated on SDS-PAGE, and immunoblots were stained with antibodies against ETS2 and HSP90. Cells with reduced or absent CDK10 kinase activity displayed increased levels of ETS2, which is normally degraded once it is phosphorylated by CDK10.
(E) Primary human fibroblasts from control and affected individuals were grown in the presence of serum over 7 days and did not show any noticeable differences.
Using a series of primers covering the CDK10 cDNA, qPCR revealed significantly reduced overall CDK10 levels, suggestive of nonsense-mediated decay of mutant transcripts (Figure 2C). The levels of ETS2, which is phosphorylated by CDK10 and subsequently targeted for degradation,4 were increased (Figure 2D), consistent with absent or reduced CDK10 kinase activity in subject-derived cells. Despite these changes, mutant and WT cells did not exhibit differential growth parameters (Figure 2E) when grown in complete (10% serum) media over 7 days. Together, these results indicate that the classical splice-site mutations uncovered here behave like loss-of-function CDK10 alleles, which do not impair cell growth ex vivo but cause a severe growth retardation syndrome during human development.
Cdk10-Knockout Mice as a Model for Al Kaissi Syndrome
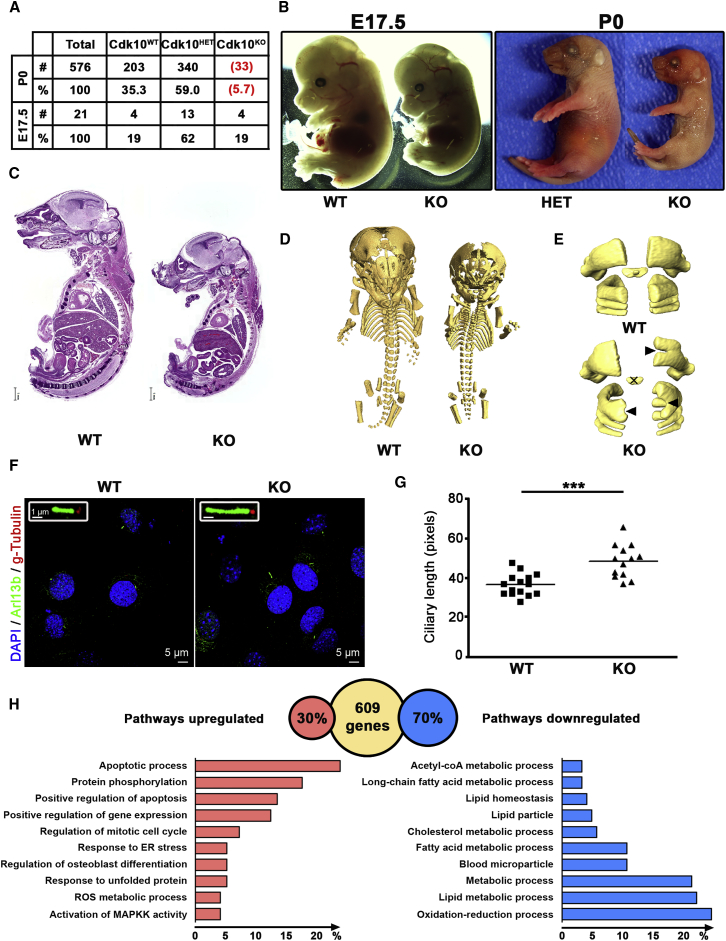
To better understand the requirement for CDK10 at the tissue and organismal levels, we generated a complete knockout of the homologous Cdk10 in mice. We accomplished this by deleting exons 2–8 of Cdk10 via standard recombineering techniques (Figure S2A). Recombination at the correct locus was verified by Southern blotting, genotyping, and qPCR analysis (Figures S2B–S2D). Even though homozygous Cdk10 mutants (Cdk10KO) were detected at a normal Mendelian ratio on E17.5, fewer than 6% of the embryos were born, indicating partial prenatal lethality (Figure 3A). Of the Cdk10KO mice born, very few survived the first day of life, suggesting that Cdk10 is an essential gene during mouse development. Similar to the individuals with CDK10 mutations (see Figure 1), all Cdk10KO embryos and mice displayed severe growth retardation (Figures 3B and 3C). The combination of growth retardation and lethality suggests that CDK10 is an essential gene like a number of other cell-cycle regulatory genes, including CDK1 (MIM: 116940)19 and MASTL (MIM: 608221).31 Therefore, we isolated MEFs from Cdk10KO and WT embryos on E13.5 to analyze their cell-cycle and proliferation characteristics. Surprisingly, the proliferation rate, cell-cycle progression determined by FACS, and timely expression of cyclin A2 and phosphorylated retinoblastoma protein (Rb) were indistinguishable between WT and Cdk10KO MEFs (Figure S5), indicating that CDK10 is not essential for cellular proliferation. We hypothetize that the growth retardation in Cdk10KO embryos is more likely driven by developmental defects rather than a strictly cell-autonomous requirement of CDK10.
Figure 3.
Mice Lacking Cdk10 as a Model for the Human Disease
(A) Heterozygous Cdk10WT/KO mice were interbred, and the genotypes of the offspring were analyzed at embryonic day 17.5 (E17.5) and birth (P0). Numbers in red indicate that pups were dead at the moment of collection.
(B) Control Cdk10WT/WT (WT), heterozygous Cdk10WT/KO (HET), and knockout Cdk10KO/KO (KO) embryos at E17.5 and pups at P0 were isolated and photographed.
(C) Hematoxylin & eosin staining of sagittal sections of the entire Cdk10WT/KO and Cdk10KO/KO P0 pups. The scale bar represents 1 mm.
(D) 3D μCT images of the mineralized part of the entire skeleton from Cdk10WT/WT and Cdk10KO/KO P0 pups. Note the shorter stature of the Cdk10KO pups and the defects in the vertebrae.
(E) 3D μCT images of C1–C4 vertebrae from Cdk10WT/WT and Cdk10KO/KO P0 mice. Note the presence of bifidity (black arrow) and the absence of mineralized dens (cross) in the Cdk10KO/KO mice.
(F) Cdk10KO and WT MEFs were starved for 4 days, and cilia were stained with Arl13b and gamma-tubulin antibodies. The inset shows magnified cilia.
(G) Quantification of the maximal length of the cilia in WT and Cdk10KO MEFs. ∗∗∗p value < 0.001 (Student’s t test).
(H) Pathway analysis from gene expression data of eight different tissues obtained from Cdk10WT/WT and Cdk10KO/KO P0 mice. From the entire dataset, 609 genes were differentially expressed in at least two tissues. In the absence of Cdk10, 30% of these genes were downregulated and 70% were upregulated. Pathway analysis for datasets of up- and downregulated genes is displayed as the percentage of genes annotated for the indicated pathway.
Because individuals with CDK10 mutations display unique skeletal defects, we analyzed newborn Cdk10KO pups by using micro-CT (Figure 3D). We observed several bone defects that affect the axial skeleton, including a reduced volume of mineralized matrix from the head, the occurrence of bifidity (clefting) at C1 (atlas) or C2 (axis), and the absence of dens (Figure 3D and 3E and Figures S3 and S4). The appendicular skeletal defects are characterized by the volume reduction of mineralized matrix from the femur, tibia, and fibula, the length reduction of the left femur, and the malformations of the mandible (Figure S3). Therefore, the Cdk10KO mouse model partly recapitulates the phenotype observed in individuals with loss-of-function CDK10 mutations.
Mice with Mutant Cdk10 Display Defects in Multiple Organs
In addition to observing growth retardation, we detected defects in several organs of the Cdk10KO mice, including in the kidney, lung, heart, spleen, liver, and muscle (Figure S6). For example, Cdk10KO mouse kidneys displayed severe tubulonephrosis. Approximately 30%–50% of available renal tubules were severely degenerated or necrotic. Most of the affected tubules were lined with severely attenuated mineralized epithelium, and their lumens contained mineralized casts. Other tubules were lined with swollen and vacuolated epithelium that occasionally displayed nuclear pyknosis (indicative of necrosis). In Cdk10KO lungs, most of the alveoli and bronchioli were collapsed and failed to open (fetal atelectasis). The interalveolar septae were markedly thickened and were composed of collapsed alveolar walls and few lymphocytes and macrophages. The few inflated (opened) alveoli had squamous debris and activated foamy macrophages in their lumens. The bronchioles were partially filled with eosinophilic fluid and squamous debris. The current pulmonary atelectasis (failure of lung to inflate at birth) was severe and was the likely cause of death of the homozygous Cdk10KO mice. The cardiac and renal lesions might also have contributed to the general weakness and demise of these mutant mice. Diffuse fetal atelectasis usually leads to severe hypoxia and death. Causes of fetal atelectasis include weakness of respiratory diaphragmatic and intercostal muscles, which can lead to insufficient respiration attempts at birth, bronchial obstruction, or absence of alveolar surfactant.
Organs with Mutant Cdk10 Show Defects in Metabolic and Cilia-Related Genes
Because the phenotype of the Cdk10KO mice and individuals with CDK10 mutations is multifaceted and affects multiple organs, we carried out unbiased gene expression analysis on eight different organs (kidney, lung, heart, brain, stomach, intestine, thymus, and liver) from WT and Cdk10KO mice (Figure 3H and Figure S7). A total of 609 transcripts, including Cdk10 as expected (Figures S7A–S7C and Table S2), were differentially expressed in at least two different organs. Approximately 70% of the genes were downregulated (Figure 3H), indicating that CDK10 directly or indirectly stimulates gene expression. Pathway analysis indicated that 175 metabolic genes were enriched (Table S3). Of the 175 metabolic genes differentially regulated, lipid metabolic processes involving HMGCS2 (MIM: 600234), LIPO1 (MIM: 613921), and LIPA (MIM: 613497) stood out (Figure S7C and Table S3). HMGCS2 is a mitochondrial enzyme that catalyzes the first and rate-limiting reaction of ketogenesis, an important process that allows alternative metabolic adaptation by using lipids as energy sources in starved cells (for a review, see Puchalska and Crawford32). Hmgcs2 was almost 20-fold upregulated in WT MEFs upon serum starvation, whereas Hmgcs2 expression remained unchanged in Cdk10KO cells (Figure S7D). These data suggest that a metabolic impairment might contribute to the stunted growth of mice and humans with Cdk10 mutations.
As expected, a mild upregulation of Ets2 expression was detected upon CDK10 depletion (Figure S7D). CDK10 is activated by cyclin M and interacts with ETS2.6, 7 To confirm the increase in ETS2 at the protein level, we performed western blots with lysates of subject-derived primary fibroblasts (see Figure 2D). The levels of ETS2 were substantially higher in CDK10 mutant cells than in control cells, which is in accordance with the finding that ETS2 is a substrate of the CDK10/cyclin M complex.7
Finally, expression levels of genes involved in ciliogenesis, including BBS4 (MIM: 600374), CEP290 (MIM: 610142), and RPGRIP1L (MIM: 610937) (Figure S7D), differed between organs with mutant CDK10 and control organs. Mutations in these genes can lead to ciliopathies such as Bardet-Biedl syndrome (MIM: 615982) or Joubert syndrome (type 5 [MIM: 610188] or type 7 [MIM: 611560]). To investigate the functional consequences of differential expression of cilia-related genes, we examined cilia growth and length in serum-starved Cdk10KO MEFs. Immunostaining, using ARL13B and gamma-tubulin antibodies, revealed significantly longer cilia in Cdk10KO MEFs than in WT cells (Figures 3F and 3G), corroborating results that were reported after the silencing of Cdk10 or cyclin M by siRNA10 (for a review, see Guen et al.33). BBS4, which interacts with CEP290,34 is a BBSome component that is essential for cilia formation,35 providing a rationale for our observation of longer cilia in Cdk10KO MEFs.
Discussion
Our work has uncovered nine individuals with germline recessive mutations in CDK10. The clinical features of these individuals display little phenotypic variability and are best summarized by severe growth retardation, spine malformations, facial dysmorphisms, developmental delays, and intellectual disability (see Figure 1, Table 1, and Table S1).
Because cyclin M binds to and activates CDK10, and mutations in FAM58A (the gene encoding cyclin M) lead to STAR syndrome (MIM: 300707),8, 9 we compared the two syndromes (see Table S5). Superficially, the two syndromes might appear related, but careful comparison reveals that they are clinically distinct. The spine malformations, developmental delays, and intellectual disability are important symptoms present in this syndrome but absent from STAR syndrome. In addition, the severity of the growth retardation is more pronounced in individuals harboring CDK10 mutations. Distinctive features of STAR syndrome, such as toe syndactyly and anogenital, renal, and urinary-tract malformation, are rarely found in the syndrome described here. Given that we observed incomplete clinical overlap between STAR and Al Kaissi syndromes, we hypothesize either that the nature of the mutation is different or that CDK10 has additional binding partners beyond cyclin M. The phenotype in the Cdk10KO mice is similar in terms of growth retardation and spine malformation, whereas other aspects have not been examined yet. The analysis of the Cdk10KO brain warrants further investigation in light of the observed intellectual ability of individuals with CDK10 mutations. In addition, Cdk10KO newborn pups displayed a number of phenotypes that have not been detected yet in individuals with CDK10 mutations. They include lung defects, kidney problems, and others. Several reasons could explain the differences observed between mice and humans: (1) all Cdk10 transcripts are lost in the mutant mice, whereas in humans only a subset of them might carry a mutation, (2) some defects might not have been documented yet in the individuals with CDK10 mutations as a result of differential disease progression, or (3) the requirement or genetic compensations in mice could be different in humans.
CDK10 has been shown to interact with the transcription factor ETS2 and to affect its transactivation activity.6 CDK10/cyclin M complexes phosphorylate ETS2 on several residues to control its half-life, and ETS2 was the first direct CDK10 substrate to be identified. In our experiments, we uncovered that in the absence of CDK10, endogenous ETS2 was present at higher levels, which confirms previous reports. This result is noteworthy given that overexpression of ETS2 has been observed in Down syndrome (MIM: 190685), and overexpression of Ets2 in mice leads to skeletal abnormalities that are comparable to those of Cdk10KO mice and individuals harboring CDK10 mutations.36 These data indicate that the spine malformations observed in individuals with CDK10 mutations and in Cdk10KO mice could be a direct consequence of elevated ETS2 levels, suggesting that inhibition of ETS2 activity could potentially reduce the severity of spine malformations in affected individuals.
Among the most surprising results from our study is the effect of CDK10 on the expression of genes involved in lipid metabolism. Our unbiased transcriptomic approach revealed across multiple organs that approximately one-third of differentially expressed genes were involved in metabolism. Among these, HMGCS2 is particularly interesting because of its function in ketogenesis.32 Likewise, LIPA and LIPJ might function in the same pathway. We have confirmed the differential expression of these genes in subject-derived dermal fibroblasts (see Figure S7E). Although we have not been able to document overtly altered lipid profiles in individuals with CDK10 mutations, it will be very important to perform a thorough metabolic phenotyping of these probands to document whether their diet might offer a therapeutic window for mitigating their severe growth retardation.
Silencing of CDK10 in cell lines was reported to lead to longer cilia,10 and indeed we detected altered expression of a number of genes involved in ciliogenesis (BBS4, CEP290, and RPGRIP1l; see Figure S7E) in mutant cells from affected individuals. Although there is no clinical resemblance between Al Kaissi syndrome and Bardet-Biedl syndrome, we detected that monocilia are longer in Cdk10KO MEFs. Together with the lung histology of Cdk10KO mice and the recurrent infections seen in individuals with CDK10 mutations, this suggests that ciliary defects might contribute to the phenotype of individuals with CDK10 mutations. Although our results indicate that CDK10 does not affect the proliferation capacity of primary fibroblasts, we cannot rule out that other cell types would be differentially affected. It is clear that loss of CDK10 is detrimental during embryonic stages but not necessarily after development is complete. In line with this, we observed that knockout of Cdk10 in adult mice (Cdk10flox/flox ROSA26-CreERT2 treated with tamoxifen) had little to no effect (data not shown). Therefore, future studies will need to address whether loss of CDK10 affects proliferation during development. Alternatively, loss of CDK10 could affect differentiation, which would also result in a net loss of differentiated cells.
In conclusion, our work has uncovered loss-of-function biallelic germline CDK10 mutations as responsible for a growth-retardation syndrome. This congenital disease of stunted growth can be modeled in Cdk10-deficient mice. Our work suggests that deregulation of metabolic pathways could in part explain the growth restriction seen in the absence of CDK10. These unanticipated effects of CDK10 on metabolism could lead to context- or lineage-dependent arrest of cell growth and could involve transduction of signals at the level of the cilia. Future work should strive to better explain the pathogenesis of Al Kaissi syndrome with the aim of creating possible therapeutic interventions.
Acknowledgments
We thank all families for partaking in this study. This work is the result of a fruitful collaboration between several research groups, and therefore it is difficult to give sufficient credit to each author. Nevertheless, the Kaldis, Reversade, and Al Kaissi laboratories contributed equally. The authors would also like to thank Isabelle Dalle Fusine and all past and current members of the Kaldis, Reversade, Windpassinger, and Al Kaissi labs for technical assistance and fruitful discussions. We acknowledge the technical expertise provided by the Advanced Molecular Pathology Laboratory at the Institute of Molecular and Cell Biology. We are grateful to Eileen Southon and Susan Reid for help in generating the Cdk10flox mice. This work was supported by the Intramural Research Program of the NIH, the National Cancer Institute, and the Center for Cancer Research (V.C. and L.T.); a Strategic Positioning Fund for the Genetic Orphan Diseases program (SPF2012/005) and an Industry Alignment Fund for the Singapore Childhood Undiagnosed Diseases program (IAF311019) from the A∗STAR (Agency for Science, Technology, and Research) Biomedical Research Council (B.R.); and the A∗STAR Biomedical Research Council (P.K.).
Published: September 7, 2017
Footnotes
Supplemental Data include seven figures and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.08.003.
Contributor Information
Bruno Reversade, Email: bruno@reversade.com.
Philipp Kaldis, Email: kaldis@imcb.a-star.edu.sg.
Accession Numbers
Microarray data were deposited in the Gene Expression Omnibus under accession number GEO: GSE98628.
Web Resources
1000 Genomes, http://www.1000genomes.org/
Database of Genomic Variants, http://projects.tcag.ca/variation/
dbSNP146, http://www.ncbi.nlm.nih.gov/snp
dCHIP software, http://www.dchip.org/
ExAC Browser, http://exac.broadinstitute.org/
GeneMatcher, https://genematcher.org/
Human Splicing Finder, http://www.umd.be/HSF/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, http://genome.ucsc.edu/
Supplemental Data
References
- 1.Lim S., Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 2.Malumbres M., Harlow E., Hunt T., Hunter T., Lahti J.M., Manning G., Morgan D.O., Tsai L.H., Wolgemuth D.J. Cyclin-dependent kinases: a family portrait. Nat. Cell Biol. 2009;11:1275–1276. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan D.O. New Science Press Ltd; 2007. The cell cycle: principles of control. [Google Scholar]
- 4.Brambilla R., Draetta G. Molecular cloning of PISSLRE, a novel putative member of the cdk family of protein serine/threonine kinases. Oncogene. 1994;9:3037–3041. [PubMed] [Google Scholar]
- 5.Graña X., Claudio P.P., De Luca A., Sang N., Giordano A. PISSLRE, a human novel CDC2-related protein kinase. Oncogene. 1994;9:2097–2103. [PubMed] [Google Scholar]
- 6.Kasten M., Giordano A. Cdk10, a Cdc2-related kinase, associates with the Ets2 transcription factor and modulates its transactivation activity. Oncogene. 2001;20:1832–1838. doi: 10.1038/sj.onc.1204295. [DOI] [PubMed] [Google Scholar]
- 7.Guen V.J., Gamble C., Flajolet M., Unger S., Thollet A., Ferandin Y., Superti-Furga A., Cohen P.A., Meijer L., Colas P. CDK10/cyclin M is a protein kinase that controls ETS2 degradation and is deficient in STAR syndrome. Proc. Natl. Acad. Sci. USA. 2013;110:19525–19530. doi: 10.1073/pnas.1306814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boczek N.J., Kruisselbrink T., Cousin M.A., Blackburn P.R., Klee E.W., Gavrilova R.H., Lanpher B.C. Multigenerational pedigree with STAR syndrome: A novel FAM58A variant and expansion of the phenotype. Am. J. Med. Genet. A. 2017;173:1328–1333. doi: 10.1002/ajmg.a.38113. [DOI] [PubMed] [Google Scholar]
- 9.Unger S., Böhm D., Kaiser F.J., Kaulfuss S., Borozdin W., Buiting K., Burfeind P., Böhm J., Barrionuevo F., Craig A. Mutations in the cyclin family member FAM58A cause an X-linked dominant disorder characterized by syndactyly, telecanthus and anogenital and renal malformations. Nat. Genet. 2008;40:287–289. doi: 10.1038/ng.86. [DOI] [PubMed] [Google Scholar]
- 10.Guen V.J., Gamble C., Perez D.E., Bourassa S., Zappel H., Gärtner J., Lees J.A., Colas P. STAR syndrome-associated CDK10/Cyclin M regulates actin network architecture and ciliogenesis. Cell Cycle. 2016;15:678–688. doi: 10.1080/15384101.2016.1147632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh C.W., Kao S.H., Cheng Y.C., Hsu L.S. Knockdown of cyclin-dependent kinase 10 (cdk10) gene impairs neural progenitor survival via modulation of raf1a gene expression. J. Biol. Chem. 2013;288:27927–27939. doi: 10.1074/jbc.M112.420265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y.J., Liao W.L., Wang C.H., Tsai L.P., Tang C.H., Chen C.H., Wu J.Y., Liang W.M., Hsieh A.R., Cheng C.F. Association of human height-related genetic variants with familial short stature in Han Chinese in Taiwan. Sci. Rep. 2017;7:6372. doi: 10.1038/s41598-017-06766-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wölfel T., Hauer M., Schneider J., Serrano M., Wölfel C., Klehmann-Hieb E., De Plaen E., Hankeln T., Meyer zum Büschenfelde K.H., Beach D. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 14.Zuo L., Weger J., Yang Q., Goldstein A.M., Tucker M.A., Walker G.J., Hayward N., Dracopoli N.C. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat. Genet. 1996;12:97–99. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- 15.Hussain M.S., Baig S.M., Neumann S., Peche V.S., Szczepanski S., Nürnberg G., Tariq M., Jameel M., Khan T.N., Fatima A. CDK6 associates with the centrosome during mitosis and is mutated in a large Pakistani family with primary microcephaly. Hum. Mol. Genet. 2013;22:5199–5214. doi: 10.1093/hmg/ddt374. [DOI] [PubMed] [Google Scholar]
- 16.Fujita P.A., Rhead B., Zweig A.S., Hinrichs A.S., Karolchik D., Cline M.S., Goldman M., Barber G.P., Clawson H., Coelho A. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin M., Wei L.J., Sellers W.R., Lieberfarb M., Wong W.H., Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 19.Diril M.K., Ratnacaram C.K., Padmakumar V.C., Du T., Wasser M., Coppola V., Tessarollo L., Kaldis P. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc. Natl. Acad. Sci. USA. 2012;109:3826–3831. doi: 10.1073/pnas.1115201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee E.-C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D.A., Court D.L., Jenkins N.A., Copeland N.G. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 22.Lewandoski M., Meyers E.N., Martin G.R. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- 23.Truett G.E., Heeger P., Mynatt R.L., Truett A.A., Walker J.A., Warman M.L. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52–54. doi: 10.2144/00291bm09. 54. [DOI] [PubMed] [Google Scholar]
- 24.Koh H.W., Swa H.L., Fermin D., Ler S.G., Gunaratne J., Choi H. EBprot: Statistical analysis of labeling-based quantitative proteomics data. Proteomics. 2015;15:2580–2591. doi: 10.1002/pmic.201400620. [DOI] [PubMed] [Google Scholar]
- 25.Herwig R., Hardt C., Lienhard M., Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 2016;11:1889–1907. doi: 10.1038/nprot.2016.117. [DOI] [PubMed] [Google Scholar]
- 26.Todaro G.J., Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berthet C., Aleem E., Coppola V., Tessarollo L., Kaldis P. Cdk2 knockout mice are viable. Curr. Biol. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 28.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford J., Ianzano L., Savino M., Whitmore S., Cleton-Jansen A.M., Settasatian C., d’apolito M., Seshadri R., Pronk J.C., Auerbach A.D. The PISSLRE gene: structure, exon skipping, and exclusion as tumor suppressor in breast cancer. Genomics. 1999;56:90–97. doi: 10.1006/geno.1998.5676. [DOI] [PubMed] [Google Scholar]
- 30.Sergère J.C., Thuret J.Y., Le Roux G., Carosella E.D., Leteurtre F. Human CDK10 gene isoforms. Biochem. Biophys. Res. Commun. 2000;276:271–277. doi: 10.1006/bbrc.2000.3395. [DOI] [PubMed] [Google Scholar]
- 31.Diril M.K., Bisteau X., Kitagawa M., Caldez M.J., Wee S., Gunaratne J., Lee S.H., Kaldis P. Loss of the Greatwall kinase weakens the spindle assembly checkpoint. PLoS Genet. 2016;12:e1006310. doi: 10.1371/journal.pgen.1006310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puchalska P., Crawford P.A. Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab. 2017;25:262–284. doi: 10.1016/j.cmet.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guen V.J., Gamble C., Lees J.A., Colas P. The awakening of the CDK10/Cyclin M protein kinase. Oncotarget. 2017;8:50174–50186. doi: 10.18632/oncotarget.15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stowe T.R., Wilkinson C.J., Iqbal A., Stearns T. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol. Biol. Cell. 2012;23:3322–3335. doi: 10.1091/mbc.E12-02-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez-Hernandez V., Pravincumar P., Diaz-Font A., May-Simera H., Jenkins D., Knight M., Beales P.L. Bardet-Biedl syndrome proteins control the cilia length through regulation of actin polymerization. Hum. Mol. Genet. 2013;22:3858–3868. doi: 10.1093/hmg/ddt241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sumarsono S.H., Wilson T.J., Tymms M.J., Venter D.J., Corrick C.M., Kola R., Lahoud M.H., Papas T.S., Seth A., Kola I. Down’s syndrome-like skeletal abnormalities in Ets2 transgenic mice. Nature. 1996;379:534–537. doi: 10.1038/379534a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.