Abstract
RAC1 is a widely studied Rho GTPase, a class of molecules that modulate numerous cellular functions essential for normal development. RAC1 is highly conserved across species and is under strict mutational constraint. We report seven individuals with distinct de novo missense RAC1 mutations and varying degrees of developmental delay, brain malformations, and additional phenotypes. Four individuals, each harboring one of c.53G>A (p.Cys18Tyr), c.116A>G (p.Asn39Ser), c.218C>T (p.Pro73Leu), and c.470G>A (p.Cys157Tyr) variants, were microcephalic, with head circumferences between −2.5 to −5 SD. In contrast, two individuals with c.151G>A (p.Val51Met) and c.151G>C (p.Val51Leu) alleles were macrocephalic with head circumferences of +4.16 and +4.5 SD. One individual harboring a c.190T>G (p.Tyr64Asp) allele had head circumference in the normal range. Collectively, we observed an extraordinary spread of ∼10 SD of head circumferences orchestrated by distinct mutations in the same gene. In silico modeling, mouse fibroblasts spreading assays, and in vivo overexpression assays using zebrafish as a surrogate model demonstrated that the p.Cys18Tyr and p.Asn39Ser RAC1 variants function as dominant-negative alleles and result in microcephaly, reduced neuronal proliferation, and cerebellar abnormalities in vivo. Conversely, the p.Tyr64Asp substitution is constitutively active. The remaining mutations are probably weakly dominant negative or their effects are context dependent. These findings highlight the importance of RAC1 in neuronal development. Along with TRIO and HACE1, a sub-category of rare developmental disorders is emerging with RAC1 as the central player. We show that ultra-rare disorders caused by private, non-recurrent missense mutations that result in varying phenotypes are challenging to dissect, but can be delineated through focused international collaboration.
Keywords: RAC1, developmental disorders, intellectual disability, microcephaly, macrocephaly, cerebellar abnormalities, neuronal proliferation, Rho GTPase, TRIO, HACE1
Main Text
Developmental disorders (DDs) are etiologically extremely heterogeneous and affect 2%–5% of individuals.1, 2 De novo mutations account for a substantial proportion of DDs and are thought to underlie approximately 400,000 new DD-affected case subjects world-wide annually.3 Recently, large-scale next generation sequencing studies have led to the identification of several DD-associated genes that lead to clinical manifestations through protein truncating or recurrent missense variants.4, 5, 6 However, rare disorders caused by private, non-recurrent missense mutations that result in varying phenotypes remain challenging to dissect.7
Several human DDs are known to result from mutations in members of the RAS superfamily of small GTPases.8 The RAS superfamily is further divided into smaller families, one of which is the 22-member Rho family. Rho GTPases cycle between active GTP-bound and inactive GDP-bound states. Their activity is regulated by guanine nucleotide exchange factors (GEFs), GTPases activating proteins (GAPs), and guanine nucleotide dissociated inhibitors (GDIs).9 Rho GTPases modulate essential cellular functions, including cell polarity, migration, vesicle trafficking, and cytokinesis and play crucial roles in neuronal development, neuronal survival, and neurodegeneration.10, 11, 12 However, no human DDs caused by mutations in genes encoding Rho GTPases are known.
One of the most widely studied Rho GTPases is the RAS-related C3 Botulinum Toxin Substrate 1 (RAC1).13 RAC1 is part of the RAC Rho GTPases subfamily that also includes RAC2, RAC3, and RhoG.14 RAC1 is an important modulator of the cytoskeleton, with a critical function in phagocytosis, mesenchymal-like migration, neuronal polarization, axonal growth, adhesion, and differentiation of multiple cell types.10, 15, 16 Additionally, it is involved in cellular growth and cell-cycle regulation via mTOR signaling.17 In mouse studies, Rac1 is required for the formation of three germ layers during gastrulation, with Rac1-knockout mice being embryonic lethal.18 Conditional forebrain-specific Rac1-knockout mice display impaired neuronal migration, abnormal dendritic growth and remodelling, disruption of lamellipodia formation, reduced neuronal proliferation, premature differentiation, and microcephaly.19, 20, 21 Here, we report de novo missense RAC1 (MIM: 602048) mutations in individuals with DD and divergent phenotypes.
All procedures followed were in accordance with the ethical standards of the institutional and national responsible committees on human or animal experimentation and that, where relevant, informed consent was obtained. Review of data from 4,293 families, who underwent trio whole-exome sequencing (WES) as part of the Deciphering Developmental Disorders study,3 led to identification of three individuals with de novo RAC1 (GenBank: NM_006908) missense mutations: individual 3 with c.218C>T (p.Pro73Leu), individual 5 with c.190T>G (p.Tyr64Asp), and individual 6 with c.151G>A (p.Val51Met) (Table 1; Figures 1 and 2). Two additional individuals were independently ascertained through family-based diagnostic WES: individual 1 with c.53G>A (p.Cys18Tyr) and individual 2 with c.116A>G (p.Asn39Ser) (Table 1; Figures 1 and 2). While functional studies were ongoing for the five individuals, two additional individuals were identified via the GeneMatcher tool22 or through international collaboration: individual 4 with c.470G>A (p.Cys157Tyr) and individual 7 with c.151G>C (p.Val51Leu) (Table 1; Figures 1 and 2).
Table 1.
RAC1 Mutations and Phenotype
|
Individual 1 |
Individual 2 |
Individual 3 |
Individual 4 |
Individual 5 |
Individual 6 |
Individual 7 |
|
|---|---|---|---|---|---|---|---|
| microcephaly | microcephaly | microcephaly | microcephaly | normal OFC | macrocephaly | macrocephaly | |
| Gender | male | male | male | male | male | male | male |
| Ethnicity | European/Egyptian | European | European | European/Armenian | European/Asian | European | African American |
| Age at examination | 13 years | 9 years | 15 years | 4.5 months | 12 years | 33 months | 4 years 5 months |
| Mutation (GenBank:NM_006908) | |||||||
| Chromosome position (Hg19) | chr7:6426860G>A | chr7:6431563A>G | chr7:6431665C>T | chr7:6441968G>A | chr7:643163T>G | chr7:6431598G>A | chr7:6431598G>C |
| cDNA change | c.53G>A | c.116A>G | c.218C>T | c.470G>A | c.190T>G | c.151G>A | c.151G>C |
| Amino acid change | p.Cys18Tyr | p.Asn39Ser | p.Pro73Leu | p.Cys157Tyr | p.Tyr64Asp | p.Val51Met | p.Val51Leu |
| Likely effect of the mutation | dominant negative | dominant negative | unknown | unknown | constitutively active | unknown | unknown |
| Growth | |||||||
| Height | 127 cm (−2.5 SD) | 128 cm (−2.5 SD) | unknown | 62 cm (−1 SD) | 134 cm (+1.03 SD) (8 years) | unknown | 109 cm (0 SD) |
| Weight | 30 kg (0 SD) | 24 kg (−0.5 SD) | unknown | 4.8 kg (−3 SD) | unknown | unknown | 23 kg (+2.5 SD) |
| Head circumference | 50 cm (−2.5 SD) | 47.7 cm (−3 SD) | 47 cm (−5 SD) | 39 cm (−2.5 SD) | 56.5 cm (+1 SD) | 57 cm (+4.16 SD) | 59.5 cm (+4.5 SD) |
| Development | |||||||
| Intellectual disability | yes | yes | yes | yes | yes | yes | yes |
| Mild/Moderate/Severe | moderate | mild-moderate | severe | unknown | severe | moderate | unknown |
| Neurological | |||||||
| Epilepsy | yes | no | unknown | yes | no | unknown | yes |
| Hypotonia | yes | no | unknown | yes | yes | unknown | yes |
| Behavioral problems | no | yes - hyperactive | unknown | unknown | yes - sleep disturbances | unknown | yes - autism |
| Stereotypic movements | no | no | unknown | no | yes | unknown | yes |
| Brain MRI Abnormalities | |||||||
| Cerebellar abnormalities | yes | yes | unknown | yes | no | no | no |
| Hypoplasia corpus callosum | yes | yes | unknown | yes | yes | no | no |
| Enlarged lateral ventricles | yes | yes | unknown | no | no | no | no |
| Enlarged fourth ventricle | no | yes | unknown | no | no | no | no |
| Thin pons brain stem | no | yes | unknown | yes | no | no | no |
| Mega cisterna magna | yes | yes | unknown | yes | no | no | no |
| Polymicrogyria | no | no | unknown | no | yes | no | no |
| White matter lesions | yes | no | unknown | no | no | yes | yes |
| Congenital Abnormalities | |||||||
| Cardiac abnormalities | yes – NS LVC; IV | no | unknown | yes – PDA, PFO, BAV | yes - VSD | unknown | no |
| Hypospadias | no | no | unknown | yes | yes | unknown | no |
| Other | |||||||
| Neonatal feeding difficulties | yes | yes | yes | no | no | unknown | no |
| Other | plagiocephaly; scoliosis; small hands and feet; hyperlaxity; brachydactyly 5th digit; SC bilateral | recurrent pneumonias; eczema | diabetes mellitus | umbilical hernia, tracheobronchomalacia; cryptorchidism | mild visual impairment; congenital sensorineural hearing impairment; abnormal creases in hands | none | non-verbal; recurrent otitis media; eczema |
Abbreviations are as follows: BAV, bicuspid aortic valve; IV, insufficiency all valves; NS LVC, non-synchronic left ventricle contractions; PDA, patent ductus arteriosus; PFO, patent foramen ovale; SC, simian crease; VSD, ventricular septum defect.
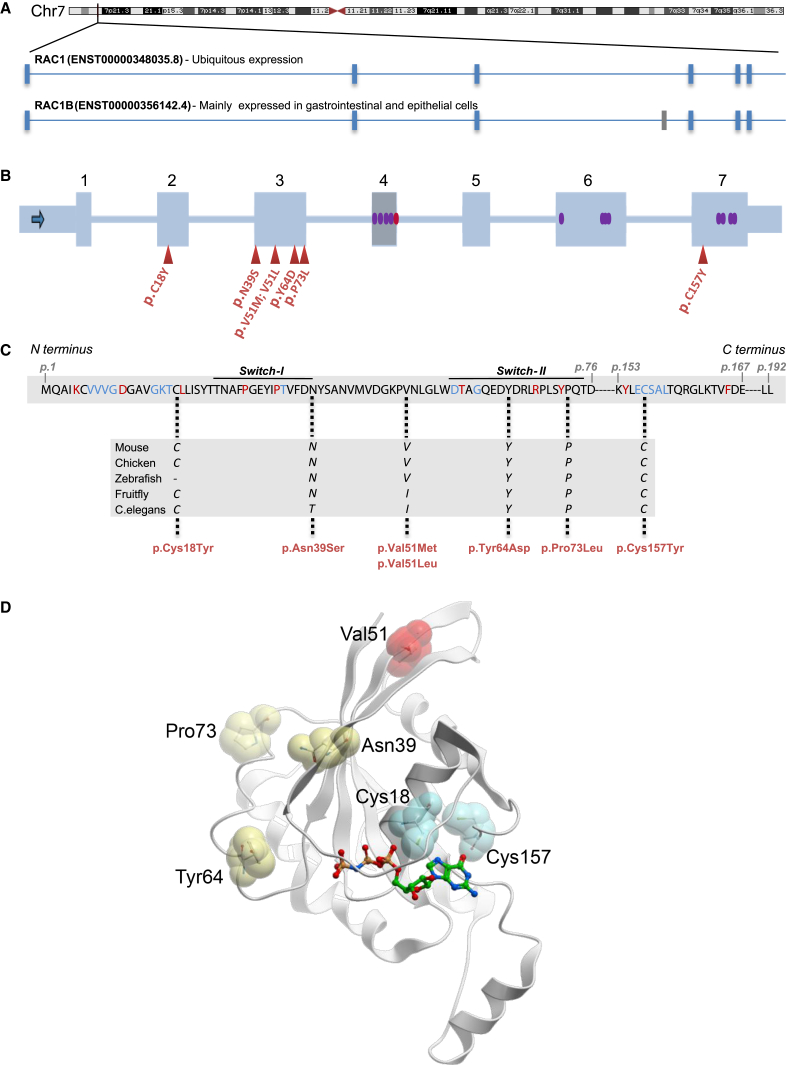
Figure 1.
Properties of Germline RAC1 Variants
(A) RAC1 is located on human chromosome 7p22.1. Exon-intron structure of the two protein-coding RAC1 transcripts is shown. The ubiquitously expressed RAC1 transcript is composed of six exons (blue vertical lines). RAC1B (mainly expressed in gastrointestinal and epithelial cells) contains an additional exon (exon 4; gray vertical line).
(B) Schematic of RAC1, with the coding exons shown as blue rectangles and the introns as blue lines. Exon 4 that is expressed only in the longer RAC1B transcript is shown as a gray rectangle. The red triangles represent the positions of the mutations identified in this study. The red oval within exon 4 shows the two splice donor variants detected in the ExAC database. The purple ovals represent the missense variants in the ExAC database.
(C) Amino acid sequence of the RAC1 across positions p.1-p.192. Red amino acids are highly conserved in >90% of the RAS family members and blue amino acids represent RAS protein G box consensus residues. Positions of the identified missense mutations (in red) are marked with dotted lines and the high conservation among different species of these amino acids is shown in the rectangle below.
(D) Crystal structure of human RAC1 showing the position of the identified missense variants presented in this study. The sites of substitution are indicated by sticks in spheres and are color coded according to their putative impact on guanine nucleotide binding (cyan), protein-protein interactions (yellow), and structural stability (red). For reference, the non-hydrolyzable GTP analog GMPPNP bound to the structure is shown in sticks. The structure was analyzed and this figure was generated using the program ICM-Pro v3.8 (Molsoft, LLC).
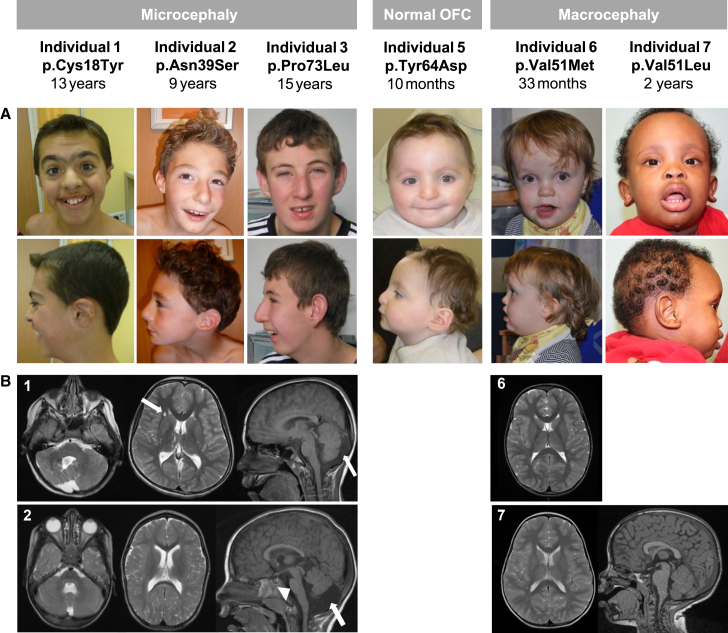
Figure 2.
Facial Dysmorphism and Brain Abnormalities Observed in Individuals with RAC1 Mutations
(A) Frontal and lateral photographs of the faces of individuals 1–3 and 5–7. Individuals 1, 2, and 3 were microcephalic, individual 5 had a normal occipital-frontal circumference, and individuals 6 and 7 had macrocephaly. Variable degrees of arched eyebrows, dysplastic ears, prominent nasal bridge, and overhanging columella were noted. Individuals with macrocephaly have prominent broad foreheads, slightly up-slanted palpebral fissures, open mouths, and “scooped out” appearance on lateral view.
(B) Panel 1 shows brain MRI images of individual 1: axial T2 images showing dysgenetic vermis and right cerebellar hemisphere (left), cystis lesions in the right frontal deep white matter (middle, arrow), and sagittal T1 images (right) showing enlarged cisterna magna (arrow). Panel 2 shows brain MRI images of individual 2: axial T2 images showing hypoplastic cerebellar vermis (left), slightly enlarged lateral ventricles (middle), and sagittal T1 images (right) showing hypoplastic corpus callosum, pons (arrow head), hypoplastic lower vermis, enlarged 4th ventricle, and cisterna magna (arrow). Panel 6 shows brain MRI image of individual 6: axial T2 image showing non-specific white matter changes in the frontal and parietal lobes. No hydrocephalus or extra-axial fluid was observed. Panel 7 shows brain MRI of individual 7: T2 weighted MRI images. Left: axial image showing very discrete bilateral patchy high intensity signal changes in the deep white matter of the frontal and parietal lobes; right: mid-sagittal section showing, except for megalencephaly, normal brain structures.
Human RAC1 (ENSG00000136238) encodes six transcripts, of which two are protein coding: RAC1 and RAC1B (Figure 1A). Of the two protein-coding transcripts, GenBank: NM_006908 (RAC1, ENST00000348035.8) lacks exon 4 and encodes the shorter RAC1 isoform of 192 amino acids, which is ubiquitously expressed in all tissues.23 RAC1 is under strict mutational constraint with only 15 missense variants observed versus 75.9 expected in ∼60,000 exomes cataloged in the ExAC database (z-score = 3.42).24 Moreover, 7 of these 15 observed missense variants are located in exon 4, which is included only in RAC1B (ENST00000356142.4) that encodes the longer isoform and is mainly expressed in gastrointestinal and epithelial tissues.25 Of particular note, there are no missense variants in exons 1, 2, 3, and 5 in the ExAC database (Figure 1B). Six out of seven mutations described here are in exons 2 and 3. The p.Cys157Tyr change (p.Cys176Tyr in the longer RAC1B) lies in a sub-region of exon 7 with no known germline human missense variants (Figure 1B). None of the identified mutations were present in any of the in-house variant databases of the four centers participating in this study. All six amino acids affected by the seven mutations are highly conserved among different species (Figure 1C). All reported RAC1 mutations are within or in proximity to G box residues and/or conserved residues present in 90% of the RAS superfamily members (Figure 1C).26 Collectively, the genetic data were strongly supportive of deleteriousness for each of the seven mutations.
Informed consent for publication of photographs was obtained from legal guardians. Detailed clinical information was collected on all affected individuals (Table 1; Figure 2; see Supplemental Note). All seven individuals (age range 4.5 months–15 years) had moderate to severe intellectual disability (ID) and variable degrees of neurological involvement including hypotonia (4/7), epilepsy (3/7), behavioral problems (3/7), and stereotypic movements (2/7). However, their occipital frontal circumferences (OFC) were remarkably different: individuals 1, 2, 3, and 4 were microcephalic (OFCs of −2.5, −3, −5, and −2.5 SD, respectively), individual 5 with p.Tyr64Asp mutation had a normal OFC (+1 SD), and individuals 6 and 7, both with mutations affecting Val51, were macrocephalic (OFCs of +4.16 and +4.5 SD, respectively) (Table 1; Figure 2). Hypoplasia of the corpus callosum and the cerebellar vermis were the commonest features observed on available magnetic resonance imaging (MRI) studies of individuals with microcephaly. However, they all had additional abnormalities (Supplemental Note; Figure 2). Individual 5 (with normal OFC) was reported to have polymicrogyria and hypoplastic corpus callosum (images not available). The two individuals with macrocephaly (individuals 6 and 7) showed periventricular white matter lesions (Table 1; Figure 2). Arched eyebrows, dysplastic ears, prominent nasal bridges, and overhanging columellae were shared between a majority of individuals without macrocephaly (Supplemental Note; Figure 2), but their facial dysmorphism does not overlap sufficiently to make this condition recognizable via their gestalt. Both individuals with macrocephaly displayed prominent broad foreheads, open mouth appearance, and scooped out appearance on lateral view (Figure 2). Collectively, the clinical data suggested a remarkable phenotypic variability in our cohort.
We mapped each of the seven identified mutations onto the available crystal structure of human RAC1 (PDB: 3TH5) (Figure 1D) to gain insights into their effect on the protein structure and function. The amino acids affected by the seven mutations could be categorized into three groups. (1) Cys18 and Cys157 are located in and adjacent to, respectively, the guanine nucleotide binding site that binds GTP/GDP. These two cysteine thiols can be oxidized by glutathionylation, a post-translational modification that alters GTP binding and exchange activities of RAC1.27 The p.Cys18Tyr and p.Cys157Tyr substitutions introduce a bulky aromatic residue in place of the thiol group and are expected to impact GTPase activity either by directly interfering with GTP binding or indirectly by abolishing the post-translational modification. (2) Asn39 is part of the switch I motif, while Tyr64 and Pro73 are within and adjacent to, respectively, the switch II motif. Both switch motifs are highly conserved regions involved in the interactions with various GEFs and GAPs (e.g., Rex1, DOCK),28 mediating the conformational changes for guanine nucleotide exchange (Figure S1), and downstream effectors. Missense mutations affecting these residues are likely to impact the protein-protein interactions directly. (3) Val51 has no obvious involvement in GTP binding or interactions with GEF/GAPs. Free energy calculations by FoldX (in the SNPeffect 4.0 server)29 revealed a reduction of protein stability for these two substitutions (ΔΔG of 2.13 and 1.94 kcal/mol for p.Val51Met and p.Val51Leu, respectively).
RAC1 is known to regulate the spreading of fibroblasts plated onto fibronectin.30 The genetic data and in silico modeling suggested that the phenotypes are unlikely to result from haploinsufficiency. We reasoned that the RAC1 mutations identified in this study could be dominant negative or acting as constitutively active. If so, these mutations should result in changes in fibroblast spreading. To test this hypothesis, we introduced selected RAC1 mutations in NIH 3T3 fibroblasts. Mammalian expression plasmids encoding GFP-Rac1, GFP-Rac1-T17N, and GFP-Rac1-Q61L were obtained from Prof. Viki Allan (Manchester). A Quikchange Lightning kit (Agilent Technologies) was used to introduce point mutations in the GFP-Rac1 expression plasmid. We generated plasmids encoding 6/7 variants identified in this study (p.Cys18Tyr, p.Asn39Ser, p.Val51Met, p.Tyr64Asp, p.Pro73Leu, and p.Cys157Tyr) along with p.Thr17Asn that is known to have a dominant-negative effect31 and p.Gln61Leu which is known to result in constitutive protein activation.32 NIH 3T3 fibroblasts were cultured in a 24-well plate and were transfected with the Rac expression plasmids 24 hr after plating using Fugene 6 reagent (Promega). 48 hr after transfection, the fibroblasts were trypsinized then replated onto fibronectin-coated coverslips. 30 min after re-plating, the coverslips were rinsed with PBS and fixed with 4% paraformaldehyde (PFA). The fixed cells were then permeabilized with 0.1% Triton X-100 in PBS followed by blocking with 1% BSA in PBS. The cells were stained with a rabbit anti-GFP (Invitrogen) antibody (1:500), followed by an Alexa 488 anti-rabbit secondary antibody (Invitrogen) and Alexa568-phalloidin (Invitrogen) before mounting in Prolong Gold (Invitrogen). Cells were imaged on a Nikon A1R confocal microscope, using a 60× 1.4NA oil objective.
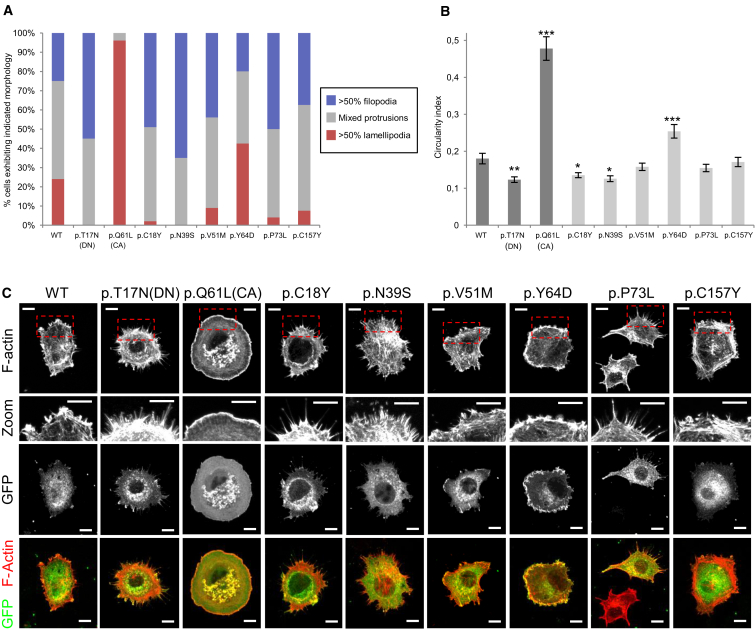
Cell circularity values were obtained using ImageJ software. Cell perimeters were identified using the ImageJ “threshold” function and then circularity index (4π × area/perimeter2) was calculated using the “analyze particles” function. Circularity datasets were statistically analyzed using one-way ANOVA with Dunnett’s correction for multiple comparisons. At least 50 cells were analyzed for each dataset except p.Asn39Ser, p.Tyr64Asp, p.Cys157Tyr (>40 cells), and p.Gln61Leu (25 cells). Data were pooled from three independent experiments and highly expressing cells were excluded. Morphology was also assessed qualitatively by classifying cells according to their predominant actin protrusion type. Three categories were used: (1) >50% of cell perimeter occupied of filopodia, (2) >50% of perimeter occupied by lamellipodia/ruffles, and (3) mixed protrusions, with neither type occupying 50% of the perimeter.
The majority (51%) of cells transfected with wild-type Rac1 exhibited mixed protrusions, with the remaining cells split almost equally between those in which >50% the perimeter was occupied by filopodia or lamellipodia/ruffles (Figure 3A). As observed previously, transfection of the known dominant-negative p.Thr17Asn variant resulted in a filopodia-rich cell perimeter and a significantly reduced circularity index (Figures 3A–3C).30 By contrast, expression of the known constitutively active p.Gln61Leu variant resulted in virtually all cells exhibiting large lamellipodia or membrane ruffles and a significantly increased circularity index.33 Expression of the p.Cys18Tyr and p.Asn39Ser variants (seen in individuals 1 and 2 with microcephaly) resulted in a phenotype reminiscent of dominant-negative Rac1, with an increase in the proportion of cells rich in filopodia and a reduction in cells rich in lamellipodia/ruffles (Figures 3A and 3C). Consistently, these mutations exhibited a significantly decreased circularity index (Figure 3B). By contrast, cells transfected with the p.Tyr64Asp substitution resulted in a phenotype more reminiscent of constitutive active Rac1, with significantly increased circularity index and a greater proportion of cells exhibiting lamellipodia or ruffles (Figures 3A–3C). Cells expressing p.Val51Met, p.Pro73Leu, and p.Cys157Tyr all showed a tendency toward increased filopodia and reduced lamellipodia but did not result in a significant change in circularity index relative to cells expressing wild-type Rac, suggesting at most a modest impact on Rac function in these assays (Figures 3A–3C). All the Rac1 mutant proteins exhibited similar cellular localization to wild-type Rac1 with the exception of p.Tyr64Asp, which appeared to localize strongly to the leading edge of ruffles and lamellipodia (Figure 3C).
Figure 3.
Distinct RAC1 Mutations Genocopy Dominant-Negative or Constitutively Active Variants
(A) NIH 3T3 fibroblasts transfected with 500 ng of DNA per coverslip expressing the indicated Rac1 mutants and were fixed 30 min after plating onto fibronectin and stained with Alexa-568 phalloidin to reveal the actin cytoskeleton and anti-GFP to reveal the expressed construct. Cells were imaged by confocal microscopy and divided into the three indicated categories. At least 50 cells were analyzed for each dataset except p.Asn39Ser, p.Tyr64Asp, p.Cys157Tyr (40 cells), and p.Gln61Leu (25 cells). Data were pooled from three independent experiments.
(B) Graph showing mean circularity index of cells expressing indicated Rac1 mutants. Error bars indicate SEM. Datasets statistically analyzed using ANOVA test with Dunnett’s correction for multiple comparisons. Asterisks indicate datasets significantly different to WT (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(C) Images of cells expressing the indicated RAC1 mutation. In each case the cell is a representative example of the most common morphological category for that mutation (see A). Top panels show Alexa-568 phalloidin staining to reveal F-actin distribution, while the panels immediately below show a magnified view of the boxed region from the top panels to highlight the morphology of actin protrusions at the cell periphery. The third row of panels show the GFP channel of the same cells to reveal the distribution of the expressed RAC1 mutation, and the bottom panels show a merge of the F-actin (red) and GFP (green) channels. All scale bars indicate 10 μm.
Next, we sought to explore the in vivo effects of RAC1 mutations in zebrafish embryos. Toward this, we identified rac1a as the sole zebrafish ortholog with highest degree of homology to the human RAC1 (90% similarity, 90% identity) and used the CRISPR/Cas9 system to introduce deletions, as a way to explore the phenotypic effects induced by loss of function of RAC1. Guide RNAs targeting the Danio rerio coding region of rac1 were generated as previously described.34, 35 We observed sequence aberrations in 30% of the evaluated rac1 clones (Figure S2A). Assessment of mosaic F0 embryos injected with a guide against exon 2 did not result in statistically different head size counts between F0 CRISPR and control embryos (Figure S2B). No overt morphological changes were observed in the genetically edited embryos.
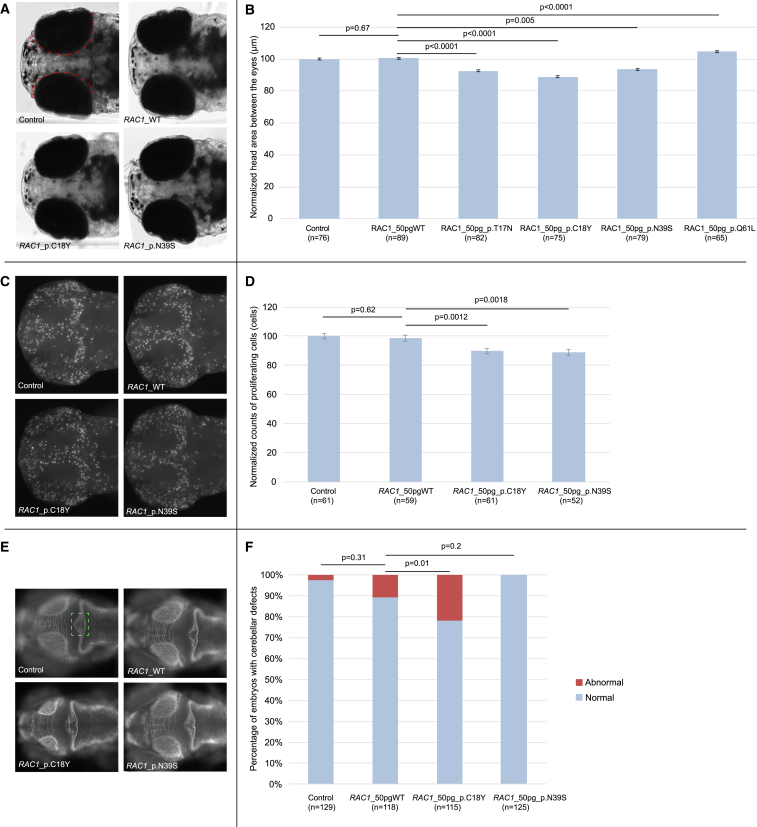
We next considered a dominant effect of the RAC1 alleles. To test this hypothesis, we cloned the human wild-type RAC1 mRNA (GenBank: NM_018890) into the pCS2+ vector and transcribed in vitro using the SP6 Message Machine kit (Ambion). The mouse fibroblast spreading assays had indicated that dominant-negative effects could be a common mechanism for RAC1 mutations. We therefore introduced the two variants (p.Cys18Tyr and p.Asn39Ser) that were shown to be the strongest genocopies of the known dominant-negative mutation using Phusion high-fidelity DNA polymerase (New England Biolabs) and custom-designed primers. Additionally, for sake of comparison, we also introduced the known dominant-negative (p.Thr17Asn) and constitutively active (p.Gln61Leu) mutants. Based on the dose-curve for the titration of the effect that WT RAC1 had on the head-size phenotype (Figure S3), we injected 50 pg of WT or mutant RNA into wild-type zebrafish embryos at the 1- to 4-cell stage. The injected larvae were grown to 5 dpf and imaged live on dorsal view. The area of the head was traced excluding the eyes from the measurements and statistical significance was calculated using Student’s t test. All experiments were repeated three times and scored blind to injection cocktail. Injection of RNA encoding p.Thr17Asn, p.Cys18Tyr, or p.Asn39Ser in zebrafish embryos induced a significant decrease in head-size (p < 0.0005) compared to controls (Figures 4A and 4B). Injection of RNA encoding p.Gln61Leu induced a significant increase in head-size (p < 0.0001) compared to controls (Figures 4A and 4B). Importantly, the head-size could not be rescued by co-injection of mutant RAC1 with WT message (Figure S4) arguing further in favor of these RAC1 mutations acting as dominant alleles in vivo, consistent with our results of mouse fibroblast spreading assay.
Figure 4.
Overexpression of Human RAC1 p.Cys18Tyr and p.Asn39Ser Cause Microcephaly, while p.Gln61Leu Causes the Mirror Phenotype in Zebrafish Embryos
Functional assessment of de novo variants in RAC1 by in vivo complementation in zebrafish larvae.
(A) Dorsal view of 5 days post fertilization (dpf) control and overexpressant larvae. For each experiment, embryos were injected with either WT or mutant RAC1 human mRNA message. The embryos were allowed to grow to 5 dpf and imaged live for head size. The midbrain area between the eyes highlighted with the dashed red line was measured for every imaged embryo, to produce a quantitative score.
(B) Bar graph of normalized values showing the quantification of the head size phenotype in control embryos and embryos injected with either WT or mutant human RAC1 message, from two plotted experiments. Statistical analyses were performed by Student’s t test.
(C) Dorsal view of 2 dpf control and overexpressant larvae, fixed in Dent’s fixative and whole-mount immuno-stained with an anti-phospho histone 3 (PH3) antibody that marks proliferating cells. For each experiment, embryos were injected with either WT or mutant RAC1 human mRNA message. The embryos were allowed to grow to 2 dpf and subsequently were fixed, stained, and imaged. Embryos were imaged dorsally and z stacks were acquired every 100 nm. The z stacks were collapsed to produce an extended depth of focus (EDF) image that was then processed for scoring. The PH3-positive cells were counted for every embryo imaged using the ITCN plugin from ImageJ, to produce a quantitative score.
(D) Bar graph of normalized values showing the quantification of the proliferating cell count phenotype, across three biological replicas. Statistical analyses for the neuronal proliferation experiments were performed by Student’s t test.
(E) Dorsal view of 3 dpf control and overexpressant larvae for RAC1 WT and mutant conditions. The area of the cerebellum consisting of the neuronal axons that cross the midline is highlighted with a green dashed box. At least 50 embryos per condition were imaged live and evaluated at 3 dpf for the depletion of axons within the area highlighted.
(F) Bar graph of cumulative plotted experiments across three biological replicas showing percentages of embryos with cerebellar defects. Statistical analyses for the cerebellar integrity assay using a χ2 test.
Error bars define 95% confidence interval.
Driven by the fact that RAC1 is involved in neuronal proliferation,20 we next assessed neuronal proliferation in the brain of embryos injected with WT or a subset of mutant RAC1 (encoding p.Cys18Tyr or p.Asn39Ser). To do this, the injected embryos were fixed overnight at 48 hr post fertilization (hpf) in Dent’s fixative (80% methanol, 20% DMSO) at 4°C. The embryos were first rehydrated in progressively decreasing methanol solutions, bleached with 10% H2O2, 0.5% KOH, and 0.1% Triton-X. After two washes in PBS, the tissue was permeabilized with proteinase K followed by post-fixation with 4% PFA. PFA-fixed embryos were washed first in PBS and subsequently in IF buffer (0.1% Tween-20, 1% BSA in PBS) for 10 min at room temperature. The embryos were incubated in the blocking buffer (10% FBS, 1% BSA in PBS) for 1 hr at room temperature. After two washes in IF buffer for 10 min each, embryos were incubated overnight at 4°C with 1:500 phospho histone 3 (PH3, a marker for proliferating cells) primary antibody (ser10)-R (sc-8656-R, rabbit, Santa Cruz) in blocking solution. After two additional washes in IF buffer for 10 min each, embryos were incubated in the secondary antibody solution (Alexa Fluor goat anti-mouse IgG [A21207, Invitrogen] and donkey anti-rabbit [A21206, Invitrogen], 1:1,000) in blocking solution for 1 hr at room temperature. Proliferating cells were quantified by counting all positive cells on a dorsal view of a 48 hpf embryos, excluding the eyes from the scored area, using the ITCN ImageJ plugin that counts cells with 10 pixel width and 5 pixel minimum distance between them in order to be considered as separate cells. Statistical significance for this assay was established using Student’s t test. This assay showed significantly reduced cellular proliferation for both mutants (p = 0.0012 for p.Cys18Tyr and p = 0.0018 for p.Asn39Ser when compared to WT RAC1; Figures 4C and 4D).
Finally, armed with prior knowledge that RAC1 has a known role in cerebellar development36 together with the observation that cerebellar abnormalities were reported in 3/7 individuals in our cohort, we sought to study the effect of the p.Cys18Tyr and p.Asn39Ser variants on cerebellar development of our zebrafish model. For the assessment of the cerebellum, embryos were fixed at 72 hpf in Dent’s fixative and subsequently whole-mount stained following the same staining protocol as for neuronal proliferation and using a primary antibody against acetylated tubulin (1:1,000; Thr7451, mouse, Sigma-Aldrich). The embryos were then scored qualitatively assaying the integrity of the cerebellum by scoring for the presence and organization of axons along the midline of the structure (highlighted with a green dashed box; Figure 4E) and statistical significance was determined using a χ2 test. Structural defects in the integrity of the cerebellum consisted of depletion of the axons that cross the midline were observed only upon overexpression of RAC1 p.Cys18Tyr (p = 0.01) but not RAC1 p.Asn39Ser (p = 0.2) (Figures 4E and 4F). The latter is likely consistent with the more pronounced cerebellar defect observed in individual 1 (who harbors the p.Cys18Tyr variant) (Figure 2B).
In summary, we report seven individuals with de novo missense RAC1 mutations and variable developmental delay with additional features. Remarkably, the OCFs observed in this study ranged from −5 SD to +4.5 SD. Previously, some chromosomal regions have been associated with both microcephaly (deletions) and macrocephaly (duplications).37, 38, 39 However, it is extremely rare that point mutations within the same gene can cause differences of such magnitude (∼10 SD) in head size of affected individuals. It is interesting to note that RAC1 is involved in mTOR signaling and other disorders in this pathway also result in significant alterations in head size.17, 40, 41 The variability of RAC1 phenotypes appear to be dependent on specific mutations, although the contribution of genetic background cannot be ruled out. In silico modeling, mouse fibroblasts spreading assays, and zebrafish experiments demonstrate that some RAC1 mutations (those encoding p.Cys18Tyr and p.Asn39Ser) genocopy a known dominant-negative mutant (p.Thr17Asn) and result in reduced neuronal proliferation, microcephaly, and cerebellar abnormalities in vivo. On the other hand, the in vitro effects of p.Tyr64Asp, seen in affected individual 5 with OFC within the normal range, are similar to the known constitutively active RAC1 mutation. Of note, in vitro expression of dominant-negative and constitutively active Rac1 have been previously shown to cause opposite effects on dendritic growth and morphology.18 However, the link between the human mutations and the resultant phenotypes is likely to be complex depending on the balance between interactors, regulators, and effectors of RAC1 signaling.42 This is likely to be especially true for other RAC1 mutations identified in this study that could not be clearly classed as being either dominant negative or constitutively active (those encoding p.Val51Met, p.Pro73Leu, and p.Cys157Tyr). Further studies will be required in the future to uncover the precise underlying mechanisms of phenotypic variability of RAC1 mutations.
Our findings show that mutations in genes encoding members of the RhoGTPases-family can cause DDs. Of note, mutations in TRIO (MIM: 601893), a gene that encodes a RAC1 GEF, have been shown to result in mild intellectual disability.43, 44 Interestingly, missense mutations in RAC-GEF domain of TRIO result in a more severe phenotype with global developmental delay, microcephaly, and reduced RAC1 activity.43 Furthermore, bi-allelic mutations in HACE1 (MIM: 610876), a known interactor of RAC1, have been recently shown to result in an autosomal-recessive syndrome with macrocephaly.45 Notably, overexpression of a WAVE mutant has been demonstrated to partially rescue axon growth in Rac1 knock-out neurons,19 suggesting that some of the conditions in this group could be potentially treatable. Overall, a potentially treatable sub-category of rare DDs appears to be emerging with RAC1 as the central player.
Finally, our results show that ultra-rare disorders caused by private, non-recurrent missense mutations, resulting in varying phenotypic effects severity and degrees of severity, are challenging to dissect but can be delineated through focused international collaborations.
Acknowledgments
We would like to thank all the families for agreeing to participate in this study. We acknowledge the Rare Disease Foundation and the BC Children’s Hospital Foundation for their support for this work (S.B., grant number 17-48). N.K. is a Distinguished George W. Brumley Professor. W.W.Y. is a researcher at the Structural Genomics Consortium, a registered charity (number 1097737; funding details online). We thank Prof. Viki Allan (University of Manchester) for providing mammalian expression plasmids encoding GFP-Rac1, GFP-Rac1-T17N, and GFP-Rac1-Q61L. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute (grant number WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83 granted by the Cambridge South REC and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network.
Published: September 7, 2017
Footnotes
Supplemental Data include Supplemental Note and three figures and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.08.007.
Contributor Information
Han G. Brunner, Email: han.brunner@radboudumc.nl.
Siddharth Banka, Email: siddharth.banka@manchester.ac.uk.
Web Resources
ExAC Browser, http://exac.broadinstitute.org/gene/ENSG00000136238
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
ImageJ, https://imagej.nih.gov/ij/
Molsoft, http://www.molsoft.com/icm_pro.html
OMIM, http://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Structural Genomics Consortium, http://www.thesgc.org/
Supplemental Data
References
- 1.Sheridan E., Wright J., Small N., Corry P.C., Oddie S., Whibley C., Petherick E.S., Malik T., Pawson N., McKinney P.A., Parslow R.C. Risk factors for congenital anomaly in a multiethnic birth cohort: an analysis of the Born in Bradford study. Lancet. 2013;382:1350–1359. doi: 10.1016/S0140-6736(13)61132-0. [DOI] [PubMed] [Google Scholar]
- 2.Ropers H.H. Genetics of early onset cognitive impairment. Annu. Rev. Genomics Hum. Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- 3.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lelieveld S.H., Reijnders M.R., Pfundt R., Yntema H.G., Kamsteeg E.J., de Vries P., de Vries B.B., Willemsen M.H., Kleefstra T., Löhner K. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 7.MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., Adams D.R., Altman R.B., Antonarakis S.E., Ashley E.A. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rauen K.A. The RASopathies. Annu. Rev. Genomics Hum. Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Aelst L., D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 10.Heasman S.J., Ridley A.J. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 11.Duquette P.M., Lamarche-Vane N. Rho GTPases in embryonic development. Small GTPases. 2014;5:8. doi: 10.4161/sgtp.29716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stankiewicz T.R., Linseman D.A. Rho family GTPases: key players in neuronal development, neuronal survival, and neurodegeneration. Front. Cell. Neurosci. 2014;8:314. doi: 10.3389/fncel.2014.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aspenström P., Fransson A., Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem. J. 2004;377:327–337. doi: 10.1042/BJ20031041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boureux A., Vignal E., Faure S., Fort P. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol. Biol. Evol. 2007;24:203–216. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahirovic S., Hellal F., Neukirchen D., Hindges R., Garvalov B.K., Flynn K.C., Stradal T.E., Chrostek-Grashoff A., Brakebusch C., Bradke F. Rac1 regulates neuronal polarization through the WAVE complex. J. Neurosci. 2010;30:6930–6943. doi: 10.1523/JNEUROSCI.5395-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wojnacki J., Quassollo G., Marzolo M.P., Cáceres A. Rho GTPases at the crossroad of signaling networks in mammals: impact of Rho-GTPases on microtubule organization and dynamics. Small GTPases. 2014;5:e28430. doi: 10.4161/sgtp.28430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saci A., Cantley L.C., Carpenter C.L. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol. Cell. 2011;42:50–61. doi: 10.1016/j.molcel.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugihara K., Nakatsuji N., Nakamura K., Nakao K., Hashimoto R., Otani H., Sakagami H., Kondo H., Nozawa S., Aiba A., Katsuki M. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene. 1998;17:3427–3433. doi: 10.1038/sj.onc.1202595. [DOI] [PubMed] [Google Scholar]
- 19.Threadgill R., Bobb K., Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 20.Leone D.P., Srinivasan K., Brakebusch C., McConnell S.K. The rho GTPase Rac1 is required for proliferation and survival of progenitors in the developing forebrain. Dev. Neurobiol. 2010;70:659–678. doi: 10.1002/dneu.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L., Melendez J., Campbell K., Kuan C.Y., Zheng Y. Rac1 deficiency in the forebrain results in neural progenitor reduction and microcephaly. Dev. Biol. 2009;325:162–170. doi: 10.1016/j.ydbio.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matos P., Skaug J., Marques B., Beck S., Veríssimo F., Gespach C., Boavida M.G., Scherer S.W., Jordan P. Small GTPase Rac1: structure, localization, and expression of the human gene. Biochem. Biophys. Res. Commun. 2000;277:741–751. doi: 10.1006/bbrc.2000.3743. [DOI] [PubMed] [Google Scholar]
- 24.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordan P., Brazåo R., Boavida M.G., Gespach C., Chastre E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene. 1999;18:6835–6839. doi: 10.1038/sj.onc.1203233. [DOI] [PubMed] [Google Scholar]
- 26.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci. STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobbs G.A., Mitchell L.E., Arrington M.E., Gunawardena H.P., DeCristo M.J., Loeser R.F., Chen X., Cox A.D., Campbell S.L. Redox regulation of Rac1 by thiol oxidation. Free Radic. Biol. Med. 2015;79:237–250. doi: 10.1016/j.freeradbiomed.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y., Xing J., Streuli M., Leto T.L., Zheng Y. Trp(56) of rac1 specifies interaction with a subset of guanine nucleotide exchange factors. J. Biol. Chem. 2001;276:47530–47541. doi: 10.1074/jbc.M108865200. [DOI] [PubMed] [Google Scholar]
- 29.De Baets G., Van Durme J., Reumers J., Maurer-Stroh S., Vanhee P., Dopazo J., Schymkowitz J., Rousseau F. SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 2012;40:D935–D939. doi: 10.1093/nar/gkr996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price L.S., Leng J., Schwartz M.A., Bokoch G.M. Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol. Biol. Cell. 1998;9:1863–1871. doi: 10.1091/mbc.9.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridley A.J., Paterson H.F., Johnston C.L., Diekmann D., Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 32.Machesky L.M., Hall A. Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J. Cell Biol. 1997;138:913–926. doi: 10.1083/jcb.138.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michiels F., Habets G.G., Stam J.C., van der Kammen R.A., Collard J.G. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- 34.Jao L.E., Wente S.R., Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perles Z., Moon S., Ta-Shma A., Yaacov B., Francescatto L., Edvardson S., Rein A.J., Elpeleg O., Katsanis N. A human laterality disorder caused by a homozygous deleterious mutation in MMP21. J. Med. Genet. 2015;52:840–847. doi: 10.1136/jmedgenet-2015-103336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulherkar S., Uddin M.D., Couvillon A.D., Sillitoe R.V., Tolias K.F. The small GTPases RhoA and Rac1 regulate cerebellar development by controlling cell morphogenesis, migration and foliation. Dev. Biol. 2014;394:39–53. doi: 10.1016/j.ydbio.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nevado J., Rosenfeld J.A., Mena R., Palomares-Bralo M., Vallespín E., Ángeles Mori M., Tenorio J.A., Gripp K.W., Denenberg E., Del Campo M. PIAS4 is associated with macro/microcephaly in the novel interstitial 19p13.3 microdeletion/microduplication syndrome. Eur. J. Hum. Genet. 2015;23:1615–1626. doi: 10.1038/ejhg.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Dijck A., van der Werf I.M., Reyniers E., Scheers S., Azage M., Siefkas K., Van der Aa N., Lacroix A., Rosenfeld J., Argiropoulos B. Five patients with a chromosome 1q21.1 triplication show macrocephaly, increased weight and facial similarities. Eur. J. Med. Genet. 2015;58:503–508. doi: 10.1016/j.ejmg.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Shinawi M., Liu P., Kang S.H., Shen J., Belmont J.W., Scott D.A., Probst F.J., Craigen W.J., Graham B.H., Pursley A. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J. Med. Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivière J.B., Mirzaa G.M., O’Roak B.J., Beddaoui M., Alcantara D., Conway R.L., St-Onge J., Schwartzentruber J.A., Gripp K.W., Nikkel S.M., Finding of Rare Disease Genes (FORGE) Canada Consortium De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosner M., Hanneder M., Siegel N., Valli A., Fuchs C., Hengstschläger M. The mTOR pathway and its role in human genetic diseases. Mutat. Res. 2008;659:284–292. doi: 10.1016/j.mrrev.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Hetmanski J.H.R., Schwartz J.M., Caswell P.T. Rationalizing Rac1 and RhoA GTPase signaling: A mathematical approach. Small GTPases. 2016:1–6. doi: 10.1080/21541248.2016.1218406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pengelly R.J., Greville-Heygate S., Schmidt S., Seaby E.G., Jabalameli M.R., Mehta S.G., Parker M.J., Goudie D., Fagotto-Kaufmann C., Mercer C., DDD Study Mutations specific to the Rac-GEF domain of TRIO cause intellectual disability and microcephaly. J. Med. Genet. 2016 doi: 10.1136/jmedgenet-2016-103942. jmedgenet-2016-103942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ba W., Yan Y., Reijnders M.R., Schuurs-Hoeijmakers J.H., Feenstra I., Bongers E.M., Bosch D.G., De Leeuw N., Pfundt R., Gilissen C. TRIO loss of function is associated with mild intellectual disability and affects dendritic branching and synapse function. Hum. Mol. Genet. 2016;25:892–902. doi: 10.1093/hmg/ddv618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hollstein R., Parry D.A., Nalbach L., Logan C.V., Strom T.M., Hartill V.L., Carr I.M., Korenke G.C., Uppal S., Ahmed M. HACE1 deficiency causes an autosomal recessive neurodevelopmental syndrome. J. Med. Genet. 2015;52:797–803. doi: 10.1136/jmedgenet-2015-103344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.