Abstract
OBJECTIVES
To establish whether two families from Malopolska and Mazovia Provinces in Poland are affected by hereditary gingival fibromatosis type 1, caused by a single-cytosine insertion in exon 21 of the Son-of-Sevenless-1 gene.
MATERIAL AND METHODS
Six subjects with hereditary gingival fibromatosis and five healthy subjects were enrolled in the study. Gingival biopsies were collected during gingivectomy or tooth extraction and used for histopathological evaluation. Total RNA and genomic DNA were purified from cultured gingival fibroblasts followed by cDNA and genomic DNA sequencing and analysis.
RESULTS
Hereditary gingival fibromatosis was confirmed by periodontal examination, X-ray and laboratory tests. Histopathological evaluation showed hyperplastic epithelium, numerous collagen bundles and abundant to moderate fibroblasts in subepithelial and connective tissue. Sequencing of exons 19–22 of the Son-of-Sevenless-1 gene did not reveal a single-cytosine insertion nor other mutations.
CONCLUSIONS
Patients from two Polish families under study had not been affected by hereditary gingival fibromatosis type 1, caused by a single-cytosine insertion in exon 21 of the Son-of-Sevenless-1 gene. Further studies of the remaining regions of this gene as well as of other genes are needed to identify disease-related mutations in these patients. This will help to unravel the pathogenic mechanism of gingival overgrowth.
Keywords: hereditary gingival fibromatosis, pedigree, SOS-1, mutation, heterogeneity
Introduction
Hereditary gingival fibromatosis (HGF, MIM 135300) is a genetically heterogeneous and rare disease with unknown prevalence, characterized by a benign, fibrous and slowly progressive overgrowth of the gingiva (Häkkinen and Csiszar, 2007). It may affect the mandible and/or the maxilla locally or manifest itself as a generalized condition. In severe cases, the overgrowth can cover entire crowns of the teeth, thus resulting in prolonged retention of primary dentition, diastemas or malposition of teeth. It may also cause phonetic, masticatory and psychological issues (Gawron et al, 2016a). Although gingival overgrowth (GO) does not directly affect the marginal alveolar bone, it may induce bacterial plaque accumulation and periodontitis resulting in alveolar bone loss and halitosis (Coletta and Graner, 2006; Gorlin et al, 1990). GO may appear as an isolated condition or co-exist with a genetic syndrome or disease. It has been identified in craniofacial dysmorphism (MIM 228560), progressive deafness (MIM 135550), Murray-Puretic-Drescher syndrome (MIM 228600), Zimmermann-Laband syndrome (MIM 135500), Rutherfurd syndrome (MIM 180900), infantile systemic hyalinosis (MIM 236490) or amelogenesis imperfecta (MIM 614253) with the incidence of one or less per million. The frequency of other syndromes i. e., hypertrichosis syndrome (MIM 135400) and Ramon syndrome (MIM 266270) that may co-exist with a periodontal phenotype is unknown and may present with or without mental deficiency. Sporadic co-existence of GO with Bardet-Biedl syndrome (MIM 209900) and multiple hamartoma syndrome (MIM 158350) was also reported (Gawron et al, 2016b). Mutation in several genes has been proposed as a candidate or a causative factor of some genetic syndromes manifesting with periodontal phenotypes (Liaw et al, 1997; El-Kamah et al, 2010; O’Sullivan et al, 2011; Denadai et al, 2012; Jaureguiberry et al, 2012; DeStefano et al, 2014; Kortum et al, 2015).
HGF is transmitted as a Mendelian trait in an autosomal dominant or, less frequently, an autosomal recessive mode (Häkkinen and Csiszar, 2007). Linkage analysis of families suffering from HGF revealed a diversified genetic background and indicated several regions on chromosomes that may potentially contain mutations contributing to this condition. Loci for non-syndromic autosomal dominant variants of HGF have been identified on chromosome 2p21-p22 (GINGF) in a Brazilian family (Hart et al, 1998), chromosome 5q13-q22 (GINGF2) (Xiao et al, 2001), 2p22.3-p23.3 (GINGF3) (Ye et al, 2005) and 11p15 (GINGF4) (Zhu et al, 2007) in several Chinese families. Sequence analysis in the candidate region 2p21-p22 underlying GINGF locus showed an insertion of a single base in the Son-of-Sevenless-1 (SOS-1) gene (MIM 182530) in a large multigeneration Brazilian family characterized by a high (autosomal dominant) penetrance of HGF. Most notably, the single cytosine insertion resulting in a truncated protein was identified in all family members affected by HGF, while it was not found in unaffected family members or in the control group (Hart et al, 2002). Therefore, a mutation in SOS-1 has been proposed as one possible etiological factor for non-syndromic HGF, and defined as type 1.
The aim of this study was to establish whether two families from Malopolska and Mazovia Provinces in Poland are affected by hereditary gingival fibromatosis type 1, caused by a single-cytosine insertion in exon 21 of the SOS-1 gene.
Material and Methods
Study participants
The study was carried out in accordance with the Declaration of Helsinki. The Bioethics Committee at the Jagiellonian University, Medical College in Krakow, Poland approved the protocol, including the clinical/periodontal examination and collection of gingival tissue biopsies (KBET/133/B/2012). All subjects read and signed a written informed consent prior to recruitment. Six affected subjects from two families (further referred to as Family 1 and Family 2) with non-syndromic HGF and five healthy subjects (control group) were recruited. Medical, dental and medication histories of the subjects were taken by the clinicians. The diagnosis was confirmed by periodontal and radiographic examination, and routine laboratory tests. Additionally, two unaffected individuals from Family 1 and three unaffected members of Family 2 underwent clinical and oral examination. The inclusion criteria were diagnosis of non-syndromic HGF and the indication for treatment of the condition (gingivectomy) and/or tooth extraction due to orthodontic indications.
Exclusion criteria included a medical condition requiring pre-medication and/or pre-treatment prior to dental procedures, five or more decayed teeth at screening (cavities), diagnosis of other diseases of the oral cavity, use of antibiotics or antimicrobial drugs 30 days prior to surgery, history of genetic diseases/syndromes or systemic diseases, i.e., cardiovascular disease or epilepsy, women during pregnancy and/or lactation, tobacco smokers, immune- compromised individuals, i.e., undergoing immunosuppression, radio- and chemotherapy, HIV/AIDS.
Sample collection
Gingival biopsies from five subjects with non-syndromic HGF from two unrelated families were collected during gingivectomy, namely, from the anterior region of the mandibular gingiva from a 32-year-old female (Fig. 1a) (Gawron et al, 2014), and the anterior region of the maxilla from a 13-year-old (Fig. 1b) and an 11-year-old females (Fig. 1c) (Gawron et al, 2016a) (Family 1), as well as from the gingiva covering the mandibular incisors from a 33-year-old female (Fig. 1d) and a 41-year-old female (Fig. 1e) (Family 2). Gingivectomy was carried out to remove excessive amount of the marginal/coronal part of the keratinized gingiva. Gingival samples from the third patient from Family 2 (a 10-year-old son of the 41-year-old female) were collected from the maxillary gingiva during selective extraction of a deciduous molar for orthodontic reasons (Fig. 1f). Control gingival biopsies were collected from five healthy volunteers (four females and one male, aged 30–38 years). Immediately after collection, tissue biopsies were rinsed with sterile phosphate-buffered saline (PBS, pH 7.2) and divided into two portions. One portion of the tissue was kept for 24 h at 4°C in 10% formalin in PBS, and then proceeded to embed in paraffin. The second portion was kept in PBS at 4°C for isolation of gingival fibroblasts.
Fig. 1. Clinical presentation of patients from two Polish families with non-syndromic hereditary gingival fibromatosis enrolled in the study: a).
a 32-year-old female (Gawron et al, 2014), b) a 13-year-old female (Gawron et al, 2016b), c) an 11-year-old female (Gawron et al, 2016a) from Family 1; d) a 33-year-old female, e) a 41-year-old female, f) a 10-year-old male from Family 2. The oral status before gingivectomy is depicted in all cases.
Histopathology
Gingival biopsies were fixed in 10% formalin in PBS for 24 h at 4°C, and then rinsed with PBS for further 24 h. Tissue was dehydrated in series of alcohols and acetone, then kept in xylene bath (2 × 1.5h), rinsed in paraffin bath (3 × 1h), and finally paraffin embedded. Gingival tissue was serially cut for the sections of 7 μm thickness and stained using haematoxylin and eosin (Sigma-Aldrich, Poland). In brief, paraffin sections of gingiva were incubated for 1 h at 37°C, then kept in xylene baths (2 × 10 minutes) for deparaffinization. Next, tissues were rehydrated in graded alcohols and rinsed in bidistilled water. Harris haematoxylin solution (Sigma-Aldrich, Poland) was used for a 2-minute staining, followed by rinsing in tap water for 30 minutes. Next, eosin H solution (Sigma-Aldrich, Poland) was added for 20 seconds. The tissues were further dehydrated in graded alcohols, kept in xylene bath (2 × 10 minutes) and mounted in Shandon Consul Mount (ThermoScientific, Poland). Histopathological evaluation of gingival tissues was carried out using a light microscope (Nikon Eclipse Ti, Japan) connected to NIS-Elements F 3.0 software (Nikon Inc.).
Primary human gingival fibroblasts isolation and culture
In brief, gingival biopsy sample was digested in dispase (Invitrogen) at 4°C for 18 h. The obtained subepithelial tissue was digested using 0.1% collagenase I (Invitrogen) at 37°C for 12 h. The cell pellet was plated in Dulbecco’s Modified Eagle Medium (DMEM, PAA GmbH) with 10% heat-inactivated fetal bovine serum (FBS, PAA GmbH), antibiotics (penicillin/streptomycin 50 u/ml, Gibco; gentamycin 50 μg/ml, ThermoFisher Scientific), nystatin (6 μg/ml, Sigma) and cultivated for 7 days. The homogeneity of the fibroblast culture was confirmed after first passage by immunofluorescent staining against vimentin and cytokeratin. Cell viability amounted to 99% using Trypan blue. The experiments were carried out between the third and sixth passage.
Sequence analysis of exons 19–22 of the SOS-1 gene
Total RNA was purified from gingival fibroblasts by TRI Reagent (Ambion), followed by DNA digestion with Turbo DNase (Life Technologies). 400 ng RNA was converted to cDNA using High-Capacity cDNA Reverse Transcription Kit (Life Technologies) in the total reaction volume of 20 μl. The mRNA of exons 19–22 of the SOS-1 gene was amplified using Phusion DNA polymerase (Thermo Scientific). Initial denaturation was conducted at 98°C for 30 sec. Target DNA was amplified at 98°C for 10 sec, 62°C for 30 sec and 72°C for 30 sec over 40 cycles, followed by an extension at 72°C for 10 min. Products of reaction were separated in agarose gel and purified using GeneJET Gel Extraction Kit (Thermo Scientific). Adenine overhangs were added after 10 min of incubation at 72°C using DreamTaq Green PCR Master Mix (2X) (Thermo Scientific) followed by purification with GeneJET PCR Purification Kit (Thermo Scientific). The obtained inserts were cloned into pTZ57R/T vector using InsTAclone PCR Cloning Kit (Thermo Scientific) and amplified in E.coli DH5α. Plasmids were then purified by GeneJET Plasmid Miniprep Kit (Thermo Scientific) and inserts were sequenced by Genomed S.A (Warsaw, Poland) using standard M13 primers for pTZ57R plasmid (Table 1). The target DNA sequences were analysed and aligned using Unipro UGENE 1.17.0 software.
Table 1.
Primers used for amplification of the SOS-1 gene fragments (Hart et al, 2002), and for sequencing, synthesized by Genomed S.A. (Warsaw, Poland).
| Forward primer | Reverse primer | |
|---|---|---|
| SOS1 mRNA exons 19–22 | ACCCTGAGGTCCTAAAAAGACATG | GCTCGAATGATCGGAATCAAATAC |
| SOS1 exon 21 | AGGGCTTTAGCAAAATAGAATGTT | ACTTGCAGATTTTAAGACTGATCT |
| Sequencing primers for pTZ57R (M13) | GTAAAACGACGGCCAGT | CAGGAAACAGCTATGAC |
Genomic DNA from gingival fibroblasts was isolated using GeneJET Genomic DNA Purification Kit (Thermo Scientific). Exon 21 of the SOS-1 gene was amplified with Phusion DNA polymerase (Thermo Scientific). After initial denaturation for 30 sec at 98°C, DNA was amplified at 98°C for 10 sec, 50°C for 30 sec, and 72°C for 30 sec over 40 cycles, followed by an extension at 72°C for 10 min. The DNA products were purified using GeneJET PCR Purification Kit (Thermo Scientific) and sequence analysis was carried out by Genomed S.A. (Warsaw, Poland) using primers identical as for amplification. The obtained sequences were analyzed using Unipro UGENE 1.17.0 software. The sequences of primers are depicted in Table 1.
Results
Clinical description of patients
Three patients (IV-2, V-1, V-2) from a five-generation family from the Malopolska Province in Poland (Fig. 2a) and three patients (III-2, II-3, IV-1) from a four-generation family from the Mazovia Province in Poland (Fig. 2b) with history of non-syndromic autosomal dominant HGF and five control donors were enrolled in the study. All participants underwent clinical, oral and radiographic examination by two experienced periodontologists. In addition, two unaffected individuals from Family 1 (IV-1, IV-3) and three from Family 2 (III-1, III-4, IV-2) underwent oral and radiographic examination (Fig. 2). As ascertained by a 32-year-old proband, apart from her and the two daughters (Fig. 1a–c) the condition was also present in her father, grandfather and great grandfather. In three affected members of Family 1 enrolled to participate in the study, the onset of GO was noted at the time of deciduous dentition and revealed diffuse involvement of mandibular and maxillary arches. The 32-year-old female underwent complete surgical excision of the lesion in 2013. Twelve months later the recurrence of GO was observed, particularly in the mandible (Gawron et al, 2014). Her 11-year-old and 13-year-old daughters underwent gingivectomy in 2015 for functional, hygienic, and psychological reasons. Control oral examination at one, three and six months after surgery revealed uneventful healing and no signs of recurrence in the 13-year-old daughter. In the case of the 11-year-old sibling, recurrence in the maxilla was noted two weeks after surgery and in the mandible at four weeks postoperatively. The recurrence involved 25% of the pre-operative tissue volume within the maxilla and 45% of the initial volume in the mandible. Gradual decrease of the recurrence was noted in both jaws in the following two months (Gawron et al, 2016a). All patients received outpatient care with control visits once every six months. Thus far, further surgical intervention has not been scheduled.
Fig. 2. Pedigree of two Polish families diagnosed with non-syndromic hereditary gingival fibromatosis: a).
Family 1, and b) Family 2. HGF-affected subjects are marked by blackened symbols, and healthy subjects by colorless symbols. Males are denoted by squares, and females by circles; a slash across a symbol indicates a deceased individual. Unknown or uncertain diagnosis is depicted by a question mark within the semi-blackened symbol. Arrows indicate the probands.
In Family 2, both sisters, aged 41 and 33 years were diagnosed with HGF at the initial examination in 2012 and 2014, respectively (Fig. 1d, 1e). Both sisters received surgical treatment in order to improve aesthetics and to initiate orthodontic treatment in the case of the older sibling. The orthodontic treatment in the older sibling aimed at alignment of slight malposition of teeth and lasted 12 months. This treatment neither clinically changed the status of gingiva nor affected the initial clinical diagnosis. Both patients underwent gingivectomy, alveolar osteotomy and osteoplasty under local anesthesia, first in the maxilla and then in the mandible. The older sister reported a recurrence of GO that was detectable four months after excision in the mandible, while the gingiva in the maxilla remained clinically normal. During the follow-up period the patient underwent orthodontic treatment with fixed orthodontic appliance. The recurrence of GO in the younger sister was observed exclusively in the mandibular anterior area. Consequently, both sisters underwent a second surgery in the anterior mandible. The surgeries were limited to gingivectomy since the alveolar bone level remained unaffected after initial osteoplasty. Both patients were then followed up for a year and no significant recurrence of GO was noted. The third patient from Family 2 (the 10-year-old son of the elder female) underwent extraction of deciduous molar tooth for orthodontic reasons (Fig. 1f). In this case, uneventful healing was observed after the surgery.
Histopathology of gingival tissue
Histopathological evaluation showed mild to moderate acanthosis of gingival epithelium with elongated or enlarged rete pegs extending into subepithelial and connective tissue (Fig. 3c, f, h, i). Abundant collagen bundles were present in subepithelial and dense connective tissue (Fig. 3b, d, g). Unlike normal gingiva (Fig. 3k, l), locally irregular, thick collagen fibers oriented in variable directions were predominantly visible within the deeper parts of connective tissue and around blood vessels (Fig. 3d, e, j). Abundant to moderate fibroblasts with numerous to scanty blood vessels were visible in gingival stroma (Fig. 3c–g). In some cases inflammatory infiltrates were observed in the subepithelial tissue and in almost entire connective tissue (Fig. 3a, b) or locally beneath extending rete pegs (Fig. 3i). Plasmocytes and histiocytes were predominant infiltrating cells, and less numerous population was represented by lymphocytes.
Fig. 3. Histological staining (hematoxylin and eosin) of gingival tissue biopsies from patients from two Polish families with non-syndromic hereditary gingival fibromatosis enrolled in the study: a, b).
a 32-year-old female (Gawron et al, 2014; Gawron et al, 2016b), c) a 13-year-old female (Gawron et al, 2016b), d, e) an 11-year-old female (Gawron et al, 2016a) from Family 1; f, g) a 33-year-old female, h) a 41-year-old female, i, j) a 10-year-old male from Family 2; k, l) control gingiva. Original magnific. 100 ×, bars = 100 μm.
Mutation analysis in exons 19–22 of the SOS-1gene
Sequencing of exon 21 of SOS-1 cDNA and genomic sequence analysis of exon 21 in HGF which affected members of the two recruited families did not show a single-cytosine insertion between nucleotides 3248 and 3249 (c.3248–3249insC) or between nucleotides 126,142 and 126,143 (g.126,142–126,143insC), respectively (Figs. 4 and 5). Sequencing analysis spanning across the exons 19, 20, 21 and 22 of SOS-1 cDNA did not show any other mutation, either.
Fig. 4. SOS-1 sequencing results from two Polish families with non-syndromic hereditary gingival fibromatosis.
Part of SOS-1 cDNA containing a potential insertion site, c.3248–3249insC in CDS, was cloned and sequenced in six individuals affected by hereditary gingival fibromatosis and five control donors. Sequences were aligned and compared to wild type and mutated variants. None of the analysed sequences contained the insertion, therefore, they are not shown individually. The mutated sequence is shown for comparison, protein sequences of the wild type gene (top) and the mutated variant (bottom) show a frame shift and an altered translation product. Inserted cytosine is denoted in a bold font, numeration of bases begins at the first letter of the start codon.
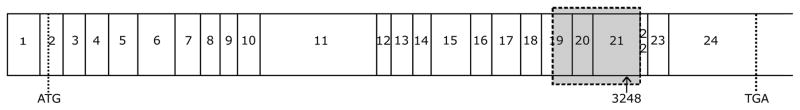
Fig. 5. Schematic presentation of SOS-1 cDNA.
Translation initiation and stop codons are marked with dotted lines as ATG and TGA, respectively. A grey box indicates sequenced region spanning over the 19, 20 and 21 exons. The arrow marks the insertion site after nucleotide 3248; numeration begins at the first letter of the start codon.
Discussion
So far, four distinct loci related to the non-syndromic variant of HGF have been identified, i.e., two map to chromosome 2 (GINGF on 2p21-p22 and GINGF3 on 2p22.3-p23.3) (Hart et al, 1998; Ye et al, 2005), one maps to chromosome 5 (GINGF2 on 5q13-q22) (Xiao et al, 2001) and one to chromosome 11 (GINGF4 on 11p15) (Zhu et al, 2007). Of these loci, only mutations in the SOS-1 gene underlying the GINGF locus (MIM 182530) have been identified. Originally, a single-cytosine insertion in the genomic sequence (g.126,142–126,143insC) and the cDNA sequence (c.3248–3249insC) of the SOS-1 gene, mapped to 2p21-p22 locus, have been identified as a causative factor of non-syndromic HGF type 1 in one family from Brasil (Hart et al, 1998; Hart et al, 2002). The single-cytosine insertion causes a frame shift and an early termination of the protein. The chimeric protein has augmented activity since it lacks the carboxyl-terminal domain that normally exerts negative allosteric regulation (Corbalan-Garcia et al, 1998; Saygun et al, 2003). In physiological conditions, SOS-1 mediates Ras activation, which stimulates several protein kinases, including the members of the MAPK, PI3K and proteins of the Rho family (Rusanescu et al, 2001; Funato et al, 2004). The consequence is transcriptional control of the genes required for cell proliferation, differentiation and migration (Endou et al, 2005; Xie et al, 2005).
In the present study, the sequence analysis of exons 19–22 of the SOS-1 gene was performed to establish whether the members of the two Polish families are affected by HGF type 1 caused by a single-cytosine insertion mutation, previously reported by Hart et al (2002). Overall, a total of ten affected individuals were identified in both families, and six of them were enrolled in the study. In both families, autosomal dominant transmission of the condition was noted. Histopathological evaluation revealed typical features of GO, such as hyperplastic epithelium with rete pegs extending into subepithelial tissue and gingival stroma, numerous collagen fibers, locally irregular and oriented in variable directions and abundant to moderate fibroblasts in subepithelial and connective tissue (Fig. 3). Additionally, inflammatory infiltrates were visible in two patients (Fig. 3a, b, i). The sequence analysis showed that neither single-cytosine insertion in exon 21 nor any other mutation in exons 19, 20, 21, and 22 of SOS-1 were involved in the manifestation of HGF in these patients. In line with these observations, Ma et al (2014) also did not detect a single-cytosine insertion in exon 21 nor any other mutation in 23 exons of SOS-1 in three multigeneration Chinese families with isolated HGF.
On the other hand, Lee et al (2006) characterized the metabolic activity of fibroblasts derived from the HGF-affected individuals harboring a single-cytosine insertion in the SOS-1 (g.126,142–126,143insC). Fibroblasts carrying the mutation in the SOS-1 gene demonstrated accelerated proliferation, higher density, altered ability to attach, enhanced propensity to form protrusions with lamellipodia and alterations in the surrounding matrix, i.e., increased collagen synthesis (Lee et al, 2006). The authors suggest that these interactions can correspond to the alterations of cell content and collagen overproduction in the gingiva of the HGF-affected patients with a single-cytosine insertion in the SOS-1. Nonetheless, enhanced collagen synthesis and accelerated proliferation were noted in fibroblasts isolated from patients analyzed herein (Gawron et al, 2016a; unpublished data). Notably, these observations corresponded to the GO recurrence, particularly in the mandibular gingiva of the 32-year-old female (Gawron et al, 2014) as well as substantial de novo production of fibrotic tissue and unusually progressive re-growth of mandibular and maxillary gingiva in the 11-year-old female (Gawron et al, 2016a). Similarly, increased cell proliferation and collagen upregulation observed in vitro, correlated with the GO recurrence in the mandible of the 33-year-old and the 41-year-old females (unpublished data). Thus, by contrast with the results reported by Lee et al (2006), highly proliferative and fibrotic phenotype of HGF-derived fibroblasts were found in individuals who did not harbor a single-cytosine insertion in exon 21 of the SOS-1 gene.
In summary, patients from two families with history of non-syndromic autosomal dominant HGF from Malopolska and Mazovia Provinces in Poland had not been affected by HGF type 1, caused by a single-cytosine insertion in exon 21 of the SOS-1 gene. Sequencing analysis spanning across the exons 19, 20, 21 and 22 of SOS-1 did not show any other mutation, either. These data highlight the importance of studies of the remaining regions of the SOS-1 gene as well as of other genes probing to identify the HGF-associated mutations in these patients. Identification of the specific genetic background of the disease in families described herein will help to unravel the pathogenic mechanism underlying GO.
Acknowledgments
Funding
This work was supported by grant from the National Science Centre, Poland (2012/07/B/NZ6/03524 to K.G.) and US NIH/NIDCR (DE 022597 to J.P.). Faculty of Biochemistry, Biophysics and Biotechnology of Jagiellonian University is a partner of the Leading National Research Center (KNOW) supported by the Ministry of Science and Higher Education.
Footnotes
Conflicts of interest: none to declare
References
- Coletta RD, Graner E. Hereditary gingival fibromatosis: a systematic review. J Periodontol. 2006;77:753–764. doi: 10.1902/jop.2006.050379. [DOI] [PubMed] [Google Scholar]
- Corbalan-Garcia S, Margarit SM, Galron D, Yang SS, Bar-Sagi D. Regulation of Sos activity by intramolecular interactions. Mol Cell Biol. 1998;18:880–886. doi: 10.1128/mcb.18.2.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denadai R, Raposo-Amaral CE, Bertola D, et al. Identification of 2 novel ANTXR2 mutations in patients with hyaline fibromatosis syndrome and proposal of a modified grading system. Am J Med Genet. 2012;158A:732–742. doi: 10.1002/ajmg.a.35228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano GM, Kurban M, Anyane-Yeboa K, et al. Mutations in the cholesterol transporter gene ABCA5 are associated with excessive hair overgrowth. PLoS Genet. 2014;10:e1004333. doi: 10.1371/journal.pgen.1004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kamah GY, Fong K, El-Ruby M, et al. Spectrum of mutations in the ANTXR2 (CMG2) gene in infantile systemic hyalinosis and juvenile hyaline fibromatosis. Brit J Derm. 2010;163:213–215. doi: 10.1111/j.1365-2133.2010.09769.x. [DOI] [PubMed] [Google Scholar]
- Endou M, Mizuno M, Nagata T, et al. Growth inhibition of human pancreatic cancer cells by human interferon-beta gene combined with gemcitabine. Int J Mol Med. 2005;15:277–283. [PubMed] [Google Scholar]
- Funato Y, Terabayashi T, Suenaga N, Seiki M, Takenawa T, Miki H. IRSp53/Eps8 complex is important for positive regulation of Rac and cancer cell motility/invasiveness. Cancer Res. 2004;64:5237–5244. doi: 10.1158/0008-5472.CAN-04-0327. [DOI] [PubMed] [Google Scholar]
- Gawron K, Łazarz-Bartyzel K, Fertala A, Plakwicz P, Potempa J, Chomyszyn-Gajewska M. Gingival fibromatosis with significant de novo formation of fibrotic tissue and a high rate of recurrence. Version 2. Am J Case Rep. 2016a;17:655–659. doi: 10.12659/AJCR.899997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawron K, Lazarz-Bartyzel K, Lazarz M, et al. In vitro testing the potential of a novel chimeric IgG variant for inhibiting collagen fibrils formation in recurrent hereditary gingival fibromatosis: chimeric antibody in a gingival model. J Physiol Pharmacol. 2014;65:585–591. [PubMed] [Google Scholar]
- Gawron K, Łazarz-Bartyzel K, Potempa J, Chomyszyn-Gajewska M. Gingival fibromatosis: clinical, molecular and therapeutic issues. Orphanet J Rare Dis. 2016b;11:9. doi: 10.1186/s13023-016-0395-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin R, Cohen M, Levi L. Syndromes of the Head and Neck. Oxford Press (Oxford Monographs on Medical Genetics); New York: 1990. [Google Scholar]
- Hart TC, Pallos D, Bowden DW, Bolyard J, Pettenati MJ, Cortelli JR. Genetic linkage of hereditary gingival fibromatosis to chromosome 2p21. Am J Hum Genet. 1998;62:876–883. doi: 10.1086/301797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart TC, Zhang Y, Gorry MC, et al. A mutation in the SOS1 gene causes hereditary gingival fibromatosis type 1. Am J Hum Genet. 2002;70:943–954. doi: 10.1086/339689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häkkinen L, Csiszar A. Hereditary gingival fibromatosis: Characteristics and novel putative pathogenic mechanisms. J Dent Res. 2007;86:25–34. doi: 10.1177/154405910708600104. [DOI] [PubMed] [Google Scholar]
- Jaureguiberry G, De la Dure-Molla M, Parry D, et al. Nephrocalcinosis (enamel renal syndrome) caused by autosomal recessive FAM20A mutations. Nephron Physiol. 2012;122:1–6. doi: 10.1159/000349989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortum F, Caputo V, Bauer CK, et al. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nature Genet. 2015;47:661–667. doi: 10.1038/ng.3282. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Jang SI, Pallos D, Kather J, Hart TC. Characterization of fibroblasts with Son of Sevenless-1 Mutation. J Dent Res. 2006;85:1050–1055. doi: 10.1177/154405910608501115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nature Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- Ma Y, Sun Z, Hu Y, Liu Y, Jin L, Zhang F. Non-syndromic hereditary gingival fibromatosis in three Chinese families is not due to SOS1 gene mutations. Cell Biochem Biophys. 2014;70:1869–1873. doi: 10.1007/s12013-014-0144-9. [DOI] [PubMed] [Google Scholar]
- O’Sullivan J, Bitu CC, Daly SB, et al. Whole-exome sequencing identifies FAM20A mutations as a cause of amelogenesis imperfecta and gingival hyperplasia syndrome. Am J Hum Genet. 2011;88:616–620. doi: 10.1016/j.ajhg.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Esteban L, Vass WC, et al. The Sos1 and Sos2 Ras-specific exchange factors: differences in placental expression and signaling properties. EMBO J. 2000;19:642–654. doi: 10.1093/emboj/19.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusanescu G, Gotoh T, Tian X, Feig LA. Regulation of Ras signaling specificity by protein kinase C. Mol Cell Biol. 2001;21:2650–2658. doi: 10.1128/MCB.21.8.2650-2658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saygun I, Ozdemir A, Gunhan O, Aydintug YS, Karslioglu Y. Hereditary gingival fibromatosis and expression of Ki-67 antigen: a case report. J Periodontol. 2003;74:873–878. doi: 10.1902/jop.2003.74.6.873. [DOI] [PubMed] [Google Scholar]
- Xiao S, Bu L, Zhu L, et al. A new locus for hereditary gingival fibromatosis (GINGF2) maps to 5q13-q22. Genomics. 2001;74:180–185. doi: 10.1006/geno.2001.6542. [DOI] [PubMed] [Google Scholar]
- Xie Y, Wang Y, Sun T, et al. Six post-implantation lethal knockouts of genes for lipophilic MAPK pathway proteins are expressed in preimplantation mouse embryos and trophoblast stem cells. Mol Reprod Dev. 2005;71:1–11. doi: 10.1002/mrd.20116. [DOI] [PubMed] [Google Scholar]
- Ye X, Shi L, Cheng Y, et al. A novel locus for autosomal dominant hereditary gingival fibromatosis, GINGF3, maps to chromosome 2p22.3-p23.3. Clin Genet. 2005;68:239–244. doi: 10.1111/j.1399-0004.2005.00488.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Zhang W, Huo Z, et al. A novel locus for maternally inherited human gingival fibromatosis at chromosome 11p15. Hum Genet. 2007;121:113–123. doi: 10.1007/s00439-006-0283-1. [DOI] [PubMed] [Google Scholar]