Summary
Clonal hematopoiesis (CH), as evidenced by recurrent somatic mutations in leukemia-associated genes, commonly occurs among aging human hematopoietic stem cells. We analyzed deep coverage, targeted next-generation sequencing (NGS) data of paired tumor and blood samples from 8,810 individuals to assess the frequency and clinical relevance of CH in patients with non-hematologic malignancies. We identified CH in 25% of cancer patients, with 4.5% harboring presumptive leukemia driver mutations (CH-PD). CH was associated with increased age, prior radiation therapy, and tobacco use. PPM1D and TP53 mutations were associated with prior exposure to chemotherapy. CH and CH-PD led to an increased incidence for subsequent hematologic cancers, and CH-PD was associated with shorter patient survival. These data suggest CH occurs in an age-dependent manner and specific perturbations can enhance fitness of clonal hematopoietic stem cells, which can impact outcome through progression to hematologic malignancies and through cell non-autonomous effects on solid tumor biology.
Keywords: Biological sciences/Cancer/Cancer genomics, Biological sciences/Cancer/Hematological cancer, Biological sciences/Genetics/Mutation, Health sciences/Diseases/Hematological diseases
Graphical Abstract
Introduction
Age-related clonal hematopoiesis (CH) was first identified through the finding of non-random X-chromosome inactivation (NRXI) in the peripheral blood of healthy elderly women. This observation led to the identification of recurrent somatic TET2 mutations in women with NRXI (Busque et al., 1996; Busque et al., 2012) suggesting TET2 mutations promote expansion of clonal populations of hematopoietic stem or progenitor cells, or CH, in the absence of overt hematologic transformation. Large-scale next generation sequencing (NGS) studies of unpaired exome data from blood samples obtained from healthy subjects identified somatic mutations in leukemia-associated genes, most commonly in DNMT3A, TET2, and ASXL1, in asymptomatic individuals without known hematologic disease(Genovese et al., 2014; Jaiswal et al., 2014; McKerrell et al., 2015; Xie et al., 2014; Young et al., 2016). These mutations lead to clonal expansion of hematopoietic cells, in the absence of clinically overt hematologic transformation and have been demonstrated to persist over time(Jaiswal et al., 2014; Young et al., 2016) suggesting that these mutations occur in hematopoietic stem cells (HSC).
CH (also referred to as “CHIP”/clonal hematopoiesis of indeterminate potential and “ARCH”/age-related clonal hematopoiesis) is thought to be analogous to other precursor clonal states, such as monoclonal gammopathy of undetermined significance (MGUS) and monoclonal B-cell lymphocytosis (MBL)(Ogawa, 2016; Steensma et al., 2015). Consistent with this notion, a subset of healthy subjects with CH subsequently develop hematologic cancers at an estimated rate of approximately 0.5–1.0% per year compared to <0.1% for non-CH controls(Genovese et al., 2014; Jaiswal et al., 2014). CH has been linked to decreased overall survival (OS)(Genovese et al., 2014; Jaiswal et al., 2014), including an increased risk for cardiovascular mortality(Jaiswal et al., 2014). CH has similarly been identified in patients with clonal cytopenias of undetermined significance(Cargo et al., 2015; Kwok et al., 2015) and aplastic anemia(Babushok et al., 2015; Yoshizato et al., 2015) with a different distribution of mutations often at higher variant allele fractions (VAF) than in studies of healthy individuals unselected for hematologic phenotypes.
Taken together, these studies suggest that CH is a common finding among aging HSCs, and that the presence of CH may impact overall outcome and subsequent risk for hematologic malignancies. However, due to the reliance on hematopoietic DNA without comparison to paired non-hematopoietic DNA, previous studies have been limited to the identification of known “hot-spot” mutations and mutations predicted to attenuate tumor suppressor gene function. Further, the prevalence and clinical implications of CH in patients with non-hematologic cancers have not been extensively studied(Xie et al., 2014). At Memorial Sloan Kettering Cancer Center (MSKCC), we have developed a high throughput capture-based NGS assay, MSK-IMPACT, encompassing the full protein-coding sequence of 341 (and more recently, 410) cancer-associated genes, for prospective clinical sequencing of matched tumor and blood DNA(Cheng et al., 2015). We hypothesized that sequencing of paired hematopoietic and non-hematopoietic DNA from patients with advanced cancer would allow us to accurately delineate the incidence of CH in this population and to assess whether the presence of CH impacts clinical outcome in patients with advanced cancers. We therefore sought to further characterize CH and its associated clinical impact in patients with non-hematologic cancers who underwent genomic profiling using paired tumor/blood sequencing.
Results
Prevalence of CH
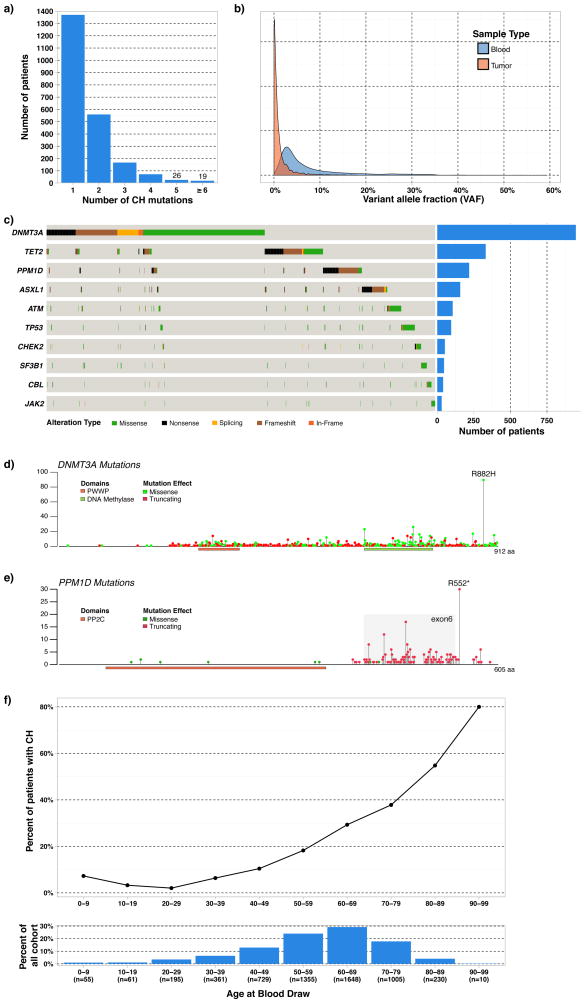
We examined MSK-IMPACT sequencing data to identify the prevalence of CH in 8,810 patients that underwent paired tumor and blood sequencing. Median coverage was 419X for blood normal and 698X for the tumor samples. Mean age at time of sample collection was 58.3 yrs (0.13–98.7 yrs). Mutational analysis of hematopoietic cells identified 3,547 presumptive somatic, non-silent mutations in 295 genes. 2,213 (25.1%) patients had at least one somatic mutation, and 38% (843 out of 2,213) patients with CH harbored more than one mutation (Figure 1A). Of the variants detected, 23% were not detected in the tumor tissue at all (variant fraction= 0) and the remaining 77% were detected at a low level in the tumor tissue though the variant fraction, which is the proportion of alleles in the sample that are mutated, was always at least twice as high in the blood. The median VAF for CH mutations was 0.044 in blood compared to 0.005 in the corresponding tumor, presumably due to the infiltration of blood cells in tumor tissue (Figure 1B). Mutations in DNMT3A were most common, followed by TET2, PPM1D, ASXL1, ATM, and TP53; these six genes accounted for 60% of all mutations detected (Figure 1C).
Figure 1.
Characteristics of clonal hematopoiesis (CH) mutations identified in the cohort. A. Number of mutations harbored per patient. B. Density of mutations by variant allele fraction in blood and tumor tissue C. Patterns of mutation co-occurrence and number of patients with mutations in ten most recurrently mutated genes. The first 1912 patients in the cohort underwent sequencing with a 341-gene assay, which did not include PPM1D but included all other genes listed. The remaining patients (N=6,898) underwent sequencing with a 410-gene assay, which included all genes shown. D. Distribution of DNMT3A mutations within cohort. Gene models are based on uniprot database. E. Distribution of PPM1D mutations within cohort. F. CH association with age and age distribution of patients in cohort.
Similar to prior CH studies, 43.8% of the mutations in DNMT3A were truncating in nature while 16% (89 out of 557) of DNMT3A missense mutations occurred at the R882 codon (Figure 1D)(Genovese et al., 2014; Jaiswal et al., 2014). 92 missense mutations led to new cysteine residues, which could inhibit protein function through the formation of novel di-sulfide bonds. Recurrent DNMT3A mutations reported in the COSMIC database were present at a higher VAF than the novel mutations in DNMT3A (median = 0.058 vs. 0.039, p<0.001). These data suggest allele-specific selective pressures underlying clonal expansion such that different mutations may confer differential fitness advantages to hematopoietic stem/progenitor cells (Figure S1). TET2 mutations were mainly protein-truncating variants similar to those seen in myeloid malignancies. The majority of TP53 mutations were missense, and were clustered in the DNA binding domain (Figure S2). PPM1D mutations were primarily protein-truncating variants in exon 6, these mutations have previously been demonstrated as gain-of-function mutations that lead impaired to TP53 function (Figure 1E)(Kleiblova et al., 2013).
Presumptive driver mutations
Comparing CH mutations to the genes most commonly altered in AML from prior published series(Papaemmanuil et al., 2016), we observed a significant degree of variation in the distribution of CH mutations (Figure S2). This was particularly apparent for DNMT3A, where the spectrum of somatic mutations observed in CH was widespread with many mutations that have not been previously characterized. These data suggest that only a subset of the mutations which initiate CH are capable of driving further clonal expansion and malignant transformation, such that this smaller subset of mutations confer a sufficient fitness advantage to further expand over time. We hypothesized that mutations in known leukemia genes with a higher minimum VAF would allow us to identify putative driver genes which predict for the development of subsequent hematologic cancers and impact overall outcome. Utilizing an adjusted schema for identifying high VAF (≥0.10) mutations in presumptive drivers (PD), we identified CH-PD in 4.5% of patients (393 of 8,810) (Figure S3A–B).
CH and clinical associations
Baseline clinical parameters: age, race, gender, and blood counts
Of 5,713 patients for whom detailed clinical review was performed for this study, 64 (1.1%) were excluded due to the presence of an active hematologic cancer before genomic analyses were performed (Table S1), for a final cohort of 5,649 patients. The majority of patients in the cohort (72%, or 4,058) had been exposed to prior chemotherapy, radiation therapy (RT), or both; the remainder (28%, 1,591) were naïve to both chemotherapy and RT. Of the 5,649 patient cohort, 1,353 (24%) patients harbored at least one CH mutation, and 246 (4.4%) had CH-PD. Median follow up among survivors was 16.3 months (interquartile range: 12.6–21.1 months).
CH demonstrated a statistically significant association with increased age (p<0.001) (Figure 1F) and race (p=0.002) with CH occurring most often in Caucasian patients. Starting at age 30, the estimated increase in the odds of CH was 6% for every 10 years (OR: 1.06 (95% CI 1.05–1.07) p <0.001). We did observe CH in pediatric cancer patients, with a similar frequency of CH in patients aged 0–9 as adults in the 30–39 age group. CH was strongly associated with current or former tobacco use (p<0.001). CH was associated with increased white blood cell (WBC) count (p=0.008), increased absolute monocyte count (AMC) (p<0.001), increased absolute neutrophil count (ANC) (p=0.023), increased mean corpuscular volume (MCV) (p<0.001), and decreased platelet count (p=0.029). There was no relationship between CH and hemoglobin, red-cell distribution width (RDW), or absolute lymphocyte count (ALC). CH-PD demonstrated similar clinical associations as the overall CH cohort (Table 1). These data demonstrate that CH is associated with alterations in hematopoiesis, similar to those observed in patients with myeloid malignancies including increased myelopoiesis and reduced megakaryopoiesis.
Table 1.
Associations between clonal hematopoiesis (CH) and clonal hematopoiesis in presumptive drivers (CH-PD) and clinical variables. Chi-square tests and Wilcoxon-rank sum tests were used to compare patient and clinical characteristics among patients with and without CH and CH-PD.
| Overall | CH | No CH | P-value | CH-PD | No CH-PD | P-value | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Number of patients | 5,649 | 1,353 | 4,296 | 246 | 5,403 | ||
|
| |||||||
| Age | 58.3 | 66.1 | 55.8 | <0.001 | 69.1 | 57.8 | <0.001 |
|
| |||||||
| Gender (%male) | 49% | 50% | 49% | 0.40 | 48% | 49% | 0.89 |
|
| |||||||
| Race: | %represented in cohort | %with CH | %without CH | 0.002 | %with CH-PD | %without CH-PD | 0.18 |
| • Caucasian | 81% | 25% | 75% | 5% | 95% | ||
| • Black | 6% | 20% | 80% | 2% | 98% | ||
| • Other | 13% | 20% | 80% | 4% | 96% | ||
|
| |||||||
| Cancer category | %represented in cohort | %with CH | %without CH | 0.06 | %with CH-PD | %without CH-PD | 0.039 |
| • GI | 22% | 24% | 76% | 4% | 96% | ||
| • GU | 18% | 25% | 75% | 3% | 97% | ||
| • Lung | 16% | 27% | 73% | 6% | 94% | ||
| • Breast | 13% | 21% | 79% | 3% | 97% | ||
| • Sarcoma | 8% | 21% | 79% | 4% | 96% | ||
| • Gyn | 5% | 24% | 76% | 4% | 96% | ||
| • Other | 19% | 24% | 76% | 5% | 95% | ||
|
| |||||||
| WBC | 7.1 | 7.4 | 7.1 | 0.008 | 7.5 | 7.1 | 0.15 |
|
| |||||||
| ANC | 5.03 | 5.23 | 4.97 | 0.023 | 5.39 | 5.02 | 0.12 |
|
| |||||||
| AMC | 0.46 | 0.50 | 0.45 | <0.001 | 0.51 | 0.46 | 0.029 |
|
| |||||||
| ALC | 1.35 | 1.36 | 1.35 | 0.51 | 1.31 | 1.35 | 0.38 |
|
| |||||||
| NLR>4 | 38% | 40% | 38% | 0.11 | 44% | 38% | 0.062 |
|
| |||||||
| Hemoglobin | 12.4 | 12.4 | 12.5 | 0.12 | 12.3 | 12.4 | 0.19 |
|
| |||||||
| RDW | 14.7% | 14.8% | 14.6% | 0.096 | 14.9% | 14.7% | 0.047 |
|
| |||||||
| MCV | 90.2 | 91.2 | 89.9 | <0.001 | 92.1 | 90.1 | <0.001 |
|
| |||||||
| Platelet count | 254 | 249 | 256 | 0.029 | 242 | 255 | 0.065 |
|
| |||||||
| RBC transfusion | 22% | 21% | 22% | 0.69 | 22% | 22% | 0.91 |
|
| |||||||
| Platelet transfusion | 6% | 4% | 6% | 0.011 | 5% | 6% | 0.83 |
|
| |||||||
| Growth factor | 28% | 27% | 28% | 0.70 | 27% | 28% | 0.79 |
|
| |||||||
| Current or former smoker | 46% | 53% | 44% | <0.001 | 55% | 46% | 0.004 |
|
| |||||||
| Prior chemotherapy | 64% | 64% | 64% | 0.89 | 60% | 64% | 0.24 |
|
| |||||||
| Prior radiation therapy | 37% | 41% | 35% | <0.001 | 44% | 37% | 0.021 |
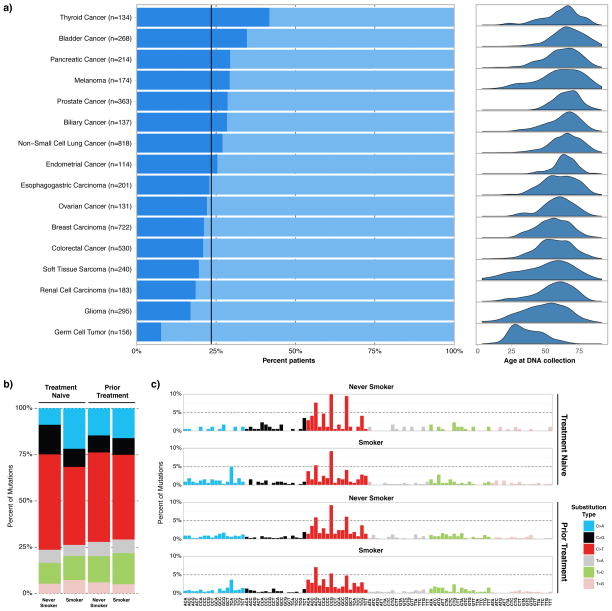
We next sought to identify any potential relationship between the incidence of CH and primary cancer type. The incidence of CH was highest in thyroid cancer patients, possibly related to radioactive iodine exposure, and lowest in patients with germ cell tumors (Figure 2A), with the low rate in germ cell tumors likely attributable to younger age of patients with this malignancy. While CH did not show any significant associations across larger cancer categories, CH-PD was slightly more common in patients with lung cancer compared to other tumor types, likely related to the enrichment for current/former smokers among lung cancer patients (Table 1). Both CH and CH-PD demonstrated a statistically significant relationship with patients having received prior RT (p<0.001 and p=0.021, respectively) but not with prior chemotherapy (p=0.89 and p=0.24, respectively) (Table 1).
Figure 2.
A. Detailed tumor types and percent of patients with clonal hematopoiesis and corresponding age distributions. This includes the 16 most frequently sequenced tumors, and patients with less common tumors are not included on this figure. Note that for detailed tumor types depicted, that Non-Small Cell Lung Cancer does not include Small Cell Lung Cancers or Mesotheliomas. B. Effect of smoking on observed clonal hematopoiesis (CH) mutations. Rates of different substitution types that lead to silent and non-silent CH mutations are compared between four groups: Treatment naïve never smokers, treatment naïve smokers, previously treated (chemotherapy, RT, or both) never smokers and previously treated (chemotherapy, RT, or both) smokers. C. Rates of different substitutions with their nucleotide context are compared amongst the same four groups of patients.
Associations for recurrently mutated genes
We next queried whether mutations in specific genes harbored unique phenotypes with respect to their effect on hematologic parameters and associations with prior toxic exposures (Table S2). Although the presence of CH in general was not associated with previous exposure to chemotherapy, mutations in TP53 and PPM1D were significantly associated with prior chemotherapy exposure (p=0.047 and p<0.001, respectively) and RT (p<0.001 for both). Additional associations regarding mutational effect on hematologic parameters are demonstrated in Table S2. Together, these results suggest that CH mutations in leukemia-associated genes may influence hematologic parameters in a gene-specific manner.
CH and mutational signatures
Overall, the most common single nucleotide substitution amongst the coding mutations were C>T transitions which have been demonstrated to accompany aging-associated processes(Alexandrov et al., 2013) (data not shown). To further elucidate whether smoking, generally characterized by C>A transversions, was an additional significant mutagenic factor leading to the spectrum of mutations observed, we assigned all silent and non-silent mutations and their trinucleotide context into previously described mutational signatures in smokers compared to never smokers(Alexandrov et al., 2013). This analysis did not show a significant difference in the proportion of mutations attributed to the smoking signature (Signature 4) (data not shown). We posited that the prior cytotoxic exposures might mask any significant effects smoking might impose. Therefore we categorized patients into treatment naïve and non-naïve groups (those with a history of prior chemotherapy, RT, or both). We observed an increased rate of C>A substitutions in treatment naïve smokers compared to treatment naïve never smokers, suggesting smoking may affect the type of CH mutations that occur (Figure 2B, Figure S3C). Further delineating the mutations based on the nucleotide context suggest smoking might affect substitutions specifically in the [TCA] context (Figure 2C).
CH and risk for subsequent hematologic cancer
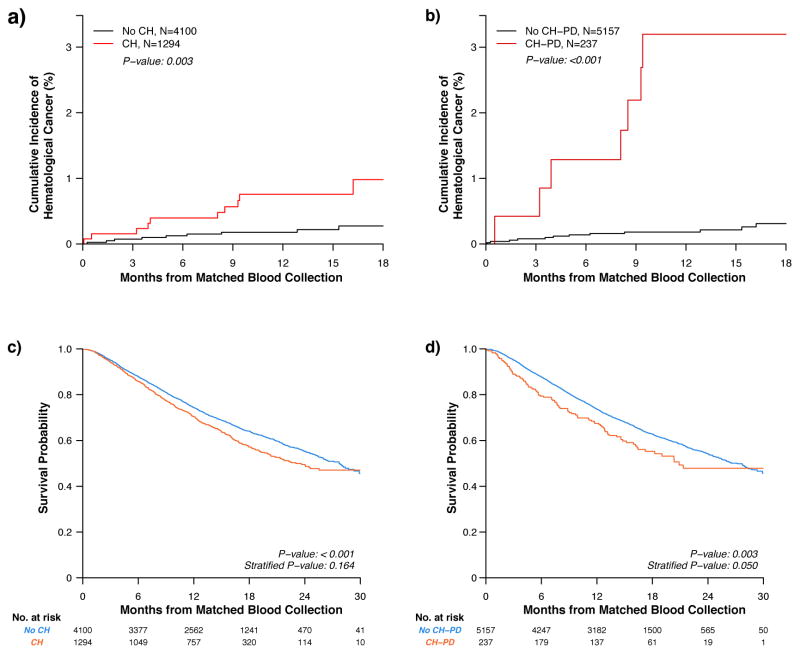
Of the 5,394 patients who were prospectively followed, 19 developed a new hematologic cancer over the study period at a mean time of 187 days following collection of matched normal blood. Over the entire study period, 10 of 1,294 patients with CH developed a hematologic cancer for an 18-month cumulative incidence estimate of 1% (0.5–1.8%), compared to 9 of 4,100 non-CH patients for an estimate of 0.3% (0.1–0.5%) (p=0.003). Seven of 237 patients with CH-PD developed a hematologic cancer, compared to 12 of 5,157 patients without CH-PD (3.2% (1.4–6.2%) and 0.3% (0.2–0.6%) estimates at 18 months, respectively; p<0.001) (Figure 3A–B).
Figure 3.
Clonal hematopoiesis (CH) and clonal hematopoiesis in presumptive drivers (CH-PD) and associations with subsequent hematologic cancers and overall survival. A. Incidence of new hematologic cancer for patients with and without CH. B. Incidence of new hematologic cancer for patients with and without CH-PD. C. OS in non-CH vs. CH, all ages. Time-point estimates for non-CH vs. CH are 12-month OS: 0.70 (0.68–0.73) vs. 0.74 (0.73–0.76); 18-month OS: 0.57 (0.54–0.60) vs. 0.64 (0.62–0.66); and 24-month OS: 0.50 (0.46–0.53) vs. 0.55 (0.53–0.57). D. OS in non-CH-PD vs. CH-PD, all ages. Time-point estimates for non-CH-PD vs. CH-PD are 12-month OS: 0.67 (0.62 vs. 0.74) vs. 0.74 (0.72–0.75); 18-month OS: 0.55 (0.49–0.63) vs. 0.63 (0.61–0.64); and 24-month OS: 0.48 (0.40–0.57) vs. 0.54 (0.52–0.56). Differences in the incidence of new hematologic cancer were compared using Gray’s test, while survival differences were assessed using the Peto & Peto modification of the Gehan-Wilcoxon test. Survival was compared both overall and stratified based on categories of age, gender, and smoking status.
CH and survival
We assessed whether the presence of CH had an effect on OS. CH was associated with inferior survival (p<0.001); however, this comparison did not remain statistically significant upon stratifying for age, gender, and smoking status (p=0.16) (Figure 3C). We next examined the relationship between CH-PD and OS. CH-PD was associated with inferior survival (p=0.003); which remained significant when stratifying by age, gender, and smoking status (p=0.050) (Figure 3D). Of note, the association between CH-PD and adverse outcome was seen at different VAF thresholds, with the most significant impact on overall survival seen in patients with the largest clonal burden (VAF>0.2 with stratified p=0.005) (Figure S4).
Deaths from hematologic cancers did not account for the shorter OS, as only 1 patient with CH died from a newly developed hematologic cancer, compared to 4 patients in the non-CH group who died of hematologic cancers (Table S3). The most common cause of death in all patients was progression of the primary non-hematologic cancer (98% of all deaths). Patients with CH did not have increased transfusion or growth factor requirements compared to patients without CH (Table 1 and Table S2). On an individual gene level, patients with DNMT3A mutations were less likely to require growth factor support (p=0.039), whereas patients with PPM1D mutations were more likely to require growth factor therapy (p<0.001) (Table S2).
Discussion
Prior studies examining CH in individuals unselected for hematologic phenotypes have utilized unpaired, lower coverage exome analysis of blood to identify putative somatic mutations(Genovese et al., 2014; Jaiswal et al., 2014). These approaches limit the spectrum of mutations that can be identified and do not allow for detection of lower prevalence subclones, which may expand and influence subsequent outcomes. Using a targeted sequencing panel, we were able to confidently detect down to 0.01 VAF the presence of somatic mutations in hematopoietic cells by utilizing concurrent sequencing of non-hematopoietic tumor specimens. We demonstrated that CH in patients with non-hematologic cancers is very common and is associated with increasing age, smoking, and prior exposure to RT. The frequency of CH in our cohort was significantly higher than in previous studies, likely due to increased sensitivity to detect smaller CH clones, comprehensive somatic mutational analysis of >300 cancer genes, and exposure to RT, tobacco, and chemotherapy in cancer patients. Subsequent studies using comparable methodologies are needed to ascertain if CH is more common in cancer patients than in the general population. Importantly, the presence of CH-PD was associated with adverse survival.
Clonal mosaicism, as evidenced in peripheral blood or buccal DNA, has been reported to be more common in individuals with cancer compared to those without cancer(Jacobs et al., 2012). Given our ability to examine sequencing data from more than one tissue (blood and tumor), our data indicate that mutations in genes such as PPM1D, which previously were reported as mosaic constitutional variant alleles that predispose to solid tumors(Ruark et al., 2013), are commonly hematopoietic in origin, which has been identified recently by other groups(Pharoah et al., 2016; Swisher et al., 2016). As the majority of detected mutations occurred in genes previously reported to cause CH(Genovese et al., 2014; Jaiswal et al., 2014; Xie et al., 2014), combined with our filtering schema, which required CH events to be enriched in blood compared to tumor, the likelihood of these events representing germline mosaic events is very low.
We have prospectively demonstrated that the risk for the subsequent development of a hematologic cancer is increased in patients with CH and/or CH-PD and a non-hematologic cancer compared to those without a mutation, similar to two recently published retrospective case-control studies from large cancer centers(Gillis et al., 2017; Takahashi et al., 2017) and in patients undergoing autologous stem-cell transplant for lymphoma(Gibson et al., 2017). In our study, death from hematologic cancer was not the primary determinant of adverse outcome in cancer patients with CH-PD. CH does not appear to limit the ability of cancer patients to receive cytotoxic chemotherapy, as CH did not increase the requirement for transfusion or growth factor therapy. In fact, it is likely that mutations in some genes, such as DNMT3A, allow for CH clones to persist following cytotoxic chemotherapy as has been shown in AML patients with residual CH following chemotherapy(Bhatnagar et al., 2016; Guryanova et al., 2016; Shlush et al., 2014). In contrast to CH in a general population without a diagnosis of advanced cancer, where cardiovascular events are the primary basis for adverse outcome(Jaiswal et al., 2014), the primary cause of death in patients in this cohort was progression of the primary, non-hematologic cancer. The association between CH and cancer progression could be due to cell-cell interactions between CH clones and cancer cells(Kleppe et al., 2015), the impact of CH on immune surveillance, or other factors which will require further clinical and functional investigation. One possibility is that some CH patients, particularly those with mutations in PPM1D and TP53, represent a subset with more extensively previously treated cancers and CH is an indirect measurement of refractoriness to therapy which could lead to inferior outcome. Longer follow-up of a larger cohort of patients will allow us to delineate the spectrum of adverse events driving poor outcome in cancer patients with CH.
We observed gene-specific interactions between previous therapies and clinical outcome in patients with CH. PPM1D and TP53 demonstrated a statistically significant association with exposure to both chemotherapy and RT. TP53 mutations have a well-established role in development of therapy-related myeloid neoplasms(Wong et al., 2015); however, the role of PPM1D mutations in therapy-induced myeloid malignancies has not been delineated though recent findings suggest an association with PPM1D variants and therapy-related myelodysplastic syndromes (MDS)(Lindsley et al., 2017). Notably, TP53 and PPM1D mutants demonstrated the strongest trend toward a relative increase in MCV, common in MDS. These data suggest that investigations of the relationship between cancer therapy, CH and risk of hematologic cancers must be assessed on a gene-specific level, and that there may be a role for assessing for the presence of CH driven by specific mutations in better informing clinicians and patients of the risk for subsequent hematologic cancers following chemotherapy and RT in the adjuvant setting. Moreover, our data suggest that further studies of the relationship between known leukemia disease alleles and response to specific therapies might identify gene-specific effects which confer therapeutic resistance to leukemia stem/progenitor cells.
A recent study utilizing error-corrected sequencing allowing detection down to VAF 0.0003 demonstrated CH in 95% of a small cohort of healthy subjects, suggesting that CH at a low level is present in almost all adults(Young et al., 2016). These data, coupled with our observations, suggest that mutant clones constantly develop in HSCs throughout life, and that the presence of clinically detectable CH represents selection of a particular mutant HSC clone with increased fitness. It is likely that the mutational spectrum that can drive CH-PD represents a subset of the mutations seen in CH, and that other factors including environmental/therapeutic exposures, immune surveillance, and others govern the likelihood of developing CH-PD. We would posit that CH represents a clinically relevant context in which investigators can elucidate the interaction between age-dependent stochasticity, specific mutations, and the local and systemic microenvironment to promote clonal outgrowth of clonal HSCs that have clinical relevance in pleiotropic contexts. These data suggest a model by which age-dependent mutations are constantly acquired in HSCs, which are then selected for by external perturbations that allow clones to expand, which has impact on the growth and therapeutic response of primary tumors and on the subsequent risk of hematologic malignancies. Most importantly, these data suggest that identification of CH can inform the use of specific interventions, including alterations in systemic/local therapies, lifestyle interventions, and targeted therapies which can be used to identify and target CH and to prevent clonal expansion and clinical sequelae.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources should be directed to and will be fulfilled by Lead Contact, Ross L. Levine, leviner@mskcc.org
Experimental model and subject details
Human subjects
The study population included patients with non-hematologic cancers at MSKCC that underwent matched tumor and blood sequencing on an institutional prospective tumor sequencing protocol ClinicalTrials.gov number, NCT01775072. This study was approved by the MSKCC Institutional Review Board (IRB). A subset of patients that underwent tumor-genomic profiling as standard of care were not directly consented, in which case an IRB waiver was obtained to allow for inclusion into this study. Associations between peripheral blood counts were determined using complete blood counts obtained at time of matched normal blood sampling (within 30 days). Associations with prior cytotoxic therapy were based on clinical review of any cancer-directed systemic therapy (excluding hormonal therapy) or radiation therapy (RT) that patients had received prior to the date of matched normal blood sampling. Patients were excluded from analysis if they had an active hematologic cancer or precursor condition such as MGUS or MBL at the time of blood sequencing (Table S1). Clinical characteristics were analyzed in all patients for whom blood sampling was performed on or prior to October 1, 2015 (N=5,649). Patients were prospectively followed for development of subsequent hematologic cancers with last date of follow up June 1, 2016 and for OS with last date of follow up October 18, 2016. Clinical data for all subjects are deposited onto the following website: https://data.mendeley.com/datasets/kcwswn7tdp/draft?a=3c797857-ddf0-4653-b2e2-d8a526431eb8. These data include information on gender, age and race of patients included in this study.
Method details
Next-generation sequencing assay
All patients underwent NGS using MSK-IMPACT, a hybridization capture-based next-generation sequencing assay encompassing all protein-coding exons of 341 or 410 cancer-associated genes (Table S4A) MSK-IMPACT is validated and approved for clinical use by New York State Department of Health Clinical Laboratory Evaluation Program and is used to sequence advanced stage cancer patients at Memorial Sloan Kettering Cancer Center (MSKCC). DNA is extracted from de-paraffinized formalin fixed paraffin embedded (FFPE) tumor tissue and patient matched blood sample using chemagic STAR instrument (Hamilton) using magnetic beads (PerkinElmer). Extracted DNA samples were normalized in TE buffer and sheared on the Covaris instrument. KAPA Biosystems library preparation kit was used to prepare barcoded DNA molecules on the Biomek FXP instrument. Libraries were pooled and DNA fragments were captured using custom designed biotinylated probes (NimbleGen). Further details are previously described in (Cheng et al., 2015) and (Zehir et al., 2017).
Variant calling methodology
Pooled libraries were sequenced on an Illumina HiSeq 2500 with 2×100bp paired-end reads. Sequencing reads were aligned to human genome (hg19) using BWA (0.7.5a). Reads were re-aligned around indels using ABRA (0.78), followed by base quality score recalibration with Genome Analysis Toolkit (GATK) (2.6–5). Median coverage in the blood samples was 419X, and median coverage in the tumors was 698X. Coverage for ten most recurrently mutated genes is described in Table S4B.
Single nucleotide alterations were called in the blood sample, using a mixture of 10 unrelated normal DNAs as a comparator. Insertions and deletions were called using Somatic Indel Detector (2.3–9) and Pindel (0.2.5a7). All called mutations were genotyped in the matching tumor sample. Mutations were annotated with Annovar. False positive (FP) calls and germline mutations were filtered out in the following manner:
Each variant is genotyped across the normals sequenced in the same pool as the blood sample where the variant is identified. If a variant is present in the 20% of these normals then it is marked as a potential sequencing artifact.
Variants that were not present in the COSMIC database (v78) but are present in more than 5 blood samples were marked as potential sequencing artifacts.
Variants that were present in more than 35% of the total reads in both the blood and the tumor sample were marked as likely germline events.
Variants where the total number of reads covering the genomic position harboring the mutation was less than 20 were marked as high risk for false positives.
Determination of Clonal Hematopoiesis (CH) events
We identified CH mutations in all genes captured in MSK-IMPACT through a two-tiered filtering schema where the variant allele fraction (VAF) in the blood was greater than twice as much the VAF in the tumor after removing false positives and germline mutations. The following criteria were used to retain mutations:
Mutations present in one of the curated leukemia/lymphoma related gene list (ASXL1, CBL, DNMT3A, GNAS, JAK2, NRAS, SF3B1, TP53, U2AF1, IDH2, BCOR, PPM1D, TET2, IDH1, IDH2, SRSF2, RUNX1, SH2B3, ZRSR2, STAT3, KRAS, MYD88, ATM, CALR, CEBPA, ETV6, EZH2, FLT3, KIT, MPL, NPM1, SRSF2, STAG2, WT1, SETD2, CREBBP) where VAF was greater than or equal to 2% and at least 8 reads supported the alternate allele.
Mutations present in non-leukemia/lymphoma genes where VAF is greater than or equal to 5% and at least 8 reads supported the alternate allele.
In samples where one CH mutation was identified, we also looked for the presence of additional mutations at lower variant frequencies, where mutations with VAF greater than or equal to 1% in leukemia/lymphoma genes and VAF greater than or equal to 3% for non-leukemia/lymphoma genes were retained.
In addition, we identified mutations in leukemia/lymphoma genes where the variant frequency was greater than 35% in the blood sample but less than 35% in the tumor tissue. As a subset of these mutations could conceivably be considered as germline events with loss of allele in tumor tissue, we included only the mutations that were present in the COSMIC database with more than 10 occurrences in the “haematopoietic and lymphoid” category into the pool of CH mutations. All CH mutations detected across all samples are provided at https://data.mendeley.com/datasets/kcwswn7tdp/draft?a=3c797857-ddf0-4653-b2e2-d8a526431eb8.
Determination of CH-Potential Driver (PD) events
We used the same filters to eliminate FP and germline mutations. Subsequently, only variants with VAF greater than or equal to 10% in the normal were considered for CH-PD. The following criteria were used to further filter CH-PD mutations:
Any variant in COSMIC occurring in the “haematopoietic and lymphoid” category greater than or equal to 5 times
Any damaging variant in DNMT3A gene within exons 7–23
Any damaging variant in these genes: ASXL1, TET2, PPM1D, TP53, RAD21, STAG2, ATM, NF1
Any inactivating mutation in CALR exon9
JAK2 V617F variant
CBL E366K, C384Y, C404S, and C416S variants
SETD2 R1625C variant
MPL W515S variant
Any oncogenic variant in Papaemmanuil et al’s 2016 NEJM study that did not satisfy the above criteria(Papaemmanuil et al., 2016)
Determination of mutations signatures
Filtering criteria above were applied to silent and intronic mutations and then combined with the non-silent mutations that were observed in COSMIC less than 20 times. This step is to ensure only background mutations are included in the analysis since they are the ones that are most likely to display the effects of mutagenic processes. 3′ and 5′ nucleotide context were used to determine the mutation signatures using an in-house developed decomposition algorithm. Briefly, the mutations were resampled 1,000 times and then subjected to decomposition analysis in which the Kullback–Leibler divergence was minimized between the signatures affecting our cohort and the approximation built from 30 signatures that had been previously described(Alexandrov et al., 2013), such that each signature was assigned a weight that corresponded to the percentage of mutations explained by each given signature. The analysis code can be found here: https://github.com/mskcc/mutation-signatures and it was recently used in analysis of > 10,000 solid tumor samples sequenced with MSK-IMPACT(Zehir et al., 2017).
Categorization of non-hematologic cancers
Refer to Methods S1 for this information.
Determination of new hematologic cancers
At date of last follow up with respect to new hematologic cancer development (June 1, 2016), MSKCC databases were queried for all patients on this study to determine whether a new hematologic cancer had developed since the date of matched normal blood sampling. This was performed by querying across institutional databases in the following ways:
-
All patients with diagnostic codes (stated as ICD-10 codes) pertaining to hematologic cancers were reviewed
C81 Hodgkin lymphoma
C82 Follicular lymphoma
C83 Non-follicular lymphoma
C84 Mature T/NK-cell lymphomas
C85 Other specified and unspecified types of non-Hodgkin lymphoma
C86 Other specified types of T/NK-cell lymphoma
C88 Malignant immunoproliferative diseases and certain other B-cell lymphomas
C90 Multiple myeloma and malignant plasma cell neoplasms
C91 Lymphoid leukemia
C92 Myeloid leukemia
C93 Monocytic leukemia
C94 Other leukemias of specified cell types
C95 Leukemia of unspecified cell type
C96 Other and unspecified malignant neoplasms of lymphoid, hematopoietic and related tissue
D45 Polycythemia vera
D46 Myelodysplastic syndromes
D47 Other neoplasms of uncertain behavior of lymphoid, hematopoietic, and related tissue
All patients who were seen as a “new visit” by providers in leukemia, lymphoma, and myeloma clinics were reviewed
All patients undergoing bone marrow aspirate/biopsy (CPT codes 38220 and 38221) were reviewed
All patients with a documented peripheral blast count >20% across all recorded CBCs at MSKCC were reviewed
All patients who underwent testing with in-house 30 gene myeloid panel were reviewed
From the patients identified by the above queries, charts were reviewed individually and date of new hematologic cancer, if present, was tabulated as the date of the pathologic report that confirmed the diagnosis. Hematologic cancers diagnosed prior to date of matched normal blood sampling were excluded from the analysis.
Known precursor conditions including monoclonal B-cell lymphocytosis (MBL) and monoclonal gammopathy of uncertain significance (MGUS) that were diagnosed following the collection of matched normal blood were not considered as new hematologic cancers.
Survival determination
Survival follow up on patients in the cohort was obtained through internal MSKCC databases based upon deaths that occurred while admitted to MSKCC hospital or by Death Notification Forms submitted from physicians’ offices. Last known follow up information was also electronically updated for all patients with activity at any MSKCC site. Additionally, vital status was updated electronically by utilizing the Social Security Death Index (SSDI) and matched against the MSKCC patient database. The SSDI update is run on a monthly basis. For Cancer Registry patients, additional processes are utilized to determine vital status. If there is no update or activity on a patient’s account over a 15 month period, the Omnipro Medicare Database is queried to determine vital status and/or data of last account activity.
Quantification and statistical analysis
Differences in VAF by mutation type were assessed using a Wilcoxon rank-sum test. Chi-square tests and Wilcoxon-rank sum tests were further used to compare patient and clinical care characteristics among patients with and without CH. Clinical characteristics were also explored for individual mutations for the five most recurrently mutated genes. The incidence of hematologic cancer, defined as the time from matched normal blood sampling to pathologically confirmed hematologic cancer development, was visually displayed using cumulative incidence functions, whereas death in the absence of a hematologic malignancy was considered a competing event. Gray’s test was used to assess differences in the cumulative incidence based on CH and CH-PD. OS, defined as the time from sample collection to death or last follow-up, was displayed using Kaplan-Meier methods. Due to evidence the proportional hazards assumption was violated for the association of CH-PD and OS, a Cox proportional hazards model was not pursued. Alternatively, differences in survival based on mutational status were assessed using the Peto & Peto modification of the Gehan-Wilcoxon test(Harrington and Fleming, 1982) The test statistic was evaluated both overall and stratified based on an age categorization (≤25, 24–45, 46–55, 56–65, 66–75, 76–85, >85), gender, and smoking status (current/prior smoker vs. never smoker). Cumulative incidence and survival estimates only included patients with follow-up beyond the date of matched normal blood collection. Statistical analyses were performed using the R statistical package (www.r-project.org).
Data and software availability
Clinical data and mutation calls for all patients analyzed during this study are included at https://data.mendeley.com/datasets/kcwswn7tdp/draft?a=3c797857-ddf0-4653-b2e2-d8a526431eb8. R code is available at https://github.com/ahmetz/CSC_CH_Paper_Code.
Additional resources
The study population included patients with non-hematologic cancers at MSKCC who participated in an institutional prospective tumor sequencing protocol: ClinicalTrials.gov number, NCT01775072.
Supplementary Material
Acknowledgments
This study was supported in part by NIH/NCI P30 CA008748 (Cancer Center Support Grant). C.C.C. is supported by a Conquer Cancer Foundation American Society of Clinical Oncology Young Investigator Award. R.L.L. is supported by a Stand up to Cancer Convergence Grant with support from the V foundation and the National Science Foundation. We gratefully acknowledge the members of the Molecular Diagnostics Service in the Department of Pathology and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. We also acknowledge the patients and their families for their participation.
Footnotes
Author Contributions
C.C.C. analyzed data, collected data, and wrote the initial draft of the manuscript. A.Z. analyzed data, performed bioinformatics analyses, and wrote the initial draft of the manuscript. S.M.D. performed biostatistical analyses and assisted with manuscript preparation (methods section). A.K. performed data collection. A.S. and P.J. performed bioinformatics analyses. D.M.H., D.B.S., M.E.R., J.B., M.E.A., M.L., and M.S.T. provided critical review and revision of the manuscript. R.L.L. and M.F.B. conceived, designed, and supervised the study, provided critical review and revision of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok DV, Perdigones N, Perin JC, Olson TS, Ye W, Roth JJ, Lind C, Cattier C, Li Y, Hartung H, et al. Emergence of clonal hematopoiesis in the majority of patients with acquired aplastic anemia. Cancer genetics. 2015;208:115–128. doi: 10.1016/j.cancergen.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar B, Eisfeld AK, Nicolet D, Mrozek K, Blachly JS, Orwick S, Lucas DM, Kohlschmidt J, Blum W, Kolitz JE, et al. Persistence of DNMT3A R882 mutations during remission does not adversely affect outcomes of patients with acute myeloid leukaemia. British journal of haematology. 2016;175:226–236. doi: 10.1111/bjh.14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busque L, Mio R, Mattioli J, Brais E, Blais N, Lalonde Y, Maragh M, Gilliland DG. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood. 1996;88:59–65. [PubMed] [Google Scholar]
- Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nature genetics. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargo CA, Rowbotham N, Evans PA, Barrans SL, Bowen DT, Crouch S, Jack AS. Targeted sequencing identifies patients with preclinical MDS at high risk of disease progression. Blood. 2015;126:2362–2365. doi: 10.1182/blood-2015-08-663237. [DOI] [PubMed] [Google Scholar]
- Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics: JMD. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. The New England journal of medicine. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CJ, Lindsley RC, Tchekmedyian V, Mar BG, Shi J, Jaiswal S, Bosworth A, Francisco L, He J, Bansal A, et al. Clonal Hematopoiesis Associated With Adverse Outcomes After Autologous Stem-Cell Transplantation for Lymphoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2017 doi: 10.1200/JCO.2016.71.6712. Jco2016716712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis NK, Ball M, Zhang Q, Ma Z, Zhao Y, Yoder SJ, Balasis ME, Mesa TE, Sallman DA, Lancet JE, et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. The Lancet Oncology. 2017;18:112–121. doi: 10.1016/S1470-2045(16)30627-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryanova OA, Shank K, Spitzer B, Luciani L, Koche RP, Garrett-Bakelman FE, Ganzel C, Durham BH, Mohanty A, Hoermann G, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nature medicine. 2016;22:1488–1495. doi: 10.1038/nm.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington DP, Fleming TR. A class of rank test procedures for censored survival data. Biometrika. 1982;69:553–566. [Google Scholar]
- Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, Hutchinson A, Deng X, Liu C, Horner MJ, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nature genetics. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. Age-related clonal hematopoiesis associated with adverse outcomes. The New England journal of medicine. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiblova P, Shaltiel IA, Benada J, Sevcik J, Pechackova S, Pohlreich P, Voest EE, Dundr P, Bartek J, Kleibl Z, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. The Journal of cell biology. 2013;201:511–521. doi: 10.1083/jcb.201210031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe M, Comen E, Wen HYLB, Blum B, Rapaport FT, Keller M, Granot Z, Socci N, Viale A, et al. Somatic mutations in leukocytes infiltrating primary breast cancers. npj Breast Cancer. 2015;1 doi: 10.1038/npjbcancer.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok B, Hall JM, Witte JS, Xu Y, Reddy P, Lin K, Flamholz R, Dabbas B, Yung A, Al-Hafidh J, et al. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood. 2015;126:2355–2361. doi: 10.1182/blood-2015-08-667063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley RC, Saber W, Mar BG, Redd R, Wang T, Haagenson MD, Grauman PV, Hu ZH, Spellman SR, Lee SJ, et al. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. The New England journal of medicine. 2017;376:536–547. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrell T, Park N, Moreno T, Grove CS, Ponstingl H, Stephens J, Crawley C, Craig J, Scott MA, Hodkinson C, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell reports. 2015;10:1239–1245. doi: 10.1016/j.celrep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S. Clonal hematopoiesis in acquired aplastic anemia. Blood. 2016;128:337–347. doi: 10.1182/blood-2016-01-636381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. The New England journal of medicine. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah PD, Song H, Dicks E, Intermaggio MP, Harrington P, Baynes C, Alsop K, Bogdanova N, Cicek MS, Cunningham JM, et al. PPM1D Mosaic Truncating Variants in Ovarian Cancer Cases May Be Treatment-Related Somatic Mutations. Journal of the National Cancer Institute. 2016;108 doi: 10.1093/jnci/djv347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruark E, Snape K, Humburg P, Loveday C, Bajrami I, Brough R, Rodrigues DN, Renwick A, Seal S, Ramsay E, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493:406–410. doi: 10.1038/nature11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher EM, Harrell MI, Norquist BM, Walsh T, Brady M, Lee M, Hershberg R, Kalli KR, Lankes H, Konnick EQ, et al. Somatic Mosaic Mutations in PPM1D and TP53 in the Blood of Women With Ovarian Carcinoma. JAMA oncology. 2016;2:370–372. doi: 10.1001/jamaoncol.2015.6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Wang F, Kantarjian H, Doss D, Khanna K, Thompson E, Zhao L, Patel K, Neelapu S, Gumbs C, et al. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: a case-control study. The Lancet Oncology. 2017;18:100–111. doi: 10.1016/S1470-2045(16)30626-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, Lamprecht TL, Shen D, Hundal J, Fulton RS, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518:552–555. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nature medicine. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, Sato-Otsubo A, Sato Y, Liu D, Suzuki H, et al. Somatic Mutations and Clonal Hematopoiesis in Aplastic Anemia. The New England journal of medicine. 2015;373:35–47. doi: 10.1056/NEJMoa1414799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nature communications. 2016;7:12484. doi: 10.1038/ncomms12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nature medicine. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.