Abstract
Background:
Peroxisome proliferator-activated receptors-λ (PPAR-λ) is a member of nuclear receptor superfamily and acts as a ligand-dependent transcription factor often found in the adrenal gland, the spleen, and adipose tissue. The Pro12Ala polymorphism of PPAR-λ has been associated with the risks of gestational diabetes mellitus (GDM); however, association studies have provided conflicting results. The aim of this Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) compliant meta-analysis is to reach a more up-to-date and accurate estimation of the relationship between Pro12Ala genetic polymorphisms and the risks of GDM.
Methods:
Eligible studies were retrieved by searching PubMed, EMBASE, Web of Science, Ovid, WanFang, and Chinese National Knowledge Databases and selected according to a pre-defined inclusion criterion. The risk of bias was assessed using the Newcastle-Ottawa quality assessment scale. The per-allele odds ratio (OR) of risk allele proline (Pro) was compared between cases and controls in each study to describe the association between the Pro allele and an individual's risk of GDM. The ORs were pooled using both the random-effects model (the DerSimonian and Laird method) and the fixed effects model (the Mantel-Haenszel method) and the 95% confidence interval (95% CI) was calculated using Woolf method.
Results:
The final meta-analysis included a total of 11 articles of 12 data sets consisting of 7054 controls and 2980 GDM cases. Our results demonstrate that the Pro allele is not associated with GDM [OR: across multiple populations, 95% CI: 0.98–1.24; P(Z) = 0.01; P(Q) = 0.003]. In the stratified analysis by ethnicity, significantly increased risks were found for the Chinese (OR = 2.36; 95% CI: 1.47–3.78) and Korean (OR = 1.39; 95% CI: 1.00–1.93) populations.
Conclusion:
These data suggest the potential role of Pro allele in the pathogenesis of GDM in Asian populations. Although the funnel plot of included studies showed assymetry, the results using the “trim and fill” method did not alter the conclusion of this study.
Keywords: genetic polymorphism, gestational diabetes mellitus, gestational diabetes, meta-analysis, peroxisome proliferator-activated receptors, Pro12Ala
1. Introduction
Gestational diabetes mellitus (GDM) is defined as the intolerance of glucose that was not present or detected before preganacy[1] and often occurs when a woman's pancreatic function is not sufficient to overcome the diabetogenic environment of pregnancy.[2] GDM is the most common metabolic disorder during pregnancy,[3] and its frequency has further increased in the past decade, with increases ranging from 10% to 100% in different groups of patients and ethnicities.[4–6] Recent trends such as the decrease in physical activity,[7] epidemic of obesity,[8] and adoption of unhealthy lifestyles may all contribute to the increasing prevalence of GDM.[9]
Although the exact disease etiology of GDM is still very much unknown, evidence to date suggests that it is a careful interplay between environmental factors and genetic background.[10] Considerable research has been devoted to identifying potential genetic factors that contribute to GDM, and many genome-wide association studies have been conducted.[11,12] The list of variants associated includes polymorphism within genes such as CDKAL1, IGF2BP2, KCNQ1, KCNF11, MTR1B, TCF7L2, PPAR, etc.[13–18]
Peroxisome proliferator-activated receptors-λ (PPAR-λ) is a member of nuclear receptor superfamily and acts as a ligand-dependent transcription factor often found in the adrenal gland, the spleen, and adipose tissue.[19–21] PPAR-λ forms heterodimers with the retinoid X receptors and regulates various genes involved in metabolism and adipocyte differentiation.[22,23] Furthermore, PPAR-λ has been shown to have diverse functions such as negatively regulates macrophage activation,[24] inhibits the production of monocytes inflammatory cytokines,[25] adipogenesis, and insulin desensitization.[26] Mutations in the PPAR-λ gene have been associated with obesity and diabetes-related phenotypes, such as improved insulin sensitivity and plasma leptin levels.[27–29] The polymorphism of a proline (Pro) substituted with an alanine (Ala) at Amino acid 12 is a common polymorphism. The Ala allele is associated with reduced activity of PPAR-λ.[27] The Pro12Ala has been heavily researched for its role in obesity and type 2 diabetes and is considered one of the most common genetic risk factors for human diabetes.[30–32] However, studies have found conflicting results in Pro12Ala's role in GDM. For example, some studies have reported such a correlation, while other studies have found otherwise. To clarify the in-conflict findings reported so far as well as heterogeneity and publication bias that exists between studies, we have conducted a meta-analysis of genetic association studies of the PPAR-λ Pro12Ala polymorphism to assess its effect on the risk of GDM.
2. Methods
2.1. Search strategy and inclusion criteria
We searched the literature hosted on PubMed, EMBASE, Web of Science, Ovid, WanFang, and Chinese National Knowledge Databases with keywords related to disease (e.g., “gestational diabetes mellitus,” “GDM”) and the gene of interest (e.g., “peroxisomal proliferator-activated receptor gamma,” “PPAR-λ,” or “PPARG”). Genetic association studies published before May 2016 were retrieved, and their references were checked to identify other relevant publications. No earlier date limit was applied. The search was conducted without any restrictions on the language used but focused on human subjects. We did not define a minimum number of patients as a criterion for a study's inclusion in this meta-analysis.
All retrieved study were screened, and all eligible studies included needed to satisfy each point of the following criteria: original papers containing independent data, pathological confirmation of GDM, case–control or cohort study, and genotype distribution information or odds ratio (OR) with its 95% confidence interval (CI) and P value. The major reasons for exclusion of studies were overlapping data; review articles, case-only studies, and family-based studies.
2.2. Ethic approval
Ethic approval was deemed unneccesary, as this study is a systematic-review.
2.3. Data extraction
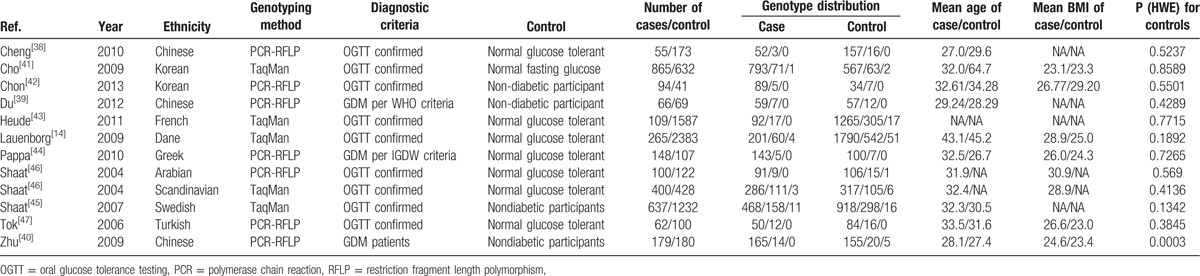
Data extraction was performed independently by 2 reviewers (WL, XP). All data were checked for internal consistency and disagreements were resolved through careful discussion between all authors. For each study, the following were extracted from each article: first author's name, publication year, diagnostic criterion, definition and numbers of cases and controls, frequency of genotypes, genotyping method, source of controls, Hardy–Weinberg equilibrium (HWE), age, body mass index (BMI), and ethnicity. Studies with different ethnic groups were considered as individual studies for our analyses.
2.4. Risks of bias between individual studies
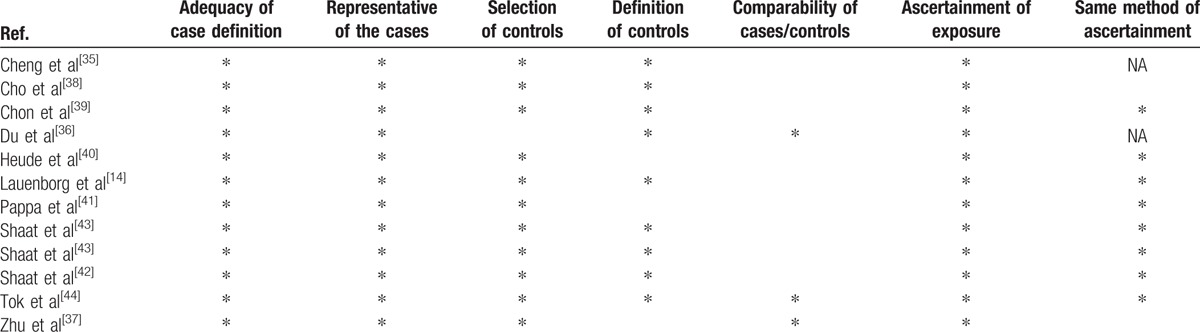
The Newcastle–Ottawa Scale (NOS) was used for quality assessment. The 3 parameters assessed by NOS in case–control studies are selection, comparability, and exposure; stars were assigned to each parameter according to criteria in the NOS manual.[33,34] Selection is evaluated by the definition and assignment of cases and controls (see Table 1).[35–44] The comparability of the article focuses on the design and analysis of the study. Potential of bias was determined by the method of ascertainment of both cases and controls. The risk of bias is considered high if a study obtained 1 or 0 stars for selection, comparability, and exposure. Two stars for selection, 1 star for compatibility, and 2 stars for exposure are the minimum requirement for a study to be considered having a medium risk. Finally, the risk of bias is recognized as low if a study was awarded 4 stars for selection, 2 stars for comparability, and 3 stars for the ascertainment of exposure.
Table 1.
The Newcastle–Ottawa quality assessment scale for studies included in this meta-analysis.
2.5. Statistical analysis
The association strength between PPAR-λ Pro12Ala polymorphism and GDM was assessed by calculating OR with 95% CI.
The χ2 test was used to evaluate whether there is a significant deviation from HWE among the control subjects of the study. The per-allele OR of risk allele proline (Pro) was compared between cases and controls in each study to quantitatively describe the presence of the Pro allele and an individual's risk of GDM. The ORs were pooled using both the random-effects model (the DerSimonian and Laird method) and the fixed effects model (the Mantel–Haenszel method) as previously described,[45,46] and 95% CI was calculated using Woolf method.[47] The results of the random effects model were reported in this article because it takes into consideration the variation between studies. A prespecified stratified analysis was conducted to explain the heterogeneity between each study and to investigate the relationship present in a subgroup. Stratified analysis was performed for ethnicity (Caucasian, Chinese, Korean, and Middle Eastern).
Heterogeneity across individual studies was examined using Cochran χ2 Q test.[48] Q test was also performed to detect the heterogeneity within each subgroup. Publication bias was assessed using the linear regression approach to measure funnel plot asymmetry on the natural logarithm of OR, as described by Egger et al.[49] All statistical analysis were carried out with Stata statistical software version 13.0 (Stata Corporation, College Station, TX). Type I error rate was set at 0.05, and all P values were for 2-sided analysis.
3. Results
3.1. Study characteristics
The search yielded a combined 69 references. Study selection process is shown in Fig. 1. The final meta-analysis included a total of 11 articles of 12 data sets.[14,35–44] The 12 data sets included 7054 controls and 2980 GDM cases. The detailed characteristics of included studies are summarized in Table 2. Of the GDM cases, 300 were Chinese, 959 were Korean, 1559 were Caucasian, and 162 were Middle Eastern.
Figure 1.
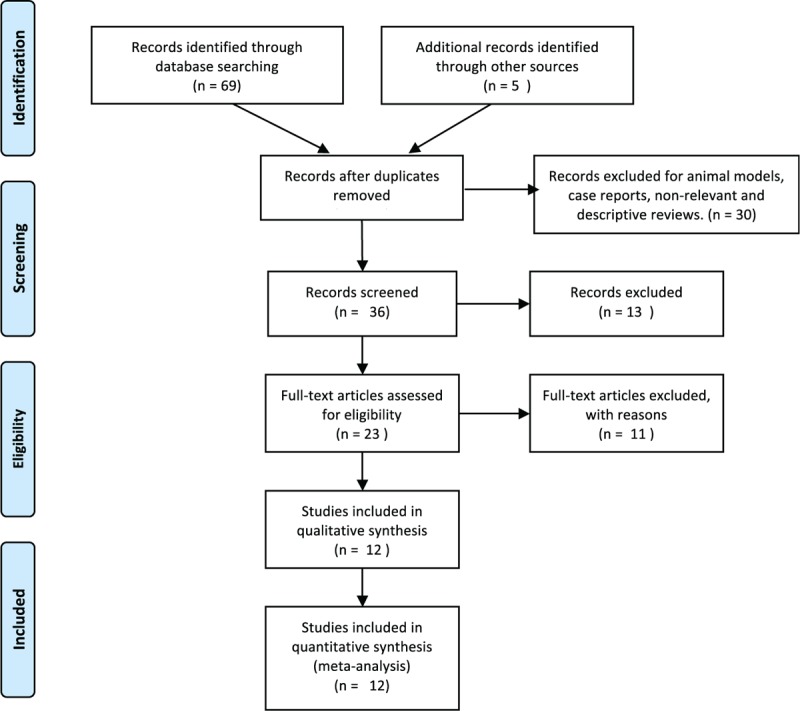
PRISMA flowchart of study selection.
Table 2.
Characteristics of included studies.
3.2. Meta-analysis results
Overall, there was no evidence of an association between the Pro12Ala variant and increased risks of GDM when all data sets were pooled together. The per-allele OR of Pro using the random effects models was 1.10 [95% CI: 0.98–1.24; P(Z) = 0.01; P(Q) = 0.003; Fig. 2]. The main results of the meta-analysis are listed in Table 3.
Figure 2.
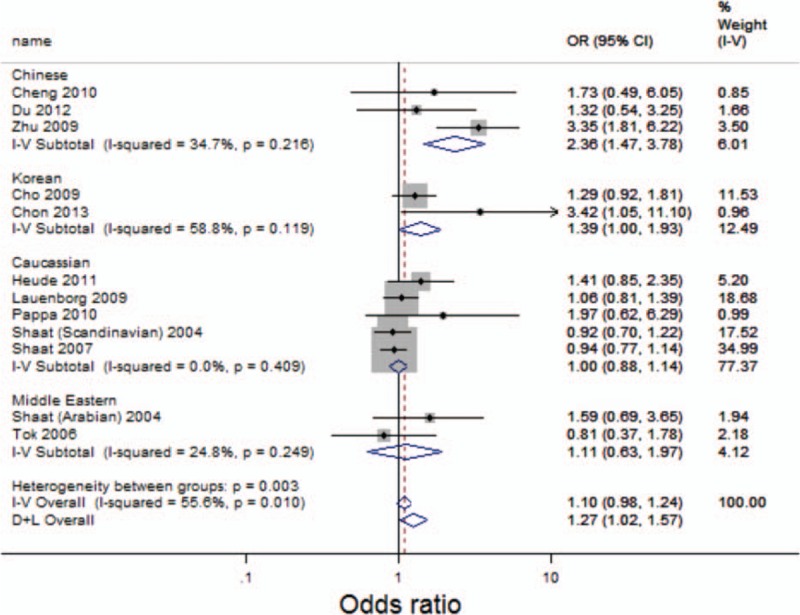
Forest plot of GDM risk associated with the Pro allele at amino acid position 12. CI = confidence interval, D + L = DerSimonian and Laird method, I-V = inverse variance, OR = odds ratio.
Table 3.
Meta analysis of the PPAR-λ Pro12Ala polymorphism and the risks of GDM.
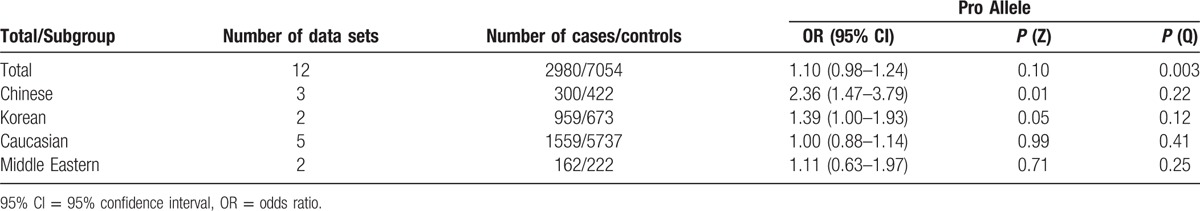
In the stratified analysis by ethnicity, significantly increased risks were found for the Chinese (OR = 2.36; 95% CI: 1.47–3.78) and Korean (OR = 1.39; 95% CI: 1.00–1.93) population (See Fig. 2). However, no significant associations were detected for the Caucasian (OR = 1.00; 95% CI: 0.88–1.14) and Middle Eastern (OR = 1.11; 95% CI: 0.63–1.97) populations.
3.3. Sensitivity analysis
Sensitivity analyses using single-study omission demonstrated that this meta-analysis was stable (Fig. 3). Statistical significance of the summary ORs was not modified (data not shown). Therefore, the results of this study are stable.
Figure 3.
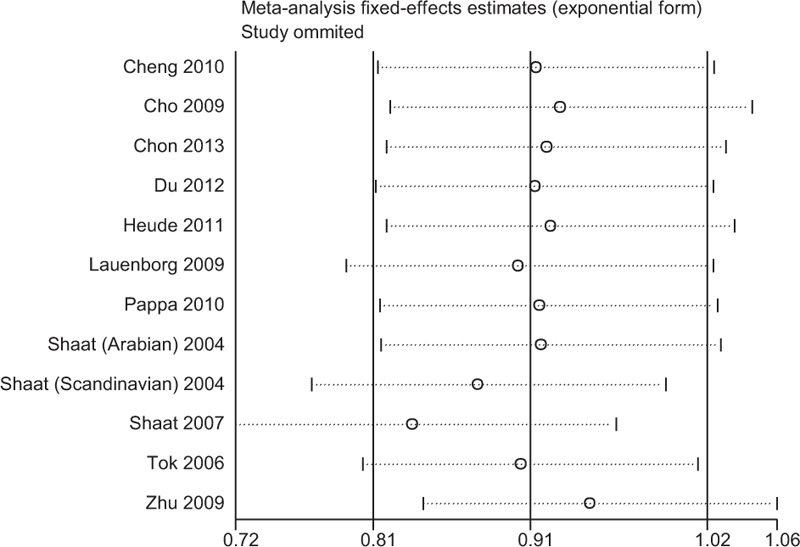
Sensitivity analysis of the Pro12Ala polymorphism and the risk of GDM.
3.4. Publication bias
Begger's and Eggar's funnel plots were constructed using the standard error and compared against the OR of each study (Figs. 4 and 5). The plots suggest the possibility of publication bias toward positive findings in smaller studies. The Duval and Tweedie nonparametric “trim and fill” method was utilized to adjust for publication bias[50] and its results did show different conclusions (data not shown). Thus, this indicates that this meta-analysis is statistically robust.
Figure 4.
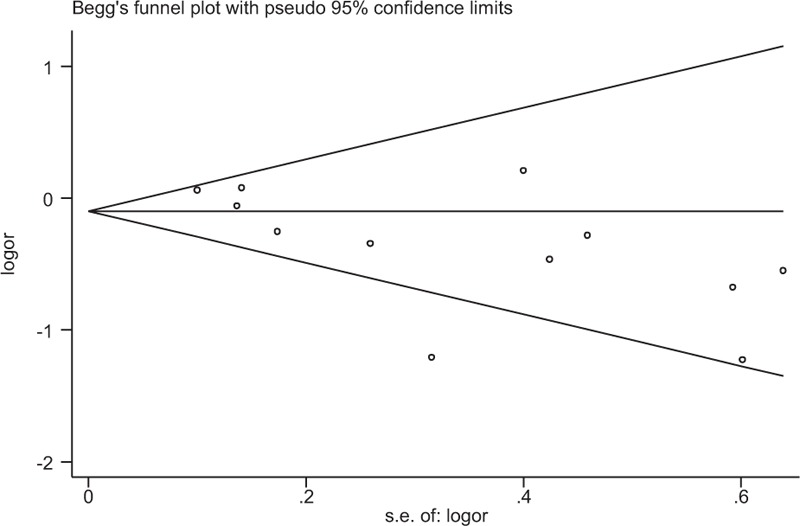
Funnel plot of the Pro12Ala polymorphism shows a possible excess of smaller studies with positive findings beyond the 95% CI. Ala = alanine, CI = confidence interval, logor = log odds ratio, Pro = proline, s.e = standard error.
Figure 5.
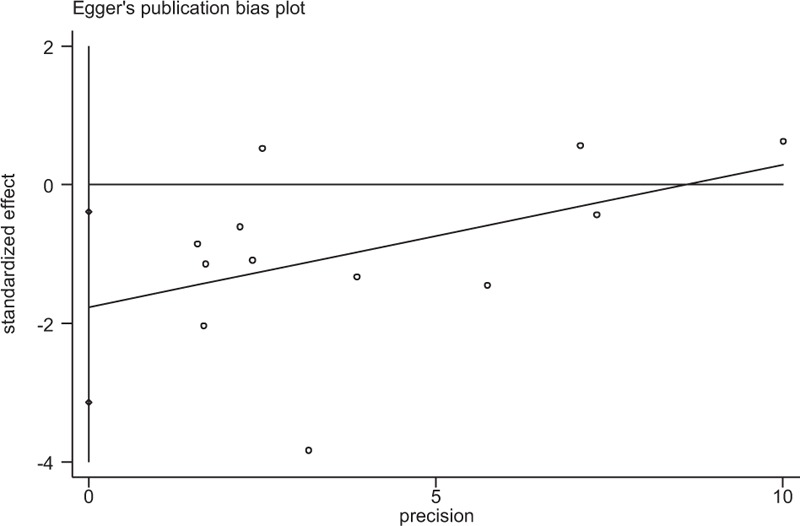
Egger's test of the Pro12Ala polymorphism (P = 0.005). Ala = alanine, Pro = proline.
4. Discussion
PPAR-λ is a ligand-dependent transcription factor involved in many body functions, including adipogenesis and also regulates immune responses.[20,25] The substitution of a Pro to Ala at site 12 is associated with reduced PPAR-λ activities[27] and has been identified as a possible polymorphism involved obesity and type 2 diabetes.[30–32]
Our up-to-date meta-analysis summarizes the evidence to date regarding the association between PPAR-λ Pro12Ala and GDM using a total of 7054 controls and 2980 GDM cases. Our study suggests that Pro12Ala is not associated with the risks of GDM.
In our stratified analysis by ethnicity, a strong association was observed for both the Chinese (OR: 2.36, 95% CI: 1.47–3.78) and Korean (OR: 1.39, 95% CI: 1.00–1.93) population but not for the Caucasian (OR = 1.00, 95% CI = 0.88–1.14) and Middle Eastern (OR = 1.11, 95% CI = 0.63–1.97) populations. These results indicate that the association of the polymorphism has a genetic and possibly environmental background factor in contributing to the pathology of GDM. Other factors such as differences in matching criteria and selection bias could also play a role in the difference between ethnic groups. It should also be noted that the analysis only included 3 Chinese studies and 2 Korean studies. This suggests the possibility that the observed differences may be due to chance. Thus, additional studies are required to increase the statistical power and validate the racial difference of the Pro12Ala polymorphism and GDM risk.
The preferential publication of studies with positive results is a significant source of bias in many meta-analyses. However, the included studies in our meta-analysis also consist of studies with negative conclusions. Although our funnel plots showed asymmetry, the results using the “trim and fill” method did not alter the conclusion of this study. This suggests that the bias may not be caused by publication bias but by potential heterogeneity between each study's population, language bias, citation bias, or simply by chance.
Several limitations should be noted in interpreting our results. We were not able to adjust for potential confounding effects conferred by gender, environmental factors, and lifestyle due to the lack of data. Our results were based on unadjusted estimates—a more precise analysis could be conducted if all raw data were available. The lack of individual health and metabolic data, such as fasting plasma glucose levels, β-cell function, and indices for insulin sensitivity also forbid us from performing a more sensitive analysis.
In conclusion, the pooled results of our meta-analysis indicate that Pro12Ala is not associated with the risks of GDM. However, in the Chinese and Korean populations, the Pro allele is strongly associated with the risks for GDM. Larger association studies with strict selection criteria are required to validate this result.
Footnotes
Abbreviations: Ala = alanine, BMI = body mass index, CI = confidence Interval, GDM = gestational diabetes mellitus, HWE = Hardy–Weinberg equilibrium, OR = odds ratio, PPAR-λ = peroxisome proliferator-activated receptors-λ, Pro = Proline.
Authorship: Conceived and designed the experiments: LW. Performed the experiments: LW, WX, and XW. Analyzed the data: LW, WX. Wrote the paper: LW, WX, and XW.
Funding/support: This work is supported by the Natural Science Foundation of Jiangsu Province in 2015 (BK20151261). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
The authors have no conflict of interest.
References
- 1.Buchanan TA, Xiang AH. Gestational diabetes mellitus. J Clin Invest 2005; 115:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilmartin ABH, Ural SH, Repke JT. Gestational diabetes mellitus. Rev Obstet Gynecol 2008; 1:129–134. [PMC free article] [PubMed] [Google Scholar]
- 3.Metzger BE, Coustan DR. Summary and recommendations of the Fourth International Workshop-Conference on Gestational Diabetes Mellitus. The Organizing Committee. Diabetes Care 1998; 21 (Suppl 2):B161–B167. [PubMed] [Google Scholar]
- 4.Ferrara A, Kahn HS, Quesenberry CP, et al. An increase in the incidence of gestational diabetes mellitus: Northern California, 1991-2000. Obstet Gynecol 2004; 103:526–533. [DOI] [PubMed] [Google Scholar]
- 5.Dabelea D, Snell-Bergeon JK, Hartsfield CL, et al. Increasing prevalence of gestational diabetes mellitus (GDM) over time and by birth cohort: Kaiser Permanente of Colorado GDM Screening Program. Diabetes Care 2005; 28:579–584. [DOI] [PubMed] [Google Scholar]
- 6.Ferrara A. Increasing prevalence of gestational diabetes mellitus: a public health perspective. Diabetes Care 2007; 30 (Suppl 2):S141–S146. [DOI] [PubMed] [Google Scholar]
- 7.Tobias DK, Zhang C, van Dam RM, et al. Physical activity before and during pregnancy and risk of gestational diabetes mellitus: a meta-analysis. Diabetes Care 2011; 34:223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mokdad AH, Serdula MK, Dietz WH, et al. The spread of the obesity epidemic in the United States, 1991-1998. JAMA 1999; 282:1519–1522. [DOI] [PubMed] [Google Scholar]
- 9.Pan XR, Yang WY, Li GW, et al. Prevalence of diabetes and its risk factors in China, 1994. National Diabetes Prevention and Control Cooperative Group. Diabetes Care 1997; 20:1664–1669. [DOI] [PubMed] [Google Scholar]
- 10.Martin AO, Simpson JL, Ober C, et al. Frequency of diabetes mellitus in mothers of probands with gestational diabetes: possible maternal influence on the predisposition to gestational diabetes. Am J Obstet Gynecol 1985; 151:471–475. [DOI] [PubMed] [Google Scholar]
- 11.Kwak SH, Kim S-H, Cho YM, et al. A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes 2012; 61:531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Nie M, Li W, et al. Association of six single nucleotide polymorphisms with gestational diabetes mellitus in a Chinese population. PLoS One 2011; 6:e26953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groenewoud MJ, Dekker JM, Fritsche A, et al. Variants of CDKAL1 and IGF2BP2 affect first-phase insulin secretion during hyperglycaemic clamps. Diabetologia 2008; 51:1659–1663. [DOI] [PubMed] [Google Scholar]
- 14.Lauenborg J, Grarup N, Damm P, et al. Common type 2 diabetes risk gene variants associate with gestational diabetes. J Clin Endocrinol Metab 2009; 94:145–150. [DOI] [PubMed] [Google Scholar]
- 15.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 2007; 39:770–775. [DOI] [PubMed] [Google Scholar]
- 16.Grarup N, Rose CS, Andersson EA, et al. Studies of association of variants near the HHEX, CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects: validation and extension of genome-wide association studies. Diabetes 2007; 56:3105–3111. [DOI] [PubMed] [Google Scholar]
- 17.Tam CHT, Ho JSK, Wang Y, et al. Common polymorphisms in MTNR1B, G6PC2 and GCK are associated with increased fasting plasma glucose and impaired beta-cell function in Chinese subjects. PLoS One 2010; 5:e11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei F-Y, Nagashima K, Ohshima T, et al. Cdk5-dependent regulation of glucose-stimulated insulin secretion. Nat Med 2005; 11:1104–1108. [DOI] [PubMed] [Google Scholar]
- 19.Tontonoz P, Hu E, Graves RA, et al. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev 1994; 8:1224–1234. [DOI] [PubMed] [Google Scholar]
- 20.Lemberger T, Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: a nuclear receptor signaling pathway in lipid physiology. Annu Rev Cell Dev Biol 1996; 12:335–363. [DOI] [PubMed] [Google Scholar]
- 21.Kliewer SA, Forman BM, Blumberg B, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A 1994; 91:7355–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee R, Jow L, Croston GE, et al. Identification, characterization, and tissue distribution of human peroxisome proliferator-activated receptor (PPAR) isoforms PPAR 2 versus PPAR 1 and activation with retinoid X receptor agonists and antagonists. J Biol Chem 1997; 272:8071–8076. [DOI] [PubMed] [Google Scholar]
- 23.Dubuquoy L, Dharancy S, Nutten S, et al. Role of peroxisome proliferator-activated receptor gamma and retinoid X receptor heterodimer in hepatogastroenterological diseases. Lancet (London, England) 2002; 360:1410–1418. [DOI] [PubMed] [Google Scholar]
- 24.Ricote M, Li AC, Willson TM, et al. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature 1998; 391:79–82. [DOI] [PubMed] [Google Scholar]
- 25.Jiang C, Ting AT, Seed B. PPAR-gamma agonists inhibit production of monocyte inflammatory cytokines. Nature 1998; 391:82–86. [DOI] [PubMed] [Google Scholar]
- 26.Fajas L, Debril MB, Auwerx J. PPAR gamma: an essential role in metabolic control. Nutr Metab Cardiovasc Dis 2001; 11:64–69. [PubMed] [Google Scholar]
- 27.Deeb SS, Fajas L, Nemoto M, et al. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet 1998; 20:284–287. [DOI] [PubMed] [Google Scholar]
- 28.Doney ASF, Fischer B, Cecil JE, et al. Association of the Pro12Ala and C1431T variants of PPARG and their haplotypes with susceptibility to Type 2 diabetes. Diabetologia 2004; 47:555–558. [DOI] [PubMed] [Google Scholar]
- 29.Meirhaeghe A, Fajas L, Helbecque N, et al. A genetic polymorphism of the peroxisome proliferator-activated receptor gamma gene influences plasma leptin levels in obese humans. Hum Mol Genet 1998; 7:435–440. [DOI] [PubMed] [Google Scholar]
- 30.Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 2000; 26:76–80. [DOI] [PubMed] [Google Scholar]
- 31.Lohmueller KE, Pearce CL, Pike M, et al. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 2003; 33:177–182. [DOI] [PubMed] [Google Scholar]
- 32.Tönjes A, Scholz M, Loeffler M, et al. Association of Pro12Ala polymorphism in peroxisome proliferator-activated receptor gamma with pre-diabetic phenotypes: meta-analysis of 57 studies on nondiabetic individuals. Diabetes Care 2006; 29:2489–2497. [DOI] [PubMed] [Google Scholar]
- 33.Wells G, Shea B, O’connel D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. 2013. Available from: http://www.ohri.ca/programs/clinical_epidemiology/nos_manual.pdf. [Google Scholar]
- 34.Lo CK-L, Mertz D, Loeb M, et al. Newcastle-Ottawa Scale: comparing reviewers’ to authors’ assessments. BMC Med Res Methodol BioMed Central 2014; 14:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Y, Ma Y, Peng T, et al. [Genotype discrepancy between maternal and fetal Pro12Ala polymorphism of PPARG2 gene and its association with gestational diabetes mellitus]. Zhonghua Fu Chan Ke Za Zhi 2010; 45:170–173. [PubMed] [Google Scholar]
- 36.Du J, Xie F, Xie X, et al. [Study on the correlation between PPARγ gene polymorphisms with gestational diabetes mellitus in Han Chinese in Hubei]. Mod J Integr Tradit Chinese West Med 2012; 21. [Google Scholar]
- 37.Zhu Y, Wu Y. [Relationship between Prol2Ala polymorphism in peroxisome proliferators-activated receptor gamma 2 gene and gestational diabetes mellitus]. Shi Yong Yi Xue Za Zhi 2009; 1963–1965. [Google Scholar]
- 38.Cho YM, Kim TH, Lim S, et al. Type 2 diabetes-associated genetic variants discovered in the recent genome-wide association studies are related to gestational diabetes mellitus in the Korean population. Diabetologia 2009; 52:253–261. [DOI] [PubMed] [Google Scholar]
- 39.Chon SJ, Kim SY, Cho NR, et al. Association of variants in PPAR(2, IGF2BP2, and KCNQ1 with a susceptibility to gestational diabetes mellitus in a Korean population. Yonsei Med J 2013; 54:352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heude B, Pelloux V, Forhan A, et al. Association of the Pro12Ala and C1431T variants of PPARgamma and their haplotypes with susceptibility to gestational diabetes. J Clin Endocrinol Metab 2011; 96:E1656–E1660. [DOI] [PubMed] [Google Scholar]
- 41.Pappa KI, Gazouli M, Economou K, et al. Gestational diabetes mellitus shares polymorphisms of genes associated with insulin resistance and type 2 diabetes in the Greek population. Gynecol Endocrinol 2011; 27:267–272. [DOI] [PubMed] [Google Scholar]
- 42.Shaat N, Lernmark A, Karlsson E, et al. A variant in the transcription factor 7-like 2 (TCF7L2) gene is associated with an increased risk of gestational diabetes mellitus. Diabetologia 2007; 50:972–979. [DOI] [PubMed] [Google Scholar]
- 43.Shaat N, Ekelund M, Lernmark A, et al. Genotypic and phenotypic differences between Arabian and Scandinavian women with gestational diabetes mellitus. Diabetologia 2004; 47:878–884. [DOI] [PubMed] [Google Scholar]
- 44.Tok EC, Ertunc D, Bilgin O, et al. PPAR-gamma2 Pro12Ala polymorphism is associated with weight gain in women with gestational diabetes mellitus. Eur J Obstet Gynecol Reprod Biol 2006; 129:25–30. [DOI] [PubMed] [Google Scholar]
- 45.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986; 7:177–188. [DOI] [PubMed] [Google Scholar]
- 46.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959; 22:719–748. [PubMed] [Google Scholar]
- 47.Woolf B. On estimating the relation between blood group and disease. Ann Hum Genet 1955; 19:251–253. [DOI] [PubMed] [Google Scholar]
- 48.Cochran WG. The combination of estimates from different experiments. Biometrics 1954; 10:101–129. [Google Scholar]
- 49.Egger M, Smith GD, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997; 315:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000; 56:455–463. [DOI] [PubMed] [Google Scholar]


