Abstract
Coinfection of blood-borne hepatitis B and hepatitis C viruses (HBV and HCV, respectively) in human immunodeficiency virus type 1 (HIV-1)-positive individuals frequently occurs in inmate population and peculiar viral strains and patterns of virological markers may be observed.
Plasma from 69 HIV-1-positive inmates was obtained from 7 clinical centers connected with correctional centers in different towns in Italy. HIV, HBV, and HCV markers were tested by commercial assays. Virus genotyping was carried out by sequencing the protease and reverse transcriptase-encoding region (PR-RT region) for HIV and a region encompassing the NS5B gene for HCV and subsequent phylogenetic analysis.
Twelve over 14 HIV-subtyped inmates were infected with HIV-1 subtype B strains. The 2 non-B strains belonged to subtype G and CRF02_AG, in an Italian and a Gambian patient, respectively. Variants carrying the K103N and Y181C resistance mutations to non-nucleoside reverse transcriptase inhibitors (NNRTIs) were found in 2 out of 9 patients naive for combined antiretroviral therapy (cART) (22.2%). Most HIV-positive patients (92.8%) showed evidence of past or present HBV and/or HCV infection. Prevalence of HBV and HCV was 81.2% for both viruses, whereas prevalence of HBV/HCV coinfection was 69.6%. A significantly higher presence of HCV infection was found in Italians [odds ratio (OR) 11.0; interval 1.7–80.9] and in drug users (OR 27.8; interval 4.9–186.0). HCV subtypes were determined in 42 HCV or HBV/HCV-coinfected individuals. HCV subtypes 1a, 3a, 4d, and 1b were found in 42.9%, 40.5%, 14.3%, and 2.4% of inmates, respectively. Low titers of HBV DNA in HBV DNA positive subjects precluded HBV subtyping.
The high prevalence of HBV and HCV coinfections in HIV-infected inmates, as well as the heterogeneity of HIV and HCV subtypes suggest the need to adopt systematic controls in prisons to monitor both the burden and the genetic forms of blood-borne viral infections, in order to apply targeted therapeutic interventions.
Keywords: HBV, HCV, HIV, inmates, Italy, virus variability
1. Introduction
Prevalence of HIV infection among inmates in Italy has been estimated to be about 7.5%, more than 25 times the estimated rate in the general Italian population in 2012.[1,2] Factors, such as overcrowding, scarce hygiene, promiscuous intercourses, reduced perception of risk of infection and absence of effective health policies, can favor transmission of HIV among inmates, as well as of other blood pathogens, such as HBV and HCV, in particular in HIV-positive inmates, who are at a greater risk of coinfection with HBV and HCV, than the general population.[3] Prevalence rates of blood-borne virus infections vary substantially among prison settings of different countries, reflecting the epidemiological characteristics of the geographic area, the risk category of people entering prisons, and the prevalence of foreign prisoners.[4] Different prevalence values were reported for HIV and viral hepatitis antibody markers, varying from 0.04% to 17%, from 21.7% to 52%, and from 4.9% to 80% for HIV, HBV, and HCV, respectively,[1,5–7] mostly depending on the prevalence of drug users.
The high prevalence of HIV, HBV, and HCV infections raises concerns for their spreading within the prison and the general population in Italy. However, efforts to contain spreading of these blood-borne pathogens in prisons are strongly limited by severe budget constraints that lead to crumbing and obsolete health facilities and by frequent inmate transfers between prisons that do not allow regular and constant clinical visits and maintenance of an effective correctional health care database.[8,9]
In HIV-positive patients, pattern modification of coinfecting virus markers may occur, possibly due to altered immune response. Specific therapeutic approaches are therefore required in presence of HBV and/or HCV coinfection and in presence of different coinfecting viral strains and genotypes.[10–12] To this regard, few data are available on genetic variants of HIV, HBV, and HCV strains circulating in the population of inmates in Italy. Prevalence of HIV subtypes in prison is strongly influenced by the frequency of foreign people in each prison.[13] As well, prevalence of HCV genotypes may be influenced by the route of infection and coinfection, regardless of the geographic variable, and may differ between risk groups and the general population.[14] Knowledge of the prevalence of different subtypes of HIV, HBV, and HCV circulating in prisons may be useful to track the epidemiology of infection and choose appropriate therapies.
Here, we describe a cross-sectional multicenter study aimed to assess epidemiological, clinical, and behavioral characteristics of HIV, HBV, and HCV infections in HIV-infected inmates, as well as to investigate circulating genotypes and subtypes of HIV and hepatitis viruses, in detention centers distributed over the Italian territory.
2. Methods
2.1. Study population
A multicenter study was performed from January to December 2013 in clinical centers connected to detention centers of 7 Italian towns (from North to South: Brescia, Genoa, Modena, Viterbo, Sassari, Bari, and Lamezia Terme). Plasma samples leftovers, previously collected and stored at −80°C in the clinical centers in the period 2011 to 2013, from a total of 69 inmates already diagnosed with HIV infection, were sent to the National AIDS Center (NAC) of the Istituto Superiore di Sanità (ISS) in Rome for determination of markers of HBV and HCV and for HIV, HBV, and HCV subtyping. All studies regarding the Italian Penitentiary System have been nationally approved by the Ethics Committee of the University of Rome Tor Vergata (Registro Sperimentazioni 73/05). For the study purposes, no ad hoc withdrawal was required and all data were collected and analyzed in an anonymous and aggregate form. Demographic, behavioral, clinical, immunological, and virological data were collected for each patient in complete anonymity in each center in a case report form provided by NAC. Molecular assays for HIV were performed at NAC, whereas the assays for HBV and HCV were carried out at the Department of Infectious Diseases of ISS. Contribution of samples from each center varied according to HIV prevalence. A number of 32 samples were obtained from Genoa, 14 from Brescia, 9 from Sassari, 7 from Viterbo, 4 from Bari, 2 from Lamezia Terme, and 1 from Modena.
2.2. Serological assays
Anti-HCV, anti-HBc (IgM and IgG), and HBsAg markers were tested by chemiluminescent assays on an automated analyzer (COBAS Elecsys e401; Roche Diagnostics, Basel, Switzerland) using Elecsys anti-HCV II, Elecsys anti-HBc, and Elecsys HBsAg II kits, the last one with a sensitivity of 20 to 30 mUI/mL. Anti-HBs antibodies were detected by Enzygnost Anti-HBs II (Siemens Healthcare Diagnostics Products, Marburg, Germany) and results evaluated according to the manufacturer's instructions.
2.3. Viral nucleic acid quantitation
HIV viral load was carried out at each center where enrollment was performed, as a common routine analysis for HIV-infected inmates, using different commercial kits.
HBV DNA quantitation was performed on plasma using the High Pure System Viral Nucleic Acid Kit (Roche Diagnostics, Basel, Switzerland). Amplification and detection were performed on a COBAS AmpliPrep/COBAS TaqMan Instrument using the COBAS TaqScreen MPX Test v2.0; this assay is currently used for blood screening and has a sensitivity of 2.1 IU/mL.
HCV RNA quantitation was performed on plasma using the High Pure System Viral Nucleic Acid Kit (Roche Diagnostics, Basel, Switzerland). Amplification and detection were carried out on a COBAS TaqMan 48 Analyzer using the COBAS TaqMan HCV Test v2.0; this test has a sensitivity of about 9.3 IU/mL and a linear range from 25 to 3.91 × 108 IU/mL (as reported in the package insert).
2.4. Virus genotyping and detection of drug resistance mutations
Patients with HIV-detectable plasma viral load (according to data reported by each center) were chosen for HIV subtyping. For this study purpose, the limit for a detectable virus RNA was set up at ≥1.69 log10 RNA copies/mL. HIV RNA was extracted from plasma using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). Extracted RNA was processed for reverse transcription and a portion of the pol gene, encompassing the protease and reverse transcriptase encoding region (the PR-RT region) was amplified by polymerase chain reaction (PCR), following a previously described protocol.[15] The resulting amplicon was purified using PCR Clean Up (Abbott Molecular, Des Plaines, IL, USA) and directly sequenced using an ABI 3730 automated sequencer.
For HCV genotyping, HCV RNA was extracted from plasma, using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). RNA was reverse transcribed using the SuperScript II reverse transcriptase protocol (Invitrogen, Life Technologies, Carlsbad, CA) and cDNA amplified by nested PCR using the FastStart High Fidelity PCR system (Roche Diagnostics, Basel, Switzerland). Primers for the first and second rounds of amplification have been previously described.[16] Nested PCR products encompassed the NS5B gene (nt 8256–8632) of the HCV genome. PCR conditions for both rounds were 94°C for 2 minutes followed by 28 cycles of denaturation at 94°C for 15 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 45 seconds, and a last extension step at 72°C for 7 minutes. HCV PCR products were purified using the High Pure PCR Cleanup Micro Kit (Roche Diagnostics, Basel, Switzerland). Both strands were sequenced using the Genome Lab DTCS Quick Start KiT (Beckman Coulter, Inc., Fullerton, CA). Sequencing reactions were run on an automated DNA sequencer (Beckman Coulter, Inc., Fullerton, CA).
HIV and HCV sequences were separately aligned by Clustal W (BioEdit package) and manually edited to maximize alignment. HIV aligned sequences were compared with reference sequences for the major HIV-1 subtypes and the circulating recombinant forms (CRFs) available at HIV Los Alamos database (http://www.hiv.lanl.gov), whereas HCV-aligned sequences were compared with 20 reference sequences representing the major known HCV genotypes/subtypes, downloaded from the HCV Los Alamos database (http://hcv.lanl.gov/content/index). Only references with confirmed genotype were downloaded.
Phylogenetic analysis of both HIV-1 PR-RT and HCV NS5B sequences was carried out by building a phylogenetic tree inferred using Neighbor-Joining (Kimura-2 parameter model) through Phylip 3.67 (evolution.genetics.washington.edu/phylip.html). The statistical robustness and the reliability of the phylogenetic tree were confirmed by bootstrap analysis using 1000 replicates.
2.5. Statistical analysis
Pearson Chi-squared test or Fisher exact test, when necessary, were used to evaluate the difference in the prevalence of viral hepatitis markers between groups based on demographic or clinical characteristics. The association between demographic and clinical determinants and the positivity for HBV and HCV markers was evaluated by crude odds ratios (ORs) and their 95% confidence intervals (95% CIs); exact CIs were used on small samples. P values less than 0.05 were considered statistically significant.
All the statistical procedures were performed using the STATA statistical package, release 13.1 (STATA Corp, TX, USA).
2.6. Accession numbers
Both HIV and HCV nucleotide sequences are in the process to be submitted to GenBank.
3. Results
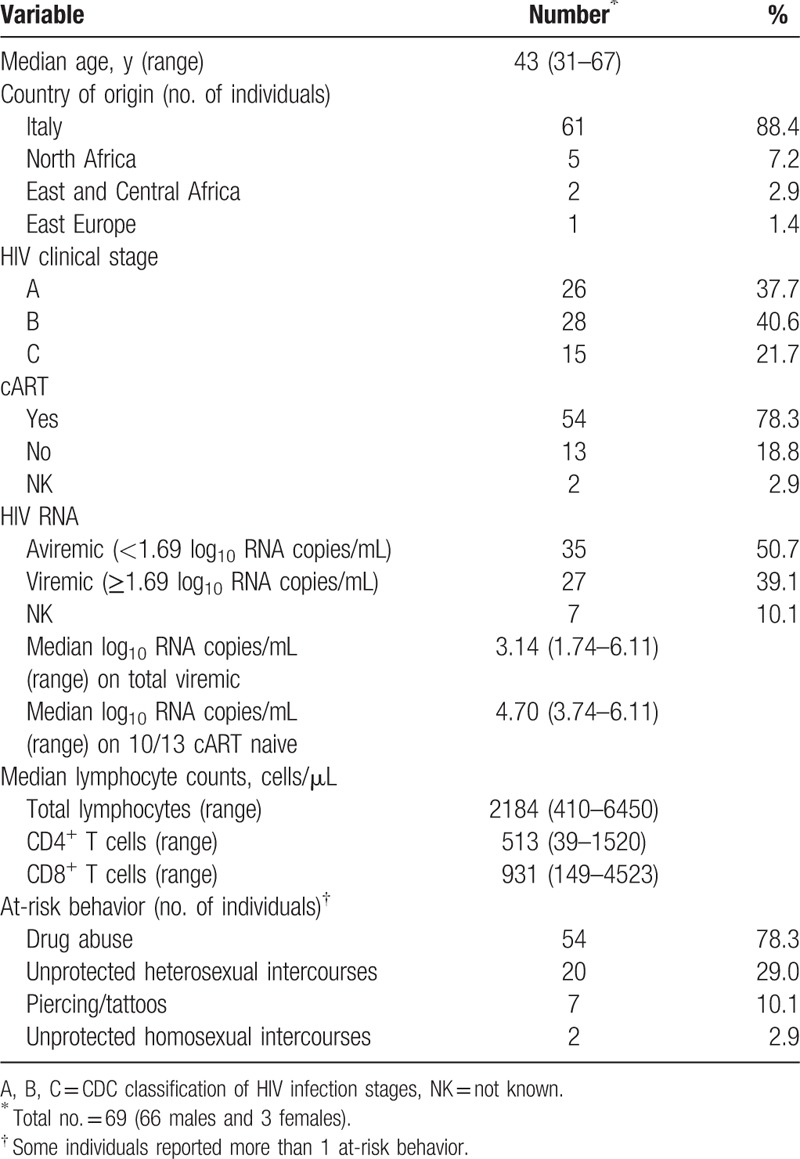
Table 1 summarizes the demographic, clinical, immunological, virological, and behavioral data reported in the case report forms of the 69 anti-HIV positive inmates. Sixty-six inmates were males and 3 females. Median age was 43 years (range 31–67). Sixty-one (88.4%) inmates were Italian and 8 (11.6%) from other countries. In particular, 5 inmates were from North Africa (Tunisia, Algeria, and Morocco), 2 from East and Central Africa (The Gambia and Nigeria), and 1 from East Europe (Moldova). Regarding HIV infection, 37.7%, 40.6%, and 21.7% were in A, B, and C clinical stages (CDC classification), respectively, at the time of sample collection. Of the participants, 54 out of 69 (78.3%) were on cART for HIV and 13 (18.8%) were naive for therapy. For 2 inmates, information on cART therapy was not reported. A total of 27 inmates had a detectable HIV plasma viral load with a median of 3.14 log10 RNA copies/mL (range 1.74–6.11); of these, 13 were naive for cART and 10 of them had a detectable HIV plasma viral load with a median of 4.70 log10 RNA copies/mL (range 3.74–6.11). Median of the total lymphocyte counts in the 69 inmates was 2184 cells/μL, while the median of CD4+ and CD8+ T cell counts were 513 and 931 cells/μL, respectively. Many inmates reported more than 1 behavior at risk for blood-borne virus infections. The most common one was the abuse of drugs (injective and/or noninjective) that was present in 78.3% of the participants. Other at-risk behaviors were unprotected heterosexual intercourses (29.0%), piercing and tattoos in a hygienically uncontrolled environment (10.1%), and unprotected homosexual intercourses (2.9%).
Table 1.
Demographic, clinical, immunological, virological, and behavioral information available for the 69 participants to the study.
HIV sequencing and subtyping was successful for 14 of the 27 HIV viremic individuals. Nine of the 14 HIV subtyped inmates were native of Italy, 3 of Tunisia, 1 of the Gambia, and 1 of Moldova. Phylogenetic analysis of the HIV PR-RT region sequence revealed the presence of several different HIV subtypes and recombinant forms, including subtype B in 11 patients, subtype G in 1 patient, and the CRF CRF02_AG in another patient (Fig. 1). All Italian individuals but one, who was infected by a subtype G strain, were infected by subtype B variants. The 3 patients from Tunisia and the 1 from Moldova were also infected by a subtype B strain, whereas the patient from The Gambia hosted the CRF02_AG variant (Fig. 1).
Figure 1.
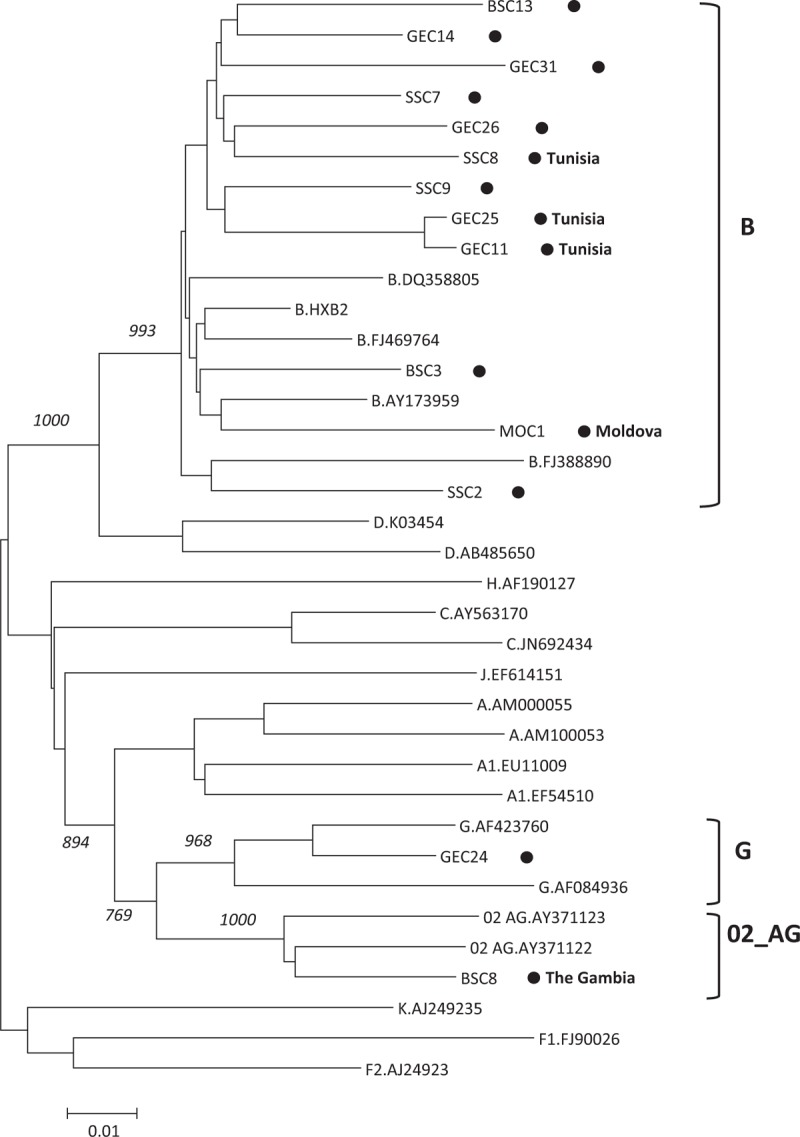
Neighbor-joining phylogenetic tree of HIV variants based on PR-RT sequence. The tree shows the phylogenetic relationships between HIV sequences obtained from 14 inmates (marked by a dot) and reference HIV sequences with a known genotype. The country of origin of non-Italian inmates is indicated. The abbreviations in capital letters before each number of the dotted sequences represent the city where the inmate resided. BS = Brescia, GE = Genoa, MO = Modena, SS = Sassari.
HIV-1 mutations that confer resistance to antiretroviral drugs in the 14 HIV subtyped individuals were found and are reported in Table 2. Major drug resistance mutations (DRMs) were observed in 1 out of 4 patients on cART. This patient was under treatment with drugs from the nucleoside reverse transcriptase inhibitor (NRTI) and protease inhibitor (PI) drugs and had a variant carrying both the M184V mutation, directed to NRTI drugs and the K103N mutation, directed to NNRTI, although the patient was not treated (nor was previously treated) with drugs of the NNRTI class. No major DRMs against PIs were observed. More important, 2 out of 9 patients naive for cART (22.2%) hosted variants that carried the K103N (in one patient) and the Y181C (in the other patient) mutations, both directed to drugs of the NNRTI class.
Table 2.
Major DRMs in the HIV-1 subtyped patients on different cART regimens.

Hepatitis B and C serological and virological markers (HBsAg, HBV DNA, anti-HBc, anti-HBs, anti-HCV, and HCV RNA) were tested in plasma from the enrolled HIV positive inmates. Table 3 reports the results. Prevalence of HBsAg and HBV DNA markers was 8.8% and 7.2%, respectively. The level of HBV DNA was very low, below 25 IU/mL (the lower limit of the linear range for quantification) in all HBV DNA positive samples. All but one HBV DNA positive individual were also HBsAg positive; the HBsAg-negative case (an Italian inmate naive for HIV cART) could be diagnosed having an occult HBV infection (OBI) (HBsAg negative, and HBV DNA <200 IU/mL in serum, but still detectable). The anti-HBc marker was detected in 69.6% of total inmates; this marker was the only one detectable (isolated anti-HBc) in 39.3% of the 61 inmates in which all HBV markers could be tested (see footnotes in Table 3). The anti-HBs marker was detected in 30.6% of subjects. Prevalence of anti-HCV and HCV RNA markers was 78.3% and 65.2%, respectively, with HCV RNA detected in 79.6% of anti-HCV positive samples. No significant trend for age was found for any tested marker. HCV infection was, instead, significantly more frequent in Italian than in foreign subjects (P < 0.001) and resulted significantly associated with Italian nationality (OR: 11.0, 95% CI: 1.7–80.9) (data not shown in the table).
Table 3.
Prevalence of HBV and HCV markers and distribution by age and nationality.
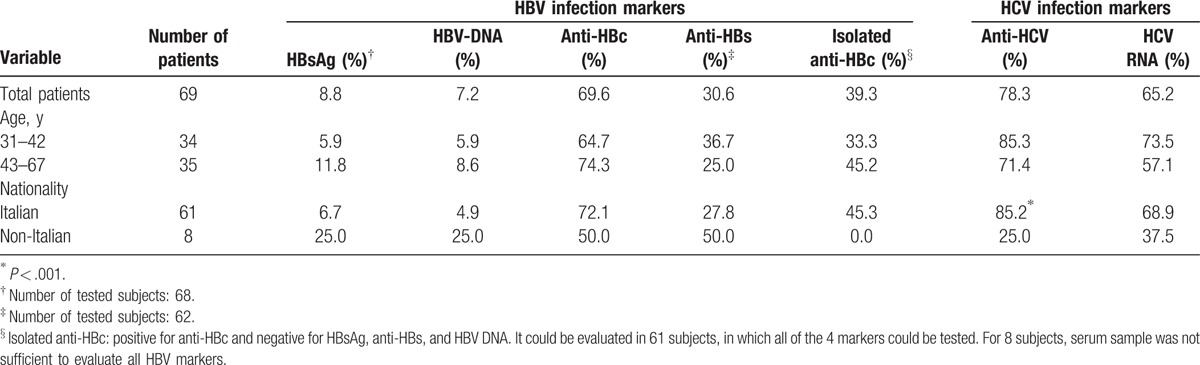
Figure 2 summarizes the overall prevalence of HBV and HCV infection in the studied population. To include both past and current infections, subjects were considered to be positive for HBV or HCV if at least 1 HBV or HCV marker was detected. The great majority of subjects (64/69, 92.8%) proved to be positive for HBV and/or HCV infection, with only a small fraction (5/69, 7.2%) negative for all the assayed markers. Among the 64 subjects positive for HBV and/or HCV, 56 were HBV positive and 56 HCV positive. Merging of HBV and HCV data showed 8 inmates to be positive for only HBV, 48 for both viruses, and 8 for only HCV (11.6%, 69.6%, and 11.6% over the total 69 inmates, respectively).
Figure 2.
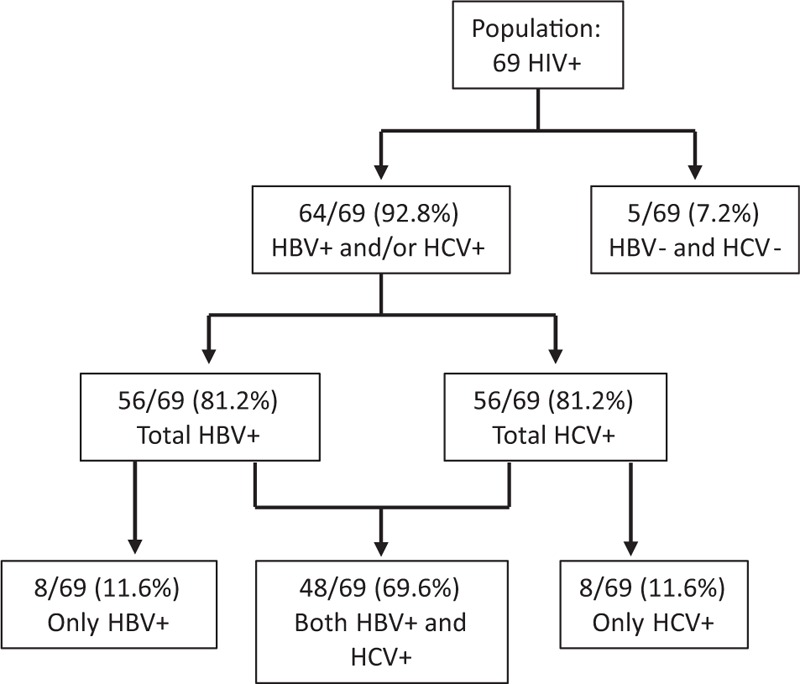
Prevalence of HBV and HCV infection in HIV-infected inmates. Subjects showing at least 1 positive marker for HBV and/or HCV were considered to be positive for past or current HBV and/or HCV infection.
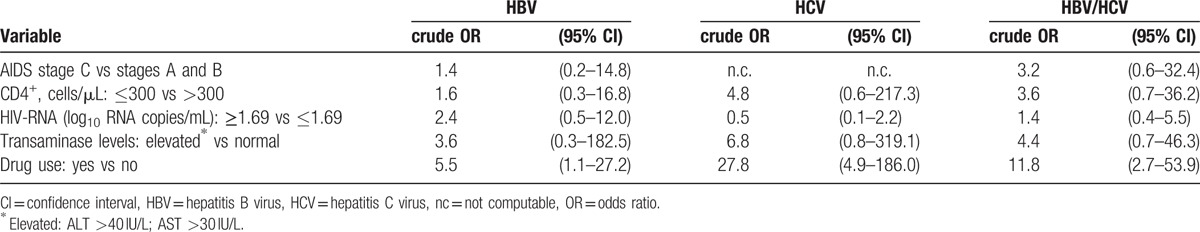
Results of the univariate analysis for the association between HBV, HCV, and HBV/HCV coinfections and clinical variables are reported in Table 4. Associations between drug use and HBV, HCV, and HBV/HCV coinfections were observed (OR: 5.5, 95% CI: 1.1–27.2; OR: 27.8, 95% CI: 4.9–186.0 and OR: 11.8, 95% CI: 2.7–53.9, respectively), whereas no association of HBV, HCV, and HBV/HCV coinfections with either AIDS stage, or CD4+ T cells, or plasma HIV RNA copies, or elevated transaminase levels was present.
Table 4.
Univariate analysis of clinical factors associated with hepatitis B and C coinfections in HIV-positive subjects.
HCV sequencing and subtyping was successful in 42 individuals. The results of the phylogenetic analysis are shown in Fig. 3. Subtypes 1a and 3a were present in 18 and 17 individuals (42.9% and 40.5%), respectively, while subtypes 4d and 1b in 6 (14.3%) and 1 (2.4%) individuals, respectively. All but 2 isolates were from Italian inmates. The 2 isolates were from Tunisian patients and belonged to 1a and 4d subtypes. The very low level of HBV DNA made unsuccessful any attempt to amplify HBV DNA for sequencing.
Figure 3.
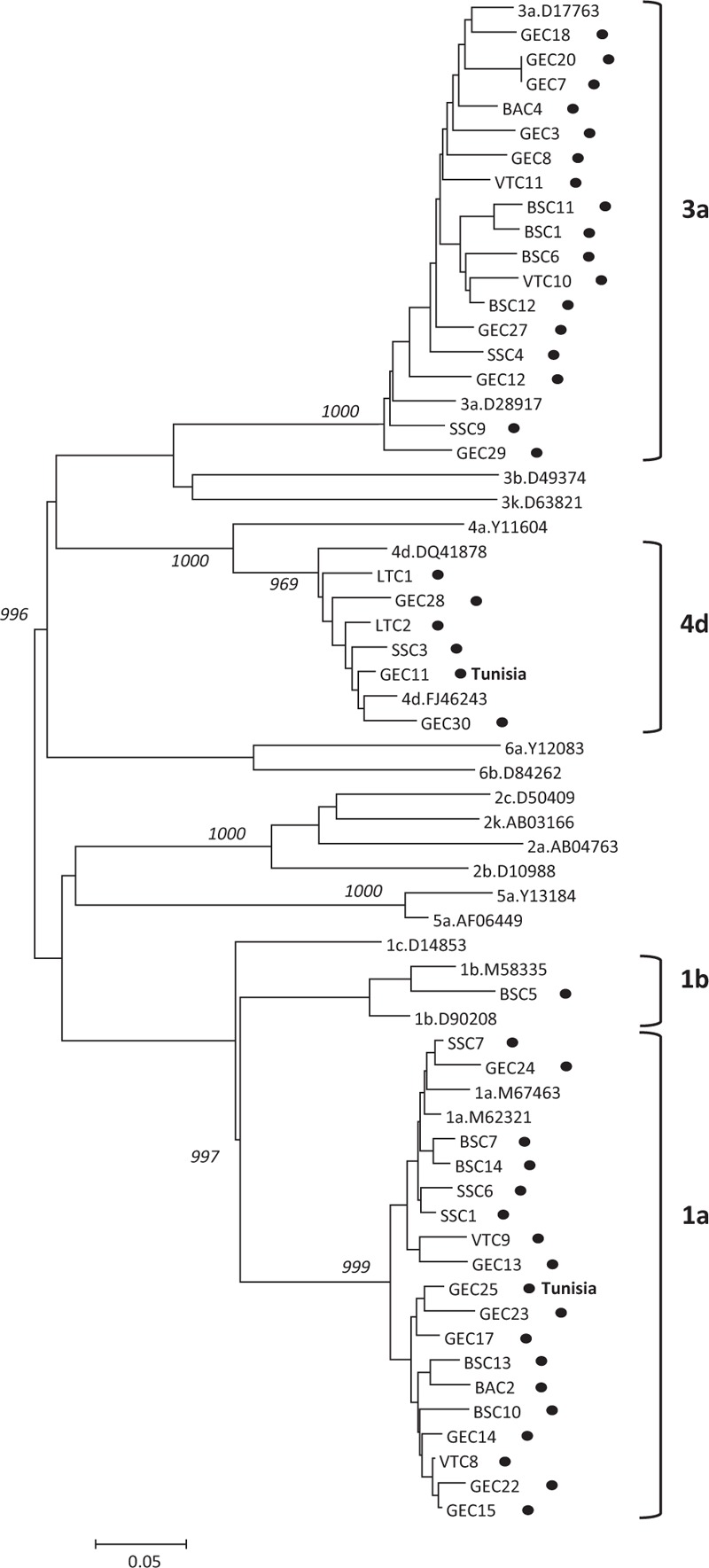
Neighbor-joining phylogenetic tree of HCV variants based on the NS5B gene sequence. The tree shows the phylogenetic relationships between HCV sequences obtained from 42 inmates (marked by a dot) and reference HCV sequences with a confirmed sub-genotype. The country of origin of non-Italian inmates is indicated. The abbreviations in capital letters before each number of the dotted sequences represent the city where the inmate resided. BA = Bari, BS = Brescia, GE = Genoa, LT = Lamezia Terme, SS = Sassari, VT = Viterbo.
4. Discussion
Most of the published prevalence data on blood-borne infections in correctional facilities of different countries are referred to all inmate population. The specific rates of HBV and HCV coinfection markers in HIV-1 positive inmates are rarely reported, often because of the scarce number of subjects. The present multicenter study, carried out in correctional facilities on the Italian territory, was focused to describe HIV and hepatitis infections in HIV-1 positive subjects.
Most of the HIV subtyped inmates were infected by HIV-1 subtype B. This subtype is predominant in North America and Western Europe[17,18] including Italy.[19] In recent years, frequency of non-B clade forms has been reported to increase in clade B restricted geographical areas and in Italy.[19,20] In the present study, 1 case of HIV-1 clade G infection was found in an Italian inmate. This finding is in line with previous studies indicating that subtype G can be present in Italian individuals, in particular in injecting drug users.[21] In fact, most of the enrolled inmates reported to be drug users, including the Italian individual infected by the subtype G strain. The recombinant form CRF02_AG was found in 1 inmate of Gambian origin who reported to have had unprotected sexual intercourses. CRF02_AG strains have been described to be highly prevalent in West Africa, including The Gambia.[22]
Sequence analysis of HIV-1 isolates showed the presence of major DRMs in HIV variants from 2 out 9 patients naive for cART (22.2%). Both patients were Italian. Mutations were the K103N in 1 case and the Y181C in the other case, both known to be major DRMs to NNRTI drugs, according to the Stanford database (http://hivdb.stanford.edu/). In particular, K103N is a nonpolymorphic mutation selected in patients receiving nevirapine and efavirenz and able to reduce both nevirapine and efavirenz susceptibility by ≥10 folds.[23] Y181C is a nonpolymorphic mutation described to reduce susceptibility to nevirapine by >50-folds.[24] Prevalence rate of DRMs in cART-naive patients has been described to vary from 0% to 15%, depending on different variables, such as the geographical area, HIV infection prevalence, routes of transmission, and infecting subtype.[25–28] The slightly higher prevalence of DRMs found here may be due to the low number of patients we were able to characterize for the presence of variants with DRMs. However, the possibility of finding strains carrying DRMs in cART-naive inmates suggests to consider adopting antiretroviral therapy (ART) regimens including drugs with a high genetic barrier to resistance and reinforcing strategies to increase adherence to therapy in prisons.
The great majority of the analyzed HIV-positive population showed laboratory evidence for past or current HBV and/or HCV infection (92.8% prevalence). The prevalence values of HBV, HCV, or HBV/HCV coinfections (81.2%, 81.2%, and 69.6%, respectively) were higher than those resulting from data reported in individual questionnaires, filled in according to medical records and interviews (36.8%, 75.0%, and 35.3% for HBV, HCV, or both viruses, respectively, data not shown). The comparative analysis between data reported in the forms and the results from sample testing showed concordance in 63.5%, 88.4%, and 54.8% of cases for HBV, HCV, and HBV/HCV coinfections, respectively, indicating either no full knowledge for presence of HBV or HCV coinfection at the time of interview, or incomplete reporting, or both. Prevalence rates obtained by laboratory markers resulted to be stable over time if compared with data from a previous similar survey conducted 10 years earlier that included 73 HIV-positive inmates from Italian prisons (prevalence of HBsAg, anti-HBc, and anti-HCV: 9.6%, 73.9%, and 80.8% in the previous study vs 8.8%, 69.6%, and 78.3% in the present study).[1] These findings suggest that measures to stop spreading of hepatitis infections among inmates in Italian prisons, in particular in individuals with behaviors at risk of blood-borne infections, such as HIV-infected people, have been either not taken or ineffective.
HCV coinfection was significantly more frequent in Italian than non-Italian inmates. In Italy, a high prevalence of HCV infection was described in injecting drug users.[29] Accordingly, in the present study, the majority of anti-HCV positive individuals reported to be drug users. In particular, the drug abuse risk factor, including both injection and other assumption practices, was present at a higher rate (78.3%) than other risk factors, such as unprotected sexual intercourses and tattooing. These data are in agreement with other surveys conducted in correctional facilities in various countries.[5,8,30] Finally, the drug abuse risk factor was found to be significantly associated not only with HCV but also with HBV and HBV/HCV coinfections.
Isolated anti-HBc positivity was observed in 39.3% individuals. This pattern in HIV-positive patients was reported to be associated with HCV positivity,[31] although the issue is still controversial.[32] In another study, isolated anti-HBc marker in HIV/HCV-coinfected subjects was associated with drug use, older age, HIV infection, and low CD4 T cell counts.[33] One possible explanation for this altered antibody pattern is that these individuals may be more likely to lose previously developed protective anti-HBs antibodies.[34]
An Italian inmate naive for HIV ART showed the key features of OBI (HBsAg negative, HBV-DNA <200 IU/mL in serum, but detectable). OBIs have been frequently reported in HIV-positive, anti-HBc-positive individuals,[35] particularly in areas highly endemic for HBV.[36,37]
Finally, in contrast with other studies, no significant associations were found between the HBV coinfection and the CD4 T cells counts and HIV-RNA titers.[38,39]
The heterogeneity of circulating genotypes and subtypes of HCV may be influenced by the route of infection and coinfection, regardless of the geographic variable, and may differ between risk groups, such as drug users and/or inmates, and general population.[14] HCV genotypes and subtypes can differ in the level of response to combination therapies, including those based on new generation antivirals. In particular, several subtypes of genotypes 1, 3, and 4 are more difficult to treat than subtypes of genotype 2.[40] In the present study, a high prevalence of 1a and 3a HCV subtypes was found (42.9% and 40.5%, respectively). These subtypes are most frequently detected among drug users in Europe and in Italy[29,41] while, to date, the HCV subtypes 1b, 2a/2c are more prevalent in the general population.[42] The application of preventative measures to avoid the spreading in the general population of subtypes that are difficult to treat is recommended. Finally, 6 inmates (5 from Italy and 1 from Tunisia) harbored HCV subtype 4d isolates (14.3%). Increased circulation of genotype 4 in Italy was recently reported.[42]
In conclusion, high prevalence of HBV and HCV coinfections was found in the HIV-1 positive inmate population of Italian correctional facilities, indicating the need for systematic controls and screening at entrance and during the permanence in prison settings. Sequence analysis should be performed to monitor the possible emergence of drug-resistant HIV variants and more aggressive viral genotypes. It should be emphasized that prisons, particularly in situations of frequent turnover of prisoners, may be considered as a sentinel site for monitoring both viral epidemiology changes and circulation of virus strains among risk groups and the general population. In addition, an accurate surveillance system is needed for implementing appropriate prevention measures and therapeutic interventions.
Acknowledgment
The authors thank S. De Menna, S. Tobelli, and F. Fedeli for administrative support and P. Arciero for his technical support.
Footnotes
Abbreviations: cART = combined antiretroviral therapy, CRF = circulating recombinant form, DRM = drug resistance mutation, HBV = hepatitis B virus, HCV = hepatitis C virus, HIV-1 = human immunodeficiency virus type 1, ISS = Istituto Superiore di Sanità, NAC = National AIDS Center, NNRTI = non-nucleoside reverse transcriptase inhibitors, NRTI = nucleoside reverse transcriptase inhibitors, OBI = occult HBV infection, PCR = polymerase chain reaction, PI = protease inhibitors, PR-RT region = protease-reverse transcriptase region.
Funding/support: The work was funded by the project 2M26 of the Italian Ministry of Health.
The authors report no conflicts of interest.
References
- 1.Babudieri S, Longo B, Sarmati L, et al. Correlates of HIV, HBV, and HCV infections in a prison inmate population: results from a multicentre study in Italy. J Med Virol 2005; 76:311–317. [DOI] [PubMed] [Google Scholar]
- 2.Camoni L, Regine V, Stanecki K, et al. Estimates of the number of people living with HIV in Italy. Biomed Res Int 2014; 2014:209619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bick JA. Infection control in jails and prisons. Health Epidemiol 2007; 45:1047–1055. [DOI] [PubMed] [Google Scholar]
- 4.Sagnelli E, Starnini G, Sagnelli C, et al. Blood born viral infections, sexually transmitted diseases and latent tuberculosis in Italian prisons: a preliminary report of a large multicenter study. Eur Rev Med Pharmacol Sci 2012; 16:2142–2146. [PubMed] [Google Scholar]
- 5.Tresó B, Barcsay E, Tarján A, et al. Prevalence and correlates of HCV, HVB, and HIV infection among prison inmates and staff, Hungary. J Urban Health 2012; 89:108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mir-Nasseri MM, Mohammadkhani A, Tavakkoli H, et al. Incarceration is a major risk factor for blood-borne infection among intravenous drug users. Hepatitis Monthly 2011; 11:19–22. [PMC free article] [PubMed] [Google Scholar]
- 7.Reekie JM, Levy MH, Richards AH, et al. Trends in HIV, hepatitis B and hepatitis C prevalence among Australian prisoners: 2004, 2007, 2010. Med J Aust 2014; 200:277–280. [DOI] [PubMed] [Google Scholar]
- 8.Babudieri S, Starnini G, Brunetti B, et al. HIV and related infections in Italian penal institutions: epidemiological and health organization note. Ann Ist Super Sanità 2003; 39:251–257. [PubMed] [Google Scholar]
- 9.Dell’Isola S, Caturelli E, Ialungo A, et al. Detention and incompatibility of HIV patients in Italy. Ann Ist Super Sanità 2013; 49:332–333. [DOI] [PubMed] [Google Scholar]
- 10.Soriano V, de Mendoza C, Fernández-Montero JV, et al. Management and treatment of chronic hepatitis B in HIV-positive patients. Ann Med 2014; 46:290–296. [DOI] [PubMed] [Google Scholar]
- 11.Coppola N, Martini S, Pisaturo M, et al. Treatment of chronic hepatitis C in patients with HIV/HCV coinfection. W J Vir 2015; 4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Almasio PL, Babudieri S, Barbarini G, et al. Recommendations for the prevention, diagnosis, and treatment of chronic hepatitis B and C in special population groups (migrants, intravenous drug users and prison inmates). Dig Liver Dis 2011; 43:589–595. [DOI] [PubMed] [Google Scholar]
- 13.Longo B, Novati S, Montieri S, et al. (ISGHP). HIV-1 diversity among inmates of Italian prisons. J Med Virol 2008; 80:1689–1694. [DOI] [PubMed] [Google Scholar]
- 14.Tresó B, Takács M, Dencs Á, et al. Molecular epidemiology of hepatitis C virus genotypes and subtypes among injecting drug users in Hungary. Euro Surveill 2013; 18:pii: 20639. [DOI] [PubMed] [Google Scholar]
- 15.Fokam J, Salpini R, Santoro MM, et al. Performance evaluation of an in-house human immunodeficiency virus type-1 protease-reverse transcriptase genotyping assay in Cameroon. Arch Virol 2011; 156:1235–1243. [DOI] [PubMed] [Google Scholar]
- 16.Lu L, Nakano T, Smallwood GA, et al. A refined long RT-PCR technique to amplify complete viral RNA genome sequences from clinical samples: application to a novel hepatitis C virus variant of genotype 6. J Virol Methods 2005; 126:139–148. [DOI] [PubMed] [Google Scholar]
- 17.Weidle PJ, Ganea CE, Irwin KL, et al. Presence of human immunodeficiency virus (HIV) type 1, group M, non-B subtypes, Bronx, New York: a sentinel site for monitoring HIV genetic diversity in the United States. J Infect Dis 2000; 181:470–475. [DOI] [PubMed] [Google Scholar]
- 18.Tatt ID, Barlow KL, Clewley JP, et al. Surveillance of HIV-1 subtypes among heterosexuals in England and Wales, 1997-2000. J Acquir Immune Defic Syndr 2004; 36:1092–1099. [DOI] [PubMed] [Google Scholar]
- 19.Lai A, Riva C, Marconi A, et al. Changing patterns in HIV-1 non-B clade prevalence and diversity in Italy over three decades. HIV Med 2010; 11:593–602. [DOI] [PubMed] [Google Scholar]
- 20.Santoro MM, Perno CF. HIV-1 genetic variability and clinical implications. ISRN Microbiol 2013; 2013:481314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciccozzi M, Montieri S, Salemi M, et al. An outbreak of HIV-1 subtype G among Italian injecting drug users. AIDS 2007; 21:1213–1215. [DOI] [PubMed] [Google Scholar]
- 22.de Silva TI, Turner R, Hué S, et al. HIV-1 subtype distribution in the Gambia and the significant presence of CRF49_cpx, a novel circulating recombinant form. Retrovirology 2010; 7:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melikian GL, Rhee SY, Varghese V, et al. Non-nucleoside reverse transcriptase inhibitor (NNRTI) cross-resistance: implications for preclinical evaluation of novel NNRTIs and clinical genotypic resistance testing. J Antimicrob Chemother 2014; 69:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhee SY, Liu T, Ravela J, et al. Distribution of human immunodeficiency virus type 1 protease and reverse transcriptase mutation patterns in 4,183 persons undergoing genotypic resistance testing. Antimicrob Agents Chemother 2004; 48:3122–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim MH, Song JE, Ahn JY, et al. HIV antiretroviral resistance mutations among antiretroviral treatment-naive and -experienced patients in South Korea. AIDS Res Hum Retroviruses 2013; 29:1617–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiamsakul A, Sirivichayakul S, Ditangco R, et al. Transmitted drug resistance in recently infected HIV-positive Individuals from four urban locations across Asia (2007–2010): TASER-S. AIDS Res Ther 2015; 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexiev I, Shankar A, Wensing AM, et al. Low HIV-1 transmitted drug resistance in Bulgaria against a background of high clade diversity. J Antimicrob Chemother 2015; 70:1874–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moura ME, da Guarda Reis MN, Lima YA, et al. HIV-1 transmitted drug resistance and genetic diversity among patients from Piauí State, Northeast Brazil. J Med Virol 2015; 87:798–806. [DOI] [PubMed] [Google Scholar]
- 29.Stroffolini T, D’Egidio PF, Aceti A, et al. Hepatitis C virus infection among drug addicts in Italy. J Med Virol 2012; 84:1608–1612. [DOI] [PubMed] [Google Scholar]
- 30.Prasetyo AA, Dirgahayu P, Sari Y, et al. Molecular epidemiology of HIV, HBV, HCV, and HTLV-1/2 in drug abuser inmates in central Javan prisons, Indonesia. J Infect Dev Ctries 2013; 7:453–467. [DOI] [PubMed] [Google Scholar]
- 31.Witt MD, Lewis RJ, Rieg G, et al. Predictors of the isolated hepatitis B core antibody pattern in HIV-infected and -uninfected men in the multicenter AIDS cohort study. Clin Infect Dis 2013; 56:606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang SH, Chen TJ, Lee SJ, et al. Risk factors of isolated antibody against core antigen of hepatitis B virus: association with HIV infection and age but not hepatitis C virus infection. J Acquir Immune Defic Syndr 2010; 54:122–128. [DOI] [PubMed] [Google Scholar]
- 33.Sun HY, Lee HC, Liu CE, et al. Factors associated with isolated anti-hepatitis B core antibody in HIV-positive patients: impact of compromised immunity. J Viral Hepat 2010; 17:578–587. [DOI] [PubMed] [Google Scholar]
- 34.Biggar RJ, Goedert JJ, Hoofnagle J. Accelerated loss of antibody to hepatitis B surface antigen among immunodeficient homosexual men infected with HIV. N Engl J Med 1987; 316:630–631. [DOI] [PubMed] [Google Scholar]
- 35.Ramezani A, Banifazl M, Mohraz M, et al. Occult hepatitis B virus infection: a major concern in HIV-infected patients: occult HBV in HIV. Hepat Mon 2011; 11:7–10. [PMC free article] [PubMed] [Google Scholar]
- 36.Chadwick D, Doyle T, Ellis S, et al. Occult hepatitis B virus coinfection in HIV-positive African migrants to the UK: a point prevalence study. HIV Med 2014; 15:189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panigrahi R, Majumder S, Gooptu M, et al. Occult HBV infection among anti-HBc positive HIV-infected patients in apex referral centre, Eastern India. Ann Hepatol 2012; 11:870–875. [PubMed] [Google Scholar]
- 38.Olawumi HO, Olanrewaju DO, Shittu AO, et al. Effect of hepatitis B virus co-infection on CD4 cell count and liver function of HIV infected patients. Ghana Med J 2015; 49:2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phuangchoei P, Chotiyaputta W, Chayakulkeeree M. Clinical characteristics of hepatitis B and C virus infections in HIV-infected patients. J Med Assoc Thai 2015; 98:226–231. [PubMed] [Google Scholar]
- 40.Cartwright EJ, Miller L. Novel drugs in the management of difficult-to-treat hepatitis C genotypes. Hepat Med 2013; 5:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyczyno M, Halota W, Nowak W, et al. Distribution of HCV genotypes in the populations of inmates in polish prison Potulice and patients hospitalised in Bydgoszcz. Hepat Mon 2014; 14:e14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marascio N, Liberto M, Barreca G, et al. Update on epidemiology of HCV in Italy: focus on the Calabria Region. BMC Infect Dis 2014; 14 (suppl 5):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]


