Abstract
Background:
Steroid therapy has been an important reason of nontraumatic osteonecrosis of the femoral head (ONFH). Steroids are metabolized by hepatic cytochrome P4503A, a low endogenous activity of this enzyme can contribute to the pathogenesis of ONFH. The aim of this study was to investigate the associations of polymorphisms of cytochrome P4503A4 (CYP3A4) gene with steroid-induced ONFH in Chinese patients.
Methods:
A total of 150 steroid-induced ONFH patients and 250 healthy controls were enrolled. We evaluated 5 single-nucleotide polymorphisms of the CYP3A4 gene in this case–control study.
Results:
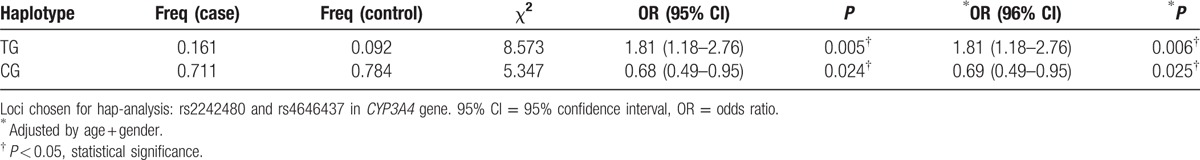
We identified rs2242480 in the CYP3A4 gene that was potentially associated with an increased risk of steroid-induced ONFH in the allele model (P = 0.023; odds ratio [OR]: 1.47; 95% confidence intervals [CI]: 1.05–2.04) and in the additive model (P = 0.028; OR: 1.44; 95% CI: 1.04–1.99) adjusted age + gender. Furthermore, we also observed a protective effect of haplotype “TG” (P = 0.025; OR: 0.69; 95% CI: 0.49–0.96) and a risk effect of haplotype “CG” (P = 0.006; OR: 1.81; 95% CI: 1.19–2.77) of the CYP3A4 gene adjusted age + gender.
Conclusion:
These findings suggested that polymorphisms of CYP3A4 gene may be associated with susceptibility to steroid-induced ONFH.
Keywords: case–control study, CYP3A4, gene, genotype analysis, single nucleotide polymorphism, steroid-induced osteonecrosis of femoral head
1. Introduction
Osteonecrosis of the femoral head (ONFH) is osteocyte death leading to the gradual disruption of the femoral head and it is characterized for impaired differentiation of mesenchymal cells, cellular toxicity, and destruction of intravascular blood flow, ultimately resulting in bone death.[1] The steroid therapy is normally specified to patients with renal transplantation, systemic lupus erythematosus, autoimmune inflammatory diseases, and nephrotic syndrome have been a central reason of nontraumatic ONFH.[2,3] The prevalence of ONFH is investigated to be annually 15,000 to 20,000 in China and 10,000 to 20,000 in the United States.[4,5] Most of the patients often need by surgery which may include osteotomy, total hip arthroplasty, or core decompression. Since not all cases with steroid therapy develop being steroid-induced ONFH, the prevention of the disorder by risk factors of individual differentiation to steroids sensitivity would be a significant tactic for cases who get steroid therapy.[6]
The cytochrome P4503A (CYP3A) is a remarkable enzyme responsible for metabolizing the steroids, and its activities varies more than 10-fold, low CYP3A activity leads to a predominant increase of steroid levels.[7,8] Previous studies have reported the importance of the CYP3A subfamily in the metabolism of statins and that genetic polymorphisms of CYP3A5 may affect the lipid-lowering responses and pharmacokinetic profiles of statins.[9,10] The cytochrome P4503A4 (CYP3A4) also manifests an approximate 40-fold degree of interindividual polymorphic variation, including CYP3A1–5 alleles, which have been associated with reduced activity of CYP3A4.[11] The CYP3A4 activity, which metabolizes steroids, was suggested to be associated with the development of osteonecrosis.[12] Meanwhile, Single nucleotide polymorphisms (SNPs) identified in CYP3A4 rs12333983 (7q22. 1), rs3735451 (7q22. 1), rs2242480 (7q22.1), rs4646437 (7q22.1), and rs2246709 (7q22.1) are associated with the incidence of ONFH in European population or in the animal model.[12–15]
This study was aimed at understanding whether the polymorphism of CYP3A4 gene was associated with a propensity to develop steroid-induced ONFH in the Chinese patients.
2. Methods
2.1. Ethics statement
The protocol in this study was strictly conformed to the principles expressed in the Declaration of Helsinki and was approved by the Ethical Committee of Zhengzhou Traditional Chinese Medicine Traumatology Hospital. Signed informed consent was obtained from each participant.
2.2. Study participants
All analyses were restricted to Chinese Han in our study population. In total, 150 steroid-induced ONFH cases were enrolled in the study from April 2015 to February 2016 in the Zhengzhou Traditional Chinese Medicine Traumatology Hospital in Zhengzhou city, China. These cases had received standard steroid therapy more than 1 year after using >2000 mg of prednisone.[16] The Arlet and Ficat classification was used for radiographic evaluation.[17] Anteroposterior and frog view X-rays of both hips were done in all of the patients. Confirmed the diagnosis of ONFH in patients without X-ray changed by using the magnetic resonance imaging. All steroid-induced ONFH cases had no previous history of prior chemotherapy or radiotherapy. All cases were recently diagnosed and confirmed to get steroid-induced ONFH.
A total of 250 healthy unrelated individuals as the controls from June 2015 to February 2016 were recruited from the medical examination at Zhengzhou Traditional Chinese Medicine Traumatology Hospital. All the participants were restricted to Chinese Han who lived in Zhengzhou city and its surrounding areas. Generally, subjects with chronic diseases and conditions involving vital organs such as brain heart, liver, and lung were excluded from this research. The aim of the above exclusion standards was to minimize the known factors that influence the variation of human complex diseases.
2.3. Genotyping
Genomic deoxyribonucleic acid (DNA) was extracted from blood samples using the phenol–chloroform extraction method,[18] and DNA concentration was measured by spectrometry (DU530 UV/VIS spectrophotometer, Beckman Instruments, Fullerton, CA). We used Sequenom MassARRAY Assay Design 3.0 Software (Sequenom Inc., San Diego, CA) to design Multiplexed SNP MassEXTEND assay.[19] SNP genotyping was performed using the Sequenom MassARRAY RS1000 (Sequenom Inc., San Diego, CA) with standard protocol recommended by the manufacturer.[19] Data management and analyses were performed using Sequenom Typer 4.0 software (Sequenom Inc., San Diego, CA) as previously described.[19,20]
2.4. Statistical analysis
The genotype frequencies of each SNP in the control subjects were checked using the Hardy–Weinberg equilibrium (HWE). Data analysis was performed using SPSS version 16.0 statistical package (SPSS, Chicago, IL) and Microsoft Excel (Chicago, IL). The significance of the difference of alleles and genotype frequencies between the groups was tested using the chi-square method.[21]P < 0.05 was considered to represent statistical significance. Differences in the distribution were analyzed using logistic regression. Odds ratios (ORs) and 95% confidence intervals (CIs) were tested using unconditional logistic regression analysis with adjustment for age and gender.[22] The 3 genetic models (Dominant, Recessive, and Additive) were applied by PLINK software (Chicago, IL) (http://pngu.mgh.harvard.edu/purcell/plink/) to assess the association of single SNPs with the risk of steroid-induced ONFH. The analyses of linkage disequilibrium (LD), and haplotype construction was used by the Haploview software package (version 4.2) (Chicago, IL).[23]
3. Results
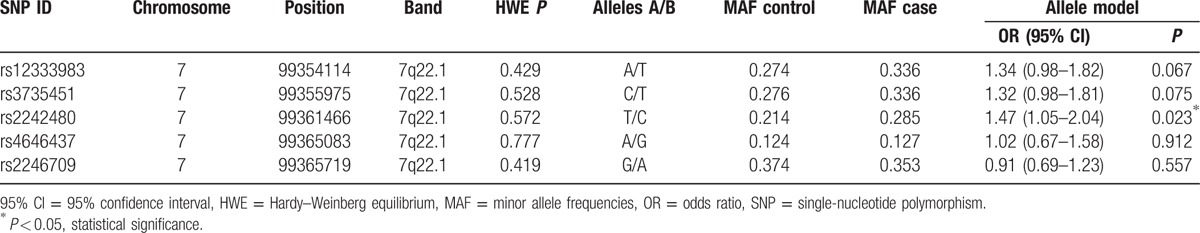
The analysis included 150 cases (89 males, 61 females; mean age 41.96 ± 11.49 years) who were receiving steroid treatment and 250 healthy controls (151 males, 99 females; mean age 43.27 ± 9.64 years). The basic characteristics of patients and control subjects are illustrated in Table 1. As listed in Table 2, a multiplexed SNP MassEXTEND assay was designed with the Sequenom MassARRAY Assay Design 3.0 Software. Chromosomal, position, band, HWE P value, alleles A/B, minor allele frequency (MAF) control, and MAF case for 5 SNPs are shown in Table 3. Meanwhile, we found that rs2242480 in the CYP3A4 gene was associated with steroid-induced ONFH as a risk factor in the allele model (P = 0.023; OR: 1.47; 95% CI: 1.05–2.04). All of the tested SNPs are in HWE in the control population of this study.
Table 1.
Basic characteristics of recruited individuals.
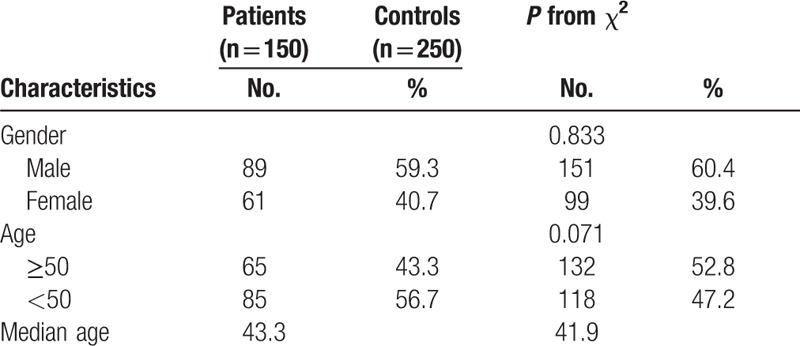
Table 2.
PCR primers in this study.
Table 3.
Examined SNPs examined in the CYP3A4 gene.
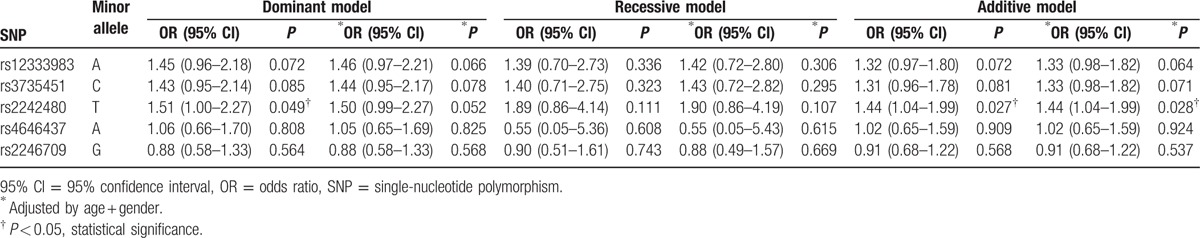
Association results between CYP3A4 SNP genotypes and the risk of steroid-induced ONFH are listed in Table 4. The significant associations were observed between the genotype “T/C” of rs2242480 and increased steroid-induced ONFH risk in the additive model (crude P = 0.027; OR: 1.44; 95% CI: 1.04–1.99; adjusted by age + gender P = 0.028; OR: 1.44; 95% CI: 1.04–1.99). The genotype “T/C” of rs2242480 was found to be associated with increased risk of steroid-induced ONFH (crude P = 0.049, OR, 1.51; 95% CI: 1.00–2.27).
Table 4.
Association between single-nucleotide polymorphism genotypes and risk of steroid-induced ONFH.
Two blocks (rs12333983 and rs3735451 in the Block1; rs2242480 and rs4646437 in the Block 2) were detected in studied CYP3A4 SNPs by haplotype analyses (Fig. 1). The results of the association between the CYP3A4 haplotype and the risk of steroid-induced ONFH are listed in Table 5. Haplotype “TG” in Block 2 was found to be associated with a risk factor of steroid-induced ONFH (crude P = 0.005, OR, 1.81; 95% CI: 1.18–2.76; adjusted by age + gender P = 0.006; OR: 1.81; 95% CI: 1.18–2.76). Haplotype “CG” in Block 2 was found to be associated with a protective factor of steroid-induced ONFH (crude P = 0.024, OR, 0.68; 95% CI: 0.49–0.95; adjusted by age + gender P = 0.025; OR: 0.69; 95% CI: 0.49–0.95).
Figure 1.
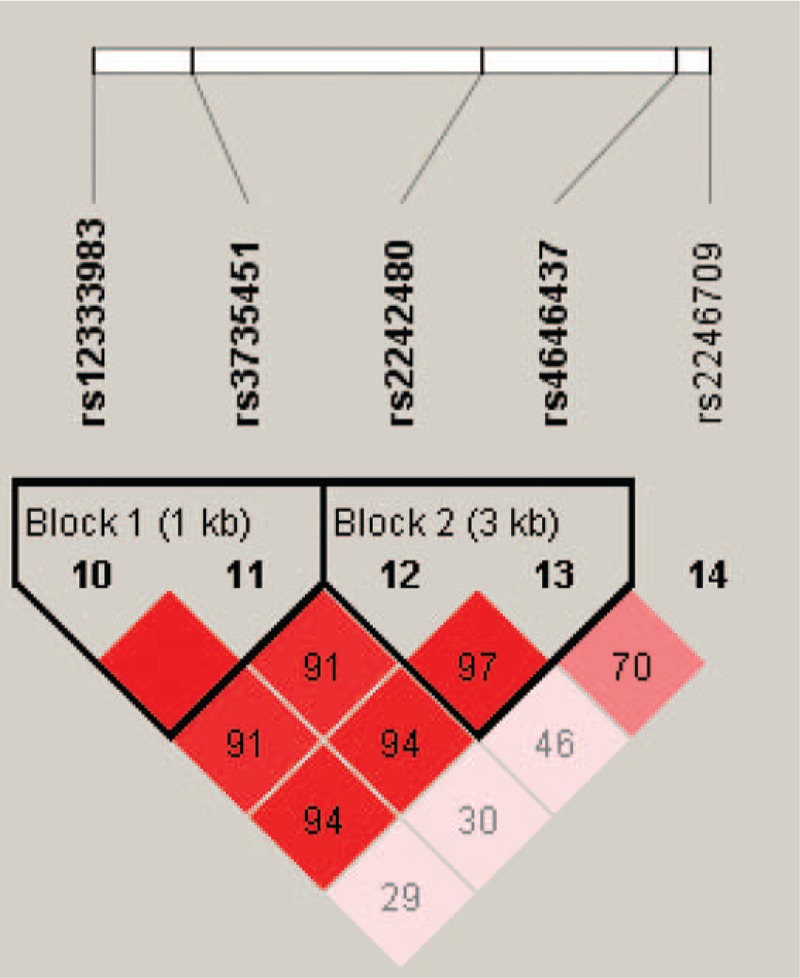
Haplotype block map for 5 single nucleotide polymorphisms (SNPs) of the cytochrome P4503A4 gene. Block 1 includes rs12333983 and rs3735451; Block 2 includes rs2242480 and rs4646437. The linkage disequilibrium between 2 SNPs is red schemes.
Table 5.
CYP3A4 haplotype frequencies associated with the risk of steroid-induced ONFH.
4. Discussion
In our case–control study, we identified rs2242480 in the CYP3A4 gene associated with an increased risk of steroid-induced ONFH in the allele model and the additive model adjusted by age + gender. A protective effect was observed for the haplotype “TG” of the CYP3A4 gene that was associated with decreased risk of developing steroid-induced ONFH. In addition, we also observed a strong effect of the “CG” haplotype, which increased the risk of developing steroid-induced ONFH.
Our results indicate a statistically significant difference between the steroid-induced ONFH and control groups regarding the CYP3A4 SNPs, suggesting a positive association between genetic polymorphism and the susceptibility of steroid-induced ONFH. In the previous study, Kitada et al[24] showed the product of the cytochrome P450 gene, CYP3A4, is considered to be the main cytochrome responsible for steroid metabolism. The cytochrome P450 family is a group of enzymes included in the oxidative and reductive metabolism of almost all lipid-soluble medicines. However, one of the potential causes of nontraumatic ONFH, lipid metabolism abnormality, is occlusion of vessels responsible for blood supply of the femoral head.[25] This might root in exposure of the femur to lower methylpredonisolone concentration for shorter duration of time by intensive metabolism in the liver by enhanced CYP3A activity. Although the exact mechanism to do harm to the osteal circulation by the high level and prolonged exposure to the exogenous steroid keep up to be researched, the CYP3A activity played an essential role that no doubt brought about development of steroid-induced ONFH.[26,27]
In the study, we found that genotype “T/C” of rs2242480 in intron 10 of the CYP3A4 gene, which is mapped to chromosome 7q22.1, was associated with the risk of steroid-induced ONFH in Chinese Han patients. The most frequent mutant allele of rs2242480 in the CYP3A4 was characterized by a G to A substitution at position 82266. The allele frequency of rs2242480 was 0.22 to 0.37 in the Chinese population.[28,29] There is an evidence of association between the mutant of rs2242480 and clopidogrel response variability in Spanish patients with coronary artery–related disease.[30] In addition, this mutant genotype was associated with a higher CYP3A metabolic activity, thus enhancing the formation of metabolites, specifically for prodrugs, which are activated via CYP3A through biotransformation.[31] According to previous researches, polymorphisms of CYP3A4 gene associated with steroid-induced ONFH had been proved in the Japanese population. It was reported that it was an allele that appeared with high frequency in Japanese and Oriental people. It is considered that the genetic polymorphism of CYP3A4 was an important cause of individual differences in drug metabolism in Oriental people. These results were consistent with the results of our present study.[12,32] Thus, the exact location and biological functions of the real causal SNPs in the CYP3A4 gene is of great interest and warrants further investigation.
Haplotype analysis suggested that steroid-induced ONFH risk was substantially elevated among individuals with specific haplotypes. In the previous study, Li et al[13] found the association between CYP3A4 rs4646437 T > C and tacrolimus pharmacokinetics. Because the rs2242480 and rs4646437 genes are both located in 7q21.1, the LD between rs2242480 and rs4646437 might influence the affect of CYP3A4 SNPs on the tacrolimus concentration (C0/D). Crettol et al[33] reported that the rs4646437-T allele was in strong LD (r2 = 0.82) with the rs2242480-G allele in Caucasian renal transplant recipients. Chau et al[34] found that genetic polymorphisms of the CYP3A4 gene included in serum concentrations were significantly correlated with finasteride metabolism. The homozygous minor allele of rs2242480 and rs4646437 in CYP3A4 were associated with lower finasteride levels. Block 2 included 2 SNPs (rs2242480 and rs4646437), and we found the haplotype association analyses showed that haplotype “TG” was associated with the increased risk of steroid-induced ONFH crude/adjusted by age + gender. Interestingly, haplotype “CG” was found to be associated with a decreased risk of steroid-induced ONFH crude/adjusted by age + gender. This explanation of results need to do the further researches.
There are several limitations to our study. First, the sample size (150 patients and 250 control subjects) was not relatively large among steroid-induced ONFH association studies. Second, steroid-induced ONFH patients and healthy controls were used in the same hospital to avoid selection bias. As we all know, population admixture was confounding factor and can caused type-I errors (false positive) in association analysis. Third, we also performed Bonferroni correction analysis and found no statistical significant associations between CYP3A4 SNPs and steroid-induced ONFH risk. However, this may be due to the weakness of Bonferroni correction itself. True important differences may be considered nonsignificant, and the likelihood of type II errors are also elevated.[35] We also performed a power analysis that showed that the power of rs2242480 was 0.76 and it was >0.75.
In conclusion, our comprehensive analysis of SNPs in the CYP3A4 gene indicates that CYP3A4 genotypes and haplotypes are associated with steroid-induced ONFH risk in Chinese Han population. In the further researches, if SNP is evaluated as a risk marker, filtrating patients at high risk of ONFH would be possible, steroid dosage could be basic on individual differences and it could prevent the development of steroid-induced ONFH.
Footnotes
Abbreviations: CIs = confidence intervals, CYP3A4 = cytochrome P4503A4, HWE = Hardy–Weinberg equilibrium, MAF = minor allele frequency, ONFH = osteonecrosis of the femoral head, ORs = odds ratios, SNPs = single nucleotide polymorphisms.
Funding/support: This study was supported by grants from the National Natural Science Foundation of China (NSFC, nos. 81160228, 81260284, and 81660378).
The authors have no conflicts of interest to disclose.
References
- 1.Lieberman JR, Berry DJ, Mont MA, et al. Osteonecrosis of the hip: management in the 21st century. Instr Course Lect 2003; 52:337–355. [PubMed] [Google Scholar]
- 2.Mont MA, Jones LC, Hungerford DS. Nontraumatic osteonecrosis of the femoral head: ten years later. J Bone Joint Surg Am 2006; 88:1117–1132. [DOI] [PubMed] [Google Scholar]
- 3.Kathiresan S, Voight BF, Purcell S, et al. Myocardial Infarction Genetics Consortium. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 2009; 41:334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones LC, Hungerford DS. Osteonecrosis: etiology, diagnosis, and treatment. Curr Opin Rheumatol 2004; 16:443–449. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Cao Y, Li Y, et al. Genetic association of the ApoB and ApoA1 gene polymorphisms with the risk for alcohol-induced osteonecrosis of femoral head. Int J Clin Exp Pathol 2015; 8:11332–11339. [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Kong X, Wang R, et al. Genetic association of the P-glycoprotein gene ABCB1 polymorphisms with the risk for steroid-induced osteonecrosis of the femoral head in Chinese population. Mol Biol Rep 2014; 41:3135–3146. [DOI] [PubMed] [Google Scholar]
- 7.Varis T, Kivisto KT, Backman JT, et al. The cytochrome P450 3A4 inhibitor itraconazole markedly increases the plasma concentrations of dexamethasone and enhances its adrenal-suppressant effect. Clin Pharmacol Ther 2000; 68:487–494. [DOI] [PubMed] [Google Scholar]
- 8.Lin YS, Lockwood GF, Graham MA, et al. In-vivo phenotyping for CYP3A by a single-point determination of midazolam plasma concentration. Pharmacogenetics 2001; 11:781–791. [DOI] [PubMed] [Google Scholar]
- 9.Wei KK, Zhang LR. Interactions between CYP3A5∗3 and POR∗28 polymorphisms and lipid lowering response with atorvastatin. Clin Drug Investig 2015; 35:583–591. [DOI] [PubMed] [Google Scholar]
- 10.He BX, Shi L, Qiu J, et al. The effect of CYP3A4∗1G allele on the pharmacokinetics of atorvastatin in Chinese Han patients with coronary heart disease. J Clin Pharmacol 2014; 54:462–467. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh KP, Lin YY, Cheng CL, et al. Novel mutations of CYP3A4 in Chinese. Drug Metab Dispos 2001; 29:268–273. [PubMed] [Google Scholar]
- 12.Masada T, Iwakiri K, Oda Y, et al. Increased hepatic cytochrome P4503A activity decreases the risk of developing steroid-induced osteonecrosis in a rabbit model. J Orthop Res 2008; 26:91–95. [DOI] [PubMed] [Google Scholar]
- 13.Li CJ, Li L, Lin L, et al. Impact of the CYP3A5, CYP3A4, COMT, IL-10 and POR genetic polymorphisms on tacrolimus metabolism in Chinese renal transplant recipients. PLoS One 2014; 9:e86206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diekstra MH, Belaustegui A, Swen JJ, et al. Sunitinib-induced hypertension in CYP3A4 rs4646437 A-allele carriers with metastatic renal cell carcinoma. Pharmacogenomics J 2016; [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 15.Gezsi A, Lautner-Csorba O, Erdelyi DJ, et al. In interaction with gender a common CYP3A4 polymorphism may influence the survival rate of chemotherapy for childhood acute lymphoblastic leukemia. Pharmacogenomics J 2015; 15:241–247. [DOI] [PubMed] [Google Scholar]
- 16.Xue Y, Zhao ZQ, Hong D, et al. MDR1 gene polymorphisms are associated with glucocorticoid-induced avascular necrosis of the femoral head in a Chinese population. Genet Test Mol Biomarkers 2014; 18:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagala J, Buraczynska M, Mazurkiewicz T, et al. Endothelial nitric oxide synthase gene intron 4 polymorphism in non-traumatic osteonecrosis of the femoral head. Int Orthop 2013; 37:1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kochl S, Niederstatter H, Parson W. DNA extraction and quantitation of forensic samples using the phenol-chloroform method and real-time PCR. Methods Mol Biol 2005; 297:13–30. [DOI] [PubMed] [Google Scholar]
- 19.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Current protocols in human genetics / editorial board, Jonathan L. Haines ... [et al.]. 2009;Chapter 2:Unit 2 12. [DOI] [PubMed] [Google Scholar]
- 20.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet 2007; 39:347–351. [DOI] [PubMed] [Google Scholar]
- 21.Adamec C. [Example of the use of the nonparametric test. Test X2 for comparison of 2 independent examples]. Cesk Zdrav 1964; 12:613–619. [PubMed] [Google Scholar]
- 22.Bland JM, Altman DG. Statistics notes. The odds ratio. BMJ 2000; 320:1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21:263–265. [DOI] [PubMed] [Google Scholar]
- 24.Kitada M, Kamataki T, Itahashi K, et al. Significance of cytochrome P-450 (P-450 HFLa) of human fetal livers in the steroid and drug oxidations. Biochem Pharmacol 1987; 36:453–456. [DOI] [PubMed] [Google Scholar]
- 25.Gangji V, De Maertelaer V, Hauzeur JP. Autologous bone marrow cell implantation in the treatment of non-traumatic osteonecrosis of the femoral head: five year follow-up of a prospective controlled study. Bone 2011; 49:1005–1009. [DOI] [PubMed] [Google Scholar]
- 26.Motomura G, Yamamoto T, Miyanishi K, et al. Combined effects of an anticoagulant and a lipid-lowering agent on the prevention of steroid-induced osteonecrosis in rabbits. Arthritis Rheum 2004; 50:3387–3391. [DOI] [PubMed] [Google Scholar]
- 27.Ichiseki T, Matsumoto T, Nishino M, et al. Oxidative stress and vascular permeability in steroid-induced osteonecrosis model. J Orthop Sci 2004; 9:509–515. [DOI] [PubMed] [Google Scholar]
- 28.Du J, Xing Q, Xu L, et al. Systematic screening for polymorphisms in the CYP3A4 gene in the Chinese population. Pharmacogenomics 2006; 7:831–841. [DOI] [PubMed] [Google Scholar]
- 29.Liu R, Zhou ZY, Chen YB, et al. Associations of CYP3A4, NR1I2, CYP2C19 and P2RY12 polymorphisms with clopidogrel resistance in Chinese patients with ischemic stroke. Acta Pharmacol Sin 2016; 37:882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, et al. Contribution of gene sequence variations of the hepatic cytochrome P450 3A4 enzyme to variability in individual responsiveness to clopidogrel. Arterioscler Thromb Vasc Biol 2006; 26:1895–1900. [DOI] [PubMed] [Google Scholar]
- 31.Fukushima-Uesaka H, Saito Y, Watanabe H, et al. Haplotypes of CYP3A4 and their close linkage with CYP3A5 haplotypes in a Japanese population. Hum Mutat 2004; 23:100. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Nakajima S, Hirota Y, et al. Genetic association of a polymorphism of the cAMP-responsive element binding protein-binding protein with steroid-induced osteonecrosis after kidney transplantation. J Bone Miner Metab 2007; 25:320–325. [DOI] [PubMed] [Google Scholar]
- 33.Crettol S, Venetz JP, Fontana M, et al. CYP3A7, CYP3A5, CYP3A4, and ABCB1 genetic polymorphisms, cyclosporine concentration, and dose requirement in transplant recipients. Ther Drug Monit 2008; 30:689–699. [DOI] [PubMed] [Google Scholar]
- 34.Chau CH, Price DK, Till C, et al. Finasteride concentrations and prostate cancer risk: results from the Prostate Cancer Prevention Trial. PLoS One 2015; 10:e0126672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perneger TV. What's wrong with Bonferroni adjustments. BMJ 1998; 316:1236–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]


