Abstract
The aim of this study was to identify and characterize the C2C2-YABBY family of genes by a genome-wide scale in common bean. Various in silico approaches were used for the study and the results were confirmed through common molecular biology techniques. Quantitative real-time PCR (qPCR) analysis was performed for identified putative PvulYABBY genes in leaf and root tissues of two common bean cultivars, namely Yakutiye and Zulbiye under salt stress condition. Eight candidate PvulYABBY proteins were discovered and the length of these proteins ranged from 173 to 256 amino acids. The isoelectric points (pIs) of YABBY proteins were between 5.18 and 9.34 and ranged from acidic to alkaline, and the molecular weight of PvulYABBYs were between 18978.4 and 28916.8 Da. Three segmentally duplicated gene couples among the identified eight PvulYABBY genes were detected. These segmentally duplicated gene couples were PvulYABBY-1/PvulYABBY-3, PvulYABBY-5/PvulYABBY-7 and PvulYABBY-6/PvulYABBY-8. The predicted number of exons among the PvulYABBY genes varied from 6 to 8 exons. Additionally, all genes found included introns within ORFs. PvulYABBY-2, -4, -5 and -7 genes were targeted by miRNAs of five plant species and a total of five miRNA families (miR5660, miR1157, miR5769, miR5286 and miR8120) were detected. According to RNA-seq analysis, all genes were up- or down-regulated except for PvulYABBY-1 and PvulYABBY-6 after salt stress treatment in leaf and root tissues of common bean. According to the qPCR analysis, six out of eight genes were expressed in the leaves but only four out of eight genes were expressed in the roots and these genes exhibited tissue- and cultivar-specific expression patterns.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0933-0) contains supplementary material, which is available to authorized users.
Keywords: C2C2-YABBY, Transcription factors, RNA-seq, In silico analysis, qPCR
Introduction
Transcription factors (TFs) play an important role in the growth and development of plants as well as all organisms in nature. To date, approximately 30 TF families have been identified and classified according to the conserved motifs that code for the DNA-binding domains and approximately half of them are considered as plant-specific TFs such as AP2/ERF, WRKY, NAC, B3, SBP and DOF families (Yamasaki et al. 2013).
The YABBY gene family is one of the plant transcription factors and is known to be specific to seed plants (Yang et al. 2016). These genes encode proteins which include two conserved domains which are N-terminal Cys2 Cys2 zinc-finger motif and C-terminal YABBY domain (Golz et al. 2004; Mackay and Crossley 1998). Previous studies have shown that some of the zinc fingers are observed as DNA-binding motifs, while many of them play roles in protein–protein interactions without binding DNA (Golz et al. 2004). The main function of YABBY genes is to specify abaxial cell fates in lateral organs in Arabidopsis (Villanueva et al. 1999). In monocots, evolution has shown that YABBY genes have taken on different functions including other processes of plant growth and development such as controlling the carpel number of tomato flower (Cong et al. 2008). Notably, WRKY TFs, regulated by YABBY, play important roles in the regulation of plant stress response both as activators and repressors (Schluttenhofer and Yuan 2015).
The YABBY comprises a small gene family, which has only six members (FIL, CRC, INO, YAB2, YAB3 and YAB5) in Arabidopsis, eight members in Orzya sativa and nine members in Lycopersicum esculentum (Huang et al. 2013). However, the molecular basis of their action has not been well understood yet (Sawa et al. 1999; Siegfried et al. 1999; Villanueva et al. 1999). In addition, YABBY is affected by Auxin, GA and other various hormones, it is estimated to be important for the regulation of the cold stress (Peng et al. 2015). The aim of this study was to identify and characterize C2C2-YABBY genes at genome-wide scale in common bean. Various in silico approaches were used and the results were confirmed through molecular biology techniques. In addition, the expression levels of eight putative PvulYABBY genes were analyzed using qPCR in leaf and root tissues of two common bean cultivars, namely Yakutiye and Zulbiye under salt stress conditions.
Materials and methods
In silico identification of YABBY proteins in Phaseolus vulgaris
Sequences of the YABBY gene family of Phaseolus vulgaris were obtained from Phytozome database v11 (www.phytozome.net) using key searching with Pfam Accession Number (PF04690), downloaded from Pfam Database (http://pfam.xfam.org/). To identify all the putative YABBY proteins in P. vulgaris, both blastp at Phytozome database v11 (www.phytozome.net) and hidden Markov model (HMM), (http://www.ebi.ac.uk) searches were performed against the P. vulgaris genome (Goodstein et al. 2012). Putative P. vulgaris YABBY proteins were used for query in blastp (NCBI) for characterization of hypothetical proteins. Redundant sequences were removed using the decrease redundancy tool (http://web.expasy.org/decrease_redundancy/) and sequences were checked for YABBY domains using HMMER (http://www.ebi.ac.uk) and Pfam databases. The physicochemical properties of YABBY proteins were calculated using ProtParam Tool (http://web.expasy.org/protparam).
Structure and physical locations of PvulYABBY genes, identification of gene duplication events and conserved motifs
The gene structure display server v2.0 (GSDS, http://gsds.cbi.pku.edu.cn/) was used (Guo et al. 2007) to obtain information on the exon–intron of PvulYABBY proteins. Both genome sequences and the coding sequences were utilized for predicting the positional information of the PvulYABBY genes using Phytozome database v11, chromosomal locations. The sizes (bp) and intron numbers of PvulYABBY genes were identified. The PvulYABBY genes were plotted on all P. vulgaris chromosomes and pictured with MapChart (Voorrips 2002). Gene duplication events were detected through the alignment of the coding nucleotide and sequences covered 70% of the longest genes and the amino acid identity between the sequences was >70% (Yang et al. 2008). To identify additional conserved motifs of PvulYABBY proteins, we used the Multiple EM for Motif Elicitation tool (MEME v4.11.1; http://meme-suite.org/) (Bailey et al. 2006).
Phylogenetic analysis and sequence alignment
The multiple sequence alignment of PvulYABBY proteins was carried out using the ClustalW (Tamura et al. 2011). The neighbor-joining (NJ) method with bootstrap value of 1000 replicates (MEGA6) was used for constructing phylogenetic trees and the tree was designed using Interactive tree of life (iTOL; http://itol.embl.de/index.shtml) (Letunic and Bork 2011).
Subcellular localization and promoter analysis of the PvulYABBY family
The 5′ upstream regions (2 kb DNA sequence of each gene of the PvulYABBY family) were obtained from Phytozome database v11 (Supplementary Table S1) and analyzed using the plantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) for a cis-element scan. Subcellular localization of the PvulYABBY proteins was determined using the WoLF PSORT (http://www.genscript.com/psort/wolf_psort.html) (Horton et al. 2007) and TargetP 1.1 (http://www.cbs.dtu.dk/services/ TargetP/) (Emanuelsson et al. 2007).
GO annotation
The functional annotation of PvulYABBY protein sequences were defined with Blast2GO (http://www.blast2go.com) (Conesa et al. 2005).
In silico prediction of miRNA targets in PvulYABBY genes
All known plant miRNA sequences were obtained from miRBase v21.0 (http://www.mirbase.org). psRNA target server (http://plantgrn.noble.org/psRNATarget) with default parameters was used for prediction of miRNAs accordingly (Zhang 2005). In silico predicted miRNA targets were identified by BLASTX with ≤1e–10 against common bean Expressed Sequenced Tags (ESTs) at NCBI database.
Comparative physical mapping of YABBY proteins between common bean and other species
To obtain a orthologous relationship among the chromosomes of common bean, Arabidopsis thaliana and Medicago truncatula species, amino acid sequences of common bean YABBYs were searched against amino acid sequences of its corresponding A. thaliana, and M. truncatula YABBYs (www.phytozome.net) using NCBI–BLASTP. The comparative orthologous relationships of YABBY genes among common bean, A. thaliana and M. truncatula, chromosomes were drawn using MapChart.
Prediction of synonymous and non-synonymous substitution rates in PvulYABBY genes
CLUSTALW software was used for the prediction of amino acid sequences of tandemly and segmentally duplicated PvulYABBY genes. The CODEML program in PAML (PAL2NAL) (http://www.bork.embl.de/pal2nal) was used to estimate the synonymous (Ks) and non-synonymous (Ka) substitution rates (Suyama et al. 2006). Time (million years ago, Mya) of duplication and divergence of each YABBY genes were determined using the following formula: T = Ks/2λ (λ = 6.56E−9) (Lynch and Conery 2003; Yang and Nielsen 2000).
Expression levels of PvulYABBY genes in transcriptome data
Illumina RNA-seq data were obtained from sequence read archive (SRA) to measure the expression levels of PvulYABBY genes. The accession numbers SRR957667 (control leaf), SRR958472 (salt-treated root), SRR958469 (control root), and SRR957668 (salt-treated leaf) were used for this purpose (Hiz et al. 2014). The analysis of transcriptomic data was performed as described by Buyuk et al. (2016). Finally, the heat maps of hierarchical clustering were constructed using the PermutMatrix (Caraux and Pinloche 2005).
Plant materials, growth conditions, and stress treatments
The seeds of drought-tolerant bean cv. ‘Yakutiye’ and drought-sensitive bean cv. ‘Zulbiye’ were supplied from ‘Transitional Zone Agricultural Research Institute, Eskişehir, Turkey’ (Guler et al. 2012). A preliminary experiment was performed to determine the salt tolerance capacity of both common bean cultivars under severe salt stress conditions (250 and 400 mM NaCl for 9 days) using malondialdehyde (MDA) analysis as an indicator test for stress. According to the preliminary data, drought-tolerant cv. ‘Yakutiye’ seemed to be more resistant to salt stress than cv. ‘Zulbiye’ exhibited (unpublished results).
For gene expression analysis, seeds of both cultivars were germinated, following the surface sterilization in a solution containing 5% (v/v) hypochlorite for 5 min, and were grown hydroponically in pots containing 0.2L of 1/10 Hoagland’s solution (Buyuk et al. 2016). Common bean seedlings were incubated in a controlled environmental growth chamber in the light with 250 mmol m−2 s−1 photosynthetic photon flux at 25 °C, 70% relative humidity. Salt stress was applied with Hoagland solution including 150 mM of NaCl (for moderate salinity stress) for 9 days after common bean seedlings reached the first trifoliate stage in growth chamber. Following the ninth day of stress application, root and leaf tissues of two different common bean cultivars were harvested to be used in qPCR analysis.
RNA isolation, cDNA synthesis and quantitative qPCR analysis
Total RNAs of tissues (leaf and root) of the bean cultivars were extracted by NucleoSpin® RNA Kit (Macherey–Nagel, Germany) as described in the manufacturer’s protocol. Nucleic acid measurements were achieved using the NanoDrop Lite UV–VIS spectrophotometer and RNAs were also confirmed by 1.5% agarose gel electrophoresis (NanoDrop Technologies, Wilmington, DE, USA). cDNAs were synthesized from 2 μg of common bean RNAs using high fidelity cDNA synthesis kit (Roche Applied Science, USA) according to the manufacturer’s recommendations. The qPCR assay was conducted using a Light Cycler® Nano System (Roche Applied Science, USA). The primers were designed using Primer3 program and the actin was used as a housekeeping gene in the qPCR analysis (Supplementary Table S2). Three biological replicates consisting of three technical replicates each were performed in all qPCR reactions. The qPCR quantifications of cDNAs were carried out with the SYBR Green PCR Master Mix (Roche Applied Science, USA) according to the manufacturer’s recommendations. The 2−∆∆CT method was used to analyze data from qPCR and one-way ANOVA was used for calculating statistics (Livak and Schmittgen 2001).
Results and discussion
Identification of YABBY gene family in common bean
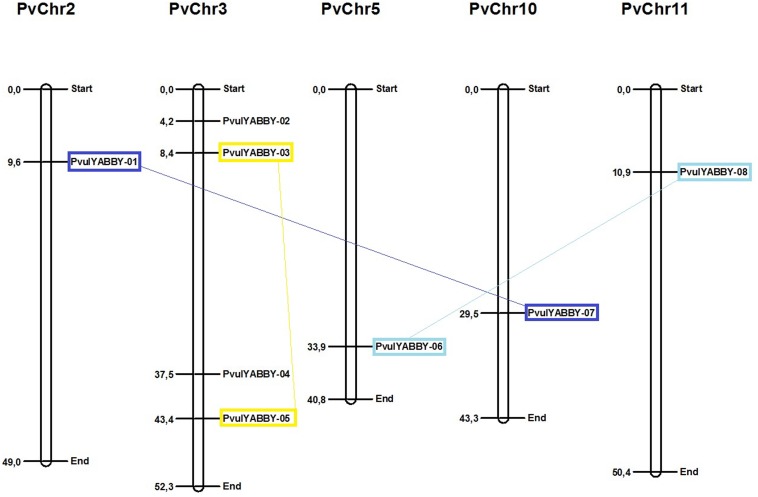
We key-searched P. vulgaris genome, acquired from the Phytozome database, using the YABBY gene family obtained from Pfam database and found eight genes with YABBY protein homology. Subsequently, YABBY domains were searched using the HMMER and Pfam databases in putative PvulYABBY proteins and redundant sequences were removed after validation. We discovered eight candidate PvulYABBY genes in the common bean genome and listed in Table 1, which shows the number of isoelectric point (pI), amino acids (length), molecular weight and NCBI annotation. All non-redundant PvulYABBY genes were distributed on chromosomes 2, 3, 5, 10, and 11 of common bean and the largest number of PvulYABBYs was on chromosome 3 (four PvulYABBY genes) (Fig. 1). The length of PvulYABBY proteins ranged from 173 (PvulYABBY-2) to 256 (PvulYABBY-1) amino acids (aa). PIs of PvulYABBY proteins were between 5.18 (PvulYABBY-1) and 9.34 (PvulYABBY-2) ranging from acidic to alkaline, and the molecular weight of PvulYABBYs were between 18978.4 Da (PvulYABBY-2) and 28916.8 Da (PvulYABBY-1).
Table 1.
The information of eight PvulYABBY proteins
| ID | Phytozome identifier | Chromosome location | Protein length | Molecular weight (Da) | pI | Stable or unstable | Instability index |
|---|---|---|---|---|---|---|---|
| PvulYABBY-1 | Phvul.002G069700.1 | 9554540..9556683 | 256 | 28916.8 | 5.18 | 46.39 | Unstable |
| PvulYABBY-2 | Phvul.003G039800.1 | 4217179..4219619 | 173 | 18978.4 | 9.34 | 39.51 | Unstable |
| PvulYABBY-3 | Phvul.003G061800.1 | 8393693..8395131 | 237 | 26412.2 | 7.94 | 52.06 | Unstable |
| PvulYABBY-4 | Phvul.003G166100.1 | 37492965..37496762 | 186 | 20894.8 | 8.4 | 41.12 | Unstable |
| PvulYABBY-5 | Phvul.003G217700.1 | 43407696..43410807 | 215 | 23982.3 | 7.13 | 54.76 | Unstable |
| PvulYABBY-6 | Phvul.005G117600.1 | 33854923..33864003 | 185 | 20704.5 | 8.95 | 41.51 | Unstable |
| PvulYABBY-7 | Phvul.010G079200.1 | 29466002..29468643 | 215 | 23915.2 | 8.58 | 48.23 | Unstable |
| PvulYABBY-8 | Phvul.011G100300.1 | 10846106..10852521 | 183 | 20458.2 | 8.22 | 48.66 | Unstable |
Fig. 1.
Distribution of PvulYABBY genes on common bean chromosomes
According to the previous studies and HMMER search, the highest number of YABBY proteins was observed in Glycine max with 17 members and Arabidopsis lyrata was found to have the lowest number of YABBY proteins with 6 among the eudicotyledons (Han et al. 2015). Musa acuminata had the highest number of YABBY proteins with 25 and Aegilops tauschii had the lowest number with 5 among the Liliopsida members. In previous studies, it was shown that seedless plants do not have YABBY genes (Han et al. 2015). It was also found that the Arabidopsis genome has six YABBY genes. Four of these genes (FIL, YAB2, YAB3 and YAB5) have a high expression level in vegetative tissues, whereas expression of other two (CRC and INO) genes is restricted to floral organs (Eckardt 2010).
The gene duplication analysis showed that three segmentally duplicated gene couples, among the eight identified PvulYABBY genes, were detected and segmentally duplicated gene couples were PvulYABBY-1/PvulYABBY-3, PvulYABBY-5/PvulYABBY-7 and PvulYABBY-6/PvulYABBY-8 (Supplementary Table S6). Ks/Ka distances and ratios were also estimated. The Ks of the segmental duplication of PvulYABBY-1 and PvulYABBY-3 was 0.598, which dated the duplication event at 4.6 Mya; PvulYABBY-5 and PvulYABBY-7 was 1.8122, dated the duplication event at 13.94 Mya and PvulYABBY-6 and PvulYABBY-8 was 0.4205, dated the duplication event at 3.23 Mya (Supplementary Table S6). The mean date of segmental duplication events showed that PvulYABBY-1/PvulYABBY-3 (4.6 Mya) and PvulYABBY-6/PvulYABBY-8 (3.23 Mya) genes were segmentally duplicated after the divergence of monocot–dicot split, which were observed at 170–235 Mya (Gupta et al. 2015). PvulYABBY-5 and PvulYABBY-7 (13.94 Mya) genes were segmentally duplicated before the divergence of the monocot–dicot split according to the Ks/Ka distances and ratios.
Phylogenetic analysis, conserved motifs, gene structure and subcellular localization of PvulYABBYs
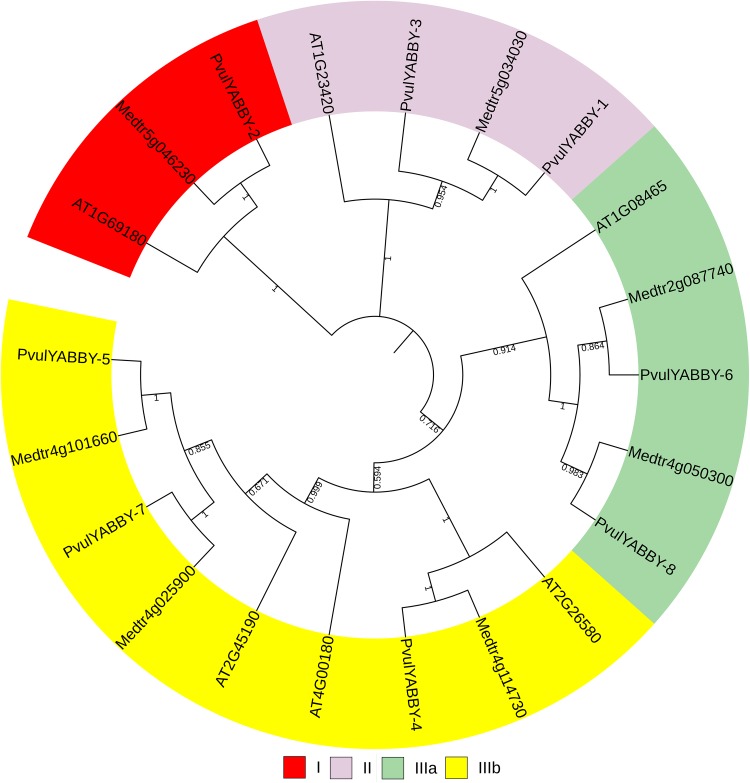
To investigate the relationships among PvulYABBY proteins, a phylogenetic tree of YABBY proteins in common bean and Arabidopsis was constructed using amino acid sequences with well-supported bootstrap values (1000 replicates) (Fig. 2) through the neighbor-joining method. PvulYABBY proteins were classified into three groups (I, II and III) and the third group could be further classified into two subgroups (IIIa and IIIb) (Fig. 2).
Fig. 2.
A phylogenetic tree of common bean YABBY proteins
The orthology analysis showed that Arabidopsis had six ortholog relationship in the common bean (AT1G08465 and YABBY 6-8; AT2G26580 and YABBY 4; AT2G45190 and YABBY 5-7; AT4G00180 and YABBY 5-7; AT1G23420 and YABBY 1-3; AT1G69180 and YABBY 2) and Medicago truncatula had seven ortholog relationship in common bean (MEDTR2G087740 and YABBY 8-6; MEDTR4G025900 and YABBY 7-5; MEDTR4G050300 and YABBY 8-6; MEDTR4G101660 and YABBY 7-5; MEDTR4G114730 and YABBY 4-6; MEDTR5G034030 and YABBY 1-3; MEDTR5G046230 and YABBY 2) (Supplementary Fig. S2).
To discover the conserved motifs in PvulYABBY proteins, MEME (v4.11.1) was used and a total of 15 conserved motifs were identified. The lengths of discovered motifs ranged from 10 to 50 amino acids. All PvulYABBY proteins included Motif 1 and Motif 2 (Supplementary Fig. S1, Table S3).
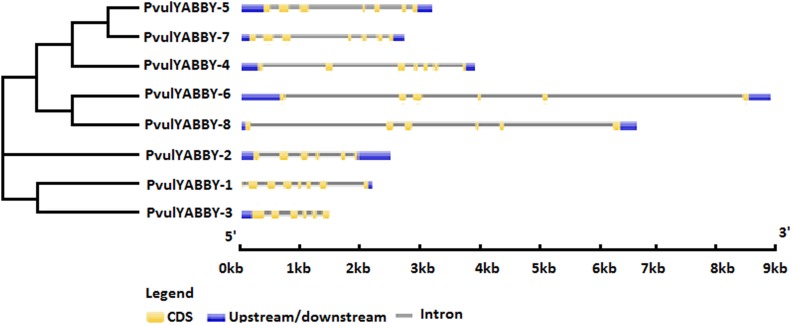
We investigated the gene structure of eight common bean YABBY genes. The predicted number of exons among the eight PvulYABBY genes varied from 6 to 8. In addition, all PvulYABBY genes included introns within ORFs (Fig. 3).
Fig. 3.
Gene structure of PvulYABBY genes
The upstream promotor region of the PvulYABBY family contained a total of 75 different cis-acting elements, which are categorized as light-responsive elements (30), development-related elements (12), environmental stress-related elements (6), hormone responsive elements (6), promoter-related elements (3), site-binding-related elements (3) and other elements (15) (Supplementary Table S4). The average number of cis-acting elements per PvulYABBY gene was 29.875 and the highest number (38) of cis-elements was found on PvulYABBY-8, while the least (23) was observed on PvulYABBY-7. GT-1 and TGACG motifs were related to salt stress in maize promotor (Hou et al. 2016). These motifs were found in PvulYABBY-4 and -7 out of eight PvulYABBY genes.
Detection of miRNAs targeting PvulYABBY genes
According to the results of psRNATarget server, the maximum expectation and accessibility of messenger RNA (mRNA) target site to miRNA ranged from 2.5 to 3.0 and 12.927 to 22.041, respectively (Supplementary Table S5). A total of four PvulYABBY genes (PvulYABBY-2, -4, -5 and -7) were targeted by miRNAs of five plant species. A total of five miRNA families (miR5660, miR1157, miR5769, miR5286 and miR8120) were detected and the PvulYABBY-2 gene was targeted by two different plant miRNAs (Supplementary Table S5).
In previous studies, several miRNAs were found against various abiotic stresses (Eldem et al. 2012; Kavas et al. 2016). A number of salt-responsive miRNAs were defined from the leaves and roots of wheat (Eren et al. 2015). Several salt stress-responsive miRNAs have been defined in diverse plant species. E.g., miR156, miR159, miR162, miR171 and miR319 were determined to be salt-responsive miRNAs in Arabidopsis, Populus, barley and rice (Eldem et al. 2013).
YABBY proteins have a zinc finger and a helix-loop-helix domain (YABBY gene family) and they are known to have crucial influence on transcriptional regulators (Juarez et al. 2004). In relation to that, miRNA-targeted zinc-finger proteins have been found as salt stress-responsive factors in plants (Fang et al. 2006). In a recent study, miR393 was found to target the helix-loop-helix domain in common bean (Kavas et al. 2016).
The miRNA-target relationship in the biological system may be more complex than was previously known (Barbato et al. 2009; Pasquinelli 2012). Specifically, miRNAs directly target the YABBY genes, however, certain miRNAs can indirectly affect the YABBY genes through a different gene. Our work presented herein, extensively covers the role YABBY genes at the genome level, shows a correlation between YABBY-related miRNAs and salt stress response. The results shown are considered to be very important for the definition of YABBY functions, which were previously unknown.
Genome-wide expression pattern of PvulYABBY genes
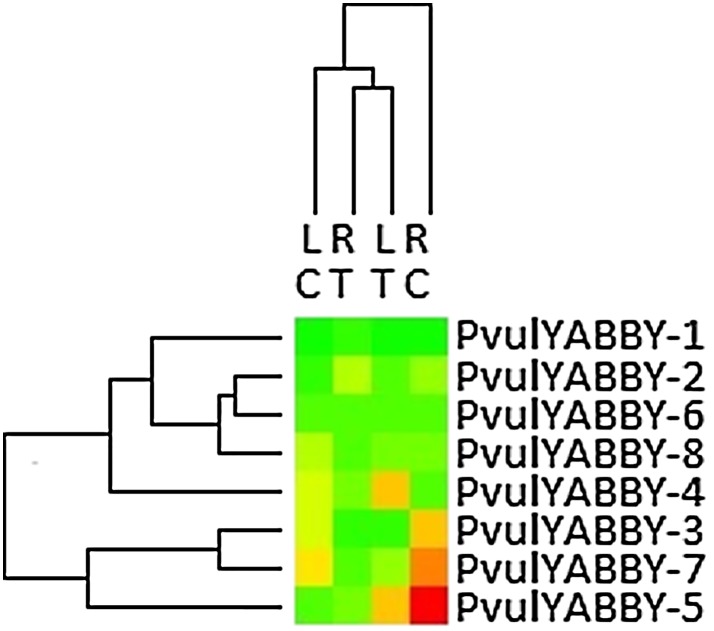
The expression profile of PvulYABBY genes, based on RNA-seq, was developed using the root and leaf tissues of common bean. Based on the RNA-seq analysis results, eight expressed PvulYABBY genes were identified through the Phytozome identifier (Table 1). According to normalized RPKM values, all PvulYABBY genes were up- or downregulated, except for PvulYABBY-1 and PvulYABBY-6, after salt stress treatment in the leaf and root tissues of common bean (Fig. 4).
Fig. 4.
Heat map of differentially expressed PvulYABBY genes under normal and salt stress conditions
Figure 4 was derived from Illumina RNA-seq data, which was reported by Hiz et al. (2014) and downloaded from Sequence Read Archive Accession numbers; SRR957667 (control leaf), SRR958472 (salt-treated root), SRR958469 (control root), and SRR957668 (salt-treated leaf) (Hiz et al. 2014). RT: Root treated, RC: Root control, LT: Leaf treated, LC: Leaf control.
Salt stress responses of PvulYABBY genes
Expression levels of all putative PvulYABBY genes were analyzed using qPCR in leaf and root tissues of Yakutiye and Zulbiye cultivars under salt stress conditions. Six out of eight genes had significant levels of expression in the leaves and these genes exhibited tissue- and cultivar-specific expression patterns. The relative expression levels of PvulYABBY-1, -4, -5, -6, -7 and -8 genes were found to be at least 1.8 times higher in leaves of salt-tolerant cv. ‘Yakutiye’ under salt stress compared to the untreated control sample (Fig. 5). The most transcribed gene in the leaves of cv. ‘Yakutiye’ was PvulYABBY-7 with approximately 1600-fold higher expression value than the control sample. On the other hand, except for PvulYABBY-4 and -6, relative expression levels of all other genes were found to be lower in the leaves of salt-sensitive cv. ‘Zulbiye’ under salt stress when compared to the control sample (Fig. 5).
Fig. 5.

qPCR data indicating the relative expression levels (Log2 values) of PvulYABBY genes in leaves and roots of Zulbiye and Yakutiye common bean cultivars under salt (150 mM NaCl) stress compared to their respective controls (unstressed tissue). Value of control sample (not shown in the histogram) is equivalent to 1
However, only four out of eight genes were found to be transcribed in the roots of cv. ‘Yakutiye’ and ‘Zulbiye’ in a tissue- and cultivar-specific manner. All of these genes (PvulYABBY-1, -5, -7 and -8) were found to be downregulated in the roots of cv. ‘Yakutiye’ and the most downregulated gene was PvulYABBY-5 with 0.02-fold lower expression value than the control sample (Fig. 5). In the roots of cv. ‘Zulbiye’, PvulYABBY-1, -7 and -8 genes were found to be upregulated, while PvulYABBY-5 gene was downregulated compared to the untreated control sample. The downregulation of PvulYABBY-5 gene was not statistically significant according to the statistical analysis. It is known that the YABBY is a quite complex gene family which consists of several members. These genes are usually found to be related with the leaf, flower, and fruit development (Dai et al. 2007). Recently, their role in formation of fruit shape has also been found in tomato (Han et al. 2015). In addition to these previous findings, this study clearly shows that YABBY genes show dynamic gene expression profile under salt stress conditions and this clue might support the idea that YABBY genes can be considered to play a role in salt stress response in common bean.
In conclusion, our identified 8 PvulYABBY genes in Phaseolus vulgaris genome were distributed on 11 different chromosomes. Gene expression analysis was carried out using all PvulYABBY genes and defined a significant variation between Yakutiye and Zulbiye variants under salt stress. The gene expression level in leaf of Yakutiye was found to be higher than those of Zulbiye. Alternatively, the expression level in root of Zulbiye was mostly higher compared to Yakutiye. The results of this study can be used for common bean breeding studies in the future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Ilker Buyuk received financial support from TUBITAK-BIDEB 2219- International Postdoctoral Research Fellowship Programme (Grant no: 1059B191600028) for his studies at Michigan State University in USA.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0933-0) contains supplementary material, which is available to authorized users.
References
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato C, Arisi I, Frizzo ME, Brandi R, Da Sacco L, Masotti A. Computational challenges in miRNA target predictions: to be or not to be a true target? Biomed Res Int. 2009 doi: 10.1155/2009/803069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buyuk İ, İnal B, İlhan E, Tanrıseven M, Aras S, Erayman M. Genome-wide identification of salinity responsive HSP70s in common bean. Mol Biol Rep. 2016;43:1251–1266. doi: 10.1007/s11033-016-4057-0. [DOI] [PubMed] [Google Scholar]
- Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;21:1280–1281. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cong B, Barrero LS, Tanksley SD. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat Genet. 2008;40:800–804. doi: 10.1038/ng.144. [DOI] [PubMed] [Google Scholar]
- Dai M, Hu Y, Zhao Y, Zhou D-X. Regulatory networks involving YABBY genes in rice shoot development. Plant Signal Behav. 2007;2:399–400. doi: 10.4161/psb.2.5.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt NA. YABBY genes and the development and origin of seed plant leaves. Plant Cell. 2010;22:2103–2104. doi: 10.1105/tpc.110.220710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldem V, Akçay UÇ, Ozhuner E, Bakır Y, Uranbey S, Unver T. Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS One. 2012;7:e50298. doi: 10.1371/journal.pone.0050298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldem V, Okay S, Ünver T. Plant microRNAs: new players in functional genomics. Turk J Agric For. 2013;37:1–21. [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Eren H, Pekmezci M, Okay S, Turktas M, Inal B, Ilhan E, Atak M, Erayman M, Unver T. Hexaploid wheat (Triticum aestivum) root miRNome analysis in response to salt stress. Ann Appl Biol. 2015;167:208–216. doi: 10.1111/aab.12219. [DOI] [Google Scholar]
- Fang Q, Xu Z, Song R. Cloning, characterization and genetic engineering of FLC homolog in Thellungiella halophila. Biochem Biophys Res Commun. 2006;347:707–714. doi: 10.1016/j.bbrc.2006.06.165. [DOI] [PubMed] [Google Scholar]
- Golz JF, Roccaro M, Kuzoff R, Hudson A. GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development. 2004;131:3661–3670. doi: 10.1242/dev.01221. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler NS, Saglam A, Demiralay M, Kadioglu A. Apoplastic and symplastic solute concentrations contribute to osmotic adjustment in bean genotypes during drought stress. Turk J Bot. 2012;36:151–160. [Google Scholar]
- Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi Chuan. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- Gupta S, Malviya N, Kushwaha H, Nasim J, Bisht NC, Singh VK, Yadav D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta. 2015;241:549–562. doi: 10.1007/s00425-014-2239-3. [DOI] [PubMed] [Google Scholar]
- Han H, Liu Y, Jiang M, Ge H, Chen H. Identification and expression analysis of YABBY family genes associated with fruit shape in tomato (Solanum lycopersicum L.) Genet Mol Res. 2015;14:7079–7091. doi: 10.4238/2015.June.29.1. [DOI] [PubMed] [Google Scholar]
- Hiz MC, Canher B, Niron H, Turet M. Transcriptome analysis of salt tolerant common bean (Phaseolus vulgaris L.) under saline conditions. PLoS One. 2014;9:e92598. doi: 10.1371/journal.pone.0092598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Van Houten J, Gonzalez G, Xiao H, van der Knaap E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol Genet Genom. 2013;288:111–129. doi: 10.1007/s00438-013-0733-0. [DOI] [PubMed] [Google Scholar]
- Hou J, Jiang P, Qi S, Zhang K, He Q, Xu C, Ding Z, Zhang K, Li K (2016) Isolation and functional validation of salinity and osmotic stress inducible promoter from the maize type-II H+-pyrophosphatase gene by deletion analysis in transgenic tobacco plants. PLoS One 11:e0154041 [DOI] [PMC free article] [PubMed]
- Juarez MT, Twigg RW, Timmermans MC. Specification of adaxial cell fate during maize leaf development. Development. 2004;131:4533–4544. doi: 10.1242/dev.01328. [DOI] [PubMed] [Google Scholar]
- Kavas M, Baloğlu MC, Atabay ES, Ziplar UT, Daşgan HY, Ünver T. Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol Genet Genom. 2016;291:129–143. doi: 10.1007/s00438-015-1095-6. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary demography of duplicate genes. J Struct Funct Genom. 2003;3:35–44. doi: 10.1023/A:1022696612931. [DOI] [PubMed] [Google Scholar]
- Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1–4. doi: 10.1016/S0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Peng X, Wu Q, Teng L, Tang F, Pi Z, Shen S. Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 2015;15:108. doi: 10.1186/s12870-015-0489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Liu YG, Shibata D, Kanaya E, Morita EH, Okada K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 1999;13:1079–1088. doi: 10.1101/gad.13.9.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluttenhofer C, Yuan L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015;167:295–306. doi: 10.1104/pp.114.251769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva JM, Broadhvest J, Hauser BA, Meister RJ, Schneitz K, Gasser CS. INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules. Genes Dev. 1999;13:3160–3169. doi: 10.1101/gad.13.23.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Seki M, Shinozaki K, Yokoyama S. DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci. 2013;18:267–276. doi: 10.1016/j.tplants.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Yang ZF, Gu SL, Wang XF, Li WJ, Tang ZX, Xu CW. Molecular evolution of the CPP-like gene family in plants: insights from comparative genomics of Arabidopsis and rice. J Mol Evol. 2008;67:266–277. doi: 10.1007/s00239-008-9143-z. [DOI] [PubMed] [Google Scholar]
- Yang C, Ma Y, Li J. The rice YABBY4 gene regulates plant growth and development through modulating the gibberellin pathway. J Exp Bot. 2016;67:5545–5556. doi: 10.1093/jxb/erw319. [DOI] [PubMed] [Google Scholar]
- Zhang YJ. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.