Abstract
We have identified a novel polymerase beta (Pol β)-like enzyme from Leishmania infantum, a parasite protozoon causing disease in humans. This protein, named Li Pol β, shows a nuclear localization that contrasts with the mitochondrial localization of Pol β from Crithidia fasciculata, a closely related parasite, the only polymerase β described so far in Trypanosomatidae. Li Pol β, that belongs to the DNA polymerase X family, displays an evolutionarily conserved Pol β-type DNA polymerase core, in which most of the key residues involved in DNA binding, nucleotide binding, dRPase and polymerization catalysis are conserved. In agreement with this, Li Pol β, overproduced in Escherichia coli, displayed intrinsic DNA polymerase activity. Cell synchronization experiments showed a correlation between both Li Pol β mRNA and protein levels along the parasite cell cycle. Analysis of these parameters at the different growth phases of the parasite, from the proliferative (non-infective) logarithmic phase to the non-dividing (highly infectious) stationary phase, showed high levels of Li Pol β at the infective phase of the parasite. The data suggest a role of Li Pol β in base excision repair in L.infantum, a parasite usually affected by oxygen stress environments into the macrophage host cells.
INTRODUCTION
Leishmaniasis is a parasitic infection caused by various species of the protozoan Leishmania. Transmitted by the bite of sand flies, it infects >15 million people and is endemic in the tropical and subtropical regions of America, Africa, the Indian subcontinent, South Asia and the Mediterranean area. The visceral form of the disease is associated with AIDS patients as an opportunistic disease (1). The disease threatens 360 million people living in the endemic areas, and large epidemics affecting hundreds of thousands still occurred in the nineties (2).
This parasitic protozoan belonging to the order of kinetoplastidae, with a single mitochondria called a kinetoplast, are among the most primitive eukaryotes (3). This ancient phylogeny results in exceptional biological features such as polycistronic transcription, trans-splicing of precursor RNAs and transcriptional editing of mitochondrial RNAs (4). The chromatin does not condense into chromosomes and the nuclear envelope remains during cell division (5). The parasite presents two forms, promastigote (extracellular) and amastigote (intracellular). The promastigote phase undergoes a process called metacyclogenesis, becoming infective before entering the host macrophage (6). This process is mimicked by the promastigote in liquid medium culture developing from a non-infectious form during logarithmic growth to a highly infectious form at the non-replicative stationary phase (7).
The polynucleotide polymerases are probably one of the earliest enzymatic activities appearing in evolution. Irrespective of the variety of DNA polymerases involved in DNA replication and repair, they display several common features. The basic mechanism of nucleotide addition involves a pair of metal ions coordinated by carboxylated amino acid residues widely conserved among DNA and RNA polymerases (8). This mechanism appears to be either evolutionarily conserved or acquired by convergent evolution (9). There are also structural similarities between the different DNA-dependent polymerases according to their crystal structures. In all structures described so far (10–17), the polymerization domain displays a hand-shaped structure with fingers, palm and thumb subdomains that define a groove to hold the DNA.
DNA polymerase beta (Pol β), a member of the family X of DNA polymerases, seems to participate in several DNA transactions in vivo, e.g. DNA replication, recombination and DNA base excision repair (BER) (18). The simple BER or ‘short patch’ BER involves the excision or replacement of a single damaged nucleotide. To carry out this process Pol β requires, in addition to the polymerization domain, an 8 kDa N-terminal domain able to excise the 5′-terminal deoxyribose phosphate residue from an incised abasic site, by a β-elimination mechanism (19). An alternative subpathway, ‘long patch’ BER, involves excision of 2–6 nt, involving DNA synthesis that may be also catalyzed by Pol δ or Pol ɛ in addition to Pol β (20–23). The only beta DNA polymerase described so far in Trypanosomatidae seems to be related to DNA synthesis processes associated with the kinetoplast DNA (24).
The ancient eukaryote Leishmania infantum, although lacking catalase (25), survives in the host macrophage not only by passive defense that relies on surface glycolipids and lipophosphoglycans but also by repairing its DNA, damaged by the macrophage oxygen burst (25). This repair is probably done by a BER mechanism. Indeed, two enzymes have been recently identified in trypanosomatidae, uracil-DNA glycosylase and A/P endonuclease (26–28), which could be involved in this type of process. However, no DNA polymerase has been so far described that may act as BER operator. In this paper we report the identification of a novel DNA polymerase from L.infantum (Li Pol β), whose three-dimensional structure modeling predicts a hand-shaped conformation similar to that described for mammalian Pol β and other DNA-dependent polymerases (29,30). The expression of Li Pol β in synchronous cell cultures appears to be regulated, correlating with the different phases of the cell cycle of the protozoon as well as with the distinct phases of the parasite, being higher at the infective stationary phase. The possible involvement of this nuclear enzyme in BER during the infective phase of the parasite, as a necessary repair mechanism to survive the hostile environment inside the host macrophage, will be discussed.
MATERIALS AND METHODS
Materials
Parasite strain (MHOM/FR/80/LEM75) was kindly provided by Dr J. Alvar (Reference Parasitology Service, Majadahonda, Madrid, Spain). Leishmania infantum promastigotes were grown at 26°C in RPMI-1640 from Gibco BRL (Scotland, UK), supplemented with 10% heat-inactivated fetal calf serum, Gibco BRL (Scotland, UK). Promastigote cultures were initiated at 1 × 106 cells/ml and harvested at logarithmic and stationary phases of growth, as defined by morphology and cell concentration (31). Escherichia coli M15 strain (pREP4) for the production of the recombinant Li Pol β was from Qiagen (Germany).
Synchronous cultures
Leishmania infantum promastigotes (107 cells/ml) from logarithmic and stationary phases were incubated for 6 h in medium containing 200 µg/ml of hydroxyurea (32). Synchronous cells were harvested by centrifugation at 2000 g for 10 min at room temperature and suspended in the same volume of fresh medium lacking hydroxyurea. Leishmania infantum promastigotes remained synchronous for ∼6 h. The mRNA and protein expression were determined every 30 min (see below).
Oligonucleotides and DNA probes
Oligonucleotides LiF (5′-CAAGCTGGAGGAGCTGGAGAG) and LiR (5′-GTCGAACACATCCTGCTCTGAAC), designed according to conserved regions of the sequences of family X DNA polymerases were used for reverse transcribed polymerase chain reaction (RT–PCR) amplification of a L.infantum DNA POL β gene fragment of 800 bp. The 5′-terminal end was amplified with the spliced leader oligonucleotide SL (5′-ATCAGTTTCTGTACTTTATTG) and the R2 oligonucleotide (5′-CTTCTGCAGCAGCTCATCCAC) of the previously determined sequence (5′-RACE). The 3′-terminal end was amplified (3′-RACE), with a specific oligonucleotide from the known sequence F1 (5′-TGTCGGCATCAAGTACTTCTACGA) and the general oligo(dT) primer [5′-GCGCCAGGAATTCGC(dT17)]. The DNA probes corresponding to the whole coding sequence of the Li Pol β were labeled with [α-32P]dCTP using the Random Primed DNA labeling kit (Boehringer Mannheim, GmbH, Germany).
Expression of L.infantum Pol β in E.coli
The open reading frame containing the DNA Pol β from L.infantum was cloned into a pQE/pREP4 bacterial expression vector, which allows the expression of recombinant proteins as fusions with a multifunctional leader peptide containing a hexahistidyl sequence for purification on Ni2+-affinity resins (33). The open reading frame of Li Pol β was PCR-amplified using oligonucleotides containing SphI and HindIII restriction sites 5′ POL SphI (5′-GCGCGCATGCTCCGCCGGAAGTTC) and 3′ POL HindIII (5′-GCGCAAGCTTAAGGGTCGCGGTTCTC). The resulting 1.15 kb PCR product was cloned at the SphI/HindIII sites of vector pQE32 (Qiagen, Germany). The construction of the recombinant expression plasmid, named pQE32-Li POL β was confirmed by DNA sequencing. Expression of the His6-tagged Li Pol β protein was carried out in the M15 E.coli strain, which contains the pREP4 repressor plasmid and the T5 promoter under the control of the isopropyl β-d-thiogalactopyranoside (IPTG)-regulated lacI gene (34). Cells were transformed with the pQE32 plasmid and grown at 37°C in Luria–Bertani (LB) medium until the OD reached 0.6 (20–30 min). Then, IPTG was added to a final concentration of 1 mM and the incubation was continued for 4 h at 37°C. Cells were collected by centrifugation at 4000 g for 20 min and lyzed in buffer A (6 M Gu–HCl, 0.1 M sodium phosphate, 10 mM Tris–HCl, pH 8.0). The suspension was cleared by centrifugation at 10 000 g for 20 min and the supernatant containing the Li Pol β recombinant protein was used for the purification process. Induction, overproduction and solubility of the recombinant protein was analyzed by 10% polyacrylamide gel electrophoresis (PAGE) in the presence of SDS and visualized by Coomassie Blue staining. Western blot was carried out as described (35,36). In all cases, unless otherwise indicated, 10 µg of protein was loaded per well.
Purification of Li Pol β
Ni2+-NTA agarose beads (Qiagen, Germany), previously equilibrated with buffer A were incubated for 30 min at room temperature with the soluble fraction containing the recombinant protein obtained as described above. The resin was loaded into a column and washed with 10 ml of buffers B (8 M urea, 0.1 M sodium phosphate, 10 mM Tris–HCl, pH 8.0) and C (8 M urea, 0.1 M sodium phosphate, 10 mM Tris–HCl, pH 6.3). The recombinant protein was eluted in 1 ml fractions with 5 ml of buffers D (8 M urea, 0.1 M sodium phosphate, 10 mM Tris–HCl, pH 5.9) and E (8 M urea, 0.1 M sodium phosphate, 10 mM Tris–HCl, pH 4.5). The fractions were analyzed by SDS–PAGE. The amount of protein was quantified by the method of Bradford (37).
In situ gel analysis of DNA polymerase activity
This assay, that allows correlation of catalytic activity with a particular polypeptide species separated by SDS–PAGE, was carried out as described (38). The assay is especially suitable to detect catalytically active species in recombinant cell extracts or affinity purified fractions that are soluble only in the presence of detergents such as SDS. The samples were electrophoresed in 10% SDS–PAGE gels containing 1.5 mg/ml activated calf thymus DNA (Pharmacia Biotech Inc., Belgium) as template-primer, followed by in situ renaturation of proteins and incubation of the gel in a DNA polymerase assay mixture. Prior to renaturation, the gel was washed twice with 50 mM Tris–HCl, pH 7.5, for 15 min at 4°C. The renaturation was allowed to occur during 3 h at 4°C in buffer F [50 mM Tris–HCl, pH 7.5, 6 mM (AcO)2Mg, or (AcO)2Mn, 40 mM KCl, 16% glycerol, 10 mM EDTA, 1 mM dithiothreitol and 400 µg/ml bovine serum albumin]. In situ polymerization was assayed with buffer G (buffer F plus 2 µM each dNTP and 1.2 nM [α-32P]dATP) for 12 h at 30°C. After washing unincorporated [α-32P]dATP from the gel, and in situ precipitation of DNA with buffer H (5% trichloroacetic acid, 1% sodium pyrophosphate), the gel was dried and the activity bands (radioactively labeled) were detected by autoradiography.
Amino acid sequence comparisons and three-dimensional structure prediction
Multiple alignment of the Li Pol β amino acid sequence with sequences of Pol β-like proteins (X family) from different sources was done using the computer program PIMA 1.4 from The Baylor College of Medicine (39) and displayed with the BOXSHADE 3.21 program from the EMBL Network-Swiss node. The alignment was manually adjusted on the basis of the secondary structure elements of the rat Pol β, as deduced from its crystal structure (29). The three-dimensional structure prediction for Li Pol β was done using knowledge-based protein modeling methods on the basis of multiple alignment of the Li Pol β sequence and those of the 3.6 and 2.6 Å resolution X-ray structures of rat (15,29) and human (40,41) Pol β, respectively. Their Cartesian coordinates were from the Brookhaven Protein Data Bank, with PDB codes 1BPD and 9ICW, respectively. Conformational calculations were carried out under the AMBER force field (42). All computations were performed on a SGI Power Challenge R10000 with the software package BIOSYM, Release 95.0 (Molecular Simulations Inc., CA).
Phylogenetic comparisons
A segment of 34 amino acids spanning the most conserved portion of the Pol β-like active site, containing two of the three catalytic aspartates, was used to generate the data matrix to infer the phyletic relationships among different members of the DNA polymerase X superfamily. This region corresponds to the rat Pol β sequence: PGLLCVACGSFRRGKVTCGDVDVLITHPDGRSHQ. Some of the proteins aligned are in the SWISS-PROT database, and the corresponding identifiers are: P06746 (Homo sapiens Pol β); P06766 (Ratus norvegicus Pol β); O57383 (Xenopus laevis Pol β); O02789 [Monodelphis domestica terminal deoxynucleotidyltransferase (TdT)]; P09838 (Mus musculus TdT); O57486 (Ambystoma mexicana TdT); P04053 (Homo sapiens TdT); P42118 (X.laevis TdT); P36195 (Gallus gallus TdT); Q92089 (Onchorrincus mykiss TdT); P06526 (Bos bovis TdT); P25615 (Saccharomyces cerevisiae Pol IV); P42494 [African swine fever virus (ASFV) Pol X]. Other sequences are available at different databases (GenBank, EMBL and dbEST), and have the following identifiers: AJ131889 (M.musculus Pol λ); AJ131890 (H.sapiens Pol λ); AW311924 (Sus scrofa Pol λ); AI703888 (R.norvegicus Pol λ); AJ393493 (G.gallus Pol λ); AJ289628 (Arabidopsis thaliana Pol λ); AJ251804 (M.musculus Pol µ); AJ131891 (H.sapiens Pol µ); AF063866 (Melanoplus sanguinipes Pol X); Z99118 (Bacillus subtilis complete genome, including Pol X); AE000838 (Methanobacterium thermotropicum complete genome, including Pol X); U19912 (Crithidia fasciculata Pol β); AF182167 (L.infantum Pol β). The multiple alignment was carried out using the CLUSTALW program (http://www.ebi.ac.uk/clustalw/). Visualization of the phylogenetic tree derived from the multiple alignment was carried out using the TreeView program, developed by Rod Page (http://taxonomy.zoology.gla.ac.uk/rod.html).
Northern blot analysis
Leishmania infantum total RNA was obtained from the distinct parasite forms and prepared as described (36). Total RNA was isolated from logarithmic and stationary growth phases using TRIzol (Gibco BRL, USA). For the analysis, 5 µg of total RNA were fractionated in formaldehyde agarose gels and transferred onto nylon membranes following standard procedures (43). The blots were hybridized with the described PCR probe for the Li Pol β coding region (see above). Blots were analyzed by autoradiography and bands were scanned and quantified by using a Molecular Dynamics Computer Densitometer and the Image Quant Software 3.3 program. To avoid sampling errors, densitometric scanning of ethidium bromide-stained gels was carried out.
Specific antibody obtention and immunofluorescence localization
New Zealand rabbit specific polyclonal antibody was obtained with 800 µg of purified recombinant Li Pol β, administered in four weekly subcutaneous shots: the first one in CFA and the following in IFA. After one additional week the rabbit was bled, the serum obtained, decomplementarized at 55°C for 1 h and subsequently stored at –20°C. The immunofluorescence was carried out with 106/ml promastigotes washed twice with phosphate-buffered saline (PBS) containing 1% fetal calf serum and 0.1% sodium azide and resuspended in the same PBS solution with identical final cell concentration. Twenty-five microliters were spotted onto 12 mm round coverslips. After 1 h at 37°C, the cells were treated with methanol/acetone (1:1) for 15 min at –20°C. The fixed cells were incubated for 15 min at room temperature in DAPI-methanol (0.1 µg/ml). After an additional washing with methanol, the cells were treated with PBS containing 0.5% Triton X-100 for 5 min and incubated in a humidity chamber for 1 h at room temperature with rabbit polyclonal anti-Li Pol β or anti-Li Topoisomerase II antibodies, diluted 1:200 in both cases. After washing in PBS, cells were incubated with anti-rabbit IgG-FITC (Dako, Denmark), diluted 1:2000, for 1 h. After washing with PBS, the slides were mounted with Mowiol (Calbiochem, USA) and observed on a Zeiss Axioplan microscope. The images were captured by a Hamamatsu CCD camera and processed in Adobe Photoshop on a Macintosh computer.
Stability of Li Pol β mRNA and response to environmental changes
Stability of the specific Li Pol β mRNA was analyzed under conditions in which RNA synthesis was inhibited. Samples of 10 µg/ml of Actinomycin D (Sigma, USA) were added to promastigote cultures at logarithmic phase and incubated for 8 h. Samples were removed every 2 h and specific mRNA molecules detected by northern blotting. Environmental changes at the promastigote phase were carried out mimicking the conditions of pH (5.5) and temperature (37°C) that correspond to the macrophage parasitophorous vacuole (44) formed after parasite infection of the mammalian host. Logarithmic phase promastigotes (5 × 106 cells/ml) were cultured at the standard conditions except that either the pH was 5.5 or the incubation was at 37°C for 8 h, and samples were removed every 2 h. The presence of the specific mRNA was detected by hybridization with the labeled specific probe described for northern analysis.
RESULTS
Identification and cloning of L.infantum POL β gene
The complete cDNA encoding sequence of the Li Pol β polypeptide was obtained by RT–PCR in two steps. First, specific oligonucleotides were designed according to conserved sequences in the family X of DNA polymerases to obtain a central region of the gene of 800 bp. In a subsequent step, the 5′ and 3′ ends were also obtained by the same procedure using specific oligonucleotides from the known sequence and oligonucleotides containing specific sequences for the ‘spliced leader’ and the mRNA poly(A) tail (5′-RACE and 3′-RACE, respectively). Additionally, a 5′ flanking sequence of 510 bp was obtained. The open reading frame 1131 bp long (GenBank accession no. AF182167), with 84% of the codons ending in G or C as described for the genes of the genus Leishmania (45), encodes a 376 amino acid protein corresponding to an estimated molecular mass of 42.7 kDa, predicted to be a new member of the family X of DNA polymerases.
Li Pol β belongs to the family X of DNA polymerases
The prediction that Li Pol β is a new member of the Pol X family was confirmed by multiple amino acid sequence alignment with Pol β, Pol λ (46), TdT (47), Pol µ (48), yeast Pol IV (49) and other Pol β-like enzymes from C.fasciculata (50,51) and ASFV (9). Figure 1 shows a multiple alignment of Li Pol β versus rat Pol β and mouse Pol λ, two of its closest homologs among all Pol X members. The 376 amino acid residues of Li Pol β presented 31 and 24% of identity in the portion aligned with Pol β and Pol λ, respectively (see Fig. 1). Interestingly, Li Pol β contained 26 of the 27 residues that are invariant in all DNA polymerases from family X, including all those involved in DNA and nucleotide binding, polymerization catalysis and conformational changes involved in the polymerization cycle. Moreover, Li Pol β also conserves the critical residues of mammalian Pol β involved in its intrinsic dRPase activity (Pol β residues H34, Y39, K60, K68, K72) that acts in concert with polymerization during BER of damaged DNA (see legend to Fig. 1).
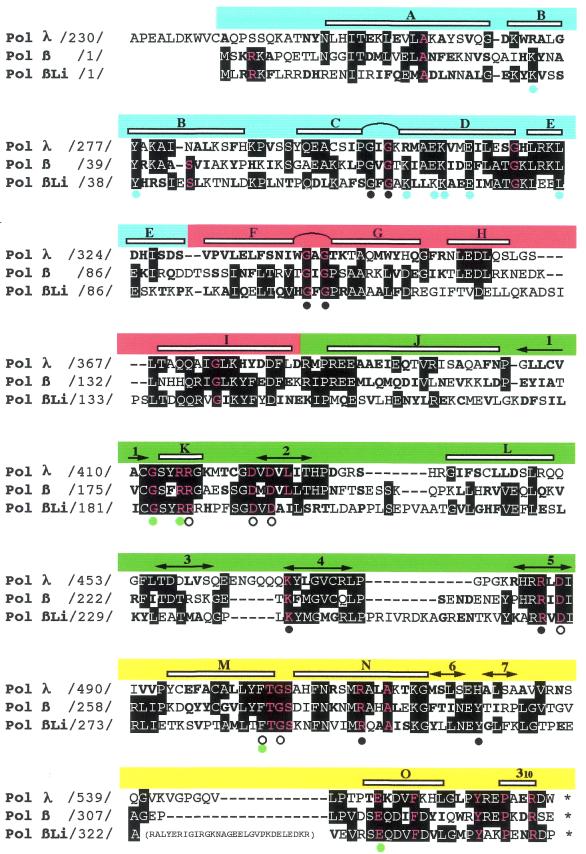
Figure 1.
Li Pol β, a Leishmania nuclear polymerase homologous to Pol β. Multiple amino acid alignment of mouse Pol λ, rat Pol β and Li Pol β. Numbers between slashes indicate the amino acid position relative to the N-terminus of each DNA polymerase. According to rat Pol β structural data (15,29), the alignment can be divided in four subdomains: ‘8 kDa’ (residues 1–84; blue segment), ‘fingers’ (residues 85–153; red segment), ‘palm’ (residues 154–276; green segment) and ‘thumb’ (residues 277–376; yellow segment). α-helices (lettered) and β-strands (numbered) are indicated at the top of the alignment. Two helix–hairpin–helix motifs are between α-helices C and D (‘8 kDa’ subdomain), and between α-helices F and G (‘fingers’ subdomain). Invariant residues among any pair of DNA polymerases are indicated with white letters (over a black background). Conservative substitutions (bold typed) were considered as follows: K, H and R; D, E, Q and N; W, F, Y, I, L, V, M and A; G, S, T, C and P. The 27 residues that are invariant among DNA polymerase X family members (8) are indicated with red letters. Colored dots at the bottom of the alignment indicate the Pol β residues (15) shown to act as DNA ligands (Gly64, Gly66, Gly105, Gly109, Lys234, Arg254, Arg283, Tyr296; black), dNTP and metal ligands (Arg183; Asp190, Asp192, Asp256, Phe272 and Gly274; white), or involved in interactions between the ‘palm’ and ‘thumb’ subdomains (Gly179/Phe272; Arg182/Glu316; green). Blue dots indicate residues involved in the dRPase activity (Lys35, Tyr39, Lys68, Glu71, Lys72, Glu75 and Lys84) present at the ‘8 kDa’ domain of Pol β (66–68). A 29 amino acid insertion in Li Pol β, located between β-strand 7 and helix O, is shown in parentheses.
The alignment shown in Figure 1 indicates that Li Pol β, as mammalian Pol β, does not contain the N-terminal extra segment, containing a BRCA1 C-terminal (BRCT) domain, present in Pol λ, and also in Pol µ, TdT and yeast Pol IV, which is likely involved in regulatory protein–protein interactions (52). However, the remaining residues could be aligned with those forming the four subdomains of Pol β: the so called ‘8 kDa’ (blue portion in Fig. 1) could be aligned with a single amino acid gap connecting α-helices A and B, and with an additional amino acid residue at α-helix B. The same occurs at the ‘fingers’ subdomain (red portion), with small differences at the connection of α-helices E and F and α-helices H and I. The regions corresponding to some stretches of secondary structure elements of both ‘palm’ (green) and ‘thumb’ (yellow) subdomains (as 1-K-2, L-3, 5-M-N-6-7 and O-310; see Fig. 1 for nomenclature) could be aligned without any gap among Li Pol β, mouse Pol λ and rat Pol β. On the other hand, Li Pol β has two main insertions, corresponding to loop regions connecting secondary structured elements, as it has been described for other members of the family X (9). One of these insertions consists of 8 amino acids (between β-strand 4 and 5), located at the ‘palm’ subdomain, whereas the other, made of 26 amino acids (between β-strand and α-helix O), is located at the ‘thumb’ subdomain.
Three-dimensional structure prediction
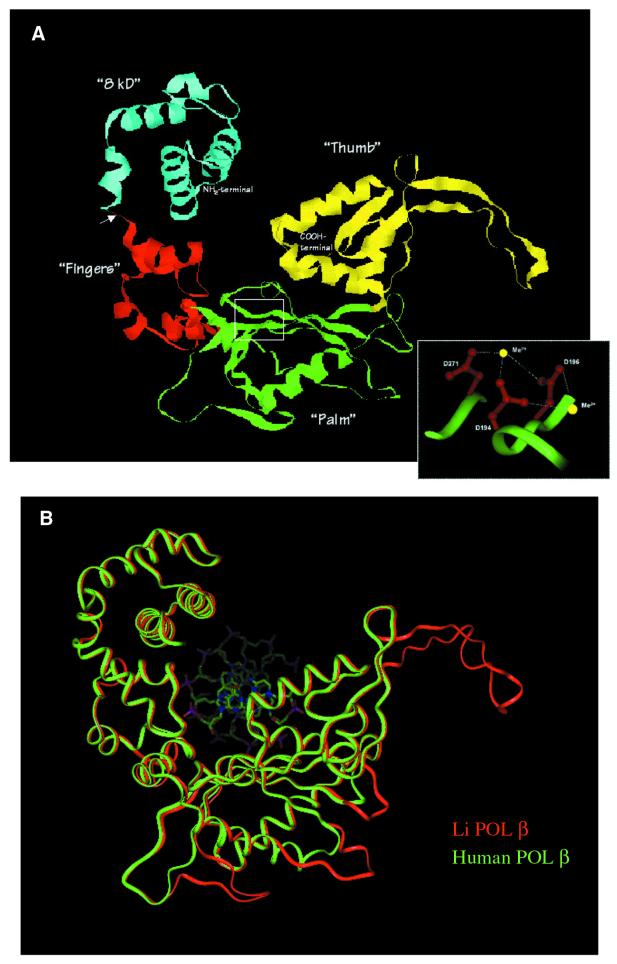
Interestingly, although the percentage of identical residues shown in the alignment of Li Pol β and rat Pol β was only 31%, the level of amino acid similarity was enough to model the structure of Li Pol β over most of the entire crystal structure of rat and human Pol β. The structural model shown in Figure 2A predicts that Li Pol β would adopt a mammalian Pol β-like fold with four distinguishable subdomains: the N-terminal protease sensitive ‘8 kDa’ domain (blue), predicted to be involved in DNA binding and deoxyribose phosphate excision, and the C-terminal 34.7 kDa region, forming the polymerization domain. This domain would contain the catalytic site with the three established subdomains present in Pol β: ‘fingers’ (red), providing contacts to handle the DNA primer strand through its helix–hairpin–helix motif, ‘palm’ (green), constituting an appropriate surface to accommodate the substrate molecules (nucleotides and DNA) and to present the pair of metal ions adequately for catalysis, and ‘thumb’ (yellow) providing template selectivity and reaction cycling via conformational changes (15,29).
Figure 2.
Three-dimensional structure prediction for L.infantum Pol β inferred from the crystal structure of rat and human Pol β. (A) A ribbon representation of the three-dimensional structure of Li Pol β, obtained as described in Materials and Methods, showing the four different subdomains described in mammalian Pol β. These subdomains are colored accordingly to the alignment shown in Figure 1: ‘8 kDa’ (blue), fingers (red), palm (green) and thumb (yellow). The inset is a close up view of the amino acid side chains of the conserved catalytic residues (Asp194, Asp196 and Asp271) that interact with the metal ions, drawn with solid sticks (red) and the polypeptide backbone as a solid ribbon (green). The arrow indicates the proteolytic cleavage site between the ‘8 kDa’ subdomain and the ‘35 kDa’ catalytic region. Ct and Nt indicate the N- and C-terminal ends, respectively. (B) A solid ribbon representation of the optimally superimposed polypeptide backbone of the model of Li Pol β (red) and human Pol β (green). The DNA from the human co-crystal is shown as solid sticks in standard colors for atom type (oxygen, red; nitrogen, blue; phosphorous, purple).
In Pol β, three catalytic aspartates (Asp190, Asp192 and Asp256) are involved in metal ion binding at the active site (8). The inset in Figure 2A shows the spatial arrangement of the corresponding residues in Li Pol β Asp194, Asp196 and Asp271. Figure 2B showed the comparison between the Li Pol β three-dimensional predicted structure (red) and the human one (green). Both structures fit quite well with each other except in the loops corresponding to the insertions that have been described above in the Li Pol β amino acid sequence (see Fig. 1).
Li Pol β overexpression in E.coli and purification
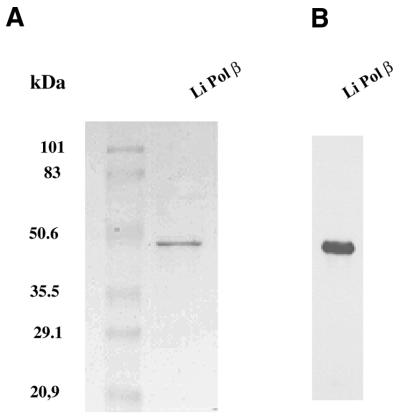
The induction of the Li Pol β was carried out in the E.coli strain M15 (data not shown). The induced new polypeptide migrated at the expected position for the Li Pol β protein (∼47 kDa, 5 kDa over the expected molecular mass due to the additional histidine chain). This protein remained soluble under the extraction conditions used. Based on the His6-tag present on its N-terminal end, the recombinant protein could be purified on a Ni2+-NTA affinity chromatography column which allowed us to obtain a single polypeptide band by SDS–PAGE (Fig. 3A) and western blotting (Fig. 3B). The yield of purified recombinant protein was 2 mg/100 ml of bacterial culture.
Figure 3.
Purification of Li Pol β. (A) SDS–PAGE analysis and Coomassie Blue staining of Li Pol β, purified through a Ni2+-NTA affinity column. (B) Western blotting of the Ni2+-NTA purified fraction, with an anti-Li Pol β specific rabbit antiserum.
Li Pol β has intrinsic DNA polymerase activity
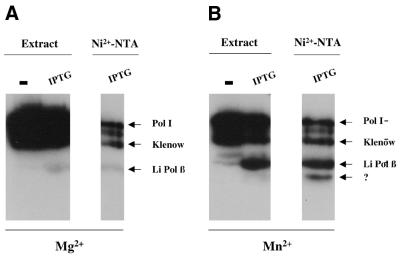
The predicted DNA polymerase activity of the recombinant Li Pol β was examined by using an in situ activity gel assay. This technique takes advantage of the high resolution of the SDS–PAGE to separate the different polymerase species. In this case a clear conclusion was expected based on the different molecular mass between the putative L.infantum Pol β polymerase (47 kDa) and those of the endogenous E.coli DNA polymerases. As shown in Figure 4, in addition to the radioactive bands corresponding to the E.coli Pol I and to its proteolytic derivative (Klenow fragment), an activity band with the electrophoretic mobility of Li Pol β was observed at both the extract and in the Ni2+-NTA fraction, corresponding to the purified protein, soluble in the presence of 0.1% of SDS. The minor activity bands, present in the Ni2+-NTA purified fraction, are due to unspecific binding of endogenous E.coli DNA polymerases to the activated beads of the Ni2+-NTA column. The sensitivity of the in situ analysis allows detection of these minor contaminants (53), that can be unambiguously distinguished from Li Pol β (47 kDa) due to their distinct molecular size. The presence in the Ni2+-NTA purified fraction of an additional activity band with a very low molecular weight (∼35 kDa) (Fig. 4B) is interesting. The technique favors the refolding of low molecular weight polypeptides, thus increasing sensitivity of truncated but active polypeptides resulting from partial proteolysis. In this case, the size of the fragment fits quite well with the predicted C-terminal catalytic fragment of the protein and may suggest that it derives from a single and preferential proteolytic cleavage of Li Pol β, as it has been described to occur also in Pol β. Moreover, an active fragment would be incompatible with the loss of the C-terminal portion of Li Pol β (see Fig. 2). The in situ activity was determined in the presence of two different divalent metal ions (Mg2+ and Mn2+) known activators of DNA-dependent DNA polymerases. Li Pol β displays a completely different metal ion requirement to that shown by the contaminating Pol I and Klenow fragments. Mn2+ was strongly required for Li Pol β (see the 47 kDa polypeptide band in Fig. 4B), the latter being residual in the presence of Mg2+. An inverse behavior was observed for the endogenous E.coli DNA activities displaying the highest activity in the presence of Mg2+ (see Fig. 4A). These results confirm that the overproduced Li Pol β corresponding to the 47 kDa band has intrinsic DNA polymerase activity. A similar preference for Mn2+ ions has recently been reported in the case of Pol µ, a DNA mutase belonging to the Pol X family (48).
Figure 4.
In situ DNA polymerase activity of Li Pol β. Autoradiography of the in situ gel analysis carried out as described in Materials and Methods. The activity was assayed in the presence of Mg2+ (A) and Mn2+ (B) as divalent metal ions. The activity was measured in complete extracts from non-induced (–) and IPTG induced (IPTG) pQE32 E.coli strain as well as in the recombinant purified fraction (Ni2+-NTA). The activity bands corresponding to the endogenous activities of E.coli: Pol I and its Klenow fragment are marked as well as the corresponding to Li Pol β. The putative 35 kDa Li Pol β active fragment is indicated by a question mark.
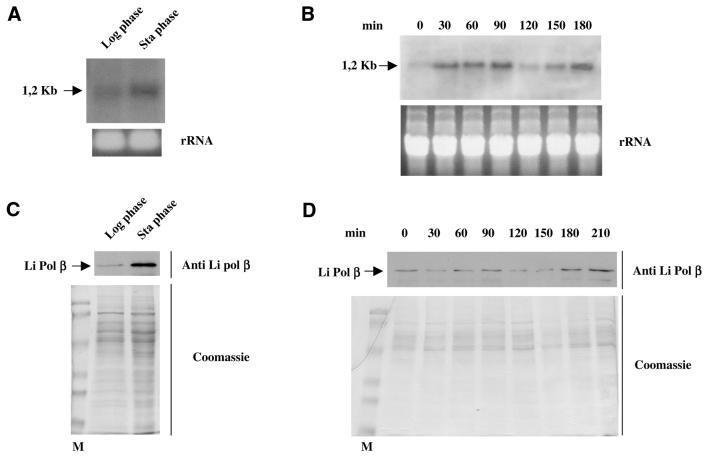
Li Pol β is differentially expressed along the cell cycle
The expression of Li Pol β was analyzed along the distinct phases of the parasite. The expression was determined following two complementary ways. First, gene expression was ascertained by analysis of the Li Pol β mRNA levels and subsequently, synthesis of the Li Pol β protein was followed by western blotting with specific antiserum. The logarithmic growth phase corresponding to the attenuated form of the parasite showed a faint hybridization band, which suggests a low level of specific mRNA. This level is clearly increased at the stationary phase corresponding to the highly infective form of the parasite (Fig. 5A). The use of hydroxyurea, which is lethal for the cells in S phase, allows synchronization of the protozoan culture. Once removed from the culture medium, the synthesis of the nuclear and kinetoplast DNA starts, and the synchronous cell cycle lasts at least for two generations. The northern blotting analysis of the samples obtained at 30 min periods showed that the levels of Li Pol β mRNA are modified along the cell cycle of the protozoon with maximum at 90 and 180 min (Fig. 5B). The rRNA control staining discounts the fact that the differences detected may be due to differences in well loading. Li Pol β protein expression was measured by western blotting of the L.infantum cell extracts obtained from samples taken at the same times than those used for the mRNA determinations. The expression of the protein was again higher at the infectious stationary phase promastigotes than at the logarithmic attenuated phase of the parasite, as indicated in Figure 5C. The protein expression along the cell cycle on synchronous cultures gave two peaks of maximum protein expression at 90 and 180–210 min with a slight delay with respect to the mRNA production at the second cell cycle, as shown in Figure 5D. Again, Coomassie-stained controls discount loading errors. The data suggest a regulatory mechanism for mRNA and Li Pol β protein expression that correlates with the protozoon cell cycle, increasing at the infective stationary phase of the parasite.
Figure 5.
Differential expression of Li Pol β mRNA. (A) Northern blot analysis using 5 µg of total RNA extracted from L.infantum promastigotes at either logarithmic or stationary phases. The Li Pol β probe was labeled with [α-32P]dCTP. The lower part shows the corresponding gel stained with ethidium bromide as loading control. (B) Northern blot analysis of the samples taken from hydroxyurea-synchronous cells culture at different times (indicated in minutes). Five micrograms of total RNA was loaded per well. The lower part shows the corresponding control gel stained with ethidium bromide. For further experimental details see Materials and Methods. (C) Analysis by western blot of the levels of Li Pol β at the logarithmic and stationary phases of the parasite; 10 µg of total protein were loaded per well. The lower part shows the SDS–PAGE gel stained by Coomassie Blue. (D) Western blot analysis of the extracts taken at different times from the synchronous culture. The lower part shows the Coomassie Blue staining of the extracts. For further experimental details see Materials and Methods.
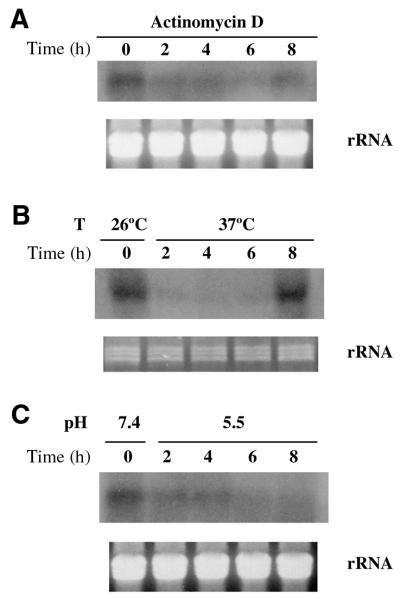
Stability of Li Pol β mRNA and response to environmental variations
Leishmania undergoes several environmental variations along its life cycle. Thus, the promastigote form grows at 26°C and neutral basic pH (7.5–8) in the sand fly gut. The intracellular amastigote form lives inside the host macrophage at mammalian temperature and at acidic pH (4.5–6). The expression of mRNA in the presence of Actinomycin D or under the temperature and pH intracellular conditions shown in Figure 6 indicates that Li Pol β messenger RNA molecules have a short life as it could be expected for a regulated molecule probably linked to DNA synthesis that should not remain in the cell for long periods. Thus, the presence of Actinomycin D leads to a rapid decay of mRNA levels that are undetectable after 2 h of treatment. The pH and temperature conditions mimicking those found by the promastigote inside the parasitophorous vacuole in the infected macrophage induce a rapid decay, as it can be seen in the Figure 6. The effect of temperature shows an interesting response with an initial rapid decay and a late increase after 8 h. This increase does not correspond to the increase of mRNA reported for a classic heat shock protein that presents an earlier response and higher messenger RNA levels (54). Taken together, the data suggest that the expression of the Li Pol β mRNA is regulated within the protozoon cell cycle and is clearly higher at the non-dividing, infective stationary phase of the parasite than at the attenuated logarithmic one.
Figure 6.
Northern blot analysis of Li Pol β mRNA expression under different environmental conditions. Extracts were prepared from samples taken at the indicated times in hours, in the presence of Actinomycin D (A), or corresponding to the different temperature (B) and pH (C) conditions. Ten micrograms of total RNA were loaded per well. A Li Pol β probe labeled with [α-32P]dCTP was used in all the cases. The lower part of the northern blots shows the corresponding agarose gel stained with ethidium bromide as a loading control. For further experimental details see Materials and Methods.
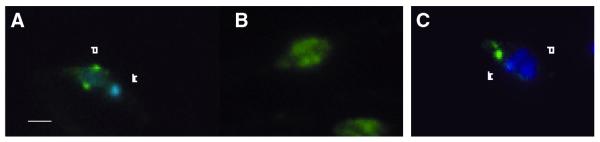
Nuclear localization of Li Pol β, detected by immunofluorescence
Immunofluorescence detection of Li Pol β in the Leishmania parasites was carried out using specific polyclonal antibodies. Simultaneous DAPI staining allowed tracing the DNA of the nucleus and the kinetoplast. The specific immunofluorescence of the protein appeared as two spots peripherally located into the nucleus, far from the DAPI staining corresponding to the cell kinetoplast (Fig. 7). This nuclear localization contrasts with that of Topoisomerase II from L.infantum used as a control and with that described for the Pol β from C.fasciculata associated to the kinetoplast (24), Conversely, it fits quite well with the nuclear localization described for the rest of Pol β-like DNA polymerases described in other species (15,18,29). This immunofluorescence pattern corresponds to the promastigote form in the logarithmic phase. The comparison between the two infective phases of the parasite is shown in Figure 7. At the stationary phase, the amount of fluorescent signal is higher, as could be expected from the previous data, and forms two strips into the nucleus along the longitudinal axis of the promastigote.
Figure 7.
Nuclear localization of Li Pol β by immunofluorescence. Leishmania infantum promastigotes were recovered from asynchronous cultures at the logarithmic and stationary phases. The DNA staining with DAPI shows the nucleus (n) and kinetoplast (k) localization at the promastigote (blue). Li Pol β and Li Topoisomerase II are localized by the respective specific antibodies and anti-rabbit IgG-FITC (green). (A) Li Pol β at logarithmic phase promastigotes. (B) Li Pol β at stationary phase promastigotes. (C) Li Topoisomerase II at logarithmic phase promastigotes. Bar 2 µm. For further experimental details see Materials and Methods.
DISCUSSION
Pol X family of DNA polymerases: structural similarities and evolutionary versions
The transfer of a nucleotide to an acceptor hydroxyl group is a central reaction in many biological processes, and is catalyzed by nucleotidyltransferases that belong to more than 10 distinct superfamilies (55). One of the most widespread superfamilies of nucleotidyltransferases is exemplified by the eukaryotic DNA Pol β, whose three-dimensional structure and detailed mechanism of nucleotidyl transference have been uncovered. A complete sequence-based classification and phyletic distribution of the whole Pol β-type nucleotidyltransferase superfamily has been previously reported (55). Among the different groups (families) of nucleotidyltransferases included in the Pol β superfamily, the Pol X family uses DNA as a substrate, mammalian Pol β and TdT being its best known members.
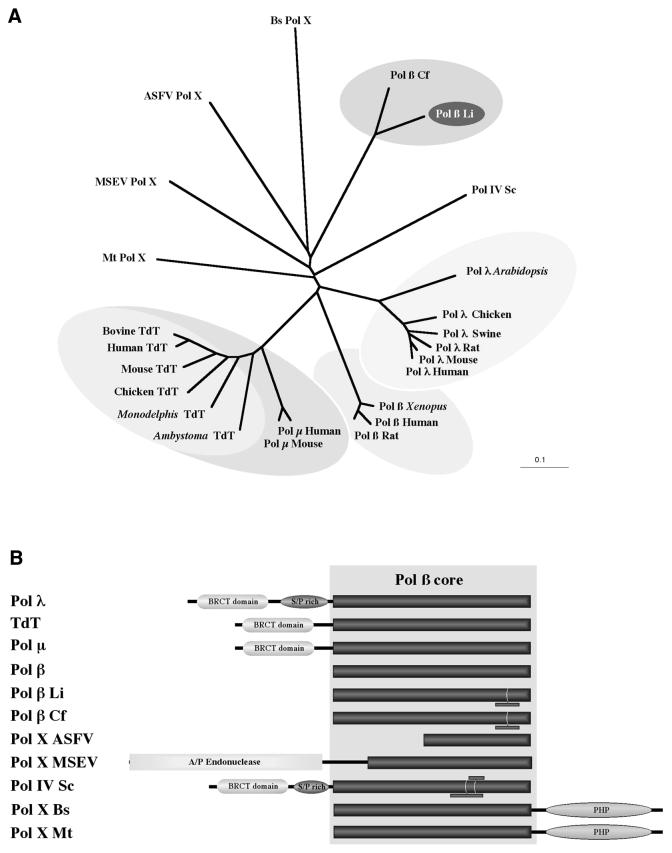
Figure 8A shows a phylogenetic unrooted tree representing the evolutionary distances of those members that are known or predicted to be DNA polymerases (Pol X family), that belong to the three divisions of life (bacteria, archea and eukaryotes). These members include the recently described Pol µ (48) and Pol λ (46), TdT (47), Pol β, viral enzymes (Pol X) from ASFV and M.sanguinipes entomopoxvirus (MSEV), Pol IV from S.cerevisiae, B.subtilis Pol X and M.thermoautotrophicum Pol X. Other bacteria possessing a Pol β polymerase (not included in Fig. 8A) are: Staphylococcus epidermidis, Aquifex eolicus, Thermoplasma acidophylum, Thermus aquaticus, Bacillus halodurans and Thermus thermophylus.
Figure 8.
The Pol X family of DNA polymerases. (A) Phylogenetic tree representing the evolutionary distances among the different members of the Pol X superfamily. The data was obtained by CLUSTALW alignment of a polypeptide segment corresponding to the most conserved segment of the Pol β core, forming the polymerization active site (see Materials and Methods). The DNA polymerases included are: Pol λ from different species of plant and animals; Pol µ from human and mouse; TdT from different vertebrates; Pol β from human, rat, X.laevis and C.fasciculata (Cf); Pol X from ASFV; Pol X from MSEV; Pol IV from S.cerevisiae (Sc); Pol X from B.subtilis (Bs); Pol X from M.thermoautotrophicum (Mt). (B) Domain organization of the different members of the Pol X family. Enzyme nomenclature is as shown in (A). The generally conserved Pol β core is shown as a dark box. The presence of additional sequences is indicated by small bars. The APE predicted at the N-terminus of the MSEV Pol X is boxed in gray. Additional domains, as the BRCT, Ser-Pro rich domain and PHP domain are also indicated.
To discard the influence of the different length of the polypeptides, due to specific deletions and insertions, or due to the presence of specific domains (described later), the phylogenetic tree was generated taking into account the information contained in a short segment of the Pol β core that has the maximal functional importance, as part of the polymerization active site (see Materials and Methods). This phylogenetic tree, that mainly represents the divergence of the Pol β core, supports the hypothesis that most of the subfamilies (Pol β, Pol λ, Pol µ, TdT, etc.) have independently and rapidly evolved from a common ancestor to occupy a particular functional niche (55).
In spite of the general conservation of the critical residues involved in metal, dNTP and DNA binding, the percentage of overall amino acid similarity among the different members of the Pol X family along the Pol β core varies from the 91% of identity between the Pol β enzymes from Crithidia and Leishmania, and the 42% of identity among Pol µ and TdT, to the 19% of identity between Li Pol β and TdT. The amino acid identity of Li Pol β with mammalian Pol β is 31%, very close to that existing between Pol λ and Pol β (32%). These amino acid similarity values support that Li Pol β, as Pol λ (46), and probably most members of the Pol X family, can be predicted to conserve a common fold, formed by the four distinct subdomains: ‘8 kDa’, ‘fingers’, ‘palm’ and ‘thumb’, described for mammalian Pol β (see Fig. 2). However, and in spite of the general conservation of the Pol β core, some members have reduced this core even at a minimum, as it occurs with ASFV Pol X (8). Further divergence of the Pol X family has been achieved by acquisition of additional domains with distinct enzymatic and regulatory activities (see Fig. 8B). Thus, Pol λ, TdT, Pol µ and yeast Pol IV possess a BRCT domain at their N-terminus. This domain, involved in protein–protein interactions, is proposed to confer responsiveness to DNA damage (52). The Pol X enzyme from M.sanguinipes entomopoxvirus has an apurinic-apyrimydinic endonuclease (APE) N-terminally connected to the Pol β core. The role of the Ser-Pro rich domain present in Pol λ and Pol IV is presently unknown. On the other hand, the Pol X enzymes from bacteria and archea have independently fused the nucleotidyltransferase domain (Pol β core) with a phosphoesterase domain (PHP in Fig. 8B), to possess both a hydrolase (nuclease) and a polymerase activity (55).
Interestingly, a few enzymes, including Li Pol β, Cf Pol β and yeast Pol IV have gained specific insertions at the Pol β core, that could serve to mediate specific protein–protein and/or protein–DNA interactions (see Fig. 2B).
Multiple functions of DNA polymerases from family X
Pol β, the paradigm of this family and one of the cellular DNA polymerases for which a role in DNA repair has been proposed (18,56), plays a key role either in short or long patch BER. This enzyme is a small nuclear DNA polymerase well suited for its proposed role, with gap-tailoring (57) and gap-filling enzymatic activities (58,59), and without proof-reading exonuclease activity (60). In spite of its small size, Pol β is a multifunctional enzyme with an 8 kDa N-terminal domain with dRP lyase activity (61) and a C-terminal domain with nucleotidyltransferase activity (57,62).
The same activities, residing in the Pol β-core, are predicted to be present also in Pol λ, although this enzyme has been proposed to be specifically involved in DNA repair processes associated with meiosis and homologous recombination (49). A similar role could be played by the S.cerevisiae Pol IV (47). On the other hand, TdT and the recently described Pol µ (48), both enzymes able to catalyze polymerization in the absence or with little template direction, are either known or proposed to be involved, respectively, in the generation of antigen receptor variability. In the latter four enzymes, the functional specificity likely relies on the protein–protein interactions mediated by the BRCT domain present at their N-terminus (Fig. 8B).
The smallest member of this family, the Pol β-like enzyme from ASFV (Pol X), is a 20 kDa enzyme proposed to be involved in viral DNA repair (8,63). The more complex enzyme of MSEV Pol X supports such a specific role in viral BER by association of the Pol β core with an AP-nuclease at its N-terminus (see Fig. 8B). In the case of bacterial and archea Pol β-like DNA polymerases, no specific role(s) has yet been proposed. The only Pol β activity described so far in Trypanosomatidae was related to the single cell mitochondria that defines the ancient order of kinetoplastidae and, therefore, specifically involved in mitochondrial DNA repair (24).
Li Pol β, a nuclear cell cycle regulated DNA polymerase
In agreement with the structural predictions, Li Pol β, overproduced in E.coli, displayed intrinsic DNA polymerase activity. Moreover, and also in agreement with the predicted Pol β-like structure for Li Pol β, this activity appears to reside in the C-terminal domain, as a fragment of the appropriate size (35 kDa) remains active after endogenous protease cleavage. As it has also been described for Pol β, this 35 kDa domain would correspond to the polymerization domain. In situ analysis of the enzymatic activity showed that polymerization by Li Pol β appears to be strongly activated by Mn2+ as metal binding ions, in comparison with the faint activation observed with Mg2+ ions. This striking preference, considering that Mg2+ is probably the divalent metal ion utilized by most polymerases for catalysis in vivo (64), must be confirmed by further enzymatic analysis of the purified protein. In this sense, it has been suggested that Mn2+ displays a mutagenic effect on polymerases promoting greater reactivity than Mg2+ at the catalytic site, allowing the nucleotidyl transfer reaction to occur with little or no regard to the template instructions (8). Therefore, it would be interesting to analyze if the Mn2+ preference displayed by Li Pol β could be related with some capacity to carry out error-prone DNA synthesis, as it has been also described for mammalian Pol β (60). Such a feature would be advantageous for L.infantum, a parasite protozoon with the ability to overcome different environmental variations and to survive in adverse conditions either natural or pharmacologically induced.
As shown here, Li Pol β is located at the nucleus of the parasite, as with mammalian Pol β, but in a clear contrast with the mitochondrial localization described for the Pol β from C.fasciculata (24). This different localization is worth noting as: (i) both Leishmania and Crithidia belong to the same group of Trypanosomatidae; (ii) it occurs irrespective of the fact that both enzymes share an N-terminal mitochondrial import tag sequence (51); (iii) both proteins have 91% amino acid identity (see Fig. 1); (iv) expression of the Crithidia Pol β protein displays a similar regulation pattern along the cell cycle (24). Therefore, it is tempting to speculate that these two closely related enzymes play a similar functional role, but targeted to different subcellular compartments. This hypothesis further stresses the notion that the evolution of the Pol β superfamily has involved rapid divergence accompanying the adaptation of distinct families to specific roles (55).
Whereas mammalian Pol β is housekeeping, in agreement with its general role in nuclear DNA repair, the Li POL β gene is cell cycle regulated, being both its mRNA and protein levels in direct correlation with DNA synthesis. It is worth noting that the maximal enzyme levels are reached during the highly infectious stationary phase. This could be related to the need for expression of new molecules involved in the developmental process of metacyclogenesis undergone by the protozoon to become infective before entering the host macrophage (6). Furthermore, preliminary results by relative quantitative PCR from our laboratory indicate that Li Pol β mRNA dramatically increases at the intracellular form of the parasite, amastigote. The nuclear location observed by immunofluorescence, with two strips along the longitudinal axis of the protozoon, implies the need for additional experiments along the metacyclogenesis process to find out a functional explanation. Meanwhile, a suitable explanation is that Li Pol β may be necessary to repair damaged DNA molecules by the macrophage oxygen burst through the BER mechanism. Alternatively Li Pol β may be used to modify newly synthesized DNA molecules that would be located at the nucleus periphery, in a way similar to that proposed for the kinetoplast DNA replicated minicircles (65).
Irrespective of different roles that could be assigned to Li Pol β by similitude with other Pol β enzymes, its role as BER operator to repair the damaged DNA under the host macrophage stress environment seems to be likely the first functional option. The cloning and characterization of Li Pol β, as well as its obtention as a recombinant active enzyme, are the first steps for a more detailed biochemical characterization, including in vitro analysis of its insertion fidelity. These studies, together with the development of a model of Li Pol β deficiency, will serve to determine the relevance of this enzyme for the development and infectivity of L.infantum.
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. A. Ollacarizqueta for her technical assistance. S.T. thanks FIS for a fellowship. M.J.R. thanks the Spanish Research Council for a fellowship. T.H. thanks the Spanish National Plan for R+D from the Spanish Ministry for Science and Technology for a fellowship. This work was supported by a grant 97/0492 from the Fondo de Investigación Sanitaria of the Spanish Health Ministry, a grant BIO1999-0853 from the Ministry of Science and Tecnology and partially by grants 08.20037/1999 and 08.1/0044.1/1998 from the CAM.
DDBJ/EMBL/GenBank accession no. AF182167
References
- 1.Jimenez M.I., Gutierrez-Solar,B., Benito,A., Aguiar,A., García,E., Cercenado,E. and Alvar,J. (1991) Cutaneous Leishmania infantum zymodemes isolated from bone marrow in AIDS patients. Res. Rev. Parasitol., 51, 91–95. [Google Scholar]
- 2.World Health Organization (1993) UNDP/World Bank/WHO8, Leishmaniasis, special program for research and training in tropical disease. Tropical Disease Research: Progress 1991–1992. Eleventh Program report, pp. 77–87.
- 3.Sogin M.L., Elwood,H.J. and Gunderson,J.H. (1986) Evolutionary diversity of eukaryotic small-subunit rRNA genes. Proc. Natl Acad. Sci. USA, 83, 1383–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donelson J.E., Gardner,M.J. and El-Sayed,N.M. (1999) More surprises from Kinetoplastida. Proc. Natl Acad. Sci. USA, 96, 2579–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solari A.J. (1995) Mitosis and genome partition in trypanosomes. Biocell, 19, 65–84. [PubMed] [Google Scholar]
- 6.Sacks D.L. and Perkins,P.V. (1984) Identification of an infective stage of Leishmania promastigotes. Science, 223, 1417–1419. [DOI] [PubMed] [Google Scholar]
- 7.Da Silva R. and Sacks,D.L. (1987) Metacyclogenesis is a major determinant of Leishmania promastigote virulence and attenuation. Infect. Immun., 55, 2802–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steitz T.A. (1998) A mechanism for all polymerases. Nature, 391, 231–232. [DOI] [PubMed] [Google Scholar]
- 9.Oliveros M., Yañez,R.J., Salas,M.L., Salas,J., Viñuela,E. and Blanco,L. (1997) Characterization of an African swine fever virus 20-kDa DNA polymerase involved in DNA repair. J. Biol. Chem., 272, 30899–30910. [DOI] [PubMed] [Google Scholar]
- 10.Ollis D.L., Brick,R., Hamlin,R., Xuong,N.G. and Steitz,T.A. (1985) Structure of large fragment of E.coli DNA polymerase I complexed with dTMP. Nature, 313, 762–766. [DOI] [PubMed] [Google Scholar]
- 11.Beese L.S., Derbyshire,V. and Steitz,T.A. (1993) Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science, 260, 352–355. [DOI] [PubMed] [Google Scholar]
- 12.Sousa R., Chung,Y.J., Rose,J.P. and Wang,B.C. (1993) Crystal structure of bacteriophage T7 RNA polymerase at 3.3 Å resolution. Nature, 364, 593–595. [DOI] [PubMed] [Google Scholar]
- 13.Jacobo-Molina A., Ding,J., Nanni,R., Clark,A.D., Lu,X., Tantillo,C., Williams,R.L., Kamer,G., Ferris,A.L., Clark,P. et al. (1993) Crystal structure of human immunodeficiency virus type I reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl Acad. Sci. USA, 90, 6320–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgiadis M.M., Jensen,S.M., Ogata,C.M., Telenitsky,A., Goff,S.P. and Hendrikson,W.A. (1995) Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase. Structure, 3, 879–892. [DOI] [PubMed] [Google Scholar]
- 15.Pelletier H., Sawaya,M.R., Kumar,A., Wilson,S.H. and Krant,J. (1994) Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer and ddCTP. Science, 264, 1891–1903. [PubMed] [Google Scholar]
- 16.Kim Y., Eom,S.H., Wang,J., Lee,D.S., Suh,S.W. and Steitz,T.A.(1995) Crystal structure of Thermus aquaticus DNA polymerase. Nature, 376, 612–616. [DOI] [PubMed] [Google Scholar]
- 17.Kiefer J.R., Mao,C., Hansen,C.J., Rasehore,S.L., Hogrefe,H.H., Braman,J.C. and Beese,L.S. (1997) Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at a 2.1 Å resolution. Structure, 5, 97–108. [DOI] [PubMed] [Google Scholar]
- 18.Wilson S.H. (1998) Mammalian base excision repair and DNA polymerase beta. Mutat. Res., 407, 203–215. [DOI] [PubMed] [Google Scholar]
- 19.Piersen C.E., Prasad,R., Wilson,S.H. and Lloyd,R.S. (1996) Evidence for an Imino Intermediate in the DNA polymerase β deoxyribose phosphate excision reaction. J. Biol. Chem., 271, 17811–17815. [DOI] [PubMed] [Google Scholar]
- 20.Kungland A. and Lindahl,T. (1997) Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J., 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim K., Biade,S. and Matsumoto,Y. (1998) Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem., 273, 8842–8848. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto Y., Kim,K. and Bogenhagen,D.F. (1994) Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: an alternative pathway of base excision DNA repair. Mol. Cell. Biol., 14, 6187–6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frosina G., Fortini,P., Rossi,O., Carrozino,F., Raspaglio,G., Cox,L.S., Lane,D.P., Abbondandolo,A. and Dogliotti,E. (1996) Two pathways for base excision repair in mammalian cells. J. Biol. Chem., 271, 9573–9578. [DOI] [PubMed] [Google Scholar]
- 24.Johnson C.E. and Englund,P.T. (1998) Changes in organization of C.fasciculata kynetoplast DNA replication proteins during the cell cycle. J. Cell. Biol., 143, 911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehlotra R.K. (1996) Antioxidant defense mechanisms in parasitic protozoa. Crit. Rev. Microbiol., 22, 295–314. [DOI] [PubMed] [Google Scholar]
- 26.Faez-Vidal M.E., Gallego,C., Ruiz-Perez,L.M. and Gonzalez-Pacanowska,D. (2000) Characterization of Uracil-DNA glycosylase activity from Trypanosoma cruzi and its stimulation by A/P endonuclease. Nucleic Acids Res., 29, 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camacho A., Hidalgo-Zarco,F., Bernier-Villamor,V., Ruiz-Perez,L.M. and Gonzalez-Pacanoswka,D. (2000) Properties of Leishmania major dUTP nucleotidehydrolase, a distinct nucleotide-hydrolizing enzyme in kinetoplastids. Biochem. J., 346, 163–168. [PMC free article] [PubMed] [Google Scholar]
- 28.Perez J., Gallego,C., Bernier-Villamor,V., Camacho,A., Ruiz-Perez,L.M. and Gonzalez-Pacanowska,D. (1999) Apurinic/apirimidinic endonuclease genes from trypanosomatidae Leishmania major and Trypanosoma cruzi confer resistance to oxidizing agents in DNA repair deficient E.coli. Nucleic Acids Res., 27, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawaya M.R., Pelletier,H., Kumar,A., Wilson,S.H. and Kraut,J. (1994) Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science, 264, 1930–1935. [DOI] [PubMed] [Google Scholar]
- 30.Davies J.F.II, Almassy,R.J., Hostomska,Z., Ferre,R.A. and Hostomsky,Z. (1994) Crystal structure of the catalytic domain of DNA polymerase β. Cell, 76, 1123–133. [DOI] [PubMed] [Google Scholar]
- 31.Zarley J.H., Britigan,B.E. and Wilson,M.E. (1991) Hydrogen peroxide-mediated toxicity for Leishmania donovani chagasi promastigotes. Role of hydroxyl radical and protection by heat shock. J. Clin. Invest., 88, 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasion S.G., Brown,G.W., Brown,L.M. and Ray,D.S. (1994) Periodic expression of nuclear and mitochondrial DNA replication genes during trypanosomatid cell cycle. Cell Sci., 107, 3515–3520. [DOI] [PubMed] [Google Scholar]
- 33.Kroll D.J., Abedel-Malek Abdel-Hafiz,H., Marcell,T., Simpson,S., Chen,C.Y., Gutierrez-Hartman,A., Lustbader,J.W. and Hoeffler,J.P. (1993) A multifunctional prokaryotic protein expression system: overproduction, affinity purification and selective detection. DNA Cell Biol., 12, 441–453. [DOI] [PubMed] [Google Scholar]
- 34.Farabaugh P.J. (1978) Sequence of the lacI gene. Nature, 274, 765–767. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Aseguinolaza G., Almazan,F., Rodriguez,F.J., Marquet,A. and Larraga,V. (1997) Cloning of the gp63 surface protease of Leishmania infantum. Differential post-translational modifications correlated with different infective forms. Biochim. Biophys. Acta, 1361, 92–102. [PubMed] [Google Scholar]
- 37.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 38.Karawya E., Swack,J.A. and Wilson,S. (1983) Improved conditions for activity gel analysis of DNA polymerase catalytic polypeptides. Anal. Biochem., 135, 318–325. [DOI] [PubMed] [Google Scholar]
- 39.Smith R.F., Wiese,B.A., Wojzynski,M.K., Davison,D.B. and Worley,K.C. (1996) BCM search launcher. An integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res., 6, 454–462. [DOI] [PubMed] [Google Scholar]
- 40.Pelletier H., Sawaya,M.R., Wolfe,W., Wilson,S.H. and Kraut,J. (1996) Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity and fidelity. Biochemistry, 35, 12742–12761. [DOI] [PubMed] [Google Scholar]
- 41.Sawaya M.R., Prasad,R., Wilson,S.H., Kraut,J. and Pelletier,H. (1997) Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry, 36, 11205–11215. [DOI] [PubMed] [Google Scholar]
- 42.Weiner S.J., Kollman,P.A., Nguyen,D.T. and Case,D.A. (1996) An all atom force field for simulations of proteins and nucleic acids. J. Comp. Chem., 7, 230–252. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Brittingham A., Morrison,C.J., McMaster,W.R., McGwire,B.S., Chang,K.P. and Mosser,D.M. (1995) Role of the Leishmania surface protease gp63 in complement fixation, cell adhesion and resistance to complement-mediated lysis. J. Immunol ., 155, 3102–3111. [PubMed] [Google Scholar]
- 45.Broccoli S., Marquis,J.F., Papadopoulou,B., Olivier,M. and Drolet,M. (1999) Characterization of a Leishmania donovani gene encoding a protein that closely resembles a type IB topoisomerase. Nucleic Acids Res., 27, 2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Diaz M., Dominguez,O., Lopez-Fernandez,L.A., de Lera,L.T., Saniger,M.L., Ruiz,F.J., Parraga,M., Garcia-Ortiz,M.J., Kirchhoff,T., del Mazo,J. et al. (2000) DNA polymerase lambda (Pol λ), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol., 301, 851–867. [DOI] [PubMed] [Google Scholar]
- 47.Kowai O., Yokota,T., Kageyama,T., Hirose,T., Yoshida,S. and Arai,K. (1986) Isolation and characterization of bovine and mouse terminal deoxynucleotidyltransferase cDNAs expressible in mammalian cells. Nucleic Acids Res., 14, 5777–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez O., Ruiz,F.J., Lain de Lera,M.T., García-Díaz,L.M., Gonzalez,M.A., Kirchhoff,T., Martinez-A,C., Bernard,A. and Blanco,L. (2000) DNA polymerase mu (Pol µ), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J., 19, 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliver S.G., van der Aart,Q.J., Agosttini-Carbone,M.L., Aigle,M., Alberghina,L., Alexandraki,D., Antoine,G., Anwar,R., Ballesta,J.P. and Benit,P. (1992) The complete DNA sequence of yeast chromosome III. Nature, 357, 38–46. [DOI] [PubMed] [Google Scholar]
- 50.Ito J. and Braithwaite,D.K. (1991) Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res., 19, 4045–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torri A.F. and Englund,P.T. (1995) A DNA polymerase beta in the mitochondrion of the trypanosomatid Crithidia fasciculata. J. Biol. Chem., 270, 3495–3497. [DOI] [PubMed] [Google Scholar]
- 52.Bork P., Hofman,K., Bucher,P., Neuwald,A.F., Altschul,S.F. and Koonin,E.V. (1997) A super-family of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J., 11, 68–76. [PubMed] [Google Scholar]
- 53.Blank A., Silber,J.R., Thelen,M.P. and Deker,C.A. (1983) Detection of enzymatic activities in sodium dodecyl sulfate-polyacrylamide gels: DNA polymerases as model enzymes. Anal. Biochem., 135, 423–430. [DOI] [PubMed] [Google Scholar]
- 54.Quijada L., Soto,M., Alonso,C. and Requena,J.M. (1997) Analysis of post-transcriptional regulation operating on transcription products of the tandemly linked Leishmania infantum hsp70 genes. J. Biol. Chem., 272, 4493–4499. [DOI] [PubMed] [Google Scholar]
- 55.Aravind L. and Koonin,E.V. (1999) DNA polymerase β-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res., 27, 4654–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dianov G.L., Prasad,R., Wilson,S.H. and Bohr,V.A. (1999) Role of DNA polymerase beta in the excision step of long patch mammalian base excision repair. J. Biol. Chem., 274, 13741–13743. [DOI] [PubMed] [Google Scholar]
- 57.Matsumoto Y. and Kim,K. (1995) Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science, 269, 699–702. [DOI] [PubMed] [Google Scholar]
- 58.Prasad R., Singhal,R.K., Srivastrava,D.K., Molina,J.T., Tomkinson,A.E. and Wilson,S.H. (1996) Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem., 271, 16000–16007. [DOI] [PubMed] [Google Scholar]
- 59.Dimitriadis E.K., Prasad,R., Vaske,M.K., Chen,L., Tomkinson,A.E., Lewis,M.S. and Wilson,S.H. (1998) Thermodynamics of human DNA ligase I trimerization and association with DNA polymerase beta. J. Biol. Chem., 273, 20540–20550. [DOI] [PubMed] [Google Scholar]
- 60.Osheroff W.P., Jung,H.K., Beard,W.A., Wilson,S. and Kunkel,T. (1999) The fidelity of DNA polymerase beta during distributive and processive DNA synthesis. J. Biol. Chem., 274, 3642–3650. [DOI] [PubMed] [Google Scholar]
- 61.Deterding L.J., Prasad,R., Mullen,G.P., Wilson,S.H. and Tomer,K.B. (2000) Mapping of the 5′-2-deoxyribose-5-phosphate lyase active site in DNA polymerase beta by mass spectrometry. J. Biol. Chem., 275, 10463–10471. [DOI] [PubMed] [Google Scholar]
- 62.Casas-Finet J.R., Kumar,A., Morris,G., Wilson,S.H. and Karpel,R.L. (1991) Spectroscopic studies of the structural domains of mammalian DNA beta-polymerase. J. Biol. Chem., 266, 19618–19625. [PubMed] [Google Scholar]
- 63.Yañez R.J., Rodriguez,J.M., Nogal,M.L., Yuste,L., Enriquez,C., Rodriguez,J.F. and Viñuela,E. (1995) Analysis of the complete nucleotide sequence of African Swine Fever virus. Virology, 208, 249–278. [DOI] [PubMed] [Google Scholar]
- 64.Kornberg A. and Baker,T.A. (1992) DNA Replication, 2nd edn. Freeman and Co., New York, NY, pp. 165–194.
- 65.Li C. and Englund,P.T. (1997) A mitochondrial DNA primase from the trypanosomatd Crithidia fasciculata. J. Biol. Chem., 272, 20787–20792. [DOI] [PubMed] [Google Scholar]
- 66.Matsumoto Y., Kim,K., Katz,D.S. and Feng,J.A. (1998) Catalytic center of DNA polymerase beta for excision of deoxyribose phosphate groups. Biochemistry, 37, 864–869. [DOI] [PubMed] [Google Scholar]
- 67.Prasad R., Beard,W.A., Chyan,J.Y., Maciejewsky,M.W., Mullen,G.P. and Wilson,S.H. (1998) Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase beta as revealed by site-directed mutagenesis. DNA binding and 5′-deoxyribose phosphate lyase activities. J. Biol. Chem., 273, 11121–11126. [DOI] [PubMed] [Google Scholar]
- 68.Prasad R., Beard,W.A., Strauss,P.R. and Wilson,S.H. (1998) Human DNA polymerase β deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J. Biol. Chem., 273, 15263–15270. [DOI] [PubMed] [Google Scholar]