Abstract
The discovery that the actinobacterium Micromonospora inhabits nitrogen-fixing nodules raised questions as to its potential ecological role. The capacity of two Micromonospora strains to infect legumes other than their original host, Lupinus angustifolius, was investigated using Medicago and Trifolium as test plants. Compatible rhizobial strains were used for coinoculation of the plants because Micromonospora itself does not induce nodulation. Over 50% of nodules from each legume housed Micromonospora, and using 16S rRNA gene sequence identification, we verified that the reisolated strains corresponded to the microorganisms inoculated. Entry of the bacteria and colonization of the plant hosts were monitored using a GFP-tagged Lupac 08 mutant together with rhizobia, and by using immunogold labeling. Strain Lupac 08 was localized in plant tissues, confirming its capacity to enter and colonize all hosts. Based on studying three different plants, our results support a non-specific relationship between Micromonospora and legumes. Micromonospora Lupac 08, originally isolated from Lupinus re-enters root tissue, but only when coinoculated with the corresponding rhizobia. The ability of Micromonospora to infect and colonize different legume species and function as a potential plant-growth promoting bacterium is relevant because this microbe enhances the symbiosis without interfering with the host and its nodulating and nitrogen-fixing microbes.
Introduction
Nitrogen-fixing nodules are unique structures, which are formed on the roots of legume and actinorhizal plants to establish a nitrogen-fixing symbiosis with either rhizobia or Frankia. One striking feature of the legume–rhizobial symbiosis is its high level of specificity in that a rhizobial strain nodulates and fixes nitrogen with usually only a limited number of host plant species. This specificity is determined by several stages of chemical signaling between the symbiotic partners1.
Nodular tissues and their carbohydrate supplies are an excellent habitat for bacteria, not only for nitrogen-fixing rhizobia, but also for many other microbes2. Numerous reports about the presence of non-rhizobial microorganisms associated with nitrogen-fixing nodules have been published. Muresu and co-workers3 described the isolation of at least 12 different taxa from surface-sterilized nodules of several wild legumes. The bacterial genera identified included Bacillus, Pseudomonas, Rhizobium, Xanthomonas, and members of the family Enterobacteriaceae. Many members of the phylum Actinobacteria have also been isolated from legume nodule tissues including Agromyces and Microbacterium spp3–5., Curtobacterium 6, and Micromonospora 7–12.
Micromonospora is a Gram-positive bacterium characterized by filamentous growth and spore production on substrate mycelium. It is aerobic and many strains produce carotenoid pigments. Micromonosporae have been mainly reported from soil and aquatic environments where they are thought to be involved in organic matter turnover, especially of cellulose13, 14. In 2007, we reported the first isolation of Micromonospora strains from nitrogen-fixing nodules of the wild legume Lupinus angustifolius 8. Since then, our research groups have focused on the ecology of Micromonospora and its interactions with plants, and we have documented the distribution of this bacterium in a wide range of legumes and actinorhizal plants10, 12, 15. Other Micromonospora strains isolated from other sources such as calcareous soil, have also been reported to promote the growth of Phaseolus vulgaris by solubilizing phosphate16.
Most studies of beneficial plant-microbe interactions focus on a single plant-microbe partnership at a time. However, simultaneous infection with rhizobia and a number of other bacteria also present in nodules enhances nodulation and plant growth in a wide variety of legumes. Current data suggest that although Micromonospora species do not induce nodules or fix nitrogen in association with a host plant, they provide many benefits to the plant by increasing the number of nodules, enhancing aerial growth and nutrient uptake17–19. Studying ways to augment plant productivity through the use of beneficial microbes will increase our knowledge of plant-microbe interactions, which has deep implications for agriculture and biotechnology.
Plant growth is promoted by various mechanisms, including improved access to and uptake of minerals and nutrients, amelioration of soil toxicity, release of growth-stimulating phytohormones as well as modulation of plant hormone production, acquisition of nitrogen and phosphate via symbioses, and/or enhancement of the effects of symbioses20. Studies based on Micromonospora strains isolated from alfalfa nodules suggest that the actinobacteria contribute to the nutritional efficiency of this legume17, and our experimental data showed that Micromonospora lupini Lupac 08 is a plant growth-promoting bacterium (PGPB). The localization of several genes in the genome that are involved in plant growth promotion, such as for the production of siderophores, phytohormones, the degradation of chitin (for biocontrol), and the biosynthesis of trehalose all contribute to the welfare of the host plant18.
Until now, most inoculation experiments to analyze the effect of Micromonospora on a host plant and its interaction with rhizobia have been carried out using the same plant species from which the strains originated. However, no information is available as to whether any specificity exists in the Micromonospora-legume interaction. Thus, the present study was designed to test the capacity of M. lupini Lupac 08 and M. saelicesensis Lupac 09T (both isolated from Lupinus angustifolius) to enter Medicago and Trifolium nodules and to obtain information about the location of Micromonospora with nodule tissues. Host-associated rhizobial strains were used for coinoculation of Micromonospora onto legume plants to induce nodulation and facilitate the entry of this microbe into plant tissues. Micromonospora strains, which were identified by 16 S rRNA gene sequencing, were re-isolated from the resulting nodules. Finally, Micromonospora localization within the plant tissues was performed using green fluorescent protein and immunogold labeling to localize strain Lupac 08 within the plant cells. The results indicate that Micromonospora has the capacity to enter and colonize additional legumes beyond the legume host from which it was isolated originally, and strongly suggest that this non-specific, but beneficial PGPB can be used to enhance productivity of a wide range of nodulating plants.
Results
In a previous study, we showed that Micromonospora strains isolated from nitrogen-fixing nodules contribute to plant development and health by acting as PGPB18. In the present work, our aim was to determine if Micromonospora when grown in vitro could re-colonize the plant from which it was isolated and also whether this process is host-specific. In addition, we searched for information about the entry and localization of Micromonospora within the nodules. To this end, three different plants, L. albus, M. sativa, and T. repens, were grown axenically and co-inoculated not only with the corresponding symbiont to induce nodulation, but also with Micromonospora lupini Lupac 08 or M. saelicesensis Lupac 09T, which were originally isolated from L. angustifolius nodules. Alfalfa and clover nodules are of the indeterminate type, as are lupine nodules. However, the latter develops several lateral meristems, which continue to undergo cell divisions and eventually form lobes that wrap around the parent root, giving the nodules a “collared” appearance21.
Recovery of M. lupini Lupac 08 and M. saelicesensis Lupac 09T from Lupinus, Medicago, and Trifolium nodules
The number of Micromonospora colonies recovered from a single nodule of the inoculated plants varied, ranging from zero to more than 300. However, on average, most of the nodules yielded between 1–4 CFUs, and about 60% of the 60 nodules screened contained Micromonospora. Regarding the success of infection, the most colonies were isolated from lupine nodules, with a slightly lower number of CFUs from the clover and alfalfa nodules. PCR with 16 S rRNA primers confirmed that the isolates were M. lupini Lupac 08 and M. saelicesensis Lupac 09T. Overall, strain Lupac 09T was more effective, with 77% of the nodules screened harboring this Micromonospora strain.
Localization of Micromonospora in lupine
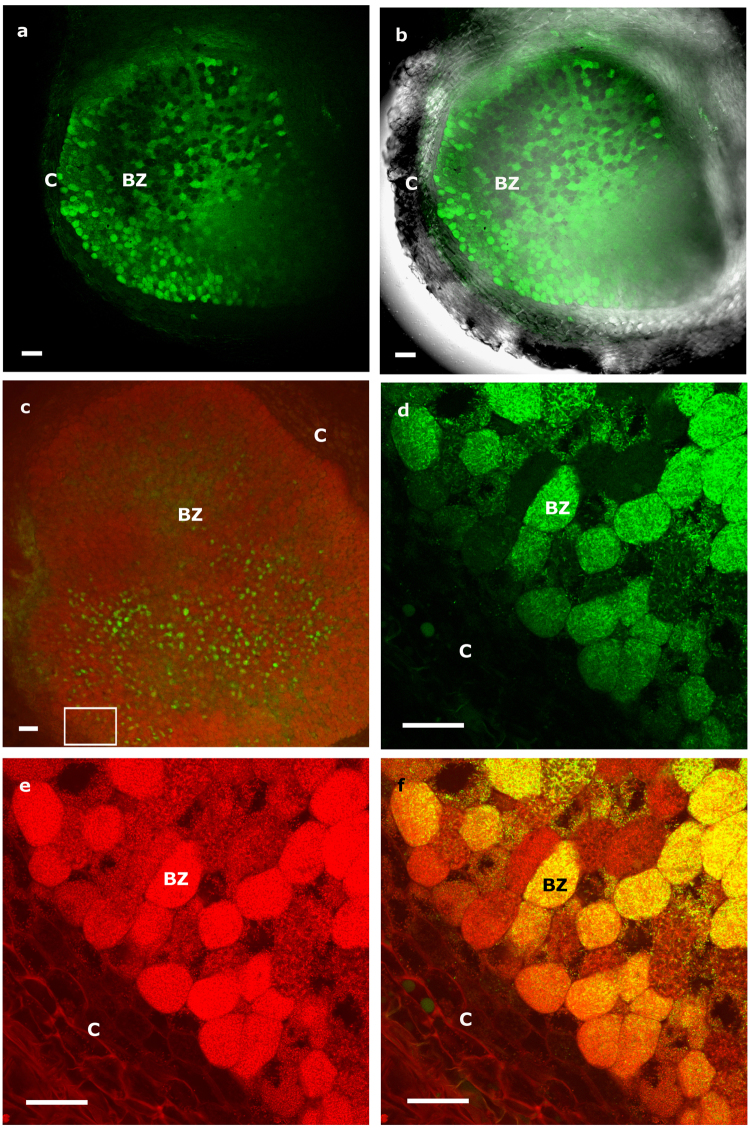
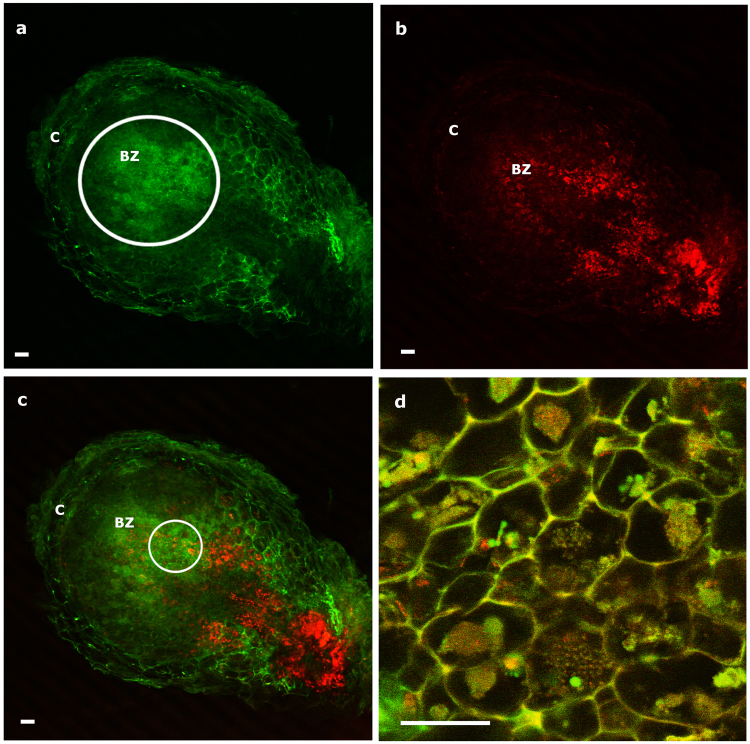
The nodules produced by all coinoculated Lupinus plants were pink in color, indicating effective nitrogen fixation. Several root nodules (~35–40 dpi) were randomly selected and longitudinally sectioned to localize M. lupini ML01-gfp cells by CLSM. The bacteria were successfully localized in several zones of the nodule, but they were especially prevalent in the infection and bacteroid zones (BZ; terminology of ref. 22) (Fig. 1a,b). Although González-Sama et al.23 reported that uninfected cells were not present in the central BZ of L. albus nodules, Micromonospora cells were observed in those host cells that appeared devoid of Bradyrhizobium. This was more obvious in longitudinal nodule sections that were counterstained with propidium iodide to differentiate Bradyrhizobium sp. CAR08 from the gfp-tagged Micromonospora (Fig. 1c–f). As expected, the bradyrhizobia occupied the majority of the cells within the nodule tissue, and were clearly seen in the infection and bacteroid zones of the nodule, whereas Micromonospora cells were observed in fewer host cells, which were interspersed among the Bradyrhizobium-infected cells (Fig. 1c). The presence of both bacteria in the same cell was detected as yellow fluorescence (Fig. 1f) due to the coincidence of the green and red fluorescence (Fig. 1d,e).
Figure 1.
Longitudinal nodule sections of Lupinus albus coinoculated with Bradyrhizobium sp. CAR08 and Micromonospora ML01-gfp (21 dpi). (a) Green fluorescence signal captured by CLSM of infected cells containing Micromonospora ML01-gfp. (b) Overlay of light and fluorescence images of the nodule section. (c) Green fluorescence localization of ML01-gfp in a nodule section stained with propidium iodide and viewed by CLSM. (d) Higher magnification image captured with the green channel. (e) Higher magnification image captured with the red channel. (f) Composite image of both channels. The white rectangle in image c shows the area where images d-f were captured. C, cortex; BZ, bacteroid zone; dpi, days post inoculation. Bars: 100 µm (a, b, c); 40 µm (d, e, f).
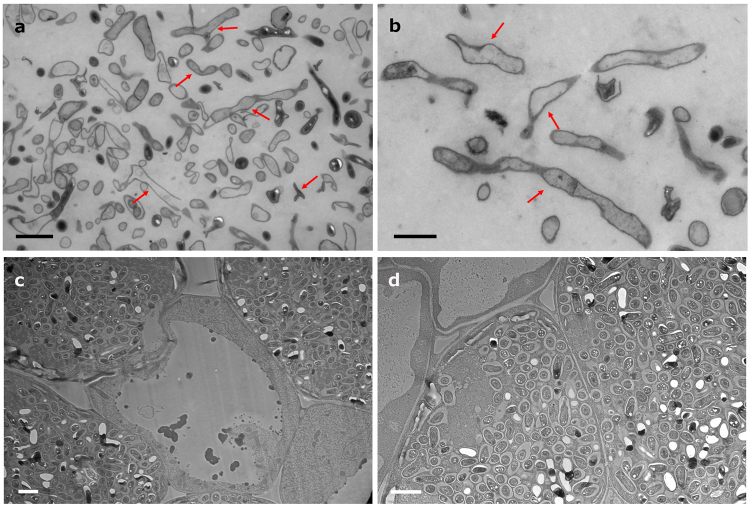
Immunogold microscopy was used to confirm the nodule occupancy of the Micromonospora cells. Pure cultures of the bacteria were observed by TEM (Jeol 1010, Japan) for comparison purposes. As expected, Micromonospora cells were seen as branched filaments or rod-shaped structures that corresponded to longitudinal and transverse sections, respectively (Fig. 2a,b). TEM preparations of nodules inoculated with Bradyrhizobium sp. CAR08 only were sectioned and served as controls. Figures 2c,d illustrate infected cells containing bacteroids within their symbiosomes as well as uninfected plant cells.
Figure 2.
Transmission electron micrographs of Micromonospora pure cultures and nodular tissue infected with Bradyrhizobium. (a,b) Micromonospora ML01-gfp pure cultures (arrows, polymorphic Micromonospora cells). (c,d) Lupine nodule tissue infected with Bradyrhizobium sp. CAR08 only. Bar: 2 µm (a, c, d); 1 µm (b).
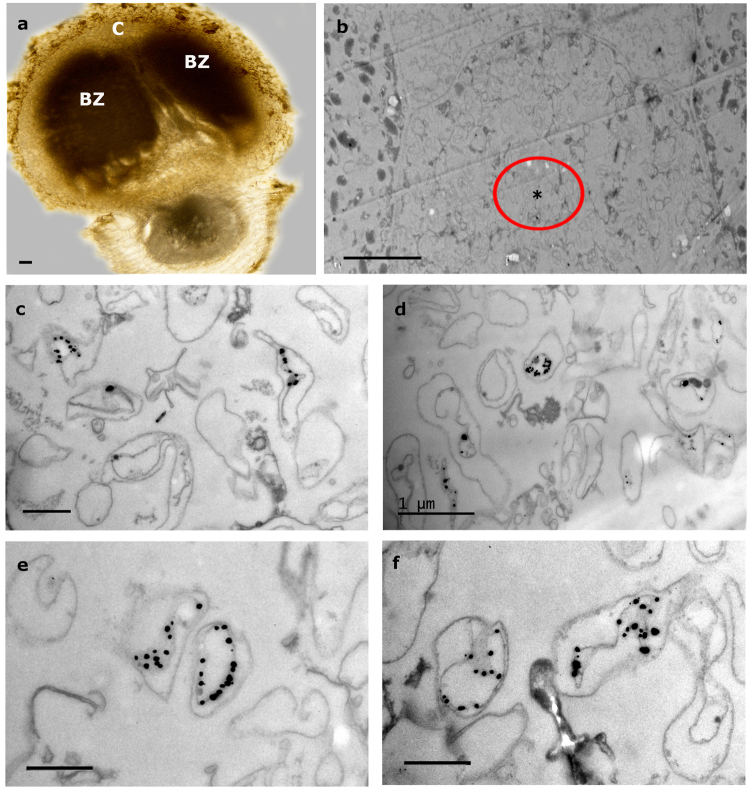
Nodule sections obtained from plants that were coinoculated exhibited a structure similar to the control nodules with zones corresponding to the nodule cortex (C) and the infection and bacteroid zones (BZ) (Fig. 3a). Within these zones, bacteria-containing plant cells were seen in addition to what appeared to be uninfected plant cells (Fig. 3b). Micromonospora hyphae were usually found in the latter cells, which at low magnification gave the impression of being “empty”. Figure 3b shows a Micromonospora-containing nodule cell, which is flanked by two host cells containing Bradyrhizobium bacteroids; the areas where the gfp-ML01 strain was found are marked with an asterisk enclosed by a circle. Within these cells, immunogold-labeled structures that resembled the cells of Micromonospora were observed (Fig. 3c–f). These structures were similar to those found in the pure culture preparations (Fig. 2a,b), but were not detected in the nodules inoculated with Bradyrhizobium only. Unlike many rhizobia that undergo physical changes such as a marked increase in cell size and morphological transformation from rod-shaped to branched cells24, the Micromonospora cells did not show any drastic morphological changes and many labeled cells resembled those observed in pure culture. In some cases, both types of bacteria were seen within the same plant cell (Fig. 1), but Micromonospora was always found in lower numbers compared to the Bradyrhizobium bacteroids.
Figure 3.
Immunoelectron microscopic images of lupine nodules infected with Bradyrhizobium CAR08 and Micromonospora ML01-gfp (21 dpi). (a) Light micrograph of a longitudinal nodule section. (b) Detail of an “empty” cell between two infected cells that contain bacteroids. (c–f) Labeled Micromonospora cells found in the area marked with an encircled asterisk in 3b. Bars: (a) 100 µm; (b) 10 µm; (d) 1 µm; 500 nm (c, e, f). C, cortex; BZ, bacteroid zone; dpi, days post inoculation.
Effect of Micromonospora on the root hairs of Medicago and Trifolium
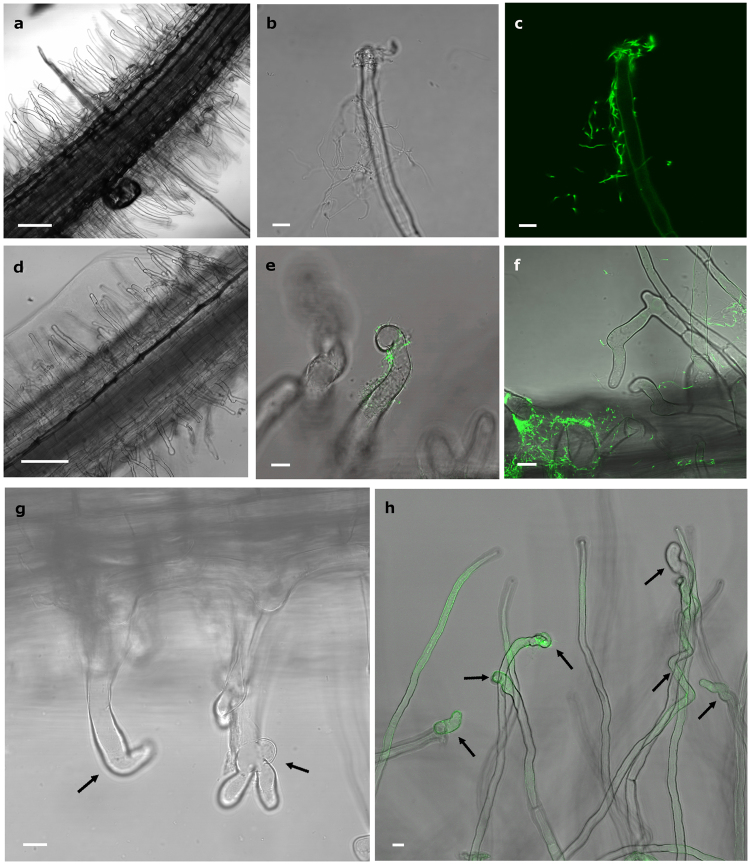
The ability of Micromonospora lupini Lupac 08 to infect legumes other than L. albus was investigated. Medicago and Trifolium plants inoculated with Micromonospora gfp-labeled ML01 were observed under light and confocal microscopy. Root hair deformations were observed as early as 2–3 days after inoculation. In both plants, Micromonospora was observed attached to the root surfaces (Fig. 4b,c,e,f). Root hairs deformed branching into Y and L shapes (Fig. 4g), developing zig-zag forms and/or swollen root hair tips (Fig. 4h). Root hairs of uninoculated Trifolium (Fig. 4a) and Medicago (Fig. 4d) control plants exhibited no deformation.
Figure 4.
Effect of Micromonospora on the root hairs of Trifolium and Medicago. (a,d) Control uninoculated plants of Trifolium and Medicago respectively. (b,c) Light and CLMS micrographs of Micromonospora cells attached to a Trifolium hair root 3 dpi. (e,f) CLMS images of Micromonospora attached to Medicago root hairs. (g) Medicago and (h) Trifolium root hairs showing different deformations (arrows) 5 dpi with Micromonospora. Bars: (a) 100 µm; (b, c, h) 8 µm; (d) 200 µm; (e, f, g) 10 µm; dpi, days post infection.
Infection of Medicago and Trifolium root nodules by Micromonospora
To determine the capacity of Micromonospora to penetrate and infect nodular internal tissues, Medicago and Trifolium plants were co-inoculated with the appropriate mCherry-tagged nitrogen-fixing rhizobia (Sinorhizobium Rm1021-mCh and Rhizobium sp. E11-mCh, respectively) as well as Micromonospora ML01-gfp. The plants were monitored by light and CLSM microscopy, every two days until approximately 25 dpi. Root tip deformations appeared 2 days after bacterial inoculation in Trifolium (Fig. 5a,b) whereas the changes in root hair deformation did not occur in Medicago until 6 dpi (Fig. 5e,f). In both sets of plants, Micromonospora surrounded the youngest regions of the root. Although green autofluorescence was detected within Medicago tissues once the nodules formed, Micromonospora cells were also clearly attached to the root hairs (Fig. 5e). Differences in root hair morphology were observed between the two plant species; most Trifolium root hairs were branched or club-shaped 2–4 dpi (Fig. 5a,b) whereas Medicago root hairs became spiral in shape, but not until 12 dpi (Fig. 5f).
Figure 5.
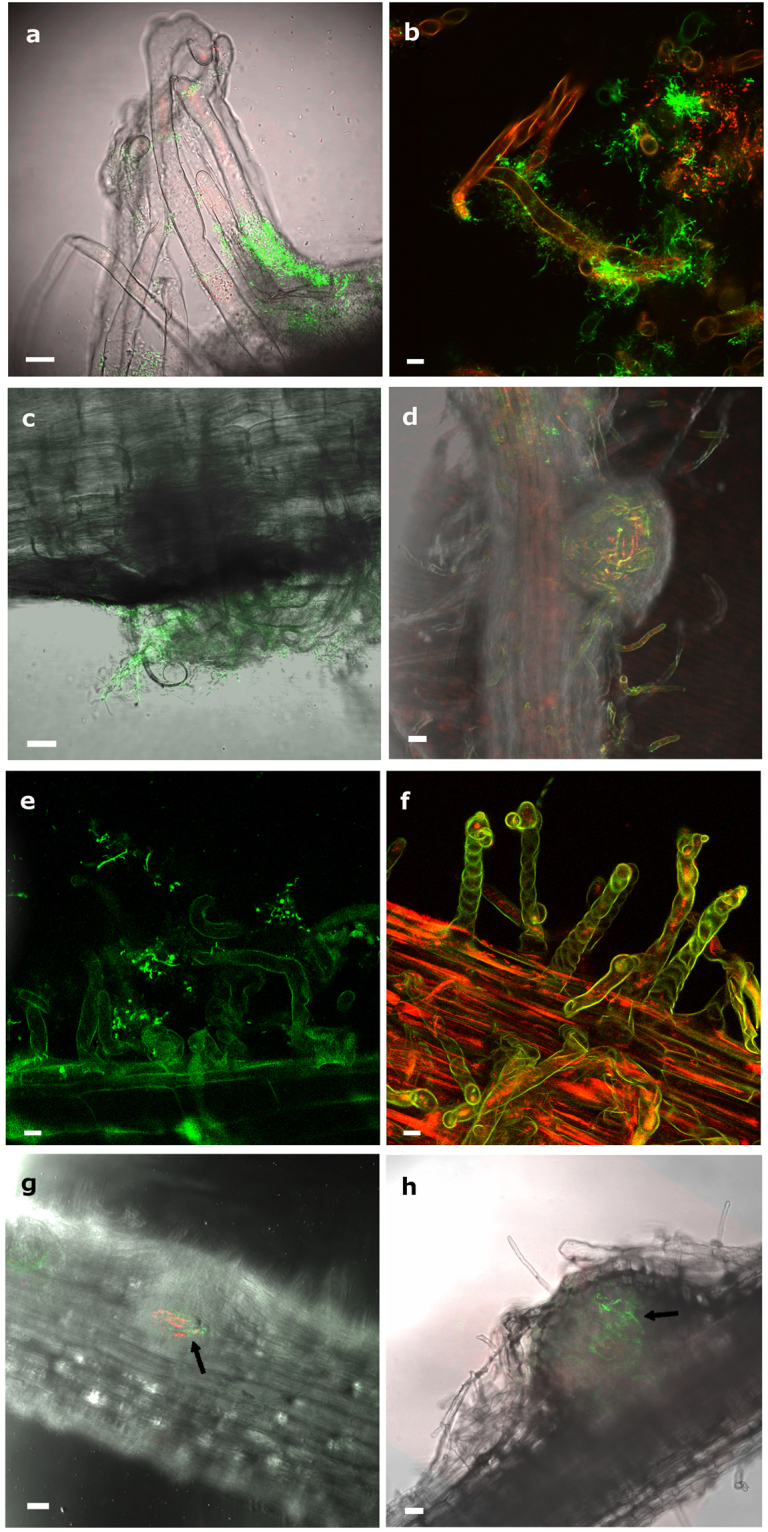
Infection and colonization of Trifolium and Medicago by Micromonospora ML01-gfp co-inoculated with strains Rhizobium sp. E11-mCh and Sinorhizobium sp. Rm1021-mCh respectively, and observed by CLSM. (a) Trifolium root tip deformations observed 3 dpi and surrounded by Micromonospora ML01-gfp. (b) Micromonospora and Rhizobium sp. co-localized on the root hairs. (c) Nodule primordium and deformed root hairs observed in Trifolium 5 dpi. (d) Young Trifolium nodule observed 11 dpi. The fluorescent bacteria are visible within the internal tissues of the nodule. (e) Attachment of Micromonospora to Medicago root tips showing deformations 6 dpi. (f) Medicago root hair tips forming spiral shapes 12 dpi. (g,h) Medicago nodules at 11 and 13 dpi with green and red fluorescence signals showing strings of bacteria. Because of the thickness of the tissue, the nodules themselves are slightly out of focus. For details see text. Bars: 8 µm (a, b); 10 µm (c, e, f); 75 µm (g, h). dpi, days post infection.
Nodule primordia were visible 3–5 dpi in Trifolium and after 7–9 days in Medicago. In both plants, the nodule primordia were covered with deformed root hairs and Micromonospora cells were attached to the hairs (Fig. 5c). Micromonospora ML01-gfp and Rhizobium E11-mCh were readily visible inside Trifolium young nodules 11 dpi, and both bacteria were co-localized as indicated by the yellow fluorescence, supporting the conclusion that both microorganisms were present (Fig. 5d). Comparably aged Medicago nodules were thicker, however, and the green and red fluorescence corresponding to Micromonospora ML01-gfp and Sinorhizobium SM1021-mCh, respectively, was detected in the intact nodules (Fig. 5g,h).
Nodules were well developed in both plants after 15–20 days and fresh samples had a pink color indicating that nitrogen fixation was taking place. Mature nodules (~20 dpi) of both plant species exhibited the typical indeterminate structure: meristematic, infection, bacteroid, and senescent zones were identified.
Longitudinal sections of 20-day old Medicago and Trifolium nodules were obtained for localizing Micromonospora by CLSM. In Trifolium, a green fluorescence signal (excitation 488-nm and 515- to 560-nm emission) was observed in the infection zone just below the meristematic area. Fluorescence expressed by Micromonospora ML01-gfp was clearly observed within the plant cells whereas the uninfected cells showed no fluorescence apart from the autofluoresence emitted by the plant (Fig. 6a). Rhizobium sp. E11-mCh cells exhibited a bright red fluorescent signal (620 nm excitation and 620–660 nm emission) in the infected, bacteroid, and senescent zones of the nodule (Fig. 6b). Co-localization of both bacteria is shown in Fig. 6c. For the most cases, the microbes were found in the infection zone, whereas in other instances, Rhizobium E11-mCh was the only occupant, especially in the senescent zone. Both bacteria were also located within the same host cell as indicated by yellow fluorescence (Fig. 6d).
Figure 6.
Trifolium longitudinal nodule section (20 dpi) showing the distribution of infected cells after co-inoculation with Micromonospora ML01-gfp and Rhizobium E11-mCh captured by CLSM. (a) Image obtained with the green channel for the localization of Micromonospora. The circled area indicates fluorescence emitted by Micromonospora concentrated in the bacteroid zone. (b) Image captured with the red channel for the localization of Rhizobium. (c) Combination of images a and b. (d) Detail of infected zone showing the co-localization of Micromonospora and Rhizobium in the host cells. The white circle in 6c shows the area where image (d) was captured. Bars: 60 µm (a–c); 20 µm (d). dpi, days post inoculation; C, cortex; BZ, bacteroid zone.
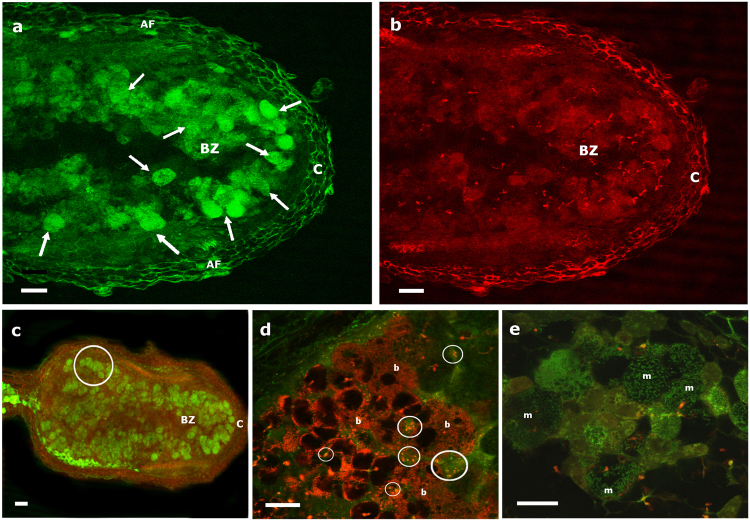
For Medicago, Micromonospora was localized across the nodule and infected cells were visible in all but the meristematic zone (Fig. 7a). As in Trifolium nodules, in some cases, both bacteria occupied the same host cells (Figs. 7c,d). Figure 7a,b show the distribution of the bacteria captured by the corresponding fluorescence channels (green for Micromonospora and red for Sinorhizobium). A close up of the nodular tissue permitted a clear visualization of Micromonospora ML01-gfp cells (Fig. 7e).
Figure 7.
Longitudinal sections of a Medicago 20 dpi nodule showing the distribution of cells infected with Micromonospora ML01-gfp and Sinorhizobium Rm1021-mCh captured by CLSM. (a) Detail of a nodule tip captured with the green channel for the localization of Micromonospora. Arrows indicate fluorescence emitted by Micromonospora to differentiate from autofluorescence emitted by the plant. (b) Detail of a nodule tip captured with the red channel for the localization of Mesorhizobium Rm1021-mCh. (c) Composite image showing the distribution of green and red fluorescence. (d) Bacteroids occupying several plant cells, small circles show areas occupied by both bacteria. (e) Micromonospora hyphae inside host plant cells. The white circle in image 7c shows the area where images d and e were captured. Bars: 75 µm (a); 30 µm (b–e). dpi, days post inoculation; C, cortex; BZ, infection zone; AF, autofluorescence; b, bacteroids; m, Micromonospora.
Discussion
Legumes are widely recognized for their agricultural importance and their capacity to form nitrogen-fixing nodules in symbiosis with bacteria, generically known as rhizobia. However, nodular tissues are an excellent habitat for microorganisms because they are richer in nutrients compared to other plant organs such as roots. Indeed the number of studies reporting the isolation of non-rhizobial bacteria from this niche is increasing2, 8, 10, 12, 25.
To our knowledge, this is the first report for a legume that describes in detail the surface interaction and colonization process of a non-rhizobial bacterium together with the nitrogen-fixing rhizobia. Our results demonstrate that Micromonspora is localized within nodules of three different legumes and strongly suggests that a non-specific relationship takes place between Micromonospora and the plant. We base this conclusion on data showing that a Micromonospora strain isolated from lupine colonizes Medicago and Trifolium as well as Lupinus, suggesting that this actinobacterial strain has a broad host range. Also, Micromonospora has been isolated from a diversity of legume nodules, including Coriaria myrtifolia 7; Pisum sativum 9, 11, and Medicago sativa 17. The ability of Micromonospora to infect different legume species contrasts with the symbiotic interactions between rhizobia and legumes and Frankia and actinorhizal plants, both of which are more restrictive26, 27. However, not all nodules housed both strains. The reason for this is not known, but the results are similar to our findings with wild, collected nodules. Further studies including additional legume species are necessary to determine the full capacity of Micromonospora to infect plants.
The plants did not show any negative effects related to the presence of Micromonospora and the nitrogen fixation process did not appear to be altered. The results also indicated that the different rhizobia were not inhibited by coinoculation with the actinobacterium and that bacteroid development proceeded normally. Indeed, the growth of the coinoculated plants was better, such that in some cases a larger number of nodules per plant resulted18. Interestingly, when Bradyrhizobium and Micromonospora were grown as a co-culture, no growth inhibition was observed by either bacterium even though strain Lupac 08 contains several genes that code for bacteriocins18. These genes may not be expressed when the two bacteria interact under the conditions tested, but more studies are needed.
The systematic isolation of Micromonospora cells from nitrogen-fixing legume and actinorhizal nodules and the application of fluorescent in situ hybridization (FISH) and TEM techniques, presented strong evidence that Micromonospora lupini Lupac 08 was a normal inhabitant of internal root nodule tissues and suggested a close interaction between the host plant and the bacterium10–12, 28, 29. In the present work, unambiguous microscopic localization of Micromonospora lupini Lupac 08 was accomplished using a combination of tagged reporter genes and immunogold labeling. Furthermore, we have demonstrated that Micromonospora not only re-enters its original host, Lupinus sp., but also interacts with other legumes such as Medicago and Trifolium. In all plant samples studied, a plant growth-promoting effect as previously reported was confirmed18, 28 and these results are in line with those reported by other researchers19, 30, 31.
By monitoring the colonization process, information about the distribution of Micromonospora in L. albus, Trifolium, and Medicago nodules was obtained. In all cases, the infection zone was the main area where Micromonospora was found and the place where both bacteria were observed occupying the same plant cell. These results strongly suggest a tripartite interaction and the coexistence of non-rhizobial bacteria within nodule tissues3, 30 although at present a specific function cannot be attributed to Micromonospora. Genomic analysis of strain Lupac 08 has revealed several features related to plant growth promotion including the production of siderophores, phytohormones and other secondary metabolites, all of which may be involved in growth enhancement18.
Furthermore, as compared to control plants inoculated with Rhizobium or Mesorhizobium only, on average, the nodules appeared and developed 1–2 days earlier on the coinoculated plants. It was previously reported that legumes coinoculated with their compatible nitrogen fixer and associated “helper” Micromonospora developed a greater number of nodules in Lupinus, Medicago, and Trifolium 18, 19. Similar results were reported for actinorhizal plants and other “helper” bacteria31–33. In the latter cases, however, these bacterial growth promoters were not considered endophytes because they were isolated from the external plant tissues or from the rhizosphere. In the present study, the Micromonospora strains tested were isolated from the internal root nodule tissues8.
Another question that remains to be answered is whether Micromonospora can enter alone or whether the presence of rhizobia is necessary for the bacterium to gain access to the internal tissues. An interaction between the bacterium and the plant’s root hairs was observed, but further studies are necessary to identify the molecules involved in this communication and whether Micromonospora enters the root hair together with the rhizobial strain or via a “crack entry” mechanism. Plant-polymer degrading enzymes are known to play an important role in infection and colonization34–36. In this respect, genomic analysis of M. lupini Lupac 08 revealed an important array of plant cell wall-degrading enzymes, which have been tested for functionality and may be involved in the infection process. Future investigations will be focused on answering some of these questions with the aim of increasing our knowledge of managing microbial communities that interact with plants. Understanding the ecology of these endophytic bacteria and their molecular interactions will have an impact in plant growth and crop yields and therefore in economics and the environment.
Materials
Strains, plant media, and culture conditions
The strains and plants used in the present study are listed in Table 1. Micromonospora strains were routinely sub-cultured on SA1 agar37. Rhizobial strains were maintained on yeast-mannitol agar38. Escherichia coli strains were grown in Tryptic Soy Broth (TSB) or Luria-Bertani agar (LB). The media were supplemented with the following antibiotics: apramycin (25 µg/ml), kanamycin (60 µg/ml), tetracycline (10 µg/ml), and streptomycin (100 µg/ml), when needed.
Table 1.
Strains used in the present study.
| Strain | Characteristics | Inoculation host |
|---|---|---|
| M. lupini Lupac 08 | Wild type | Lupinus, Medicago, Trifolium |
| M. saelicesensis Lupac 09T | Wild type | Lupinus, Medicago, Trifolium |
| S. meliloti Rm 1021 | Wild type | Medicago |
| Rhizobium sp. E11 | Wild type | Trifolium |
| Bradyrhizobium sp. CAR08 | Wild type | Lupinus |
| M. lupini ML01-gfp | M. lupini Lupac 08 egfp reporter gene | Medicago, Trifolium, Lupinus |
| Sinorhizobium Rm1021-mCh | S. meliloti Rm 1021(pM7604) carrying an mCherry reporter gene | Medicago |
| Rhizobium sp. E11-mCh | Rhizobium sp. E11 (pM7607) carrying an mCherry reporter gene | Trifolium |
Construction of reporter plasmid pSET152-eGFP
The strain Micromonospora lupini Lupac 08 was selected as the recipient for introducing a green fluorescent protein (gfp) gene for localizing the actinobacteria within the plant. The site-specific integration vector pSET152 containing aac(3)IV(Aprar), ϕC31 int, attP, and oriT was selected as the transconjugation vector for M. lupini Lupac 08 because it has been shown to integrate into the genome of various actinobacteria including Micromonospora 39. The presence of the insertion site in the genome of M. lupini Lupac 08 was confirmed by PCR and sequencing using the primers attBF 5′-GTCGACCCCGGGCCCTGGT-3′ and attBR 5′-GTCGACCACCTACCGGCCGG-3′ under the following PCR conditions: 7 min at 95 °C, 30 cycles of 1 min at 94 °C, 1 min at 65 °C and 3 min at 72 °C, followed by 10 min final extension at 72 °C. Sequencing was carried out on an ABI377 sequencer (Applied Biosystems) using a BigDye terminator v3.0 cycle sequencing kit as supplied by the manufacturer and using the primers attBF and attBR.
The construct eGFP-apramycin promoter aac(3)IV was cloned into the plasmid at the EcoRI and NotI restriction sites. The final insert pSET152-eGFP-aac(3)IV(Aprar) was inserted by electroporation into Escherichia coli S17.1, the donor strain.
Conjugation procedure
Intergeneric conjugation between E. coli S17.1 and M. lupini Lupac 08 was performed as described previously39, 40. Briefly, an overnight culture of E. coli S17.1 grown in TSB was diluted into fresh medium and incubated for 3–5 h. The cells were harvested, washed twice, and concentrated 10-fold in TSB. Strain Lupac 08 was grown in TSB for 5 days, harvested by centrifugation, washed, and re-suspended in TSB (2:1 v/v). Recipient cells were mixed with E. coli donor cells (2:1 v/v) and 150 µl were plated on MR0.1 S medium41. The plates were incubated at 28 °C for 20 h and then the medium was covered with 1 ml water containing 500 µl of nalidixic acid to inhibit further growth of E. coli and 1 mg apramycin to select M. lupini exconjugants. Incubation at 28 °C continued for 2–3 weeks to allow outgrowth of the exconjugants.
Transformation of rhizobial strains with mCherry constructs
The strains Sinorhizobium meliloti Rm1021 and Rhizobium sp. E11 were transformed with the plasmids pMP7604 and pMP760742 by conjugation according to standard methods43. Conjugation of plasmids was accomplished by mixing the donor E. coli DH5α containing pMP7604, the helper E. coli DH5α strain containing pRK2013, and the recipient strains, either S. meliloti Rm1021 or Rhizobium sp. E11.
Seed germination and infection assays
Lupinus albus, Medicago sativa, and Trifolium repens seeds were germinated axenically. L. albus seeds were surface-sterilized in 2% (v/v) sodium hypochlorite for 12 min followed by rinsing with sterile distilled water, then placed in Petri dishes containing sterilized moist filter papers, and incubated in the dark. After germination (~3–5 days), seedlings were transferred to pots containing sterile vermiculite and watered with nitrogen-free nutrient solution44 as needed. For the Medicago and Trifolium seeds, these were sequentially immersed in 70% (v/v) ethanol and 2.5% HgCl2 (w/v) for 30 s and 2 min respectively, followed by several rinses with sterile distilled water, and then placed on tap-water agar plates in the dark. After germination, seedlings were placed on square Petri dishes (120 × 120 mm) containing nitrogen-free Rigaud and Puppo nutrient agar.
All seedlings were kept in a plant growth chamber with mixed incandescent and fluorescent lighting programmed for a 16 h photoperiod, day-night cycle, with a constant temperature of 21–22 °C, and 50–60% relative humidity. Upon the appearance of the first leaves, the plants were inoculated with the appropriate bacterial suspensions (108 cells per ml). Overall, three different treatments were used: a) plants inoculated with GFP-tagged Micromonospora lupini (strain ML01); b) co-inoculation of Micromonospora ML01 and the appropriate Bradyrhizobium, Rhizobium or Sinorhizobium strain to induce nodulation (see Table 1), and c) uninoculated plants, which served as negative controls.
Monitoring bacterial colonization by confocal microscopy
Infected plant roots of Medicago and Trifolium plants were observed with fluorescence (Nikon Eclipse 80i) and confocal scanning laser (CLSM, Leica TCS model) microscopes 2 days after inoculation and monitored every other day until the nodules were fully developed. Localization of gfp and mCherry fluorescence in the root and nodule tissues was done using standard filter settings (488 nm excitation and 515 to 560 nm emission for gfp expression, and 620 nm excitation and 620–660 nm emission for mCherry). Autofluorescence was evaluated by comparing the gfp image with the red fluorescence channel (543 nm excitation and > 570 nm emission) and also by comparing the image with uninoculated plants. Coinoculated Lupinus plants were grown for 4–5 weeks. For observation, nodules were longitudinally sectioned on a cryostat (Thermo HM560), mounted on glass slides, and viewed by CLSM as described above.
Immunoelectron microscopy
Strain ML01 was localized in L. albus nodules via a pre-embedding immunogold technique with antibodies raised against GFP, following the procedures of45 and46. A vibratome (Leica V1000) was employed to obtain 60 µm-thick sections from fixed (4% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M phosphate buffer [PB]), agarose-embedded nodules. Floating sections were blocked with 10% normal goat serum (NGS) in 0.1 M Tris-buffer containing 0.9% NaCl (TBS) for 1 h. The sections were then labeled with the primary antibody raised against GFP in guinea pig (0.5–2 µg/ml diluted in 0.1 M TBS with 1% NGS; Frontiers Institute, Japan), and incubated at 4 °C overnight. After washing, 1.4 nm gold particles conjugated to goat anti-guinea pig antibodies (diluted 1:100 in TBS buffer containing 2% NGS; Nanoprobes, NY) were added and incubated for 2 h. After several PB washes, sections were post-fixed in 1% glutaraldehyde prepared in the same buffer for 10 min. They were washed in double distilled water, followed by silver enhancement of the gold particles with an HQ Silver kit (Nanoprobes). Finally, the gold-silver-labeled sections were processed for electron microscopy as described previously45.
Re-isolation of Micromonospora from legumes
Following the same procedure as above, five plants from each legume (Lupinus, Medicago, and Trifolium) were used to re-isolate the non-transformed Micromonospora strains Lupac 08 and Lupac 09T after 3 weeks incubation. After counting the nodules from each plant, ten randomly chosen nodules (from the pool of 5 plants) were selected for bacterial isolation. Washed nodules were sterilized and processed as described previously and then the bacterial suspension was inoculated on yeast mannitol agar for isolation10. After counting the number of colonies having a Micromonospora-like appearance, several purified colonies were selected for DNA extraction and 16S rRNA gene sequencing for identification10. The sequences obtained were compared to the sequences deposited in the public databases (Lupac 08, AJ783992; Lupac 09T, AJ783993).
Acknowledgements
The authors would like to thank Dr. Javier Hierro and Dr. Emilio Cervantes for their excellent support in using the CLSM and for useful discussions. Dr. Lagendijk is also greatly acknowledged for providing the plasmids. MET acknowledges financial support from the Ministerio de Economía y Competitivad (MICINN) under project CGL2014–52735-P. PB is grateful for an FPI grant from the MICINN. Support for Micromonospora research in the AMH lab came from a grant from UC-MEXUS/CONACYT.
Author Contributions
M.E.T. conceived and designed the experiments. P.B., P.A.-V., M.E.T., R.L. and C.A. performed experiments. J.A. provided material for conjugation of Micromonspora and advised during the process. M.E.T. and A.H. analysed results and wrote the manuscript. All authors reviewed the final draft.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang D, Yang S, Tang F, Zhu H. Symbiosis specificity in the legume: rhizobial mutualism. Cell Microbiol. 2012;14:334–342. doi: 10.1111/j.1462-5822.2011.01736.x. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Hidalgo, P. & Hirsch, A. M. The nodule microbiome; N2-fixing rhizobia do not live alone. Phytobiomes 1–52 (2017). doi:10.1094/PBIOMES-12-16-0019-RVW
- 3.Muresu R, et al. Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol Ecol. 2008;63:383–400. doi: 10.1111/j.1574-6941.2007.00424.x. [DOI] [PubMed] [Google Scholar]
- 4.Zakhia F, et al. Diverse bacteria associated with root nodules of spontaneous legumes in Tunisia and first report for nifH-like gene within the genera Microbacterium and Starkeya. Microb Ecol. 2006;51:375–393. doi: 10.1007/s00248-006-9025-0. [DOI] [PubMed] [Google Scholar]
- 5.Hoque MS, Broadhurst LM, Thrall PH. Genetic characterization of root-nodule bacteria associated with Acacia salicina and A. stenophylla (Mimosaceae) across south-eastern Australia. Int J Syst Evol Microbiol. 2011;61:299–309. doi: 10.1099/ijs.0.021014-0. [DOI] [PubMed] [Google Scholar]
- 6.Sturz AV, Christie BR, Matheson BG, Nowak J. Biodiversity of endophytic bacteria which colonize red clover nodules, roots, stems and foliage and their influence on host growth. Biol Fertil Soils. 1997;25:13–19. doi: 10.1007/s003740050273. [DOI] [Google Scholar]
- 7.Trujillo ME, Kroppenstedt RM, Schumann P, Carro L, Martinez-Molina E. Micromonospora coriariae sp. nov., isolated from root nodules of Coriaria myrtifolia. Int J Syst Evol Microbiol. 2006;56:2381–2385. doi: 10.1099/ijs.0.64449-0. [DOI] [PubMed] [Google Scholar]
- 8.Trujillo ME, Kroppenstedt RM, Fernandez-Molinero C, Schumann P, Martinez-Molina E. Micromonospora lupini sp. nov. and Micromonospora saelicesensis sp. nov., isolated from root nodules of Lupinus angustifolius. Int J Syst Evol Microbiol. 2007;57:2799–2804. doi: 10.1099/ijs.0.65192-0. [DOI] [PubMed] [Google Scholar]
- 9.Garcia LC, Martinez-Molina E, Trujillo ME. Micromonospora pisi sp. nov., isolated from root nodules of Pisum sativum. Int J Syst Evol Microbiol. 2010;60:331–337. doi: 10.1099/ijs.0.012708-0. [DOI] [PubMed] [Google Scholar]
- 10.Trujillo ME, et al. The genus Micromonospora is widespread in legume root nodules: the example of Lupinus angustifolius. ISME J. 2010;4:1265–1281. doi: 10.1038/ismej.2010.55. [DOI] [PubMed] [Google Scholar]
- 11.Carro L, Spröer C, Alonso P, Trujillo ME. Diversity of Micromonospora strains isolated from nitrogen fixing nodules and rhizosphere of Pisum sativum analyzed by multilocus sequence analysis. Syst Appl Microbiol. 2012;35:73–80. doi: 10.1016/j.syapm.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Carro L, Pujic P, Trujillo ME, Normand P. Micromonospora is a normal occupant of actinorhizal nodules. J. Biosci. 2013;38:685–693. doi: 10.1007/s12038-013-9359-y. [DOI] [PubMed] [Google Scholar]
- 13.de Menezes AB, Lockhart RJ, Cox MJ, Allison HE, McCarthy AJ. Cellulose degradation by Micromonosporas recovered from freshwater lakes and classification of these actinomycetes by DNA gyrase B gene sequencing. Appl. Environ. Microbiol. 2008;74:7080–7084. doi: 10.1128/AEM.01092-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Menezes AB, McDonald JE, Allison HE, McCarthy AJ. Importance of Micromonospora spp. as colonizers of cellulose in freshwater lakes as demonstrated by quantitative reverse transcriptase PCR of 16S rRNA. Appl Env. Microbiol. 2012;78:3495–3499. doi: 10.1128/AEM.07314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niner BM, et al. Analysis of partial sequences for 16S rRNA of actinomycetes isolated from Casuarina equisetifolia nodules in México. Appl Env. Microbiol. 1996;2:3034–3036. doi: 10.1128/aem.62.8.3034-3036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-tarabily, K. A., Nassar, A. H. & Sivasithamparam, K. Promotion of growth of bean (Phaseolus vulgaris L.) in a calcareous soil by a phosphate-solubilizing, rhizosphere-competent isolate of Micromonospora endolithica. 39, 161–171 (2008).
- 17.Martinez-Hidalgo P, Galindo-Villardon P, Trujillo ME, Igual JM, Martinez-Molina E. Micromonospora from nitrogen fixing nodules of alfalfa (Medicago sativa L.). A new promising plant probiotic bacteria. Sci Rep. 2014;4:6389. doi: 10.1038/srep06389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trujillo ME, et al. Genome features of the endophytic actinobacterium Micromonospora lupini strain Lupac 08: on the process of adaptation to an endophytic life style? PLoS One. 2014;9:e108522. doi: 10.1371/journal.pone.0108522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solans M, Vobis G, Wall LG. Saprophytic actinomycetes promote nodulation in Medicago sativa-Sinorhizobium meliloti symbiosis in the presence of high N. J. Plant Growth Regul. 2009;28:106–114. doi: 10.1007/s00344-009-9080-0. [DOI] [Google Scholar]
- 20.Francis I, Holsters M, Vereecke D. The Gram-positive side of plant-microbe interactions. Environ. Microbiol. 2010;12:1–12. doi: 10.1111/j.1462-2920.2009.01989.x. [DOI] [PubMed] [Google Scholar]
- 21.Guinel FC. Getting around the legume nodule: II. Molecular biology of its peripheral zone and approaches to study its vasculature. Botany. 2009;87:1139–1166. doi: 10.1139/B09-075. [DOI] [Google Scholar]
- 22.Tang CX, Robson AD, Dilworth MJ, Kuo J. Microscopic evidence on how iron-deficiency limits nodule initiation in Lupinus-angustifolius L. New Phytol. 1992;121:457–467. doi: 10.1111/j.1469-8137.1992.tb02946.x. [DOI] [PubMed] [Google Scholar]
- 23.González-Sama A, Lucas MM, De Felipe MR, Pueyo JJ. An unusual infection mechanism and nodule morphogenesis in white lupin (Lupinus albus) New Phytol. 2004;163:371–380. doi: 10.1111/j.1469-8137.2004.01121.x. [DOI] [PubMed] [Google Scholar]
- 24.Pawlowski K, Bisseling T. Rhizobial and actinorhizal symbioses: what are the shared features? Plant Cell. 1996;8:1899–1913. doi: 10.1105/tpc.8.10.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stajkovic O, De Meyer S, Lilicic B, Willems A, Delic D. Isolation and characterization of endophytic non-rhizobial bacteria from root nodules of alfalfa (Medicago sativa L.) Bot. SERBICA. 2009;33:107–114. [Google Scholar]
- 26.Pawlowski K, Demchenko KN. The diversity of actinorhizal symbiosis. Protoplasma. 2012;249:967–979. doi: 10.1007/s00709-012-0388-4. [DOI] [PubMed] [Google Scholar]
- 27.Andrews, M. & Andrews, M. E. Specificity in legume-rhizobia symbioses. (2016). doi:10.20944/preprints201608.0005.v1 [DOI] [PMC free article] [PubMed]
- 28.Trujillo ME, Riesco R, Benito P, Carro L. Endophytic actinobacteria and the interaction of Micromonospora and nitrogen fixing plants. Front. Microbiol. 2015;6:1–15. doi: 10.3389/fmicb.2015.01341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valdés M, et al. Non- Frankia actinomycetes isolated from surface-sterilized roots of Casuarina equisetifolia fix nitrogen. Appl. Environ. Microbiol. 2005;71:460–466. doi: 10.1128/AEM.71.1.460-466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokala RK, et al. Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum) Appl. Environ. Microbiol. 2002;68:2161–2171. doi: 10.1128/AEM.68.5.2161-2171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solans M, Vobis G, Cassán F, Luna V, Wall LG. Production of phytohormones by root-associated saprophytic actinomycetes isolated from the actinorhizal plant Ochetophila trinervis. World J. Microbiol. Biotechnol. 2011;27:2195–2202. doi: 10.1007/s11274-011-0685-7. [DOI] [Google Scholar]
- 32.Knowlton S, Dawson J. Effect of Pseudomonas cepacia and cultural factors on the nodulation of Alnus rubra root by Frankia. Can. J. Bot. 1983;61:2877–2882. doi: 10.1139/b83-320. [DOI] [Google Scholar]
- 33.Knowlton S, Berry A, Torrey JG. Evidence that associated soil bacterias may influence root hair infection of actinorhizal plants by Frankia. Can. J. Microbiol. 1980;26:971–977. doi: 10.1139/m80-165. [DOI] [PubMed] [Google Scholar]
- 34.Lugtenberg, B. J. J., Chin-a-woeng, T. F. C. & Bloemberg, G. V. Microbe – plant interactions: principles and mechanisms. 373–383 (2002). [DOI] [PubMed]
- 35.Compant S, et al. Endophytic colonization of Vitis vinifera L. by plant growth- promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 2005;71:1685–1693. doi: 10.1128/AEM.71.4.1685-1693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinhold-Hurek B, Maes T, Gemmer S, Van Montagu M, Hurek T. An endoglucanase is involved in infection of rice roots by the not-cellulose-metabolizing endophyte Azoarcus sp. strain BH72. Mol. Plant. Microbe. Interact. 2006;19:181–188. doi: 10.1094/MPMI-19-0181. [DOI] [PubMed] [Google Scholar]
- 37.Trujillo ME, et al. Micromonospora mirobrigensis sp. nov. Int J Syst Evol Microbiol. 2005;55:877–880. doi: 10.1099/ijs.0.63361-0. [DOI] [PubMed] [Google Scholar]
- 38.Vincent, J. A Manual for the Practical Study of Root Nodule Bacteria. (Blackwell Scientific, 1970).
- 39.Anzai Y, et al. Production of rosamicin derivatives in Micromonospora rosaria by introduction of D-mycinose biosynthetic gene with PhiC31-derived integration vector pSET152. J Ind Microbiol Biotechnol. 2009;36:1013–1021. doi: 10.1007/s10295-009-0579-y. [DOI] [PubMed] [Google Scholar]
- 40.Anzai Y, Ishii Y, Yoda Y, Kinoshita K, Kato F. The targeted inactivation of polyketide synthase mycAV in the mycinamicin producer, Micromonospora griseorubida, and a complementation study. FEMS Microbiol Lett. 2004;238:315–320. doi: 10.1016/j.femsle.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 41.Inouye M, Takada Y, Muto N, Beppu T, Horinouchi S. Characterization and expression of a P-450-like mycinamicin biosynthesis gene using a novel Micromonospora-Escherichia coli shuttle cosmid vector. Mol.Gen. Genet. 1994;245:456–464. doi: 10.1007/BF00302258. [DOI] [PubMed] [Google Scholar]
- 42.Lagendijk EL, Validov S, Lamers GE, de Weert S, Bloemberg GV. Genetic tools for tagging Gram-negative bacteria with mCherry for visualization in vitro and in natural habitats, biofilm and pathogenicity studies. FEMS Microbiol Lett. 2010;305:81–90. doi: 10.1111/j.1574-6968.2010.01916.x. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J. & Russel, D. Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor Laboratory Press, 2001).
- 44.Rigaud J, Puppo A. Indole-3-acetic acid catabolism by soybean bacteroids. J Gen Microbiol. 1975;88:223–228. doi: 10.1099/00221287-88-2-223. [DOI] [Google Scholar]
- 45.Lujan R, Nusser Z, Roberts JD, Shigemoto R, Somogyi P. Perisynaptic location of metabotropic glutamate receptors mGluR1 and mGluR5 on dendrites and dendritic spines in the rat hippocampus. Eur. J. Neurosci. 1996;8:1488–1500. doi: 10.1111/j.1460-9568.1996.tb01611.x. [DOI] [PubMed] [Google Scholar]
- 46.Chen W, James EK, Prescott AR, Kierans M, Sprent JI. Nodulation of Mimosa spp. by the beta-proteobacterium Ralstonia taiwanensis. Mol. Plant Microbe Interact. 2003;16:1051–1061. doi: 10.1094/MPMI.2003.16.12.1051. [DOI] [PubMed] [Google Scholar]