Abstract
Pleistocene climatic oscillations have greatly influenced the evolutionary history and distribution pattern of most extant species. However, their effects on species on the Qinghai-Tibet Plateau (QTP) are not well understood. To investigate the effects of past climatic shifts, particularly the Last Glacial Maximum (LGM), on plateau fish, we analysed the phylogeographic structure and demographic history of five closely related taxa of the subfamily Schizothoracinae, a representative endemic taxon of the QTP, from nine endorheic lakes on the central QTP and three peripheral exorheic rivers using the mitochondrial control region (D-loop) sequence and 12 microsatellite (SSR) markers. Phylogram from D-loop haplotypes revealed two well-supported lineages (North and South) separated by the Tanggula Mountains. The results from the D-loop and SSR revealed that endorheic populations possess high genetic diversity and a unique genetic structure. The most recent demographic expansion occurred post-LGM for most endorheic populations and in the last interglacial period for Siling Co and all exorheic populations. Phylogeographic structure, together with species distribution modelling, supports the scenario of multiple glacial refugia on the QTP during the LGM and suggests that Siling Co (4540 m asl) is a cryptic glacial microrefugia for plateau fish, which would be the highest glacial microrefugia known.
Introduction
The advances and retreats of glaciations through the Pleistocene, especially most recently the Last Glacial Maximum (LGM, 19–26 thousand years ago, kya), have significantly affected the demographic history, abundance and distribution pattern of modern species1–3. During these glaciations, many species were excluded from large parts of their ranges and forced into refugia by the expanding ice sheets, followed by recolonization and population expansion as the glaciers retreated1. For example, in Europe, palaeontological records and genetic analyses have revealed that many species were retreated southward to the three Mediterranean peninsulas (Balkans, Italian and Iberian) during the LGM and expanded northward into their current distribution areas in the subsequent warming period2, 3. Recently, biogeographic studies have led to the widespread acceptance of northern extra-Mediterranean refugia from the Atlantic coast of France in the west to the Black Sea region in the east, for example, the Carpathian Basin and even northern Central Europe4. Using phylogeographic studies, increasing numbers of previously unknown, or cryptic, refugia are identified3, 5.
The Qinghai-Tibetan Plateau (QTP) is the world’s highest and largest plateau, covering an area of 2.5 million km2 and has an average altitude of 4500 m above sea level (asl). The QTP and adjacent mountains are the largest glaciated tracks outside the polar region6. Both plateau uplifting and Pleistocene climatic oscillations have greatly influenced the biotic pattern on the QTP7, 8. Although the uplifting progress of the QTP remains controversial, most studies agree that the QTP attained its current state at least 150 kya before the last interglacial period (130–25 kya)7, 9. The present biotic pattern on the QTP was mainly shaped by Pleistocene climatic oscillations6. Unlike Europe and North America, where continental glaciations occurred, only mountain and valley glaciers formed on the QTP during the LGM10, 11. Therefore, in the common “edge refugia” model, the low-altitude southern and eastern edge of the QTP served as the glacial refugia for the plateau species; this model has proved insufficient for a comprehensive understanding of the effects of glaciations on the biota of the QTP12–14.
The influence of Pleistocene glaciations on the QTP have been studied in various species, but several basic questions, such as the locations of glacial refugia and the process of postglacial colonization, remain poorly understood13–15. Some studies of endemic birds and plants have suggested that the south-eastern edge of the QTP acts as the glacial refugia15–18, while others have failed to detect “edge refugia” but suggest multiple refugia on the platform of the QTP12, 19, 20. Correspondingly, two main scenarios for the biota on the QTP have been proposed, i.e. postglacial recolonization from peripheral refugia or the survival of the LGM in situ, with several refugia on the platform. For both scenarios, evidence for the location of refugia and postglacial colonizing routes is scant13, 15. Previous studies have mainly focused on plants, birds and amphibians in the periphery of the QTP. Therefore, more studies about additional endemic species on the central QTP are necessary for a thorough understanding of the influence of Pleistocene glaciations on the QTP biota.
Freshwater fishes are regarded as the ideal subjects to explore the effects of glaciations because of their restriction in glacial refugia that are peripheral to ice sheets and the restrictive aquatic requirements for postglacial dispersal21. Schizothoracine fishes (Teleostei: Cyprinidae) account for the largest component of the QTP ichthyofauna and are widely distributed in local lakes and rivers22, 23. They have evolved a number of adaptations to cold-water environments and are regarded as the most diverse vertebrates on the central QTP24, 25. Many lakes are located on the central QTP, and most are endorheic lakes26. As the migratory range of fish was limited by watercourses, it is likely that fishes in central QTP lakes recolonized postglacially from local refugia rather than “edge refugia”. Therefore, these lakes are ideal sites for investigating the potential of the central QTP to harbour glacial refugia.
In this study, we analysed the phylogeographic structure and demographic history of schizothoracine fishes on the central QTP and their closely related species in peripheral exorheic rivers using mitochondrial DNA (mtDNA) control region (D-loop) sequence and nuclear microsatellite markers (SSR). Species distribution modelling (SDM) was also used to predict the suitable habitat ranges for these schizothoracine fishes during the LGM. We sought to (i) assess the genetic diversity, phylogeographic structure and demographic history of those populations; (ii) infer whether the present schizothoracine fishes in the central QTP endorheic waters were recolonized from low-altitude peripheral rivers or persisted on the plateau platform through the LGM; and (iii) determine whether cryptic glacial refugia on the central QTP exist.
Results
Sequence characteristics and population genetic diversity
We sampled the only two taxa of schizothoracine fishes (Gymnocypris namensis and Schizopygopsis spp.) in nine endorheic lakes on the east Changtang Plateau, which is the central part and the earliest uplifted block of the QTP27, as well as their closely related species (Schizopygopsis younghusbandi, Schizopygopsis thermalis and Herzensteinia microcephalus), in peripheral exorheic rivers (upper Salween, Yangtze River and Yarlung Tsangpo). A total of 839 specimens belonging to five closely related taxa were collected from 19 sites in 2010 and 2015, ranging from 23–89 individuals per site (Fig. 1 and Table 1).
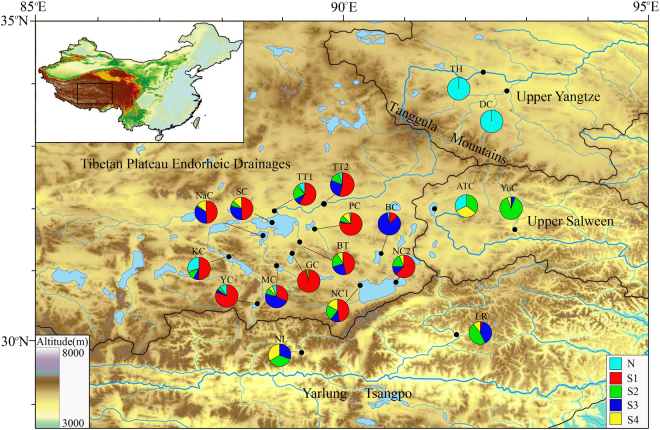
Figure 1.
Location and haploclade composition of each sampling site in this study. Pie charts exhibit the frequency of lineages or sublineages in each population. Five colours (cyan, red, green, blue and yellow) correspond to one North lineage (N1) and four South sublineages (S1, S2, S3 and S4) in the D-loop phylogram, respectively. The boundaries of the main drainage basins are also shown by black lines. Whole names for each site are listed in Table 1. The map was drawn using ArcGIS v10.0 and Adobe Illustrator CS5 v15.1.0.
Table 1.
Sample size and geological information of each site in this study.
| Species | Site | Code | Drainages | Location | Altitude(m asl) | N(m) | N(n) |
|---|---|---|---|---|---|---|---|
| SS | Tsachu Tsangpo1 | TT1 | Siling Co | 32°03′N 89°08′E | 4561 | 53 | 30 |
| Tsachu Tsangpo 2 | TT2 | 32°24′N 90°44′E | 4710 | 57 | 30 | ||
| Siling Co | SC | 31°47′N 88°48′E | 4540 | 38 | 37 | ||
| Bochu Tsangpo | BT | 31°34′N 89°19′E | 4608 | 57 | 55 | ||
| Nagtsang Co | NaC | 31°36′N 88°41′E | 4568 | 49 | 41 | ||
| Mokiu Co | MC | 31°03′N 89°01′E | 4680 | 34 | 34 | ||
| Kyaring Co | KC | 31°16′N 88°06′E | 4654 | 55 | 52 | ||
| Yueqia Co | YC | 30°28′N 88°37′E | 4810 | 41 | 41 | ||
| Pongok Co | PC | Pongok Co | 31°41′N 89°36′E | 4545 | 58 | 50 | |
| Gomang Co | GC | Gomang Co | 31°15′N 89°13′E | 4636 | 26 | 26 | |
| Bam Co | BC | Bam Co | 31°22′N 90°33′E | 4565 | 38 | 35 | |
| GN | Nam Co1 | NC1 | Nam Co | 30°55′N 90°56′E | 4729 | 23 | 23 |
| Nam Co2 | NC2 | 30°55′N 90°56′E | 4729 | 32 | 31 | ||
| SY | Namulin | NL | Yarlung Tsangpo | 29°41′N 89°05′E | 4005 | 34 | 30 |
| Lhasa River | LR | 31°11′N 91°20′E | 4048 | 32 | 30 | ||
| ST | Amdo Tsonak Co | ATC | Upper Salween | 31°56′N 91°29′E | 4599 | 89 | 30 |
| Yu Chu | YuC | 31°12′N 91°48′E | 4704 | 56 | 30 | ||
| HM | Dri Chu | DC | Upper Yangzte | 33°52′N 92°22′E | 4569 | 31 | 31 |
| Tuotuo He | TH | 34°13′N 92°26′E | 4542 | 36 | 35 |
SS = S.Spp, GN = G. namensis, SY = S. younghusbandi, ST = S. thermalis, HM = H. microcephalus, N(m), number of specimens used for D-loop sequence analysis, N(n), number of specimens used for nuclear microsatellite analysis.
Using sequencing, we obtained 839 bp aligned sites of D-loop sequences, including 81 variable sites, 47 of which were parsimony-informative, and 163 haplotypes. Three common haplotypes (Hap10, Hap47 and Hap83) shared by 455 individuals were identified. The most common haplotype Hap10 (shared by 220 individuals) was private to endorheic populations. In total, 20 haplotypes were shared across drainages (Fig. 2). Overall, both haplotype diversity (h) and nucleotide diversity (π) were high across all populations, with average values of 0.7445 ± 0.17416 and 0.0045 ± 0.0041 (mean ± s.d.), respectively. The populations of three exorheic rivers (NL, LR, ATC, YuC, DC and TH) showed higher π and h than endorheic populations. The three highest h sites (ATC, DC and TH) were located at the transition zone between exorheic and endorheic drainages (Table 2). All the haplotypes were submitted to GenBank with accession numbers MF685038-MF685200.
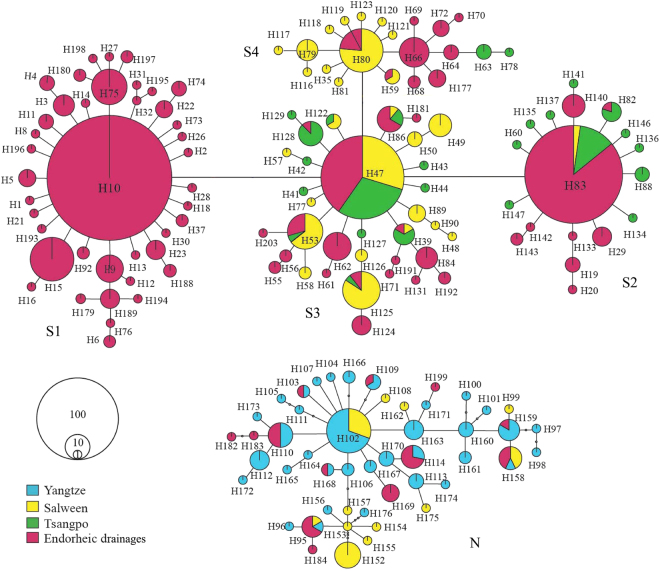
Figure 2.
Median-joining network of mitochondrial D-loop haplotypes for the five taxa of schizothoracine fish in this study. Filled circles correspond to unique haplotypes, and their sizes are proportional to haplotype frequency. The five parts of the network correspond to one lineage and four sublineages. The four colours correspond to the four main drainages in our study area (here we treat all endorheic drainages as one). Small open dots indicate missing intermediate haplotypes.
Table 2.
Summary of genetic diversity, neutrality and mismatch analyses of each population.
| Site | D-loop | SSR | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | h | π | Fu’s Fs | Tajiama’s D | SSD (P) | Ri | Tau | Growth began, kya (95% CI) | Ai | HO | HE | |
| TT1 | 16 | 0.656 | 0.0015 | −1.80622 | −1.32888 | 0.00216(0.84) | 0.03146 | 2.14 | 115.3(195.1–63.2) | 2.60 | 0.643 | 0.850 |
| TT2 | 17 | 0.844 | 0.0061 | −2.62032 | −0.31492 | 0.00151(0.18) | 0.04900 | 1.79 | 2.39 | 0.591 | 0.855 | |
| SC | 15 | 0.889 | 0.0124 | −5.68972 | −0.7133 | 0.01536(0.10) | 0.05168 | 2.88 | 2.66 | 0.682 | 0.882 | |
| BT | 19 | 0.821 | 0.003 | −4.26003 | −1.95308 | 0.01527(0.12) | 0.07584 | 2.26 | 2.49 | 0.632 | 0.871 | |
| PC | 13 | 0.586 | 0.0024 | −4.17854 | −2.19562 | 0.03185(0.13) | 0.08640 | 0.67 | 21.8(28.3–17.6) | 2.44 | 0.615 | 0.873 |
| NaC | 22 | 0.771 | 0.0025 | −3.61855 | −1.06759 | 0.0133(0.46) | 0.05150 | 3.23 | 77.2(148.6–47.8) | 2.38 | 0.589 | 0.858 |
| MC | 12 | 0.676 | 0.0022 | −2.28126 | −0.79545 | 0.04338(0.22) | 0.16480 | 3.29 | 58.7(74.3–18.3) | 2.54 | 0.652 | 0.851 |
| KC | 26 | 0.872 | 0.0134 | −0.38542 | 1.28883 | 0.04091(0.04) | 0.01300 | 6.52 | 43.8(77.1–10.5) | 2.45 | 0.612 | 0.843 |
| YC | 10 | 0.459 | 0.0063 | −0.80531 | −1.21009 | 0.01708(0.10) | 0.17679 | 0.59 | 36.2(69.5–19.8) | 2.49 | 0.632 | 0.848 |
| GC | 3 | 0.465 | 0.0009 | 3.41636 | −0.70289 | 0.26121(0.00) | 0.21524 | 0.48 | — | 2.44 | 0.615 | 0.773 |
| NC1 | 10 | 0.758 | 0.0025 | −8.71945 | −1.31482 | 0.02172(0.27) | 0.06494 | 3.23 | 55.8(95.6–21.8) | 2.48 | 0.615 | 0.843 |
| NC2 | 12 | 0.792 | 0.0021 | −4.7548 | −1.0241 | 0.02341(0.24) | 0.05471 | 3.06 | 2.45 | 0.608 | 0.832 | |
| BC | 5 | 0.334 | 0.0011 | −1.76001 | −1.33196 | 0.03082(0.22) | 0.35734 | 2.0 | 42.9(46.3–12.8) | 2.39 | 0.596 | 0.811 |
| ML | 16 | 0.722 | 0.0016 | −10.3530 | −1.32417 | 0.00891(0.08) | 0.09448 | 3.75 | 101.3(133.5–70.5) | 2.42 | 0.604 | 0.863 |
| LR | 15 | 0.873 | 0.002 | −10.1344 | −1.69583 | 0.00245(0.54) | 0.06768 | 3.62 | 2.52 | 0.613 | 0.881 | |
| ATC | 32 | 0.913 | 0.0129 | −3.75926 | 1.01608 | 0.04675(0.18) | 0.02066 | 4.88 | 137.2(252.6–48.7) | 2.46 | 0.605 | 0.865 |
| YuC | 13 | 0.878 | 0.0032 | −2.7032 | 0.34479 | 0.00064(0.33) | 0.03037 | 3.42 | 2.38 | 0.601 | 0.849 | |
| DC | 18 | 0.930 | 0.0032 | −10.3569 | −1.02339 | 0.00064(0.89) | 0.03386 | 2.69 | 130.7(188.6–89.1) | 2.41 | 0.582 | 0.893 |
| TH | 22 | 0.899 | 0.0032 | −13.4682 | −1.17882 | 0.00294(0.55) | 0.02884 | 2.91 | 2.31 | 0.567 | 0.888 | |
N, the number of D-loop haplotypes; h, haplotype diversity; π, nucleotide diversity; Ai, the average number of alleles per individual at a locus; HO, observed heterozygosity; HE, expected heterozygosity; SSD and Ri, Sum of squares deviations and Raggedness index of mismatch distribution; Tajiama’s D and Fu’s Fs are statistics of neutrality tests; Significant values (P ≤ 0.05) are indicated in bold. When estimate the growth began data, we treat each river as one site, and Siling Co and directly connected rivers as one site.
For SSR analyses, 671 specimens were used (Table 1). All 12 SSR loci showed a high level of polymorphism, and 318 alleles were amplified. The number of alleles per locus ranged from 20 to 39 (Table S1, Supplementary information), with an average of 2.46 ± 0.08 alleles per individual at each locus (Ai). The expected heterozygosity (HE) was high for most populations (0.854 ± 0.028), while the observed heterozygosity (HO) was lower (0.6134 ± 0.026). For all endorheic populations, Siling Co showed the highest h, HO and HE (Table 2).
Phylogenetic analyses and genetic structure
Phylogenetic analyses of D-loop haplotypes were conducted using Bayesian inferences (BI) and maximum likelihood (ML). The BI and ML methods obtained similar topologies; therefore, we only present the BI tree (Fig. S1, Supplementary information). Two well-supported lineages, North (N) and South (S), were identified, separated by the intervening Tanggula Mountains. Lineage N was simple, mainly comprising all upper Yangtze River haplotypes, while lineage S was complex, mainly comprising four obviously sublineages (S1, S2, S3 and S4) in spite of low bootstrap values. Congruent with the phylogenetic tree, the haplotype network (median-joining) analysis showed two isolated networks that corresponded to the two lineages. The lineage corresponding to lineage S was composed of four clusters, with cluster S3 (composed of all three drainages) at the centre and connected to cluster S1 (composed of only endorheic drainage), cluster S2 (mainly composed of endorheic and Tsangpo drainages) and cluster S4 (mainly composed of endorheic and Salween drainages). Each cluster was arrayed in a star-like structure around a central haplotype, which was regarded as the ancestral haplotype (Hap10, Hap47, Hap80, Hap83 and Hap102, Fig. 2).
In accordance with the phylogenetic analyses, spatial genetic analyses of mtDNA haplotypes revealed high values of uncorrected pairwise distance (Fst) between the upper Yangtze River (UY) and other populations (0.849 ± 0.09), suggesting that two defined groups were significantly differentiated. Fst values among populations south of the Tanggula Mountains were low (0.185 ± 0.117). Most of the Fst values among drainages (97%) were significantly higher than zero, suggesting a distinct genetic structure among drainages (Table S3, Supplementary information). An analysis of molecular variance (AMOVA) revealed that a majority of D-loop sequence variation was explained by drainage (59.29%), whereas habitat (different sites within drainage) explained only 8.75%. In addition, a significant amount of variation (31.95%) existed within populations (Table 3).
Table 3.
Analysis of molecular variance (AMOVA) of D-loop sequences.
| Source of variation | d.f. | Sum of squares | Variance components | % variation |
|---|---|---|---|---|
| Among drainages | 7 | 1383.504 | 2.64554 Va | 59.29 |
| Among sites within drainages | 11 | 214.091 | 0.39043 Vb | 8.75 |
| Within sites | 820 | 1161.631 | 1.4.2562 Vc | 31.95 |
| Total | 838 | 2759.226 | 4.46203 | 100 |
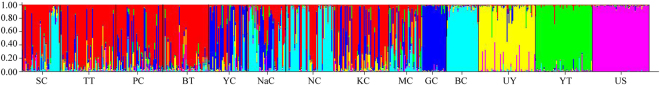
The SSR marker analyses also revealed clear genetic differentiation among drainages. When conducting the STRUCTURE analysis, LnP (D) showed no peak and increased gradually up to K = 14, but the increase slowed after K = 6. In addition, ∆K declined severely from K = 2 to K = 4 and then showed a clear peak at K = 6 (data not shown). Therefore, K = 6 was considered as the most likely number of clusters. At K = 6, the drainages were separated from each other, except for Pongok Co (PC) and Nam Co (NC), which comprised a complex cluster with populations of Siling Co drainage (Fig. 3). The SSR analyses revealed a lower Fst than did the D-loop analyses. Based on SSR markers, the mean Fst between UY and other populations was 0.144 (±0.007) and among populations south of the Tanggula Mountains was 0.037 (±0.002). Significant Fst values occurred in 56 of 91 tests, ranging from 0.024 to 0.185 (Table S3, Supplementary information).
Figure 3.
Population structure estimated from 12 nuclear microsatellite loci using STRUCTURE at K = 6. Each thin vertical line corresponds to a single individual, and the six colours represent the six genetic population clusters. Here, we treat each river as a single site. YT, Yarlung Tsangpo; US, upper Salween; UY, upper Yangtze; whole names of other codes are listed in Table 1.
Demographic history
For neutrality tests, non-significant negative values of Tajima’s D were obtained for most populations, and significantly negative values of Fu’s Fs were obtained for more than half populations (Table 2), indicating departure from neutrality. Compared with Tajima’s D, Fu’s Fs is more sensitive in detecting population expansion28. Commonly, partial population reduction, population subdivision, secondary contact among lineages or a recent bottleneck can cause discrepancy in detecting population expansion with these two neutrality test statistics29. The non-significant negative values of Fu’s Fs occurred in population of small lakes, which are easy effected by accidental events. Mismatch distribution analysis showed similar results. The values of sum of squares deviations (SSD) and the raggedness index (Ri) calculated by goodness-of-fit tests were not significant (P > 0.05) for most populations (Table 2). Most populations exhibited an observed unimodal mismatch frequency distribution, except for the Pongok Co and Gomang Co (bimodal), indicating that the model of sudden expansion could not be rejected (Fig. S2, Supplementary information). The historical population size inferred by Bayesian skyline plots (BSP) showed three growth patterns (Fig. 4). Gomang Co has been decreasing since the last interglacial period. The three exorheic rivers (Tsangpo, upper Salween and Yangtze River) and Siling Co showed a similar tendency. They began to grow at approximately 100 kya, corresponding to the last interglacial period (130–25 kya), and continued throughout the LGM. For other endorheic populations (BC, NC, MC, YC, PC, KC, and NaC), the rapid growth period began approximately 20 kya, corresponding to post LGM.
Figure 4.
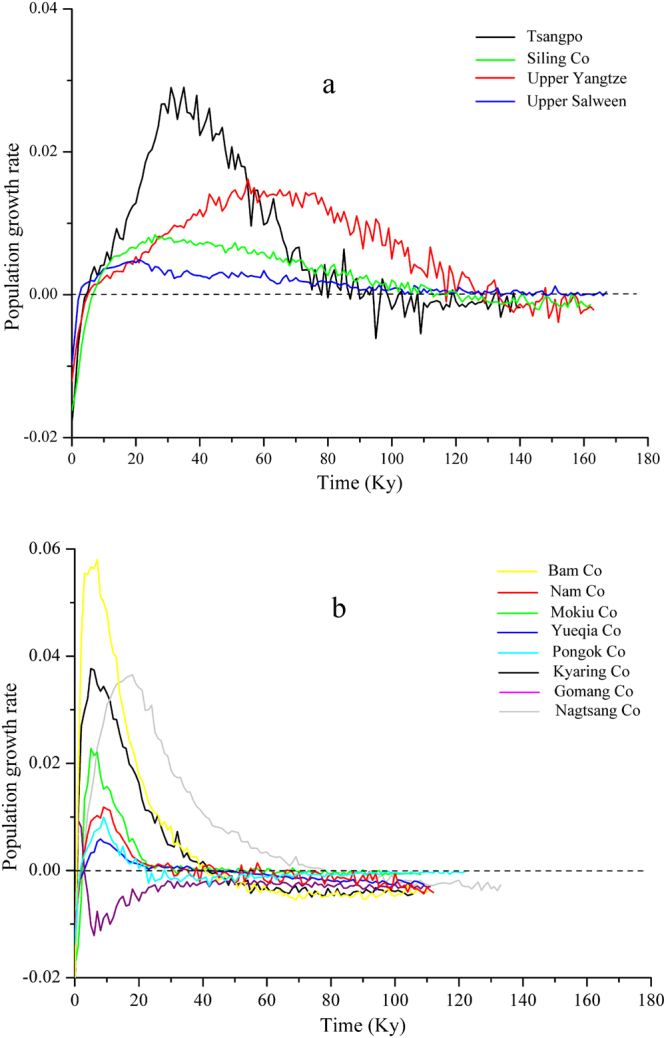
Variation of the instantaneous population growth rate reconstructed by Bayesian skyline plot based on D-loop sequences. The populations of three exorheic rivers and Siling Co (a) showed longer expansion history than other endorheic lakes (b). The x-axis is the timescale before present, and the y-axis is the estimated population instantaneous growth rate (per millennium) calculated by the equation r = (Nt2 − Nt1)/ Nt1.
Species distribution modelling
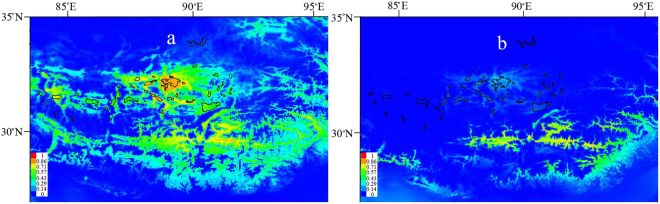
We used MaxEnt v3.3.3 to predict the potential range of the five taxa in the present and during the LGM based on 97 location records (Table S4). These five taxa have similar ecological requirements and occupied similar habitats and were therefore pooled together in our range prediction analyses. The average testing of the area under the curve (AUC) of 5 replications was 0.881 ± 0.030, implying that our model had good predictive power. The mean temperature of the warmest quarter was estimated as the main limiting environmental variable during the LGM. The predicted present distribution range was consistent with the extant distribution of those five taxa22. The community climate system model (CCSM) and the model for interdisciplinary research on climate (MIROC) predicted similar suitable habitat ranges during the LGM; therefore, we only show the result from the CCSM (Fig. 5). During the LGM, suitable habitats on the central QTP were largely reduced and mainly restricted at the south-eastern edge of the QTP. Notably, the area near Siling Co also showed a certain degree of habitat suitability, indicating that schizothoracine fishes may have persisted in this area during the LGM.
Figure 5.
Potential range of the five taxa of schizothoracine fishes on the Qiangtang Plateau in present day (a) and during the LGM (b). The predicted range during the LGM was based on a community climate system model (CCSM). The colour gradient from blue to red indicates the suitability levels from low to high. The maps were drawn using ArcGIS v10.0 and Adobe Illustrator CS5 v15.1.0.
Discussion
The results from the phylogeny and genetic structure analyses of schizothoracine fishes on the central QTP were inconsistent with the hypothesis of “edge refugia”. Commonly, glacial refugia tend to harbour more ancestral haplotypes and higher genetic diversity than recolonized regions, and populations derived from separated refugia show clear genetic differentiation due to drifts3, 5. In addition, postglacial expansion would create clines of decreasing genetic diversity along the expansion route, with increasing distance from refugia because of founder effects and bottlenecks3, 30. This phenomenon has been widely observed in European biota along a south–north gradient31. If the south-eastern and (or) eastern edge of the QTP acted as refugia for fish on the central QTP, the endorheic populations would not have a distinct genetic lineage, and the genetic diversity would have gradually decreased from peripheral populations to the interior. This was not the case in our study.
In our study, phylogenetic analyses and network analyses showed that the sublineage S1 of D-loop haplotypes was generally private to endorheic populations (Figs 1 and 2). Secondly, although exorheic populations showed high genetic diversity, there was no clear cline of genetic diversity decrease from the periphery to the interior. Instead, for all endorheic lakes, the most interior Siling Co harboured the highest genetic diversity, and we detected a clear tendency of decreasing genetic diversity from Siling Co to the outer lakes, for example, from Siling Co to Mokiu Co and to Yueqia Co (Table 2). Thirdly, four ancestral haplotypes (Hap10, Hap47, Hap80 and Hap83) were uncovered in lineage S using network analysis (Fig. 2). All four ancestral haplotypes were detected in the Siling Co area (Table S2, Supplementary information). It is unlikely that these ancestral haplotypes arrived by postglacial dispersal from exorheic rivers, as only two of them were found in Yarlung Tsangpo (Hap47 and Hap83) and three in Salween (Hap47, Hap80 and Hap83, Fig. 2). A logical explanation is that the ancestral haplotypes existed in the glacial refugia of this region over a long period and survived the LGM in situ. The above results suggest that schizothoracine fishes on the Qiangtang Plateau survived the LGM in local refugia and that Siling Co, the largest lake in Tibet (2 200 km2), which is lower than most surrounding lakes32 and is likely a cryptic glacial refugia for local fishes.
The results from demographic history reconstruction, SDM and previous geological studies support our speculation. Glacial refugia provide protection from glaciations; therefore, populations in refugia would have a longer demographic history than would postglacially expanded populations3. The demographic history reconstruction revealed that the most recent population growth of three exorheic rivers and Siling Co began approximately 100 kya, corresponding to the last interglacial period (130–25 kya). For most other endorheic populations, fast growth began approximately 20 kya, corresponding to post-LGM (Fig. 4). The Nagtsang Co population also showed earlier expansion than most endorheic populations, possibly because it is located just beside the Siling Co and is well connected. A similar demographic history was also suggested by He et al.33. The SDM also demonstrated that while most suitable habitats disappeared on the Qiangtang Plateau during the LGM, the Siling Co area showed a certain degree of suitability for those five taxa to live (Fig. 5). In addition, the geological studies suggest that the equilibrium line around the Siling Co basin (approximately 5600 m asl at present) descended by approximately 300 m during the LGM6, 34; therefore, Siling Co (4540 m asl) was approximately 800 m lower than the equilibrium line during the LGM. In our sampling sites, Yueqia Co (YC, 4810 m asl) was approximately 800 m lower than the equilibrium line. Now that schizothoracine fishes can currently complete their life history in Yueqia Co, they were likely able to complete their life history in Siling Co during the LGM. Using phylogeographic analyses, Wang et al.35 and Wang et al.36 discovered the highest microrefugia (just above 4000 m asl) ever reported. With an elevation of 4540 m asl, Siling Co is the highest glacial microrefugia known.
Phylogram from D-loop haplotypes revealed two well-supported lineages (lineages N and S) separated by the high intervening Tanggula Mountains. Lineage S was further divided into four main sublineages. Freshwater fish have restrictive aquatic requirements for dispersal. However, we noticed that the bootstrap values of the four sublineages are low (Fig. S1, Supplementary information). Except for sublineage S1, which was generally private to endorheic drainages, each of the other three sublineages (S2, S3 and S4) was composed of individuals from at least two drainages, and many haplotypes were shared by at least two drainages (Figs 1 and 2). In addition, the Fst values among the drainages south of Tanggula Mountains were low (Table S3, Supplementary information). These results indicated a certain degree of recent migration among drainages37. In total, four or five glaciations have occurred on the QTP since the Pleistocene, the last being the LGM7, 38. During the warm and wet interglacial periods, increased rainfall and melting water expanded the lake’s area and generated several temporary channels among drainages39. For example, during the last interglacial period, the QTP experienced a “high lake level period” (40–25 kya), in which the temperature was 2–4 °C higher and precipitation was 40–100% higher than today40. During that period, many presently isolated lakes, such as Nam Co, Siling Co and Pangkog Co, were expanded several times and connected with each other. In addition, the endorheic and exorheic drainages were also connected at certain times by temporary watercourses40–42. These temporary connections propelled gene flow among drainages, reducing their phylogenetic and genetic differentiation. They also served as migration routes for glacial retreat and postglacial dispersion.
The combined use of mtDNA sequences and nuclear SSR gives a more precise view of species evolutionary history and thus increases the performance of phylogenetic studies43. Compared with mtDNA sequences, SSR markers presented higher variability and faster mutation rate and provided more information to detect recent divergence44. In this study, most results from mtDNA data are consistent with those from nuclear microsatellites, except for the Bam Co and Gomang Co populations. In mtDNA analyses, haplotypes from Bam Co and Gomang Co group with those from other endorheic populations (Fig. 1), but the nuclear microsatellite analysis showed that they have a relatively independent population structure (Fig. 3). The specific geographic feature of those two lakes may contribute to this discordance. Gomang Co is located in a valley, with a small basin area but a high watershed with other lakes (60 m above its present level). For Bam Co, the watershed is much lower in the east than in other directions. During the “high lake level period”, when many endorheic lakes were connected, Gomang Co was still isolated from other lakes, and Bam Co was connected with eastern lakes rather than the Siling Co lake group in the west45. These recent divergence could have been detected by SSR analyses but not mtDNA analyses.
In summary, our study provides a comprehensive evaluation of how Pleistocene climatic oscillations have influenced the genetic variation of schizothoracine fishes on the central QTP and gives one example of such processes for the QTP freshwater fishes. Phylogeographic structure and SDM support the scenario of multiple glacial refugia on the plateau platform during the LGM and suggest Siling Co (4540 m asl) to be a cryptic microrefugia for schizothoracine fish on the central QTP that survived the LGM. Our study also illustrates how postglacial lake expansion has influenced the genetic structure among the drainages of those species, especially for the endorheic drainages.
Materials and Methods
Ethics statement
The methods involving animals in this study were conducted in accordance with the Laboratory Animal Management Principles of China. All experimental protocols were approved by the Ethics Committee of the Institute of Hydrobiology, Chinese Academy of Sciences.
Sample collection and DNA extraction
Specimens were sampled with gill nets and electrofishing. Fish identification was according to Fauna Sinica22. Measurements were taken immediately after collection. The fin tissue of each specimen was collected and preserved in 95% ethanol. Total genomic DNA was extracted using a standard phenol–chloroform method46. DNA concentration was estimated using a NanoDrop 2000 supermicro spectrophotometer (Thermo Fisher Scientific Inc., USA) and adjusted to 50 ng /μL for polymerase chain reaction (PCR).
MtDNA amplification and genetic diversity analysis
The mtDNA D-loop sequence was amplified using the pair of primers DL (5′-ACCCCTGGCTCCCAAAGC-3′) and DH (5′-ATCTTAGCATCTTCAGTG-3′) in a 60 μL mixture containing 6 μL 10 × PCR buffer (600 μM Tris-HCl, pH 8.3, 3 mM KCl, 90 μM MgCl2), dNTPs at 150 μM, forward and reverse primers at 10 pM, 1.0 unit Taq polymerase (TaKaRa) and 50 ng genomic DNA. PCR was performed with a preliminary denaturation at 94 °C for 3 min; followed by 30 cycles of strand denaturation at 94 °C for 40 s, annealing at 55 °C for 50 s and primer extension at 72 °C for 45 s; and a final extension at 72 °C for 10 min. The PCR products were sequenced on an ABI 3730 capillary sequencer from the Sangon Biotech Company (Shanghai, China). Sequences were checked with Bioedit v. 7.2.547 and corrected manually.
The D-loop sequences were initially aligned with the program Clustal × 2.148. All sequences were collapsed into unique haplotypes, and variable sites were counted with DnaSP v5.1049. Basic statistics (haplotype number (N), mean number of pairwise differences (p), haplotype (h) and nucleotide (π) diversity) were calculated for each population with Arlequin v 3.5.150.
MtDNA phylogenetic and population genetic analyses
Phylogenetic analyses of D-loop haplotypes were conducted using BI and ML, by MrBayes v3.2.351 and PhyML 3.052 respectively. Two D-loop sequences of Gymnocypris przewalskii and Gymnocypris eckloni were designated as the outgroup. The best nucleotide substitution model (GTR + G + I) was selected among 88 alternative models by the Bayesian Information Criteria (BIC) using jModelTest v2.1.453. For Bayesian analyses, two independent runs with four (one cold and three incrementally heated) Markov Chain Monte Carlo (MCMC) were conducted with the GTR + G + I substitution model, run for 20 000 000 generations and 1/1000 sample frequency. Based on convergence diagnostics, the first 4 000 trees (standard deviation of split frequencies <0.01) were discarded as burn-in. Nodal support was calculated using the mean posterior probability (BBP) values of each node of the resulting consensus tree after burn-in. For ML analysis, the node support was assessed using the nonparametric bootstrap method with 1000 replicates. The tree was visualized with FigTree v1.4.254.
A statistical parsimony network of haplotypes was constructed using TCS1.2.155 with a 95% criterion for a parsimonious connection. Excessive connections in the network were removed based on coalescent theory56. Genetic differentiation among populations was assessed by pairwise genetic distance (Fst) values. In this section, we treated every river as a single site. The genetic structure within and among drainages was further estimated by AMOVA. Populations within one drainage were pooled into one group. Both analyses were implemented in Arlequin v3.5.150.
MtDNA demographic history analyses
To assess the historical changes in effective population size, we used multiple approaches. First, we calculated Tajima’s D 57 and Fu’s Fs 58 tests to detect population expansions. The significance of both values was calculated from 10 000 simulated samples using a coalescent algorithm. Second, we performed pairwise mismatch distribution analysis (MDA) to compare theoretical distributions under sudden expansion with observed data. Goodness-of-fit was evaluated by the SSD and Raggedness index (Ri) between observed and expected mismatch distributions. The above two analyses were conducted using Arlequin 3.5.150. Finally, we generated the BSP for each population using a Bayesian MCMC algorithm in BEAST 1.8.2 and BEAUti 1.8.2 to convert the molecular sequences of each lake into the input file59. The best nucleotide substitution model was selected by jModelTest 2.1.4., using the Akaike Information Criterion (AIC).
An evolutionary rate of 0.91% per million years for the cyt b sequence of the genus Gymnocypris has been calibrated by the isolation between the upper reaches of the Yellow River and Qinghai Lake, which is a reliable geological event25. We calculated the divergence rate of cyt b and D-loop sequences between H. microcephalus in Tuotuo He and Schizopygopsis pylzovi in the Yellow River using MEGA 6.060. Based on the observed divergence rate of 1.98% for cyt b and 2.32% for the D-loop, we estimated an evolutionary rate of 1.07% per million years for the D-loop sequence. We used a strict clock model and four independent runs of 200 million generations, sampling every 2 000 runs. Tracer 1.6 was used to confirm stationary and convergence, determine the burn-in value (10%) and create the BSP61. All runs were combined using LogCombiner 1.8.2 and created a consensus tree using TreeAnnotator 1.8.2, from the BEAST package.
SSR genotyping and population structure analyses
In total, 33 published SSR primer pairs of Schizopygopsis younghusbandi 62, 63 were used to screen polymorphisms in 40 individuals from four populations (SC, LR, TH and ATC). PCR was performed as previously described, except for some variations in annealing temperature. The amplified fragments were separated on a 12% non-denaturing polyacrylamide gel. We chose 12 pairs of highly polymorphic SSR primers that amplified reliably across four populations for further genetic analyses (Table S1). Each forward primer for the 12 pairs was labelled with fluorescent dyes (FAM or HEX). PCR products were visualized on an ABI 3730xl DNA Analyser (Beijing Tianyi Huiyuan Bioscience & Technology Corporation). Every individual presented up to 4 alleles, indicating that the samples were tetraploids. Therefore, the genotypes of samples were assessed by the fluorescence height of the alleles according to the Microsatellite DNA Allele Counting Peak Ratios (MAC-PR) method64.
The number of alleles per locus (Na), allelic richness (Ai) and observed (HO) and expected heterozygosity (HE) were calculated using the program AUTOTET65. Fst values among populations were estimated by POLYSAT66. The population genetic structure was assessed by a Bayesian model-based method in STRUCTURE 2.3.467. We chose the admixture model for the ancestry of individuals and the correlated model for allele frequencies. The length of burn-in was set to 10 000, with 100 000 MCMC replication runs after burn-in. To avoid bias, we performed the program from K = 1 to 14, with 30 iterations at each value of K. The optimum value of clusters (K) was determined by comparison of mean log probability Ln P (D) and by calculating the ∆K value for each K68.
Species distribution modelling
We used SDM to predict the potential range of these five taxa in the present and during the LGM. Models were generated with MaxEnt v3.3.369, a program for maximum entropy modelling of the geographical distributions of species. Overall, 97 location records were collected, including 19 from the present study and 78 from our previous survey from 2000 to 2015 (Table S4). We downloaded 19 commonly used bioclim-variables from WorldClim (http://www.worldclim.org), with a spatial resolution of 2.5 arc-minutes. The altitude data for these sites were recorded by GPS. Highly correlated environmental variables were removed using multicollinearity test by ENMtools v1.370, and six bioclim-variables (annual mean temperature, minimum temperature of coldest month, mean temperature of warmest and coldest quarter, annual precipitation and precipitation of wettest quarter) and altitude were ultimately used.
Both the CCSM and MIROC were used to generate the environmental variables for the LGM. Those data layers were also downloaded from WorldClim. We chose the climatic-topographic models (climate model) for both time periods and set the convergence threshold and number of maximum iterations at default values of 0.00001and 500 respectively. We assigned 75% of the occurrence point as training data and the remaining 25% as testing data. The number of maximum iterations and convergence threshold were set at default values of 500 and 0.00001, respectively. We ran five fold cross-validation for each model and produced a single map per time period using the logistic output format, with suitability values ranging from 0 to 167. Model evaluation was based on the AUC71.
Electronic supplementary material
Acknowledgements
We thank Dr. Xiaoyun Sui, Mr. Chunlong Liu and Mr. Juan Tao for their help with sample collection and writing. We thank Dr. Xiu Feng, Mr. Kang Chen, Ms. Yu Cheng and Ms. Jie Zhao for their experimental assistance. This work was supported by the Natural Science Foundation of China (Nos 41030208, 31572248), the Strategic Priority Research Program and the Knowledge Innovation Project of the Chinese Academy of Sciences (Nos XDPB020403, KSCX2-YW-Z-1023), and the National Science and Technology Basic Special (Nos 2014FY210700, 2014FY210200).
Author Contributions
Y.L. contributed to molecular experiment and data analyses and drafted the manuscript. D.H. contributed to sampling, molecular experiments and data analyses. Y.J. and H.S. contributed to sampling and data analyses. Y.C. contributed to research design and the writing of the manuscript. All authors have reviewed and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11198-w
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 2.Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Phil. Trans. R. Soc. Lond. B. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Provan J, Bennett KD. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 2008;23:564–571. doi: 10.1016/j.tree.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Robin V, Nadeau MJ, Grootes PM, Bork HR, Nelle O. Too early and too northerly: evidence of temperate trees in northern Central Europe during the Younger Dryas. New Phytol. 2016;212:259–268. doi: 10.1111/nph.13844. [DOI] [PubMed] [Google Scholar]
- 5.Keppel G, et al. Refugia: identifying and understanding safe havens for biodiversity under climate change. Global Ecol. Biogeo. 2012;21:393–404. doi: 10.1111/j.1466-8238.2011.00686.x. [DOI] [Google Scholar]
- 6.Lehmkuhl F, Owen LA. Late Quaternary glaciation of Tibet and the bordering mountains: a review. Boreas. 2005;34:87–100. doi: 10.1080/03009480510012908. [DOI] [Google Scholar]
- 7.Zheng BX, Xu QQ, Shen YP. The relationship between climate change and Quaternary glacial cycles on the Qinghai-Tibetan Plateau: review and speculation. Quatern. Int. 2002;97-8:93–101. doi: 10.1016/S1040-6182(02)00054-X. [DOI] [Google Scholar]
- 8.Favre A, et al. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. 2015;90:236–253. doi: 10.1111/brv.12107. [DOI] [PubMed] [Google Scholar]
- 9.Li JJ, Fang XM. Uplift of the Tibetan Plateau and environmental changes. Chinese Sci. Bull. 1999;44:2117–2124. doi: 10.1007/BF03182692. [DOI] [Google Scholar]
- 10.Clark PU, et al. The Last Glacial Maximum. Science. 2009;325:710–714. doi: 10.1126/science.1172873. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y. Characteristics of late Quaternary monsoonal glaciation on the Tibetan Plateau and in East Asia. Quatern. Int. 2002;97-8:79–91. doi: 10.1016/S1040-6182(02)00053-8. [DOI] [Google Scholar]
- 12.Opgenoorth L, et al. Tree endurance on the Tibetan Plateau marks the world’s highest known tree line of the Last Glacial Maximum. New Phytol. 2010;185:332–342. doi: 10.1111/j.1469-8137.2009.03007.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou W, et al. River islands, refugia and genetic structuring in the endemic brown frog Rana kukunoris (Anura, Ranidae) of the Qinghai-Tibetan Plateau. Mol. Ecol. 2013;22:130–142. doi: 10.1111/mec.12087. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, et al. Phylogeography of Nanorana parkeri (Anura: Ranidae) and multiple refugia on the Tibetan Plateau revealed by mitochondrial and nuclear DNA. Sci. Rep. 2015;5:9857. doi: 10.1038/srep09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhan XJ, Zheng YF, Wei FW, Bruford MW, Jia CX. Molecular evidence for Pleistocene refugia at the eastern edge of the Tibetan Plateau. Mol. Ecol. 2011;20:3014–3026. doi: 10.1111/j.1365-294X.2011.05144.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Q, Chiang TY, George M, Liu JQ, Abbott RJ. Phylogeography of the Qinghai-Tibetan Plateau endemic Juniperus przewalskii (Cupressaceae) inferred from chloroplast DNA sequence variation. Mol. Ecol. 2005;14:3513–3524. doi: 10.1111/j.1365-294X.2005.02677.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen S, et al. Potential refugium on the Qinghai-Tibet Plateau revealed by the chloroplast DNA phylogeography of the alpine species Metagentiana striata (Gentianaceae) Bot. J. Linn. Soc. 2008;157:125–140. doi: 10.1111/j.1095-8339.2008.00785.x. [DOI] [Google Scholar]
- 18.Cun YZ, Wang XQ. Plant recolonization in the Himalaya from the southeastern Qinghai-Tibetan Plateau: Geographical isolation contributed to high population differentiation. Mol. Phylogenet. Evol. 2010;56:972–982. doi: 10.1016/j.ympev.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Qu YH, Ericson PGP, Lei FM, Li SH. Postglacial colonization of the Tibetan plateau inferred from the matrilineal genetic structure of the endemic red-necked snow finch, Pyrgilauda ruficollis. Mol. Ecol. 2005;14:1767–1781. doi: 10.1111/j.1365-294X.2005.02528.x. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann S. Population genetic structure and geographic differentiation in the hot spring snake Thermophis baileyi (Serpentes, Colubridae): indications for glacial refuges in southern-central Tibet. Mol. Phylogenet. Evol. 2012;63:396–406. doi: 10.1016/j.ympev.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Harris LN, Taylor EB. Pleistocene glaciations and contemporary genetic diversity in a Beringian fish, the broad whitefish, Coregonus nasus (Pallas): inferences from microsatellite DNA variation. J. Evolution. Biol. 2010;23:72–86. doi: 10.1111/j.1420-9101.2009.01858.x. [DOI] [PubMed] [Google Scholar]
- 22.Chen, Y. & Cao, W. Schizothoracinae. In Fauna Sinica, Osteichthyes: Cypriniformes III (ed. Yue, P.) 275-390 (Science Press, Beijing, 2000, in Chinese).
- 23.He D, Chen Y. Biogeography and molecular phylogeny of the genus Schizothorax (Teleostei: Cyprinidae) in China inferred from cytochrome b sequences. J. Biogeogr. 2006;33:1448–1460. doi: 10.1111/j.1365-2699.2006.01510.x. [DOI] [Google Scholar]
- 24.Cao, W., Chen, Y., Wu, Y. & Zhu, S. Origin and evolution of Schizothorachine fishes in relation to the upheaval of the Xizang Plateau. In Studies on the period, amplitude and type of the uplift of the Qinghai-Xizang Plateau (ed. Tibetan expedition team of the Chinese Academy of Science) 118-130 (Science Press, Beijing, 1981, in Chinese).
- 25.He D, Chen Y, Chen Y, Chen Z. Molecular phylogeny of the specialized schizothoracine fishes (Teleostei: Cyprinidae), with their implications for the uplift of the Qinghai-Tibetan Plateau. Chinese Sci. Bull. 2004;49:39–48. doi: 10.1007/BF02901741. [DOI] [Google Scholar]
- 26.Chen, C. & Fan, Y. The river, lake and water resource on Qiangtang Plateau. Resource Science2, 38-43 (1983, in Chinese).
- 27.Tapponnier P, et al. Oblique stepwise rise and growth of the Tibet plateau. Science. 2001;294:1671–1677. doi: 10.1126/science.105978. [DOI] [PubMed] [Google Scholar]
- 28.Liao PC, et al. Historical spatial range expansion and a very recent bottleneck of Cinnamomum kanehirae Hay.(Lauraceae) in Taiwan inferred from nuclear genes. BMC Evol. Biol. 2010;10:124. doi: 10.1186/1471-2148-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maruyama T, Fuerst PA. Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics. 1985;111:675–689. doi: 10.1093/genetics/111.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comes HP, Kadereit JW. The effect of quaternary climatic changes on plant distribution and evolution. Trends Plant Sci. 1998;3:432–438. doi: 10.1016/S1360-1385(98)01327-2. [DOI] [Google Scholar]
- 31.Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol.J. Linn. Soc. 1996;58:247–276. doi: 10.1111/j.1095-8312.1996.tb01434.x. [DOI] [Google Scholar]
- 32.Wan W, et al. Monitoring lake changes of Qinghai-Tibetan Plateau over the past 30 years using satellite remote sensing data. Chinese Sci. Bull. 2014;59:1021–1035. doi: 10.1007/s11434-014-0128-6. [DOI] [Google Scholar]
- 33.He D, et al. Comparative phylogeography and evolutionary history of schizothoracine fishes in the Changtang Plateau and their implications for the lake level and Pleistocene climate fluctuations. Ecol. Evol. 2016;6(3):656–674. doi: 10.1002/ece3.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi Y, Zheng B, Yao T. Glaciers and Environments during the Last Glacial Maximum (LGM) on the Tibetan Plateau. J. Glaciol. Geocryol. 1997;19:97–113. [Google Scholar]
- 35.Wang H, et al. Phylogeographic structure of Hippophae tibetana (Elaeagnaceae) highlights the highest microrefugia and the rapid uplift of the Qinghai-Tibetan Plateau. Mol. Ecol. 2010;19:2964–2979. doi: 10.1111/j.1365-294X.2010.04729.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang GN, He XY, Miehe G, Mao KS. Phylogeography of the Qinghai-Tibet Plateau endemic alpine herb Pomatosace filicula (Primulaceae) J. Syst. Evol. 2014;52:289–302. doi: 10.1111/jse.12089. [DOI] [Google Scholar]
- 37.Aldenhoven JT, Miller MA, Corneli PS, Shapiro MD. Phylogeography of ninespine sticklebacks (Pungitius pungitius) in North America: glacial refugia and the origins of adaptive traits. Mol. Ecol. 2010;19:4061–4076. doi: 10.1111/j.1365-294X.2010.04801.x. [DOI] [PubMed] [Google Scholar]
- 38.Shi Y. A suggestion to improve the chronology of Quaternary glaciations in China. J. Glaciol. Geocryol. 2002;24:687–692. [Google Scholar]
- 39.Fan Q, Ma H, Hou G. Late Pleistocene lake and glaciation evolution on the northeastern Qinghai-Tibetan Plateau: a review. Environmental Earth Sci. 2012;66:625–634. doi: 10.1007/s12665-011-1271-x. [DOI] [Google Scholar]
- 40.Shi YF, Yu G, Liu XD, Li BY, Yao TD. Reconstruction of the 30-40 ka BP enhanced Indian monsoon climate based on geological records from the Tibetan Plateau. Palaeogeogr. Palaeocl. 2001;169:69–83. doi: 10.1016/S0031-0182(01)00216-4. [DOI] [Google Scholar]
- 41.Bing-yuan L, Li-ping Z. “Greatest lake period” and its palaeo-environment on the Tibetan Plateau. J. Geogr. Sci. 2001;11:34–42. doi: 10.1007/BF02837374. [DOI] [Google Scholar]
- 42.Zhu, D. et al. Evolution and climatic change of Nam Co of Tibet and an ancient large lake in the northern Tibetan Plateau since the late Pleistocene. Geology in China31, (2004).
- 43.Seifertova M, Bryja J, Vyskocilova M, Martinkova N, Simkova A. Multiple Pleistocene refugia and post-glacial colonization in the European chub (Squalius cephalus) revealed by combined use of nuclear and mitochondrial markers. J. Biogeogr. 2012;39:1024–1040. doi: 10.1111/j.1365-2699.2011.02661.x. [DOI] [Google Scholar]
- 44.Ellegren H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 45.Guan, Z. et al. Rivers and Lakes in Tibet 115-216 (Science Press, Beijing, 1984, in Chinese).
- 46.Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular cloning: a laboratory manual (Cold Spring Harbor Laboratory Press, New York, 1989).
- 47.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 48.Larkin MA, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 49.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 50.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 51.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 53.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rambaut, A. FigTree v1.4.2: Tree figure drawing tool. Available at: http://tree.bio.ed.ac.uk/software/figtree (2014).
- 55.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 56.Cosacov A, Sersic AN, Sosa V, Johnson LA, Cocucci AA. Multiple periglacial refugia in the Patagonian steppe and post-glacial colonization of the Andes: the phylogeography of Calceolaria polyrhiza. J. Biogeogr. 2010;37:1463–1477. [Google Scholar]
- 57.Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rambaut, A., Suchard, M. A., Xie, W. & Drummond, A. J. Tracer v.1.6.0: MCMC Trace Analysis Tool. Available at: http://tree.bio.ed.ac.uk/software/tracer/ (2013).
- 62.He D, Tang W, Li C, Chen Y. Development of eighteen polymorphic microsatellite markers in Schizopygopsis younghusbandi Regan, 1905 (Ostariophysi: Cyprinidae) in Tibet, China. Conserv. Genet. Resour. 2013;5:895–897. doi: 10.1007/s12686-013-9927-3. [DOI] [Google Scholar]
- 63.Zheng H, et al. Isolation and characterization of fifteen polymorphic tetranucleotide microsatellite loci in Schizopygopsis younghusbandi Regan. Conserv. Genet. Resour. 2013;5:1147–1149. doi: 10.1007/s12686-013-9984-7. [DOI] [Google Scholar]
- 64.Esselink GD, Nybom H, Vosman B. Assignment of allelic configuration in polyploids using the MAC-PR (microsatellite DNA allele counting-peak ratios) method. Theor. Appl. Genet. 2004;109:402–408. doi: 10.1007/s00122-004-1645-5. [DOI] [PubMed] [Google Scholar]
- 65.Thrall PH, Young A. AUTOTET: A program for analysis of autotetraploid genotypic data. J. Hered. 2000;91:348–349. doi: 10.1093/jhered/91.4.348. [DOI] [PubMed] [Google Scholar]
- 66.Clark LV, Jasieniuk M. POLYSAT: an R package for polyploid microsatellite analysis. Mol. Ecol. Resour. 2011;11:562–566. doi: 10.1111/j.1755-0998.2011.02985.x. [DOI] [PubMed] [Google Scholar]
- 67.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 69.Phillips SJ, Dudik M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31:161–175. doi: 10.1111/j.0906-7590.2008.5203.x. [DOI] [Google Scholar]
- 70.Warren DL, Glor RE, Turelli M. ENMTools: a toolbox for comparative studies of environmental niche models. Ecography. 2010;33:607–611. doi: 10.1111/j.1600-0587.2009.06041.x. [DOI] [Google Scholar]
- 71.Liu CR, Berry PM, Dawson TP, Pearson RG. Selecting thresholds of occurrence in the prediction of species distributions. Ecography. 2005;28:385–393. doi: 10.1111/j.0906-7590.2005.03957.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.