Abstract
Objective:
To determine the molecular factors contributing to progressive cavitating leukoencephalopathy (PCL) to help resolve the underlying genotype-phenotype associations in the mitochondrial iron-sulfur cluster (ISC) assembly system.
Methods:
The subjects were 3 patients from 2 families who showed no inconsistencies in either clinical or brain MRI findings as PCL. We used exome sequencing, immunoblotting, and enzyme activity assays to establish a molecular diagnosis and determine the roles of ISC-associated factors in PCL.
Results:
We performed genetic analyses on these 3 patients and identified compound heterozygosity for the IBA57 gene, which encodes the mitochondrial iron-sulfur protein assembly factor. Protein expression analysis revealed substantial decreases in IBA57 protein expression in myoblasts and fibroblasts. Immunoblotting revealed substantially reduced expression of SDHB, a subunit of complex II, and lipoic acid synthetase (LIAS). Levels of pyruvate dehydrogenase complex-E2 and α-ketoglutarate dehydrogenase-E2, which use lipoic acid as a cofactor, were also reduced. In activity staining, SDH activity was clearly reduced, but it was ameliorated in mitochondrial fractions from rescued myoblasts. In addition, NFU1 protein expression was also decreased, which is required for the assembly of a subset of iron-sulfur proteins to SDH and LIAS in the mitochondrial ISC assembly system.
Conclusions:
Defects in IBA57 essentially regulate NFU1 expression, and aberrant NFU1 ultimately affects SDH activity and LIAS expression in the ISC biogenesis pathway. This study provides new insights into the role of the iron-sulfur protein assembly system in disorders related to mitochondrial energy metabolism associated with leukoencephalopathy with cavities.
Progressive cavitating leukoencephalopathy (PCL) is an autosomal recessive neurodegenerative disorder, characterized by leukoencephalopathy with cavities showing episodic clinical deterioration, and vascular permeability in the corpus callosum, centrum semiovale, or cerebellum on brain MRI.1 The pathogenesis is currently not well understood. White matter abnormalities occur in patients harboring mutations in genes encoding complex I–III assembly factors acting in mitochondrial energy metabolism.2,3 Iron-sulfur clusters (ISCs) participate in electron transfer in the mitochondrial respiratory chain and act as sulfur donors in the synthesis of lipoic acid (LA) by mitochondrial lipoic acid synthetase (LIAS).4–8 The [2Fe-2S] cluster, synthesized on the ISC unit, is converted by a protein complex consisting of ISCA1, ISCA2, and IBA57 into the [4Fe-4S] form, which is inserted into the apoprotein via complexes I and II, LIAS, and aconitase encoded by NFU1, BOLA3, and IND1.9–14
Neuroradiologically diverse phenotypes of white matter abnormalities, associated with ISC systems, have been reported. Patients with BOLA3 or NFU1 mutations present with encephalopathy and neurologic regression, leading to death in infancy.15–18 Patients with IBA57 mutations show severe encephalopathy to hereditary spastic paraplegia.19–21 However, the genotype-phenotype correlation remains elusive with respect to ISC biogenesis-associated factors. We evaluated the factors that might play a role in ISC biogenesis pathway regulation, resulting in cavitating leukoencephalopathy.
METHODS
Patients.
Patients with PCL were selected based on clinical and radiologic examinations. They showed episodic, acute onset of irritability or neurologic deficits between 11 months and 2 years of age, followed by an intermittent increase in lactate and clinical deterioration. Brain MRI showed patchy leukoencephalopathy with cavities and diffusion restriction in the affected region (figure 1). Patients 1 (II-2a) and 2 (II-3a) are siblings. All clinical materials used in this study were obtained with written informed consent. All experiments were approved by the Ethical Committee of the National Center of Neurology and Psychiatry.
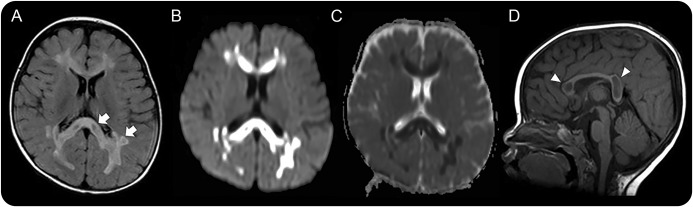
Figure 1. Illustrations of the characteristic MRI pattern in patients with progressive cavitating leukoencephalopathy.
MRIs of patient 1 (A–D) obtained at the disease onset of 11 months. Axial fluid-attenuated inversion recovery hyperintensity images (A), diffusion-weight images (B), apparent diffusion coefficient map images (C), and sagittal T1-weighted images (D) are shown. Abnormal high intensities are noted in the cerebral periventricular and subcortical white matter, which contained multiple cavities (A and B; arrow), as well as in the corpus callosum (D; arrowhead). Diffusion restriction is present in the solid part of the abnormal white matter (C).
Exome sequencing and bioinformatics analysis.
To identify the genetic cause of recessive inherited PCL, we performed whole-exome sequencing on 2 affected (II-2a and II-3a) and 3 nonaffected members (I-1a, I-2a, and II-1a) of family A, and 1 affected (II-1b) and 2 nonaffected members (I-1b and I-2b) of family B (figure 2A). Genetic and bioinformatics analysis was performed according to the procedure as described.22–24 Candidate mutations were validated by Sanger sequencing on an ABI Prism 3130 DNA Analyzer (Applied Biosystems). Within-family IBA57 variant segregation was analyzed using primers designed to amplify exons corresponding to the sequence under accession number NG_042231—PCR conditions, and primer sequences are available upon request.
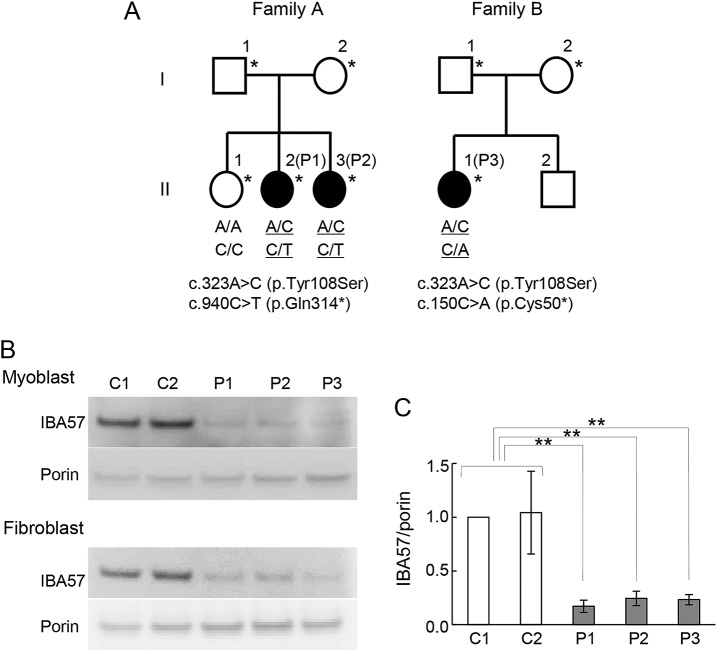
Figure 2. Genetic analysis of recessively inherited leukoencephalopathy and IBA57 protein expression.
(A) Pedigree structures for recessive leukoencephalopathy family A and family B, with filled symbols indicating affected individuals. *Individuals who were analyzed by exome sequencing. IBA57 genotypes are shown below the individuals in the pedigree. (B) IBA57 expression was analyzed by immunoblotting. C1, C2: controls; P1–3: patients. Mitochondrial fractions prepared from patient-derived myoblasts and fibroblasts were analyzed via immunoblotting. IBA57 levels were significantly lower in all patients than in the controls. Porin was used as a loading control. The experiments were performed in triplicate and normalized to porin levels. (C) IBA57 protein expression in mitochondrial fractions prepared from myoblasts. Error bars indicate SDs (**p < 0.005, Student t test).
Cell culture.
Patient-derived cells were cultured in Dulbecco modified Eagle medium/F-12 (GIBCO BRL, USA) supplemented with heat-inactivated fetal bovine serum (20% [vol/vol] for myoblasts and 10% for fibroblasts; GIBCO BRL).
Establishment of immortalized patient-derived myoblasts.
Patient-derived and control myoblasts were transfected with pEF321-T plasmid (a kind gift from Dr. Sumio Sugano, University of Tokyo) and serially cultured for more than 10 population doublings, until morphological alteration was observed.25
Preparation of mitochondrial fractions.
Mitochondrial fractions from skeletal muscle and cultured patient-derived cells were prepared as reported,26 with slight modification.
Immunoblotting.
Mitochondrial fractions were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (PAGE). Thirty micrograms of mitochondrial protein was separated on 4%–12% Bis-Tris gradient gels (Life Technologies, Carlsbad, CA) and transferred to polyvinylidene fluoride membranes as described.27 The membranes were incubated with primary antibodies against IBA57 (Sigma, St. Louis, MO), VDAC1 (Abcam, Cambridge, UK), LA (Calbiochem, San Diego, CA), NDUFS1, NDUFS8, NDUFV1, SDHB, and UQCRFS1 (all from GeneTex, Irvine, CA). Protein levels of IBA57 and ISC were quantified with ImageJ software (National Institutes of Health, Bethesda, MD).
Measurement of respiratory enzymatic activity.
Enzymatic activities of mitochondrial respiratory complexes I–V and citrate synthase (CS) were measured in mitochondrial fractions prepared from the patients' skeletal muscle and patient-derived fibroblasts. The assays for complexes I–IV and CS were conducted as described.27 The assay for complex V was performed according to Morava et al.28
High-resolution clear native PAGE and enzyme activity staining.
High-resolution clear native PAGE and activity staining were performed as described.29 In brief, isolated mitochondria were solubilized with Native PAGE Sample Buffer (Invitrogen) containing 0.3% n-dodecyl-β-d-maltoside (Dojindo, Kumamoto, Japan). Thirty micrograms of protein was applied to NativePAGE Novex 3%–12% Bis-Tris Gel (Invitrogen). Native PAGE Buffer (Invitrogen) was used as the anode buffer, and Native PAGE Buffer containing 0.02% n-dodecyl-β-d-maltoside and 0.05% deoxycholate was used as the cathode buffer.
For in-gel catalytic activity assays, the gels were incubated as described.29
Blue native PAGE and immunoblotting.
Blue native (BN)-PAGE and immunoblotting were performed as described.28 In brief, isolated mitochondria were solubilized with 0.5% (w/v) n-dodecyl-β-d-maltoside. Protein (30 μg) was applied to NativePAGE Novex 3%–12% Bis-Tris Gel. Immunoblotting was conducted as described above. Primary antibodies targeted NDUFA9 as complex I, SDHA as complex II, UQCRC2 as complex III, MTCO1 as complex IV, and ATP5B as complex V.
Expression vector construction and transfection.
For mammalian expression vector construction, full-length IBA57 was amplified from complementary DNA (cDNA) prepared from the control subject by PCR. The PCR product was cloned into pEBMulti-Pur (Wako, Osaka, Japan) and verified by Sanger sequencing. Mammalian expression vector, pEBMulti-Pur construct containing IBA57 cDNA, was transfected into immortalized myoblasts using Lipofectamine LTX Reagent (Thermo Fisher Scientific, Waltham, MA).
RESULTS
Clinical findings.
The detailed clinical and laboratory findings are summarized in table e-1 at Neurology.org/ng. Characteristic symptoms were intermittent episodic clinical deterioration and an infection-associated increase in lactate. Psychomotor development was normal until clinical onset between 11 months and 2 years of age. The patients showed marked irritability and lost acquired skills due to repeated infections. These episodes led to psychomotor regression with axial hypotonia accompanied by spasticity of the legs. Treatment with a mitochondrial cocktail recovered some of the lost skills during periods of remission. Plasma and CSF lactate levels were slightly increased intermittently, and serum amino acid and urine organic acid levels were normal based on gas chromatography/mass spectrometry. Muscle biopsy showed normal histology (data not shown). The last neurologic examination at the age of 6–7 years varied between normal and mild cognitive and acquisitive deficiency, assessed as a score of 78–82 and 73–85, respectively, according to the Kaufmann Assessment Battery for Children, Second Edition.
MRI abnormalities.
Brain MRI in all 3 patients revealed progressive leukoencephalopathy with extensive signal abnormalities in the periventricular cerebral white matter and corpus callosum (figure 1, figure e-1). The abnormal white matter in the genu and splenium of the corpus callosum showed partial cavitation. The basal ganglia, cerebellum, and brain stem were speared. Magnetic resonance spectroscopy showed an increased lactate concentration in the white matter. Diffusion restriction with low apparent diffusion coefficient values was observed in the noncavitated abnormal white matter.
Genetic analyses.
In total, 13 candidate genes from family A and 20 candidate genes from family B were assessed based on the assumption of autosomal recessive inheritance and with reference to the dbSNP135, 1000 Genomes Project database, NHLBI Exome Variant Server, and human genetic variation database for Japanese Genetic Variants databases. Among all extracted variants, we identified only 1 associated gene, IBA57, common to both families, with compound heterozygous mutations of c.323A>C and c.940C>T in family A and c.323A>C and c.150C>A in family B (RefSeq accession number NM_001010867). These variants in IBA57, encoding the mitochondrial iron-sulfur protein assembly factor, are predicted to lead to a missense and a nonsense mutation (figure 2A). The mutations were confirmed by Sanger sequencing and the compound heterozygosity by transcript analysis. In silico analyses of mutation function demonstrated that p.Tyr108Ser is predicted as damaging based on scale-invariant feature transform and Polymorphism Phenotyping v2. The amino acids involved are moderately to highly conserved.
Functional studies.
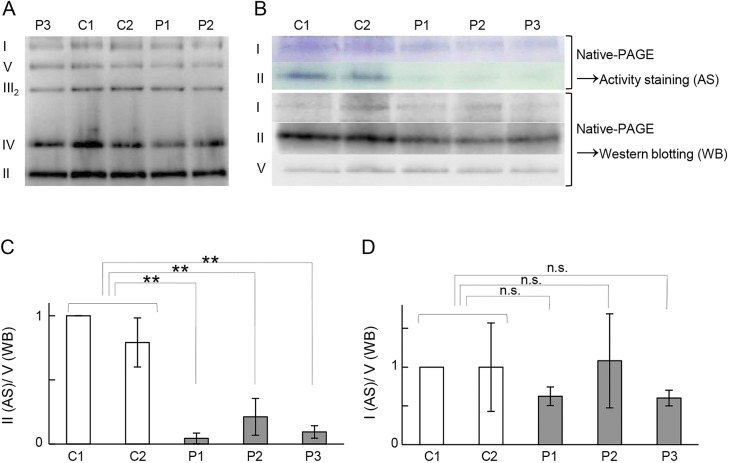
IBA57 protein expression was evaluated by immunoblotting using mitochondrial fractions prepared from patient-derived myoblasts and fibroblasts. IBA57 protein was substantially decreased in the patients; a small amount of full-length IBA57, but no truncated IBA57, was detected in the patients (figure 2, B and C). This result indicated that the mutations severely affect the level of IBA57 protein. Given that IBA57 is a mitochondrial iron-sulfur protein assembly factor, we first tested the effects of the mutations on the respiratory complexes by measuring enzyme activity and analyzing complex assembly by BN-PAGE (figure 3A) in myoblast mitochondrial fractions. Although the activities of these complexes were slightly lowered, the differences were not significant, and there were no differences in their amounts (figure 3A). As we did not detect clear decreases in activity or assembly, we next used activity staining to determine any change in the respiratory complexes. The staining intensity for SDH was weaker in all patients than in controls, indicating a defect in SDH activity (figure 3, B and C). By contrast, there was no clear decrease in complex I activity staining (figure 3, C and D).
Figure 3. Blue native polyacrylamide gel electrophoresis of respiratory chain complexes in the 3 patients (P1, P2, and P3) and controls (C1 and C2).
(A) Blue native polyacrylamide gel electrophoresis (PAGE) was performed using mitochondrial fractions prepared from myoblasts. No patient showed severely reduced assembly of respiratory chain complexes as compared to the control. (B) In-gel activity staining and Western blotting of complexes I and II. In-gel activity staining of complex II after separation by high-resolution clear native (hrCN)-PAGE using mitochondrial fractions prepared from the myoblasts. In parallel, complexes I, II, and V were detected by Western blotting after their separation by hrCN-PAGE. All patients showed clearly reduced complex II activity when compared with the control, whereas no such decrease was detected by Western blotting. The activity and amount of assembled complex I were not significantly different from those of controls. (C, D) The experiments were performed in triplicate and normalized to complex V (n.s. = not significant, **p < 0.005, Student t test).
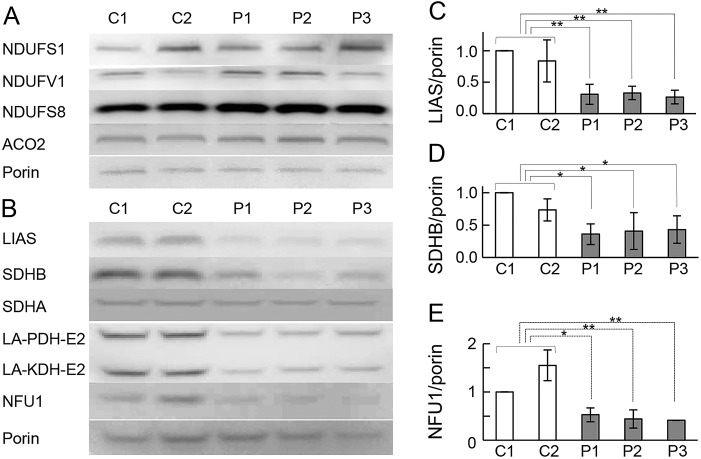
To examine the effects of the IBA57 mutations further, we assessed the expression level of each mitochondrial [4Fe-4S] protein, including 5 subunits of mitochondrial respiratory chain complexes (NDUFS1, NDUFS7, NDUFS8, and NDUFV1 in respiratory complex I, and SDHB in respiratory complex II), ACO2, and LIAS. Protein expression of the 4 subunits in complex I and ACO2 were not substantially reduced in patient vs control myoblasts (figure 4A). The levels of LIAS and the SDHB, but not the SDHA subunit of complex II, were lower in patient myoblasts (figure 4, B–D). To examine the effect of the defect in IBA57 on LIAS function, an anti-LA antibody was used to detect LA bound to the key mitochondrial enzymes pyruvate dehydrogenase (PDH) complex and α-ketoglutarate dehydrogenase (KDH), which use LA as a cofactor to form LA-PDH-E2 and LA-KDH-E2, respectively. LA-PDH-E2 and LA-KDH-E2 proteins were decreased in all patient-derived myoblasts (figure 4B), suggesting that a defect in LIAS significantly decreases LA synthesis in patient myoblasts. To evaluate the potential effects of IBA57 deficiency on other ISC components, we assessed NFU1, which inserts [4Fe-4S] into the apoprotein after [2Fe-2S] is converted to [4Fe-4S] by IBA57. NFU1 protein was significantly decreased in all patients, although no mutation in NFU1 was detected in any of the cases (figure 4, B and E).
Figure 4. Expression of mitochondrial [4Fe-4S] proteins.
(A, B) Mitochondrial [4Fe-4S] protein expression levels were analyzed by immunoblotting. Mitochondrial fractions prepared from myoblasts were analyzed. Levels of mitochondrial [4Fe-4S] proteins in all patients were comparable with those in controls (A), except lipoic acid synthetase (LIAS) and SDHB, which were clearly lower than in controls. Immunoblotting for lipoylated residues in mitochondrial fractions prepared from the myoblasts. All patients showed a severely reduced signal for the bands corresponding to the lipoylated residues pyruvate dehydrogenase (PDH)-E2 and α-ketoglutarate dehydrogenase (KDH)-E2 (B). NFU1 protein expression in mitochondrial fractions prepared from myoblasts as analyzed by immunoblotting (B). NFU1 protein level was clearly lower in all patients than in controls. The experiments were performed in triplicate and normalized to porin (C–E). Error bars indicate SDs (**p < 0.005, *p < 0.05, Student t test).
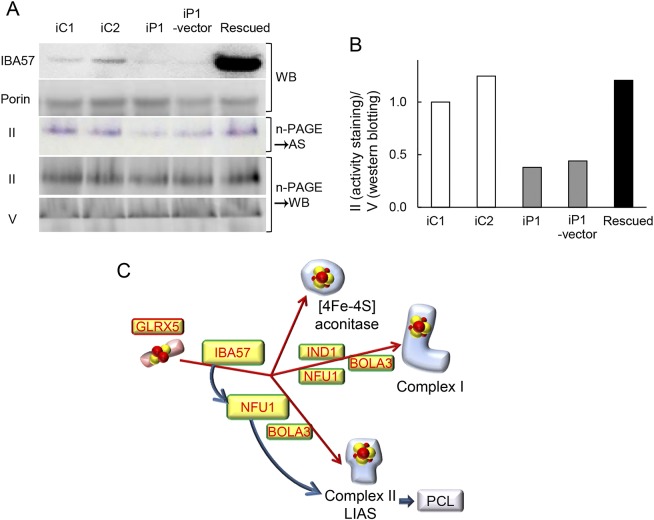
A phenotype rescue experiment was used to test whether forced expression of wild-type IBA57 in immortalized patient-derived myoblasts would complement the SDH activity defect. We confirmed that IBA57 protein expression was lower in patient-derived than in control myoblasts than in those of controls, and then transduced an empty expression vector, pEBMulti-Pur (Wako), or a pEBMulti-Pur construct containing full-length, wild-type IBA57 cDNA into the immortalized patient-derived myoblasts. IBA57 protein expression was confirmed in mitochondrial fractions prepared from the rescued myoblasts (figure 5A), and SDH activity staining showed nearly the same intensity as observed in controls (figure 5B).
Figure 5. IBA57 protein expression and activity staining of complex II in genetically rescued cells.
(A) Immortalized patient-derived myoblasts were transduced with empty vector or a wild-type IBA57-expressing construct, and rescued cells were established. iC1, iC2 = immortalized control myoblasts; iP1 = immortalized patient 1–derived myoblasts; iP1-vector = immortalized patient 1–derived myoblasts transfected with empty vector; Rescued = immortalized patient 1–derived myoblasts expressing wild-type IBA57; AS = activity staining; WB = Western blotting. IBA57 protein expression was confirmed by immunoblotting in mitochondrial fractions prepared from rescued cells. Porin was used as a loading control. Activity staining of complex II was performed to examine its restoration in rescued cells after high-resolution clear native (hrCN) polyacrylamide gel electrophoresis (PAGE). (B) In parallel, complexes II and V were detected by Western blotting after hrCN-PAGE. The band intensity of complex II was normalized to that of complex V. (C) Third step of the mitochondrial iron-sulfur cluster (ISC) biosynthesis pathway. Dedicated maturation of mitochondrial [4Fe-4S] proteins by specific ISC-targeting factors. Yellow boxes highlight ISC proteins whose genes are mutated in human ISC-related disease. LIAS = lipoic acid synthetase; PCL = progressive cavitating leukoencephalopathy.
DISCUSSION
Mitochondrial ISC assembly systems are highly conserved from yeast to humans, and their proper functioning is of vital importance to all eukaryotic organisms.4–8 All of our patients harbored compound heterozygous mutations in IBA57, which encodes a protein involved in [4Fe-4S] cluster assembly in the ISC assembly machinery. To date, 3 phenotypes linked to IBA57 mutations have been described.19–21 Homozygous mutations in IBA57 were first described in association with lethal encephalopathy, lactic acidosis, and profound respiratory complex I and II deficiencies in skeletal muscle, and brain MRI showed frontoparietal polymicrogyria with white matter abnormalities and hypoplasia of the medulla oblongata.19 A mild phenotype associated with homozygous mutations in IBA57 was reported in a highly consanguineous family with hereditary spastic paraplegia accompanied by optic atrophy and peripheral neuropathy, with complex I and II deficiencies in lymphoblastoid cells; MRI indicated mild corpus callosum and cerebellar atrophy, but no involvement of the white matter except for cystic cavitation.20 A third case of homozygous mutations in IBA57 was described as severe leukodystrophy, characterized by neurologic deterioration with complex I and II deficiencies, leading to death at 17 months.21 These previous reports suggest that IBA57 deficiency affects the mitochondrial respiratory chain complexes I and II.
Defects in the ISC biosynthesis pathway lead to abnormal function of oxidative phosphorylation as well as deficiencies in many proteins involved in the enzyme-bound cofactor lipoate and intermediary metabolic pathways.11–14 The ISC biosynthetic process is initiated by the synthesis of [2Fe-2S] clusters on a scaffold protein, followed by the generation of [4Fe-4S] clusters; their insertion into appropriate apoproteins is facilitated by the coordinated action of additional ISC maturation factors, including targeting factors such as NFU1 and BOLA3. Along with IBA57, the ISC proteins, ISC1 and ISC2, help to convert [2Fe-2S] into [4Fe-4S] clusters. Defects in IBA57 have been reported to lead to a general mitochondrial [4Fe-4S] protein deficiency, such as in respiratory complexes I and II.13,14 In our cases, there were no differences in the respiratory complexes in terms of overall activity and complex assembly, although complex activities were slightly lower in patients. However, SDH activity was substantially weaker in all patients than controls. These results suggested that IBA57 defect affected SDH function, whereas there was no effect on protein expression involved in ISC biosynthesis. Therefore, IBA57 defect might reduce the efficiency of the ISC biosynthesis process. Furthermore, detailed assessment of complex II function of mitochondrial [4Fe-4S] proteins by Western blotting revealed that the SDHB subunit of complex II was the only factor decreased in patient myoblasts, with no severe reductions in the subunits of complexes I and III. These results suggest that defects in IBA57 mainly affect the expression of SDHB.
Defects in the ISC biosynthesis pathway have a negative effect not only on the respiratory complexes but also on lipoate-containing protein, likely as a result of deficient maturation of LIAS. LIAS contains 2 [4Fe-4S] clusters that are essential for the synthesis of LA, a cofactor of PDH and α-KDH. In PDH and α-KDH, which comprise certain subunits, measurement of the lipoylation status of the E2 subunits serves as an indirect but reliable estimate of the iron-sulfur maturation efficiency of LIAS.30–34 In our cases, detection of PDH and α-KDH revealed a severe decrease in LA in patient myoblasts, providing further evidence of disturbance in the ISC biosynthesis pathway. However, the mitochondrial citric acid cycle enzyme ACO2, another reported target of IBA57 that contains 1 [4Fe-4S] cluster, was not reduced.8,9,19–21 This biochemical profile, i.e., an observed defect in LA synthesis without a corresponding defect in ACO2, appears to be inconsistent with the changes to mitochondrial [4Fe-4S] proteins identified in previous reports on IBA57 deficiencies.19–21 The present profile is more reminiscent of the phenotype related to alterations in NFU1, rather than that related IBA57 mutations.
NFU1 is characterized as a late-targeting factor of the ISC assembly machinery and is specifically required for the proper assembly of a subset of [4Fe-4S] proteins to respiratory complexes I, II, and LIAS.16–18 We determined the effect of IBA57 mutation on NFU1 to evaluate potential links of IBA57 deficiency to other aspects of the ISC assembly machinery. The NFU1 protein level was also decreased in patients, corroborating that IBA57 regulates the expression of NFU1, which subsequently affects the expression of lipoate-containing protein as well as respiratory complex II (figure 5C). There have been 5 reports describing a total of 22 patients with NFU1 mutations (table e-2). Seven of 9 cases with brain MRI data presented with leukoencephalopathy in the periventricular white matter and corpus callosum, with partial cystic degeneration or cavitation, characterized by neurologic regression, lactic acidosis, and hyperglycinemia, leading to death within 2.5 years.16,18,35–38 Navarro-Sastre et al.,18 Nizon et al.,38 and Invernizzi et al.36 reported that the activity of complex II was significantly decreased, while that of complex I was just below normal. Biochemical profiles of patients with mutations in NFU1 also suggest that NFU1 mainly affects complex II activity. The neuroimaging features and biochemical profiles in our cases are reminiscent of those reported in patients harboring mutations in NFU1. This suggests that the IBA57 mutation affects NFU1 protein expression and function. To date, the exact role of IBA57 and its functional relationship to NFU1 have remained unclear. Our results provide new insights into the pathogenic mechanisms in that defects in IBA57 essentially cause decreased respiratory complex II and LIAS activities through the aberrant function of NFU1, leading to white matter abnormalities with disturbed mitochondrial energy metabolism.
This study demonstrated that IBA57 plays essential roles in the biosynthesis of ISC proteins to regulate mitochondrial energy metabolism. IBA57 gene mutations resulted in decreased expression of SDHB as well as deficiency of LIAS through aberrant NFU1 function. This new understanding of the molecular basis of PCL pathophysiology should contribute to a better definition of the phenotypic spectrum and provide critical information to develop potential therapeutic strategies for PCL.
Supplementary Material
GLOSSARY
- BN
blue native
- cDNA
complementary DNA
- CS
citrate synthase
- ISC
iron-sulfur cluster
- KDH
ketoglutarate dehydrogenase
- LA
lipoic acid
- LIAS
lipoic acid synthetase
- PAGE
polyacrylamide gel electrophoresis
- PCL
progressive cavitating leukoencephalopathy
- PDH
pyruvate dehydrogenase
Footnotes
Supplemental data at Neurology.org/ng
AUTHOR CONTRIBUTIONS
Akihiko Ishiyama and Chika Sakai: data acquisition, analysis and interpretation, and manuscript drafting and editing. Yuichi Matsushima, Satoru Noguchi, and Satomi Mitsuhashi: bioinformatics analysis. Yukari Endo and Yukiko K. Hayashi: exome data interpretation. Yoshiaki Saito, Eiji Nakagawa, Hirofumi Komaki, and Kenji Sugai: patient clinical information collection. Masayuki Sasaki and Noriko Sato: MRI review and clinical and imaging data analysis. Ikuya Nonaka, Yu-ichi Goto, and Ichizo Nishino: study supervision and manuscript preparation and editing. All authors were responsible for data acquisition and analysis.
STUDY FUNDING
This study was supported partly by the Intramural Research Grant (29-4 to Ichizo Nishino) for Neurological and Psychiatric Disorders of NCNP and Research and Development Grants for Practical Research Project for Rare/Intractable Diseases (17ek0109285h0001 to Ichizo Nishino) from the Japan Agency for Medical Research and Development, AMED.
DISCLOSURE
A. Ishiyama and C. Sakai report no disclosures. Y. Matsushima has received research support from the Japan Society for the Promotion of Science (JSPS). S. Noguchi holds patent WO 2010/131712A1; has received research support from Novartis Pharmaceuticals, Japan; has received governmental support from the Japan Agency for Medical Research and Development (AMED); and has received institutional support from the National Center of Neurology and Psychiatry (NCNP). S. Mitsuhashi and Y. Endo report no disclosures. Y.K. Hayashi serves as an editorial board member of Neuromuscular Disorders and Journal of Physiological Sciences and received institutional support from the NCNP, the Japan Society for the Promotion of Science (JSPS), The Promotion and Mutual Aid Corporation for Private Schools of Japan, and Tokyo Medical University. Y. Saito reports no disclosures. E. Nakagawa received institutional support from the NCNP. H. Komaki has received research support from Sanofi, Daiichi Sankyo, Taiho, Pfizer, and Nippon Shinyaku and has served as a consultant for PTC Therapeutics. K. Sugai has served on the editorial board of the Journal of the Japanese Epilepsy Society. M. Sasaki, N. Sato, and I. Nonaka report no disclosures. Y.-i. Goto serves on the editorial board of Mitochondrion; has received governmental support from the Japan Agency for Medical Research and Development (AMED) and the Ministry of Health, Labour and Welfare of Japan; and has received institutional support from the NCNP. I. Nishino has received travel funding/speaker honoraria from Sanofi, Daiichi Sankyo, and Japan Blood Products Organization; serves on the editorial boards of Neuromuscular Disorders, Therapeutic Advances in Neurological Disorders, Journal of the Neurological Sciences, Journal of Neuromuscular Diseases, Skeletal Muscle, and Neurology and Clinical Neuroscience; holds a patent for Efficacious agents to pathologic condition due to GNE protein dysfunction; has served on the scientific advisory boards of Ultragenyx and Novartis Pharmaceuticals; and has received research support from Sanofi, Daiichi Sankyo, and Astellas, in addition to governmental support from the AMED and institutional support from the NCNP. Go to Neurology.org/ng for full disclosure forms.
REFERENCES
- 1.Naidu S, Bibat G, Lin D, et al. Progressive cavitating leukoencephalopathy: a novel childhood disease. Ann Neurol 2005;58:929–938. [DOI] [PubMed] [Google Scholar]
- 2.Morató L, Bertini E, Verrigni D, et al. Mitochondrial dysfunction in central nervous system white matter disorders. Glia 2014;62:1878–1894. [DOI] [PubMed] [Google Scholar]
- 3.Van der Knaap MS, Valk J. Classification of myelin disorders. In: Van der Knaap MS, Valk J, editors. Magnetic Resonance of Myelination and Myelin Disorders. Berlin, Heidelberg: Springer-Verlag; 1989:20–24. [Google Scholar]
- 4.Lill R, Hoffmann B, Molik S, et al. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta 2012;1823:1491–1508. [DOI] [PubMed] [Google Scholar]
- 5.Booker SJ, Cicchillo RM, Grove TL. Self-sacrifice in radical S-adenosylmethionine proteins. Curr Opin Chem Biol 2007;11:543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem 2005;74:247–281. [DOI] [PubMed] [Google Scholar]
- 7.Beinert H. Iron-sulfur proteins: ancient structures, still full of surprises. J Biol Inorg Chem 2000;5:2–15. [DOI] [PubMed] [Google Scholar]
- 8.Lill R. Function and biogenesis of iron-sulphur proteins. Nature 2009;460:831–838. [DOI] [PubMed] [Google Scholar]
- 9.Sheftel AD, Wilbrecht C, Stehling O, et al. The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol Biol Cell 2012;23:1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mühlenhoff U, Richter N, Pines O, et al. Specialized function of yeast Isa1 and Isa2 proteins in the maturation of mitochondrial [4Fe-4S] proteins. J Biol Chem 2011;286:41205–42116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouault TA, Tong WH. Iron-sulfur cluster biogenesis and human disease. Trends Genet 2008;24:398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rouault TA. Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis Model Mech 2012;5:155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stehling O, Wilbrecht C, Lill R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie 2014;100:61–77. [DOI] [PubMed] [Google Scholar]
- 14.Ye H, Rouault TA. Human iron-sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry 2010;49:4945–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Qi W, Cowan JA. Iron-sulfur cluster biosynthesis: functional characterization of the N- and C-terminal domains of human NFU. Biochemistry 2009;48:973–980. [DOI] [PubMed] [Google Scholar]
- 16.Cameron JM, Janer A, Levandovskiy V, et al. Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am J Hum Genet 2011;89:486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haack TB, Rolinski B, Haberberger B, et al. Homozygous missense mutation in BOLA3 causes multiple mitochondrial dysfunctions syndrome in two siblings. J Inherit Metab Dis 2013;36:55–62. [DOI] [PubMed] [Google Scholar]
- 18.Navarro-Sastre A, Tort F, Stehling O, et al. A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe-S proteins. Am J Hum Genet 2011;89:656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ajit Bolar N, Vanlander AV, Wilbrecht C, et al. Mutation of the iron-sulfur cluster assembly gene IBA57 causes severe myopathy and encephalopathy. Hum Mol Genet 2013;22:2590–2602. [DOI] [PubMed] [Google Scholar]
- 20.Lossos A, Stümpfig C, Stevanin G, et al. Fe/S protein assembly gene IBA57 mutation causes hereditary spastic paraplegia. Neurology 2015;84:659–667. [DOI] [PubMed] [Google Scholar]
- 21.Debray FG, Stümpfig C, Vanlander AV, et al. Mutation of the iron-sulfur cluster assembly gene IBA57 causes fatal infantile leukodystrophy. J Inherit Metab Dis 2015;38:1147–1153. [DOI] [PubMed] [Google Scholar]
- 22.Yukari Endo Y, Noguchi S, Hara Y, et al. Dominant mutations in ORAI1 cause tubular aggregate myopathy with hypocalcemia via constitutive activation of store-operated Ca2+ channels. Hum Mol Genet 2015;24:637–648. [DOI] [PubMed] [Google Scholar]
- 23.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim DW, Uetsuki T, Kaziro Y, et al. Use of the human elongation factor 1 alpha promoter as a versatile and efficient expression system. Gene 1990;91:217–223. [DOI] [PubMed] [Google Scholar]
- 26.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2007;2:287–295. [DOI] [PubMed] [Google Scholar]
- 27.Shimazaki H, Takiyama Y, Ishiura H, et al. A homozygous mutation of C12orf65 causes spastic paraplegia with optic atrophy and neuropathy (SPG55). J Med Genet 2012;49:777–784. [DOI] [PubMed] [Google Scholar]
- 28.Morava E, Rodenburg RJ, Hol F, et al. Clinical and biochemical characteristics in patients with a high mutant load of the mitochondrial T8993G/C mutations. Am J Med Genet A 2006;140:863–838. [DOI] [PubMed] [Google Scholar]
- 29.Mitsuhashi S, Hatakeyama H, Karahashi M, et al. Muscle choline kinase beta defect causes mitochondrial dysfunction and increased mitophagy. Hum Mol Genet 2011;20:3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayr JA, Feichtinger RG, Tort F, et al. Lipoic acid biosynthesis defects. J Inherit Metab Dis 2014;37:553–563. [DOI] [PubMed] [Google Scholar]
- 31.Tort F, Ferrer-Cortes X, Ribes A. Differential diagnosis of lipoic acid synthesis defects. J Inherit Metab Dis 2016;39:781–793. [DOI] [PubMed] [Google Scholar]
- 32.Mochel F, Knight MA, Tong WH, et al. Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am J Hum Genet 2008;82:652–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsson A, Lind L, Thornell LE, Holmberg M. Myopathy with lactic acidosis is linked to chromosome 12q23.3-24.11 and caused by an intron mutation in the ISCU gene resulting in a splicing defect. Hum Mol Genet 2008;17:1666–1672. [DOI] [PubMed] [Google Scholar]
- 34.Gelling C, Dawes IW, Richhardt N, et al. Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Mol Cell Biol 2008;28:1851–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seyda A, Newbold RF, Hudson TJ, et al. A novel syndrome affecting multiple mitochondrial functions, located by microcell-mediated transfer to chromosome 2p14-2p13. Am J Hum Genet 2001;68:386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Invernizzi F, Ardissone A, Lamantea E, et al. Cavitating leukoencephalopathy with multiple mitochondrial dysfunction syndrome and NFU1 mutations. Front Genet 2014;5:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahting U, Mayr JA, Vanlander AV, et al. Clinical, biochemical, and genetic spectrum of seven patients with NFU1 deficiency. Front Genet 2015;6:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nizon M, Boutron A, Boddaert N, et al. Leukoencephalopathy with cysts and hyperglycinemia may result from NFU1 deficiency. Mitochondrion 2014;15:59–64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.