SUMMARY
We report analytic and consensus processes that produced recommendations for clinical stage groups (cTNM) of esophageal and esophagogastric junction cancer for the AJCC/UICC cancer staging manuals, 8th edition. The Worldwide Esophageal Cancer Collaboration (WECC) provided data on 22,123 clinically staged patients with epithelial esophageal cancers. Risk-adjusted survival for each patient was developed using random survival forest analysis from which (1) data-driven clinical stage groups were identified wherein survival decreased monotonically and was distinctive between and homogeneous within groups and (2) data-driven anatomic clinical stage groups based only on cTNM. The AJCC Upper GI Task Force, by smoothing, simplifying, expanding, and assessing clinical applicability, produced (3) consensus clinical stage groups. Compared with pTNM, cTNM survival was “pinched,” with poorer survival for early cStage groups and better survival for advanced ones. Histologic grade was distinctive for data-driven grouping of cT2N0M0 squamous cell carcinoma (SCC) and cT1-2N0M0 adenocarcinoma, but consensus removed it. Grouping was different by histopathologic cell type. For SCC, cN0-1 was distinctive for cT3 but not cT1-2, and consensus removed cT4 subclassification and added subgroups 0, IVA, and IVB. For adenocarcinoma, N0-1 was distinctive for cT1-2 but not cT3-4a, cStage II subgrouping was necessary (T1N1M0 [IIA] and T2N0M0 [IIB]), advanced cancers cT3-4aN0-1M0 plus cT2N1M0 comprised cStage III, and consensus added subgroups 0, IVA, and IVB. Treatment decisions require accurate cStage, which differs from pStage. Understaging and overstaging are problematic, and additional factors, such as grade, may facilitate treatment decisions and prognostication until clinical staging techniques are uniformly applied and improved.
Keywords: esophageal malignancy, staging, statistics, squamous cell carcinoma, adenocarcinoma
INTRODUCTION
At its inception, cancer staging was limited “to cases not previously treated and that the extent of the disease must be determined and recorded on clinical examination only.”1 Lack of effective clinical staging modalities, coupled with conclusiveness of pathologic assessment of resection specimens, led to the adoption of pathologic stage groups after esophagectomy alone (pTNM) for clinical stage groups before treatment (cTNM). Today, with the advent of new staging modalities, this practice is in question.2 Post-7th edition, AJCC instructions and goals of the Worldwide Esophageal Cancer Collaboration (WECC) were to develop, if indicated, separate clinical staging recommendations for cancer of the esophagus and esophagogastric junction for the 8th edition AJCC cancer staging manual.3
Data provided by WECC4 served as substrate for a machine-learning analysis from which (1) data-driven clinical stage groups based on anatomic (cTNM) and non-anatomic cancer characteristics and (2) anatomic clinical stage groups based only on cTNM categories were produced. The AJCC Upper GI Task Force reviewed these and, by smoothing, simplifying, expanding, and assessing clinical applicability, produced (3) consensus clinical stage groups.
This manuscript reports these data-driven clinical stage groups, data-driven anatomic clinical stage groups, and consensus clinical stage groups. Finally, to aid in consistent prognostication and decision-making, the consensus pTNM stage group with equivalent survival to each consensus cTNM stage group was identified.
PATIENTS AND METHODS
Patients
At 33 WECC institutions (Supporting Information Appendix S1), 22,654 patients were treated for epithelial esophageal cancers.4 Of these, 22,123 were clinically staged before treatment decision. Patient, cancer, and survival summary data have been published.4
Endpoint
The endpoint was all-cause mortality from first management decision. Median potential follow-up was 8.9 years5; however, median actual follow-up for surviving patients was 2.5 years, with 25% followed more than 5.1 years and 10% more than 8.4 years.4
Data
Under individual institutional data use agreements and Institutional Review Board approval, data were collected using a common format with standardized definitions (Supporting Information Table S1). The Case Cancer Institutional Review Board of Case Western Reserve University and Cleveland Clinic Institutional Review Board approved the overall use of these data for research.
DATA ANALYSIS
Analytic strategy
Risk-adjusted survival for each patient was developed. From this, data-driven groups were formulated such that (1) survival decreased monotonically with increasing group number, and (2) survival of each group was distinctive, with no more than a 5% difference. cTNM categories comprising each group were then exposed. Homogeneity of survival within groups was determined.
Risk-adjusted survival
Random survival forest (RSF) methodology was used to estimate nonparametric risk-adjusted and cross-validated survival for each patient.6,7 Thirty-nine dichotomous, polytomous, ordinal, and continuous variables were used, representing patient characteristics, anatomic and non-anatomic cancer characteristics, and institutional characteristics (Supporting Information Table S2), in separate analyses for squamous cell carcinoma (SCC) and adenocarcinoma.
All computations used open source randomForestSRC R-software under default settings.8 Missing data were pre-imputed without outcome information by random forest imputation methodology, missForest.9,10 One thousand survival trees were grown using log-rank splitting. Each tree was constructed using an independent bootstrap sample of size 22,123; on average, each bootstrap sample contained 63% of the patients. The remaining unused patients (37%) were referred to as out-of-bag (OOB) observations. Each tree and its corresponding OOB observations were used to calculate a patient-specific risk-adjusted OOB (cross-validated) survival function for stage grouping and patient-specific mortality value (integrated cumulative hazard for each patient) for analysis of homogeneity.6
Data-driven clinical staging
From the individual survival estimates, 18 subsets of SCC patients and 15 for adenocarcinoma (Supporting Information Table S3 and Figs. S1 and S2) were produced with constraints such that only subsets with a sample size of 20 patients or greater were permitted. Stage groups were then formed by iteratively merging these subsets by closeness, and individual patient survival by closeness, defined as the root-mean-squared-error difference between OOB survival functions from 3 to 10 years, until no more than a 5% difference between adjacent groups, above and below, was observed.
Data-driven anatomic clinical staging
To meet UICC requirements for pure cTNM groupings, regrouping of estimated survival curves produced data-driven anatomic clinical stage groupings with grade and location used in risk adjustment.
Consensus clinical staging
The AJCC Upper GI Task Force reviewed cancer categories and both data-driven analyses. The consensus process of merging, splitting, and smoothing data-driven groups while maintaining clinical relevance and minimizing drift from data-driven groupings produced consensus clinical stage groups.
Homogeneity
Within each data-driven anatomic and consensus stage group, homogeneity was assessed as follows. OOB patient survival was analyzed with respect to the 39 independent variables using RF regression. Regression forests of 1,000 random regression trees were grown using mean-squared-error splitting. From each RF regression, variable importance (VIMP) of each variable was calculated by measuring the increase in OOB mean-squared error when the variable was removed from the analysis,7 and standardized by dividing VIMP by the variance of OOB patient mortality multiplied by 100. Standardized VIMP measured the relative importance each independent variable had in predicting OOB patient mortality within a stage group. If a group is homogeneous, standardized VIMP for all cancer facts should be near zero. Values under 5% were deemed to be non-significant.11
Equivalence of survival between consensus pTNM and consensus cTNM stage groups
For each consensus clinical stage group (cTNM), the consensus pathologic (pTNM) stage group with equivalent survival profile was identified.
RESULTS
Data-driven clinical stage groups
Only four data-driven stage groups, presented simply as cardinal numbers, were identified for SCC (Table 1) and six for adenocarcinoma (Table 2). Risk-adjusted survival within these groupings for SCC (Fig. 1A) and adenocarcinoma (Fig. 1B) monotonically decreased with increasing cardinal numbers and was distinctive between groups. Homogeneity was excellent for all groups (Supporting Information Figs. S3A and S4A).
Table 1.
Data-driven clinical stage groups: squamous cell carcinoma
| Analysis group | cT | cN | cM | cGrade |
|---|---|---|---|---|
| 1 | Tis | N0 | M0 | N/A |
| T1 | N0-1 | M0 | Any | |
| T2 | N0 | M0 | G1 | |
| 2 | T2 | N0 | M0 | G2 |
| T2 | N1-2 | M0 | Any | |
| T3 | N0 | M0 | Any | |
| 3 | T2 | N0 | M0 | G3 |
| T3 | N1-2 | M0 | Any | |
| T4a | N0-2 | M0 | Any | |
| 4 | T4a | N3 | M0 | Any |
| Any | Any | M1 | Any |
Table 2.
Data-driven clinical stage groups: adenocarcinoma
| Analysis group | cT | cN | cM | cGrade |
|---|---|---|---|---|
| 1 | Tis | N0 | M0 | N/A |
| T1 | N0 | M0 | G1-2 | |
| 2 | T1 | N0 | M0 | G3 |
| T1 | N1 | M0 | Any | |
| T2 | N0 | M0 | G1 | |
| 3 | T2 | N0 | M0 | G2 |
| 4 | T2 | N0 | M0 | G3 |
| T2 | N1 | M0 | Any | |
| T3 | N0 | M0 | Any | |
| T4a | N0-1 | M0 | Any | |
| 5 | T3 | N1 | M0 | Any |
| 6 | T3 | N2 | M0 | Any |
| Any | Any | M1 | Any |
Fig. 1.
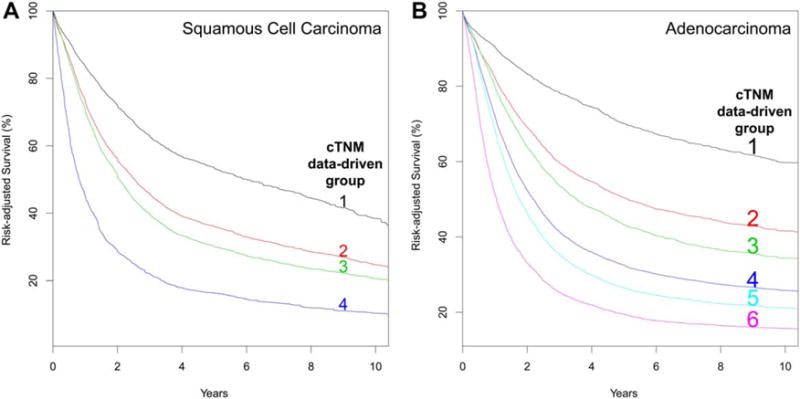
Risk-adjusted survival of data-driven clinical stage groups. A, Squamous cell carcinoma. B, Adenocarcinoma.
Squamous cell carcinoma
Location of cancer along the esophagus (upper, middle, lower) played no role in data-driven clinical stage grouping. Grade was a defining category for cT2N0M0 cancers and importantly revealed its contribution to all but group 4. Group 1 comprised a disparate collection of patients with the spectrum of cT confined to the esophagus, some with regional lymph node metastasis (cN1): cTis, cT1N0-1M0, and cT2N0M0G1. Group 2 comprised a wide range of cT2 cancers (cT2N0M0G2 and cT2N1-2M0) plus cT3N0M0. Group 3 had the largest complement of patients, with component categories being cT2N0M0G3, cT3N1-2M0, and cT4aN0-2M0. cT4aN3M0 and cM1 cancer comprised group 4. There were too few cT1N2M0 and cT1-3N3M0 cancers to group these categories.
Adenocarcinoma
Grade played an important role in stage grouping both cT1N0M0 and cT2N0M0 cancers. Similar to SCC, cT2N0M0 cancers contributed to three groups, distinguished by grade. Group 1 comprised cTis and cT1N0M0G1-2. T1N0M0G3, cT2N0M0G1, and cT1N1M0 comprised group 2. cT2N0M0G2 was the sole member of group 3, distinguished from other cT2N0M0 subcategories by grade. Group 4 had the largest complement of patients and consisted of cT2N0M0G3, cT2N1M0, cT3N0M0, and cT4aN0-1M0 cancers. cT3N1M0 cancers solely comprised group 5. Group 6 had 2 members: cT3N2M0 and cM1. There were too few cT1-2N2, cT4aN2M0, cT4bN0-2, and cTanyN3M0 adenocarcinomas to be grouped.
Data-driven anatomic clinical stage groups
Four data-driven stage groups, presented simply as cardinal numbers, were identified for SCC (Table 3A) and six for adenocarcinoma (Table 3B). Risk-adjusted survival for SCC (Fig. 2A) and adenocarcinoma (Fig. 2B) monotonically decreased, was distinctive between groups, and was homogeneous within groups (Supporting Information Figs. S3B and S4B).
Table 3.
Data-driven anatomic clinical, consensus, and equivalent pTNM stage groups
| (A) Squamous cell carcinoma
| |||||
|---|---|---|---|---|---|
| Analysis group | cT | cN | cM | Consensus stage group | pTNM equivalent group† |
| 1 | Tis | N0 | M0 | 0 | IIA |
| T1 | N0-1 | M0 | I | ||
| 2 | T2 | N0-1 | M0 | II | |
| T3 | N0 | M0 | |||
| X | T1 | N2 | M0 | III | IIIA |
| T2 | N2 | M0 | |||
| 3 | T3 | N1-2 | M0 | ||
| T4a | N0-2 | M0 | IVA | IIIB | |
| 4 | T4a | N3 | M0 | ||
| X | T4b | N0-2 | M0 | ||
| X | T1-3 | N3 | M0 | ||
| Any | Any | M1 | IVB | IVA | |
| (B) Adenocarcinoma
| |||||
|---|---|---|---|---|---|
| Analysis group | cT | cN | cM | Consensus stage group | pTNM equivalent group† |
| 1 | Tis | N0 | M0 | 0 | IB |
| T1 | N0 | M0 | I | IC | |
| 2 | T1 | N1 | M0 | IIA | IIA |
| 3 | T2 | N0 | M0 | IIB | IIB |
| 4 | T2 | N1 | M0 | III | IIIA |
| T3 | N0 | M0 | |||
| T4a | N0-1 | M0 | |||
| 5 | T3 | N1 | M0 | ||
| 6 | T3 | N2 | M0 | IVA | IIIB |
| X | T1-2 | N2 | M0 | ||
| X | T4a | N2 | M0 | ||
| X | T4b | Any | M0 | ||
| X | Any | N3 | M0 | ||
| Any | Any | M1 | IVB | IIIB | |
Note: X= not in data-driven analysis.
Equivalent survival with pTNM consensus groups.
Fig. 2.
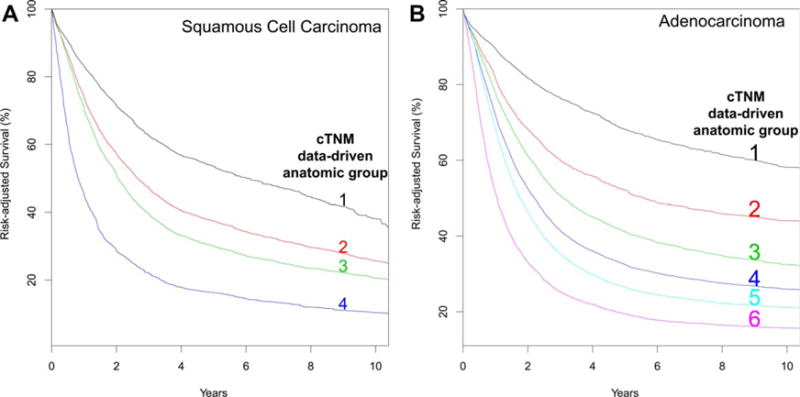
Risk-adjusted survival of data-driven anatomic clinical stage groups. A, Squamous cell carcinoma. B, Adenocarcinoma.
Squamous cell carcinoma
Group 1 comprised cTis and cT1N0-1M0. Loss of grade obliterated the distinctiveness of grade for cT2N0M0 cancers (Table 3A). cT2N0-2M0 and cT3N0M0 were in group 2. Group 3 comprised cT3N1-2M0 and cT4N0-2M0. cT4aN3M0 cancer and all cM1 cancers were placed ingroup 4.
Adenocarcinoma
Loss of grade obliterated the distinctiveness of grade for cT1-2N0M0 cancers (Table 3B). Group 1 comprised cTis and cT1N0M0. Groups 2 and 3 had single category members: group 2, cT1N1M0, and group 3, cT2N0M0. Group 4 comprised cT2N1M0, cT3N0M0, and cT4aN0-1M0 cancers. Group 5 comprised cT3N1M0. cT3N2M0 and all cM1 cancers were placed into group 6.
Consensus clinical stage groups
Risk-adjusted survival for SCC (Fig. 3A) and adenocarcinoma (Fig. 3B) monotonically decreased with increasing stage group number and was distinctive between groups. Homogeneity was excellent for all groups (Supporting Information Figs. S3C and S4C).
Fig. 3.
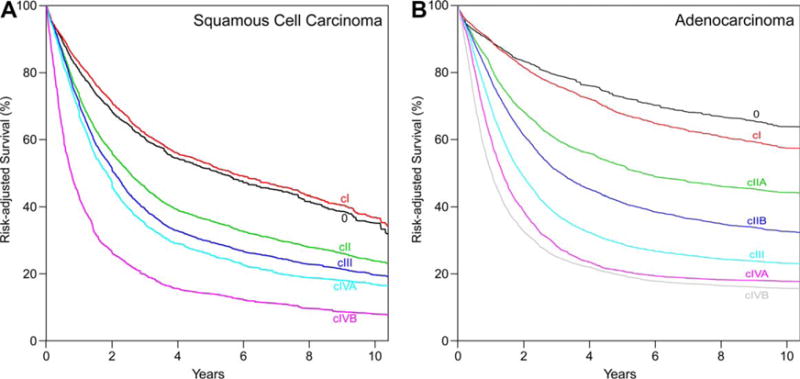
Risk-adjusted survival of consensus clinical stage groups. A, Squamous cell carcinoma. B, Adenocarcinoma.
Squamous cell carcinoma
The consensus process, adhering to prior staging rules, assigned cTis to cStage 0. Thus, cStage I was limited to cT1N0-1M0. cT2N0-1M0 and cT3N0M0 comprised cStage II. cStage III included cT1-2N2M0 and cT3N1-2M0 cancers. cStage IVA represented the most locally advanced cancers, including any T4 cancer regardless of subcategory of invasion of adjacent structure (cT4a-T4bN0-2M0) and cTanyN3M0. Adherence to staging rules placed any cM1 cancer into cStage IVB.
Adenocarcinoma
The consensus process, adhering to prior staging rules, assigned cTis to cStage 0. Thus, cStage I was limited to cT1N0M0. Stage II was the only cStage group that was subgrouped, with cT1N1M0 placed into cStage IIA and cT2N0M0 into cStage IIB. cT2N1M0 and cT3-4aN0-1M0 comprised cStage III. cStage IVA comprised the most locally advanced cancers, including cT1-4aN2M0, cT4bN0-2M0, and cTany N3M0. Adherence to staging rules placed any cM1 cancer into cStage IVB.
Equivalence of survival between consensus pTNM and cTNM stage groups
Squamous cell carcinoma
For SCC, survival of cStage groups 0 and I was equivalent to pStage group IIA, survival for cStage group II was equivalent to pStage group IIB, survival for cStage group III was equivalent to pStage group IIIA, survival for cStage IVA was equivalent to pStage IIIB, and survival for cStage IVB was equivalent to pStage IVA.
Adenocarcinoma
For adenocarcinoma, survival for cStage group 0 was equivalent to pStage group IB, survival for cStage group I was equivalent to pStage group IC, survival for cStage group IIA was equivalent to pStage group IIA, survival for cStage group IIB was equivalent to pStage IIB, survival for cStage group III was equivalent to pStage group IIIA, and survival for pStage IVA-B was equivalent to pStage group IIIB.
DISCUSSION
Principal findings
Despite the importance of histologic grade in data-driven stage grouping, the anatomic and consensus processes eliminated grade from clinical staging. Stage grouping for SCC followed a different pattern from that of adenocarcinoma. For cT1 and cT2 SCCs, there was no distinctiveness between cN0 and cN1 as there was for cT3. Stage I and II SCCs include T1-2N1M0 and T3N0M0 cancers, which are advanced-stage (cStage III) adenocarcinomas; cT1N0-1M0 is cStage I, and cT2N0-1M0 plus cT3N0M0 is cStage II. “Relatively good prognosis” advanced-stage SCCs are Stage III, cT3N1M0 and cT1-3N2, mirroring pStage IIIA-B. The most advanced cancers are subgrouped, but cT4 cancers are not subcategorized.
For adenocarcinoma, groups containing cT1-4a are distinguished by cN0-1 categorization. Subgrouping is necessary for cStage II, IIA is cT1N1M0, and IIB is cT2N0M0. Unlike SCC, cT2N1M0 and cT3-4aN0M0 are considered locally advanced cancers with relatively good survival (cStage III), closely mirroring pStage III. Also unlike SCC, there is subcategorization of T4 and merging of cN2-3.
Interpretation
Compared with pTNM survival, cTNM survival is less distinctive between cStage groups, and the survival curves are pinched: poorer survival of early cStage cancers and better survival of advanced cStage cancers. This effect is (1) much greater for SCC than adenocarcinoma and (2) less marked with risk adjustment than that seen in the presentation of raw data.4 In part this is the result of understaging early cStage cancers and overstaging advanced cStage cancers. Low accuracy of clinical staging must be considered in treatment decisions and prognostication using cStage.
The differences in clinical stage groupings between histopathologic cell types are marked. Continent was the most important variable in determining stage group of SCC, but not of adenocarcinoma.12 However, cT and cG were of similar overall importance and of greater importance than cM and cN for both histologic cell types in this analysis. The role histopathologic cell type plays in these survival differences in cStaged patients is debatable, and to a great extent differences may be mostly explained by staging variability among continents and institutions.
Assignment of treatment and coarse prognostication was set out in the 1st edition of the AJCC cancer staging manual and holds true today for cStage: “The various TNM classifications can be gathered together to represent three groups of patients: (1) those patients with a fairly good prognosis when dealt with by present-day therapeutic techniques, (2) those whose course is fulminating and rapidly fatal, and (3) those whose course lies between, including those who have little or no chance of cure but who may often live for various periods.”13 This gross partitioning holds true for clinical staging today.
Strengths and limitations
Modalities used to determine clinical stage were not available for institutions or individual patients in the WECC. Risk adjustment for staging modality was not possible.
Misconceptions abound about the best endpoint for cancer survival analyses. Cancer-specific survival is believed by many to represent the ultimate disease-specific survival. However, this soft endpoint is plagued by our ability to detect persistence of cancer after treatment. This is determined by the interval of follow-up and the sensitivity of clinical detection tools. It has been demonstrated that risk-adjusted all-cause mortality, using a rich set of patient demographics and comorbidities, is a better survival endpoint and has been used in the analysis.14–16
Random forest analysis is capable of superior risk adjustment, limited only by the depth and volume of data. The present WECC database, with multiple patient variables, permits accounting for factors other than esophageal cancer characteristics that can cause deaths in these patients. After risk adjustment, all the residual information about death may be attributed to esophageal cancer, the last remaining risk.
Although the strength of WECC isworldwide representation, it also imposes limitations related to institutional, country, and continent heterogeneity of etiology, diagnosis, clinical characterization of esophageal cancer, and treatment decisions. Imputation was necessary for missing data in some risk-adjustment variables, and some patients were excluded who did not have complete pTNM data or survival information. Although we attempted to incorporate patients who underwent less invasive treatments than esophagectomy, the numberof such patientswas small.
Clinical implications
Clinical staging is chiefly based on imaging and thus is limited by resolution of each individual technique. The shortcoming of each should be taken into account during interpretation of clinical staging. Worldwide recommendations and standardization are necessary and at the present time should include esophagogastroduodenoscopy, biopsy or endoscopic mucosal resection of the primary cancer, endoscopic esophageal ultrasonography, endoscopic esophageal ultrasonography-directed fine-needle aspiration of regional lymph nodes and distant sites in direct continuity with the upper GI tract, and CT-PET. The modalities used to determine cStage must be recorded.
cN+ was shown to be a high-risk finding for adenocarcinoma and should accordingly direct treatment decisions and crude prognostication. However, for cStage I–II SCC, the finding of cT1N1M0 or cT2N1M0 should bring into question overstaging and a concerted effort to prove that regional nodal metastases are present. If histologic proof of cN1 is found, it should lead to reconsideration of cStage and appropriate alteration of treatment.
Identification of high-risk associated findings must be included in decision-making and prognostication. Although eliminated from stage grouping, grade is important for treatment decisions. The identification of G3 in T1-2N0M0 adenocarcinoma and T2N0M0 SCC is a high-risk finding and should be used in decision-making and prognostication for these patients. Grade can and should be determined on evaluation of preoperative specimens, and it has been shown to be important also in prognostication.17,18 Clinical grade may be the only determination of grade in cancers that are obliterated with neoadjuvant therapy (ypT0N0M0 cancers).
Conclusions
This first attempt at clinical stage grouping is most useful and reliable for adenocarcinoma and should facilitate the process and uses of clinical staging. The groupings for SCC reflect variable staging to some degree and emphasize the need for universal staging protocols, uniform staging of esophageal cancer patients, identification of additional risk factors in clinically staged patients, and new staging modalities. Much work is needed to improve clinical staging for the 9th edition, but the stage has been set.
Supplementary Material
Acknowledgments
Funding: Funded in part by (1) the International Society for Diseases of the Esophagus (ISDE), (2) the Daniel and Karen Lee Chair in Thoracic Surgery at Cleveland Clinic (TWR), (3) the Kenneth Gee and Paula Shaw, PhD, Chair in Heart Research at Cleveland Clinic (EHB), and (4) the Clinical and Translational Science Collaborative of Cleveland, UL1TR000439 from the National Center for Advancing Translational Services (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the ISDE, Cleveland Clinic, or the NIH.
Footnotes
Author contributions: Conception or design of the experiment(s), or collection and analysis or interpretation of data: all authors. Drafting the manuscript or revising its intellectual content: Rice, Ishwaran, and Blackstone. Approval of the final version of the submitted manuscript: all authors.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
References
- 1.International Union Against Cancer (UICC) TNM Classification of Malignant Tumors. Geneva: UICC; 1968. [Google Scholar]
- 2.Strong VE, D’amico TA, Kleinberg L, Ajani J. Impact of the 7th edition AJCC staging classification on the NCCN clinical practice guidelines in oncology for gastric and esophageal cancers. J Natl Compr Canc Netw. 2013;11:60–6. doi: 10.6004/jnccn.2013.0009. [DOI] [PubMed] [Google Scholar]
- 3.Rice TW, Blackstone EH. Esophageal cancer staging: past, present, and future. Thorac Surg Clin. 2013;23:461–9. doi: 10.1016/j.thorsurg.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Rice TW, Apperson-Hansen C, DiPaola LM, et al. Worldwide Esophageal Cancer Collaboration: clinical staging data. Dis Esophagus. 2016;7:707–14. doi: 10.1111/dote.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldman AI. Eventcharts: visualizing survival and other timed-event data. Am Stat. 1992;46:13–8. [Google Scholar]
- 6.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann Appl Stat. 2008;2:841–60. [Google Scholar]
- 7.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 8.Ishwaran H, Kogalur UB. RandomForestSRC Random forest for survival, regression, and classification (RF-SRC) R-package version 2.1.0. 2016 http://cran.r-project.
- 9.Stekhoven DJ, Buhlmann P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28:112–8. doi: 10.1093/bioinformatics/btr597. [DOI] [PubMed] [Google Scholar]
- 10.Tang F, Ishwaran H. Random forest missing data algorithms. Stat Methods Med Res. doi: 10.1002/sam.11348. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rice TW, Rusch VW, Ishwaran H, Blackstone EH. Cancer of the esophagus and esophagogastric junction: data-driven staging for the 7th edition of the AJCC/UICC cancer staging manuals. Cancer. 2010;116:3763–73. doi: 10.1002/cncr.25146. [DOI] [PubMed] [Google Scholar]
- 12.Rice TW, Ishwaran H, Hofstetter WL, et al. Recommendations for pathologic staging (pTN) of cancer of the esophagus and esophagogastric junction for the 8th edition AJCC/UICCC staging manuals. Dis Esophagus. doi: 10.1111/dote.12533. forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manual for Staging of Cancer. American Joint Committee for Cancer Staging and End-results Reporting. Chicago: AJCC; 1977. [Google Scholar]
- 14.van Leeuwen PJ, Kranse R, Hakulinen T, et al. Disease-specific mortality may underestimate the total effect of prostate cancer screening. J Med Screen. 2010;17:204–10. doi: 10.1258/jms.2010.010074. [DOI] [PubMed] [Google Scholar]
- 15.Black WC, Haggstrom DA, Welch HG. All-cause mortality in randomized trials of cancer screening. J Natl Cancer Inst. 2002;94:167–73. doi: 10.1093/jnci/94.3.167. [DOI] [PubMed] [Google Scholar]
- 16.Lauer MS, Blackstone EH, Young JB, Topol EJ. Cause of death in clinical research: time for a reassessment? J Am Coll Cardiol. 1999;34:618–20. doi: 10.1016/s0735-1097(99)00250-8. [DOI] [PubMed] [Google Scholar]
- 17.Agoston AT, Zheng Y, Bueno R, Lauwers G, Odze RD, Srivastava A. Predictors of disease recurrence and survival in esophageal adenocarcinomas with complete response to neoadjuvant therapy. Am J Surg Pathol. 2015;39:1085–92. doi: 10.1097/PAS.0000000000000420. [DOI] [PubMed] [Google Scholar]
- 18.Davison JM, Landau MS, Luketich JD, et al. A model based on pathologic features of superficial esophageal adenocarcinoma complements clinical node staging in determining risk of metastasis to lymph nodes. Clin Gastroenterol Hepatol. 2016;14:369–77. doi: 10.1016/j.cgh.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


