Abstract
CYP2D6 is one of the most studied enzymes in the field of pharmacogenetics. The CYP2D6 gene is highly polymorphic with over 100 catalogued star (*) alleles, and clinical CYP2D6 testing is increasingly accessible and supported by practice guidelines. However, the degree of variation at the CYP2D6 locus and homology with its pseudogenes make interrogating CYP2D6 by short-read sequencing challenging. Moreover, accurate prediction of CYP2D6 metabolizer status necessitates analysis of duplicated alleles when an increased copy number is detected. These challenges have recently been overcome by long-read CYP2D6 sequencing; however, such platforms are not widely available. This review highlights the genomic complexities of CYP2D6, current sequencing methods and the evolution of CYP2D6 from allele discovery to clinical pharmacogenetic testing.
Keywords: : CYP2D6, CYP450-2D6, genotyping, long-read sequencing, pharmacogenetics, pharmacogenomics, Sanger sequencing, short-read sequencing
CYP2D6: discovery & pharmacogenetic implications
The CYP450 superfamily of enzymes is directly involved in the oxidative metabolism of numerous drugs, xenobiotics and other endogenous substances. The subfamily member, CYP2D6 accounts for only approximately 1–4% of all hepatic CYP450 enzymes, yet it metabolizes approximately 25% of commonly prescribed drugs, making it one of the most studied enzymes in the field of pharmacogenetics [1–3]. Moreover, it is also implicated in approximately 25% of the medications currently listed on the US FDA pharmacogenomic biomarkers in drug-labeling table [4], including antiarrhythmics, anticancer agents, tricyclic antidepressants, serotonin-selective reuptake inhibitors, antiemetics, antihistamines, antipsychotics, antiviral agents, β-blockers and opioids.
The discovery of CYP2D6 began with the 1969 identification of the enzyme responsible for nortriptyline plasma concentration variability [5]. The gene was later cloned, including the discovery of its involvement in debrisoquine and sparteine metabolism and the roles of variant alleles on the recessive ‘poor metabolizer’ trait [6–8]. This enzyme was characterized as CYP2D6 and its gene localized to chromosome 22q13.1 [9–13], and sequencing studies later revealed the presence of two highly homologous neighboring pseudogenes (CYP2D7 and CYP2D8) [12] (Figure 1). The CYP2D6 gene is highly polymorphic, and its variant allele frequencies can vary among different ethnic and ancestral populations [1,14]. To date, there are more than 100 variant star (*) alleles catalogued by the Human CYP450 allele nomenclature database [15,16]. Based on an individual’s CYP2D6 genotype, four different CYP2D6 metabolism phenotypes can be inferred: ultrarapid (UM), normal (previously referred to as ‘extensive’), intermediate and poor (PM) metabolizer [1,17–18]. Importantly, individuals with the more extreme UM or PM phenotypes are at higher risks for increased toxicity or reduced efficacy depending on whether a drug is bioactivated or eliminated by CYP2D6.
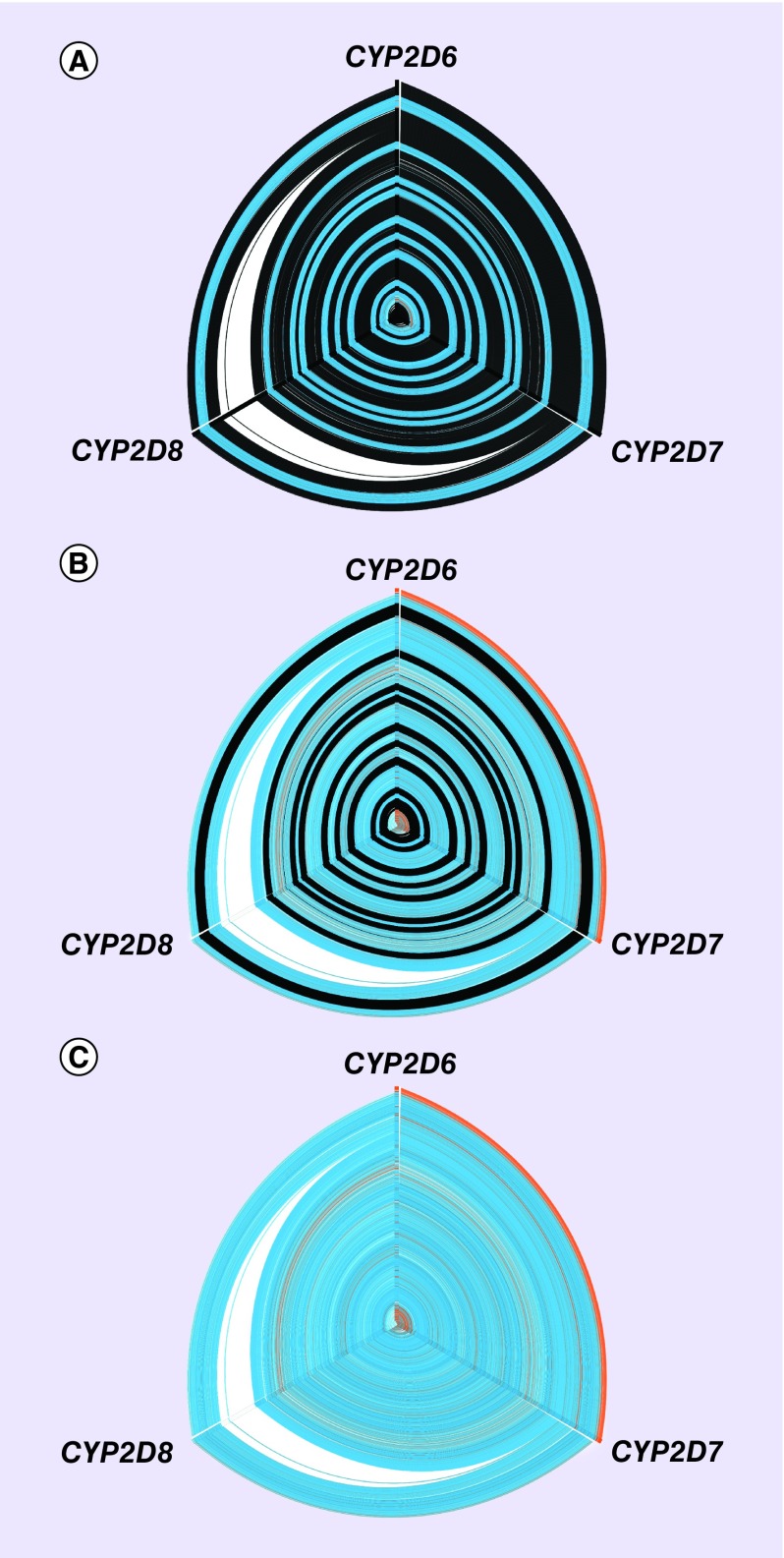
Figure 1. . Hive panel displaying multiple sequence alignment of CYP2D6, CYP2D7 and CYP2D8.
The hive plot edges display sequence similarity between CYP2D6-CYP2D7, CYP2D7-CYP2D8 and CYP2D8-CYP2D6 (clockwise from top). Three principle axes (0, 120 and 240°) of the hive plots represent the nucleotide composition of the multiple sequence alignment for the indicated gene: (A) exonic sequences (intronic sequences shown in black), (B) intronic sequences (exonic sequences shown in black) and (C) exonic and intronic sequences.
ClustalW was used to align the three genes (plus flanking 300 bp for each gene). Blue: aligned sequence is identical across the three genes; orange: aligned sequences are identical between the labeled genes; white: sequence gaps created by inserted nucleotides unique to the principle axis colored white.
The importance of CYP2D6 in human drug metabolism and increasing evidence for potential clinical utility has prompted practice guidelines on CYP2D6 for selected drugs by the Clinical Pharmacogenetics Implementation Consortium (CPIC) [19–22] and the Dutch Pharmacogenetics Working Group (DPWG) [23]. However, the CYP2D6 gene is also notable for its complex molecular architecture and pseudogene homology, which together result in technical challenges with targeted genotyping, full gene sequencing and genotype interpretation. This review highlights the genomic complexities of CYP2D6, current genotyping and sequencing methods to interrogate CYP2D6, and the evolution of CYP2D6 from allele discovery to phenotype prediction and clinical interpretation.
Genomic architecture of the CYP2D6 locus
CYP2D6 gene locus & sequence variation
Capra et al. [24] estimate that CYP2D6 arose approximately 361.2 million years ago and that the CYP2D locus gene duplication event occurred in a common ancestor of Hominini and great apes [25]. The canonical RefSeq CYP2D6 gene spans approximately 4400 nucleotides and includes 9 exons that are encoded on the minus strand at chromosome, 22q13.2. GENCODE release 25 recognizes three protein-coding transcript isoforms (ENST00000360608.5, ENST00000389970.3 and ENST00000359033.4) (Figure 2), one nonsense-mediated decay transcript (ENST00000360124.5), and one retained intron transcript (ENST00000488442.1). The 1000 Genomes Project identified more than 140 single-nucleotide variants and seven insertion/deletion variants in the CYP2D6 exonic regions across approximately 2500 individuals from 26 different populations [26]. The high degree of variation in CYP2D6 is further exemplified by the 680 PASS variants (i.e., those with a Variant Quality Score Recalibration score >99.95; 244 variants not passing quality filters) identified in >65,000 exomes by the Exome Aggregation Consortium (Supplementary Table 1) [27]. A number of repetitive and low-complexity sequences have also been annotated within and nearby CYP2D6, which give rise to the structural rearrangements characteristic of the CYP2D region, including the CYP2D6 gene duplication [28].
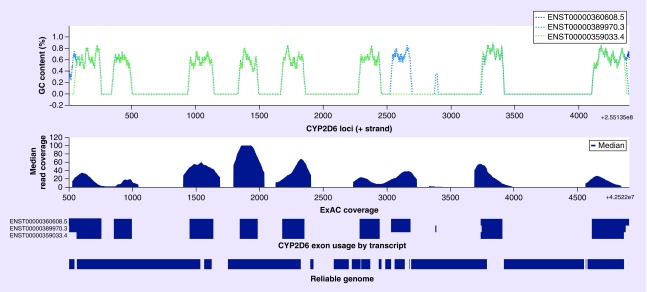
Figure 2. . Illustration of the CYP2D6 genomic region highlighting GC content across ten nucleotide-sliding windows of three functional CYP2D6 transcript isoforms (CYP2D6 loci: chr22, 42522500–42526907, GENCODE v19, hg19).
Median read coverage of ExAC exomes; base pair resolution of three functional CYP2D6 transcript isoforms; and base pair resolution of the intersection with 'reliable genome' intervals.
ExAc: Exome Aggregation Consortium.
Data taken from [29].
In addition to sequence variation in the CYP2D6 coding region, noncoding regulatory variants have recently been studied in an effort to more comprehensively inform CYP2D6 metabolism phenotype prediction. Genome-wide association studies have identified cis-regulation and more complex trans-regulation of CYP450 expression and activity, including noncoding variants implicated in CYP2D6 regulation [30]. In addition, the normal functional CYP2D6*2 allele, which is largely believed to encode an enzyme with normal activity, has recently been scrutinized based on the identification of two linked variants within an enhancer region approximately 115 kb downstream from the gene that interact with the CYP2D6 promoter through a long-range haplotype [31]. Although these regulatory variants require additional validation prior to incorporation into CYP2D6 star (*) allele nomenclature, recent functional studies suggest that the rs5758550 enhancer variant is predictive of CYP2D6 expression and may facilitate more precise CYP2D6 activity score and phenotype prediction, particularly among individuals who carry the common CYP2D6*2 variants (i.e., rs16947 [2850C > T; p.Arg296Cys] and rs1135840 [4180G > C; p.Ser435Thr]) [31,32].
CYP2D6 copy number variation
It is estimated that approximately 12% of the human genome contains copy number variants (CNVs) ranging from <1 kb to several megabases [33], which are the result of either nonallelic homologous recombination, nonhomologous end joining, fork stalling and template switching, or microhomology-mediated break-induced replication [34]. CYP2D6 CNVs include full-gene duplication and deletion, and complex rearrangements with CYP2D7, which can significantly influence the interpretation of CYP2D6 genotyping, sequencing and phenotype prediction [35–38], particularly as not all duplication alleles encode functional enzymes (e.g., reduced function CYP2D6*17xN and *36xN; no function CYP2D6*4xN). The frequencies of gene deletion (i.e., CYP2D6*5) and increased function duplication (e.g., *1xN, *2xN) alleles across worldwide populations range from approximately 2 to 6% and approximately 2 to 12%, respectively, with the CYP2D6 duplication being more prevalent among individuals of Middle Eastern descent [14].
CYP2D6 gene conversion & tandem alleles
A common polymorphism in several CYP2D6 star (*) alleles is an intron 1 CYP2D7 gene conversion at nucleotide positions 214–245 of the M33388.1 GenBank reference sequence. Although this noncoding conversion does not affect CYP2D6 transcription or translation, coding region conversion alleles have also been detected. Specifically, CYP2D6*68A is converted to CYP2D7 from intron 1 onward, the *13 series all contain an exon 1 conversion [39], *82 contains an exon 2 conversion and the exon 9 conversion is found in *4N, *36, *57 and *83.
Tandem CYP2D6 alleles have two different copies of CYP2D6 on a single chromosome, which interestingly are most commonly found together with CYP2D6/2D7 conversion (or ‘hybrid’) alleles. These tandem alleles require thorough interrogation for proper detection, and the most notable example is the *36+*10 tandem allele that is prevalent among Asians [40]. The importance of detecting these reduced function alleles is underscored by the fact that nonspecific copy number assessment of these samples would identify a gene duplication; however, these tandem alleles are not consistent with the UM phenotype [41].
CYP2D6 interrogation & allele discovery
CYP2D6 Sanger sequencing & targeted genotyping
The CYP2D6 gene and its pseudogenes were discovered by cloning and Sanger sequencing [12]; however, the subsequent availability of long-range PCR enabled Sanger sequencing of targeted exons across full-length CYP2D6 amplicons (∼2–7 kb) [42]. The long-range amplicon strategy can be coupled with nested mutation scanning techniques, such as, PCR-single strand conformation polymorphism (SSCP) and Sanger sequencing, which lead to the identification of many of the initial CYP2D6 star (*) alleles [43]. Although still a gold-standard for many molecular genetics applications, Sanger sequencing is increasingly being replaced by high-throughput second-generation sequencing. However, the CYP Nomenclature Committee still requires that all novel CYP2D6 variant alleles identified by high-throughput sequencing be confirmed by this classic technique.
Targeted genotyping is an inexpensive and common method to interrogate specific CYP2D6 variants (Table 1 & Figure 3) [44]; however, the limitations include potentially missing clinically relevant variants that were not genotyped, structurally rearranged CYP2D6 alleles and inaccurate star (*) allele haplotyping from the targeted variants. Although some CYP2D6 star (*) alleles can be identified by a single functional variant (e.g., CYP2D6*9 [2615_2617delAAG], rs5030656, p.Lys281del), many require more thorough haplotyping for identification. As noted, CYP2D6 long-range amplicons can also be used as templates for multiplexed genotyping methods, such as, TaqMan, RFLP and high-resolution melting analyses and/or allele-specific primer extension bead arrays.
Table 1. . Commercially available CYP2D6 genotyping and sequencing tests.
| Assay | Star (*) allele haplotypes interrogated | Company |
|---|---|---|
| xTAG CYP2D6 Kit v3 |
*2, *3, *4, *5, *6, *7, *8, *9, *10, *11, *15, *17, *29, *35, *41, *xN |
Luminex† |
| Ion AmpliSeq Pharmacogenomics Research Panel |
*2, *2A, *3, *4, *5, *6, *7, *8, *9, *10, *11, *12, *14, *15, *17, *20, *29, *35, *41, *29/*70, *xN |
ThermoFisher/ Ion Torrent‡ |
| DMET Plus |
*2, *3, *4, *5, *6, *7, *8, *9, *10, *11, *12, 14A, *14B, *15, *17, *18, *19, *20, *21, *29, *38, *40, *41, *42, *44, *56A, *56B, *64 |
ThermoFisher/ Affymetrix |
| PharmacoScan |
*2, *3, *4, *5, *6, *7, *8, *9, *10, *11, *12, *14A, *14B, *15, *17, *18, *19, *20, *21, *29, *38, *40, *41, *42, *44, *56A, *56B, *64, plus copy number variation |
ThermoFisher/ Affymetrix |
| iPLEX CYP2D6 Panel |
*1, (*2;*28;*32;*55;*59), (*2A;*31;*51), *2D, (*2L;*45B;*46), *2M, *3, *4, *4B, *4J, *4K, *4M, *4N;P, *53,*6, *6C, *7, *8, *9, (*10A;*37;*54), (*10B;*47;*49;*52;*72), *11, *12, *14A, *14B, *15, *17, *18, *19, *20, *21ª, *21B, *27, *29, *30, *34, *35, *36, *38, *39, *40, *41, *42, *44, *45A, *56A, *56B, *57, *58, *63, *64, *65, *68, *69, *70, *71, *82, *83, *84 |
Agena Bioscience |
| iPLEX PGx Pro Panel |
*1A, (*2A;*31;*51), (*2L;*35;*71), *3, *4, *4M, *6, *7, *8, *9, (*10;*36;*37; *47;*49;*52;*54;*57;*65;*72), *11, *12, *14A, *14B, *15, *17, *18, *19, *20, *21A, *21B, *30, *40, *41, *42, *44, *56A, *56B, *58, *64, *69 |
Agena Bioscience |
| GenoChip Tamox |
*3, *4, *5, *6, *7, *8, *9, *10, *11, *17, *29, *41, *xN |
Akabiotech |
| INFINITI CYP450 2D6I |
*2, *2A *3, *4, *5, *6, *7, *8, *9, *10, *12, *14, *17, *29, *41, *xN |
AutoGenomics |
| VeraCode ADME Core Panel |
*2A, *3, *4, *5, *6, *7, *8, *9, *10, *11, *12, *14, *15, *17, *18, *19, *20, *21, *38, *41, *42, *44, *56 |
Illumina |
| GenoChip CYP2D6 | *3, *4, *5, *6, *7, *8, *9, *10, *11, *17, *29, *41, *xN | PharmGenomics |
†The xTAG CYP2D6 Kit v3 assay is the only CYP2D6 platform approved by the US FDA for in vitro diagnostic testing.
‡The Ion AmpliSeq Pharmacogenomics Research Panel is the only commercial sequencing assay presented in the table.
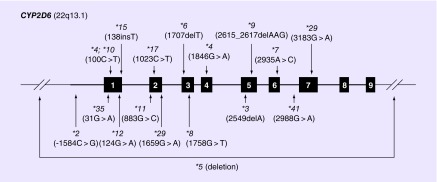
Figure 3. . Gene diagram of CYP2D6 (and chromosome cytoband location) highlighting the location of variant star (*) alleles that are commonly included in targeted genotyping assays, including the deletion allele (*5).
Note that variants are denoted by their common nucleotide nomenclature from M33388.1 GenBank reference sequence.
The detection of CYP2D6 CNVs represents another challenge, but can be enabled by targeted genotyping using CYP2D6-specific quantitative real-time PCR. In addition, assessing copy number by targeted quantitative real-time PCR at both the 5′ and 3′ regions of CYP2D6 can facilitate the identification of the CYP2D6*36+*10 tandem allele, as the 3′′ CYP2D7 gene conversion of *36 results in two detectable copies of the 5′ region (e.g., intron 2) and only one copy of the 3′ region (e.g., exon 9) on this chromosome [45].
CYP2D6 allele-specific interrogation
Allele-specific genotyping and sequencing techniques have been developed to more accurately characterize CYP2D6 haplotypes by determining the cis and trans configurations of identified variants. For example, CYP2D6*29 consists of four nonsynonymous variants in cis (1659G > A; 2850C > T; 3183G > A; 4180G > C), which encode an enzyme with reduced activity [46,47], one of these variants also occur on other star (*) allele haplotypes. In addition, individuals with a CYP2D6*1/*4 diplotype (normal metabolizer phenotype) carry the defective 100C > T (p.Pro34Ser) and 1846G > A (c.506-1G > A) variants in cis; however, if these two variants are in trans, the diplotype would be CYP2D6*4M/*10 (intermediate metabolizer phenotype) [48]. Allele-specific PCR directly amplifies specific haplotypes by anchoring PCR primers at specific variant nucleotides, which has been employed for both CYP2D6 [49] and CYP2C19 [50].
CYP2D6 second-generation short-read sequencing
Short-read sequencing produces read lengths of up to 300 bp and is predominantly derived from commercial platforms that use either light-based detection of fluorescently labeled nucleotides (Illumina) or electrical detection of proton release during nucleotide chain synthesis (Ion Torrent). Given the restrictions on read length for these platforms, the accuracy of short-read sequencing is reduced in genomic regions with low sequence complexity (e.g., tandem repeats, homopolymers), regions of dense polymorphism and/or repetitive elements (e.g., Alu, HERVs). Paired-end short-read sequencing, where reads are generated from both sides of a DNA fragment, can improve the accuracy of read mapping and variant calling by increasing the uniqueness of sequence alignments. However, repetitive elements with lengths that exceed the mean length of the sequenced DNA fragments will compromise the accuracy and efficiency of the sequencing results. Current Illumina and Ion Torrent sequencing protocols yield read lengths of approximately 100 bp with per-base error rates <1% from 100 to 1000 bp DNA fragments [51].
The CYP2D6 gene is a challenging region for short-read technologies due to repetitive elements, CNV, pseudogenes, and a high density of polymorphisms. A recent analysis of Illumina whole genome sequencing data partitioned the genome into regions of high and low reliability, which classified > 20% of CYP2D6 as ‘unreliable’ (Figure 2) [29]. This is consistent with the 1000 Genomes Project assignment of the CYP2D6 gene as ‘inaccessible’ [26]. Moreover, simulation studies using error-free standard paired-end CYP2D6 reads, which are 100 bp from DNA fragments of 500 bp, have underscored the difficulty of uniquely assigning reads to exon two of CYP2D6 versus CYP2D7 or CYP2D8 [52]. Longer read lengths and longer spacing resolve alignment ambiguity, but repetitive regions located at the upstream and downstream regions of CYP2D6 still remain challenging. Single-end reads longer than 3 kb can eliminate multiple alignment, which indicates the potential advantage of long-read CYP2D6 sequencing.
Both Illumina and Ion Torrent have high sequencing accuracy (>99 and >97%, respectively); however, the Ion Torrent chemistry has difficulty discriminating successive proton cleavage events in homopolymeric sequences [53], which are prevalent throughout CYP2D6 (Supplementary Table 2). However, Ion Torrent does have a commercial amplicon short-read sequencing panel that includes CYP2D6 (Table 1). As noted above, a principal issue for these short-read sequencing platforms when interrogated CYP2D6 (and other genes with highly homologous pseudogenes) is the ability to specifically and accurately call variants in the targeted region. Commonly used target enrichment techniques (i.e., oligonucleotide capture, amplification) are likely not specific enough to anneal to CYP2D6 and not CYP2D7 or CYP2D8, and the subsequent short-read alignment, whether specifically enriched or not, may also misalign to CYP2D7 and/or CYP2D8. In addition, the low-cycle amplification step commonly employed during Illumina sequencing library preparation may result in hybrid fragments between CYP2D6 and CYP2D7/CYP2D8. All of these issues result in off-target short-read sequencing, which subsequently leads to skewed and potentially inaccurate CYP2D6 variant calling. Illumina sequencing of the CYP2D6 region by whole-genome, whole-exome, targeted capture using PGRNseq [54] and long-read Pacific Biosciences (PacBio) sequencing is illustrated in Figure 4.
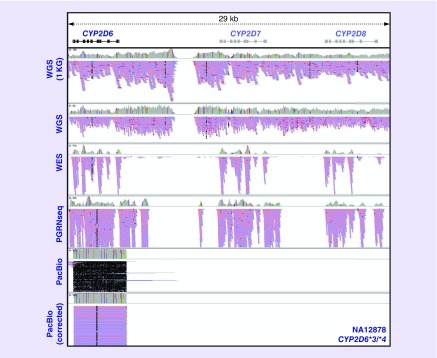
Figure 4. . Paired-end short-read sequencing (Illumina) and long-read sequencing (Pacific Biosciences) of the CYP2D6 gene region visualized with the Integrative Genomics Viewer.
Results for NA12878 (CYP2D6*3/*4) are displayed from top to bottom panels for WGS from the 1000 Genomes Project, in-house WGS, WES, targeted capture with the PGRNseq platform, targeted PacBio CYP2D6 sequencing, and ALEC-corrected targeted PacBio CYP2D6 sequencing. Of note, discrepant and skewed allele frequencies in several loci from the WGS data indicate potential read misalignment errors. Moreover, the common CYP2D6 capture strategies (e.g., WES, PGRNseq) coupled with short-read Illumina sequencing result in significant read assignment to the CYP2D7 and CYP2D8 pseudogenes. These reads indicate a lack of specificity for CYP2D6 by these target enrichment approaches and/or informatic errors related to read misalignment. Targeted PacBio sequencing results in CYP2D6-specific sequencing and no misalignment to CYP2D7 or CYP2D8, but random errors throughout the sequencing reads are characteristic to this technology. These random errors can be minimized by circular consensus sequencing read analysis; however, further correction prior to variant calling can also be accomplished by available informatics tools (e.g., Amplicon Long-read Error Correction [ALEC]).
1KG: 1000 Genomes Project; PacBio: Pacific Biosciences; WES: Whole-exome sequencing; WGS: Whole-genome sequencing.
CYP2D6 third-generation long-read sequencing
The difficulties with pseudogenes and CNVs that are inherent to short-read CYP2D6 sequencing prompted the recent development of third-generation long-read CYP2D6 sequencing using Oxford Nanopore and PacBio. Both platforms employ real-time sequencing but with different detection chemistries. The maximum read length recently achieved using Nanopore MinION was approximately 230 kb with an average of approximately 10.7 kb [55], and the latest P6-C4 chemistry from PacBio has a maximum read length of approximately 65 kb with an average of approximately 10–15 kb [56].
The recent Nanopore study sequenced approximately 5.0 kb CYP2D6 amplicons on the MinION, and variant calling and haplotype determination using the 2D consensus reads [57]. The 1D reads were discarded due to their high error and short mapping rates. BLASR was used for alignment and variants were called if they were detected in a third of reads. CYP2D6 haplotypes were inferred by interrogating nine targeted variants; however, the variant calling thresholds resulted in an ambiguous NA12878 CYP2D6 diplotype with three distinct haplotypes (*2, *3 and *4), which the authors hypothesized was due to PCR template switching or sample contamination [57].
The recent PacBio study also sequenced approximately 5.0 kb CYP2D6 amplicons using the RS II instrument, yet differed from the Nanopore study as it sequenced both ‘downstream’ and ‘upstream’ (i.e., duplicated) CYP2D6 copies by the use of specific primer sets [45]. Circular consensus sequencing reads were aligned using BWA-MEM followed by a novel error correction procedure (ALEC [58]), and variants called using GATK HaplotypeCaller (Figure 4)[45]. Sequencing of previously genotyped controls identified expected star (*) alleles, but also enabled suballele resolution, diplotype refinement and discovery of novel CYP2D6 alleles. Importantly, targeted PacBio sequencing of upstream and downstream CYP2D6 gene copies characterized the duplicated allele in control samples with CYP2D6 CNVs. This study concluded that PacBio CYP2D6 sequencing has the capacity to interrogate the entire gene in a single sequencing read as well as specifically characterize duplicated alleles when present, which ultimately could facilitate improved CYP2D6 metabolizer phenotype prediction for both research and clinical testing applications.
CYP2D6 phasing & star (*) allele haplotyping
Genetic phasing can be accomplished through analysis of parental genotypes (i.e., phasing by transmission), statistical comparison of observed variants with known haplotypes, statistical comparison of read sequences, analysis of RNAseq data and/or long-read DNA sequencing. Although phased CYP2D6 haplotypes have traditionally been determined by manual interrogation, the recent availability of high-throughput sequencing has prompted the need for more robust and automated haplotype/diplotype inference tools with logic derived from CYP2D6 star (*) allele definitions and translation tables. However, one of the current challenges is reconciling the historical star (*) allele nomenclature with current genome reference assemblies and a more informed understanding of the extent of variation in the genome [59]. In addition, a cumbersome informatics issue is converting identified CYP2D6 variants from the genome assembly used for sequencing (e.g., GRCh37/hg19) to the M33388.1 GenBank reference sequence used to define CYP2D6 star (*) alleles. This is typically accomplished by manual curation and/or with the CYP2D6 haplotype tables at PharmGKB [60], but recently has been automated by the freely available CYP2D6 VCF Translator [61]. Some of the currently available CYP2D6 phasing and diplotyping tools are explained in the following sections (Cypiripi, Constellation/Astrolabe, PharmCAT).
Cypiripi
The Cypiripi algorithm infers CYP2D6 diplotypes from short-read sequencing data, and is able to properly resolve complicated configurations, including CYP2D6/2D7 hybrids as well as CYP2D6 deletions and duplications [62]. The pipeline is composed of CYP2D6 star (*) allele library preparation (extracted from the nomenclature database); read alignment to the CYP2D genes by BLAST-like alignment tool (BLAT) and custom multimapping tools; read, variation and allele filtering; and a combinatorial optimization step that infers CYP2D6 genotype and copy numbers by integer linear programming [62].
Constellation/Astrolabe
Astrolabe (previously referred to as ‘Constellation’) was developed to impute diplotypes and assign metabolizer phenotypes for CYP2D6 and CYP2C19 from paired-end whole-genome sequencing data [52]. In the Astrolabe algorithm, identified variants are compared with all possible CYP2D6 diplotypes, which were generated using the defining variants from 119 established CYP2D6 star (*) alleles. A similarity coefficient is calculated for each possible diplotype, and the diplotype with the highest score is called for each sample. Astrolabe had a 97% sensitivity and 95% specificity and importantly, all extreme phenotypes (i.e., UM and PM) were accurately identified in the development cohort. In addition, simulation studies underscored the benefit of paired-end CYP2D6 sequencing over single-end reads as well as the increased specificity observed with long-read (e.g., >1 kb) sequencing, particularly across the highly homologous exon 2 region [52].
Pharmacogenomics Clinical Annotation Tool (PharmCAT)
The Pharmacogenomics Clinical Annotation Tool (PharmCAT) is a collaboration between the PGRN Statistical Analysis Resource, PharmGKB, the Clinical Genome Resource (ClinGen) and CPIC [63]. It extracts all CPIC level ‘A’ gene variants (except for G6PD and HLA) from a vcf file (including CYP2D6), interprets the variant alleles, infers diplotypes and generates an interpretation report based on CPIC guidelines [64]. The PharmCAT tool developers assemble and maintain the translation tables that underlie the tool, which could facilitate clinical implementation and more uniform pharmacogenetic sequencing interpretation.
CYP2D6 phenotype prediction & clinical pharmacogenetic testing
Predicting CYP2D6 metabolizer phenotype from diplotype is challenging and an imperfect inference [65]. Although there are over 100 variant CYP2D6 star (*) alleles catalogued by the Nomenclature Committee, most targeted genotyping platforms employed by clinical laboratories only interrogate a small subset of variants with established functional effect (Figure 3). Consequently, the CYP2D6*1 haplotype is assigned by default in the absence of any detected variant alleles. This commonly used system results in some alleles being incorrectly classified as CYP2D6*1 when they actually carry a low-frequency functional variant allele that was not directly genotyped. Full-gene sequencing techniques, such as, CYP2D6 SMRT sequencing [45] will result in the identification of more precise haplotypes and diplotypes, but the increased identification of rare and/or novel star (*) alleles could lead to an increased frequency of ‘indeterminate’ or ‘unclear’ CYP2D6 metabolizer phenotypes. As such, most clinical laboratories do not currently employ full-gene CYP2D6 sequencing.
A related phenotype classification system has been proposed for CYP2D6 metabolizer phenotype prediction, which has been adopted by recent CYP2D6 CPIC guidelines and is based on a continuum of ‘activity scores’ for different CYP2D6 alleles [18,66]. All CYP2D6 star (*) alleles are assigned an ‘activity score’ value when functional status is known (e.g., 0, 0.5, 1 and 2), and the sum of the maternal and paternal allele scores directly informs the CYP2D6 metabolizer phenotype (Table 2) [18]. Despite these advances in predicting CYP2D6 phenotype from genotype, some of the notable challenges still include interpreting reduced function and other low-frequency star (*) alleles with uncertain activities as well as determining which allele is duplicated when an increased CYP2D6 copy number is detected.
Table 2. . Example variant CYP2D6 alleles and the ‘activity score’ framework. † .
|
CYP2D6 diplotype |
Total CYP2D6 activity score‡ | Metabolizer phenotype§ | |
|---|---|---|---|
| Allele 1 (AS) | Allele 2 (AS) | ||
|
*1 (1) |
*1xN (2) |
3 |
UM |
|
*2 (1)† |
*35 (1) |
2 |
NM |
|
*1 (1) |
*4 (0) |
1 |
NM |
|
*4 (0) |
*10 (0.5) |
0.5 |
IM |
| *4 (0) | *7 (0) | 0 | PM |
†Please note that the recently described CYP2D6 upstream enhancer element may influence the expression of several star (*) allele haplotypes that include the common rs16947 (2850C > T) and rs1135840 (4180G > C) variants (e.g., *2, *41), which could further refine the activity scores of these alleles.
‡The activity score values assigned to CYP2D6 alleles are: 0 for *3, *4, *4xN, *5, *6, *7, *16, *36, *40, *42, *56B; 0.5 for *9, *10, *17, *29, *41, *45, *46; 1 for *1, *2, *35, *43, *45xN; and 2 for *1xN, *2xN, *35xN.
§The CYP2D6 metabolizer phenotype classification based on the diplotype activity score is: >2 = UM, 1–2 = NM, 0.5 = IM and 0 = PM.
AS: Activity score; IM: Intermediate metabolizer; NM: Normal metabolizer; PM: Poor metabolizer; UM: Ultrarapid metabolizer.
Despite the role of CYP2D6 in the metabolism of numerous medications, a clinically relevant effect of increased or decreased CYP2D6 activity has only been actualized in a subset of medications and drug classes. Clinical utility, cost–effectiveness and third-party reimbursement issues for CYP2D6 pharmacogenetic testing are beyond the scope of this review; however, clinical CYP2D6 testing is increasingly accessible and being adopted by clinicians to inform pharmacotherapy. Proficiency testing for CYP2D6 is available by the College of American Pathologists [67], and Coriell Institute Biorepository reference materials for CYP2D6 test validation have been developed by the CDC-based Genetic Testing Reference Material Coordination Program [68]. Clinical laboratory guidelines for CYP2D6 genotyping in the context of tamoxifen response testing have been published by the American College of Medical Genetics and Genomics [48], and support for healthcare practitioners on how to interpret clinical CYP2D6 diplotype and CYP2D6 metabolizer phenotype results can be found in the practice guidelines published by the CPIC [19–22] and DPWG [23].
Conclusion
Since the discovery of the polymorphic CYP2D6 gene, it has been one of the most widely studied genes in the field of pharmacogenetics due to its direct role in the metabolism of many commonly prescribed medications. Despite this extensive body of research over the past 25 years, interrogating CYP2D6 has proven challenging due to its pseudogene homology and the extent of structural variation that eventually was discovered at this locus. As such, it is likely that many early clinical research studies that only genotyped isolated CYP2D6 variants and limited star (*) alleles did not thoroughly capture the true diversity of CYP2D6 variation in their cohorts, which ultimately could have confounded result interpretation and study conclusions. Long-range PCR facilitated more accurate assessment of the CYP2D6 gene sequence; however, it has become clear that parallel copy number and sequence interrogation is necessary to properly define CYP2D6 diplotype and predicted metabolizer phenotype in any one individual. As third-generation long-read CYP2D6 sequencing becomes more commonly used by both research and clinical laboratories, it is likely that these platforms will facilitate a more precise and informed ability to infer interindividual CYP2D6-mediated drug response. These technical advancements will complement the refinement of phenotype prediction, which ultimately could further enable the reporting and implementation of clinical CYP2D6-based practice guidelines.
Future perspective
The accessibility of clinical CYP2D6 pharmacogenetic testing and interpretation practice guidelines coupled with the growing evidence for clinical validity and utility of CYP2D6 genotype-directed pharmacotherapy indicate that clinical genetic testing for this important pharmacogene will increasingly be incorporated into routine clinical care. This is further supported by the deployment of pre-emptive pharmacogenetic testing programs at selected academic medical centers [69] and the continued federal support for genomic medicine research aimed at returning ‘actionable’ sequence variants to study participants. Although CYP2D6 is an ideal gene for clinical implementation in all of these contexts, thorough full-gene sequencing and structural characterization is necessary for accurate metabolizer phenotype prediction. Long-read full-gene CYP2D6 sequencing platforms, including both allele- and duplication-specific assays, are available; however, most platforms used for clinical CYP2D6 testing are limited to only a small number of targeted CYP2D6 variants with known functional effect. As such, in order to facilitate the clinical implementation of more thorough CYP2D6 sequencing strategies, it will be imperative to develop rapid functional assays that assess novel and rare CYP2D6 variants as they are identified. In addition to facilitating more accurate metabolizer phenotype prediction for CYP2D6, a strategy of coupling long-read full-gene sequencing with functional assays is likely to have utility for other genes involved in interindividual drug response variability as well as other clinically actionable traits and Mendelian disorders.
Executive summary.
CYP450-2D6: discovery & pharmacogenetic implications
The CYP2D6 subfamily member accounts for only approximately 1–4% all hepatic CYP450 enzymes, yet it metabolizes approximately 25% of commonly prescribed drugs.
The CYP2D6 gene is implicated in approximately 25% of the medications currently listed on the US FDA pharmacogenomic biomarkers in drug-labeling table.
Genomic architecture of the CYP2D6 locus
The CYP2D6 gene is highly polymorphic with more than 100 variant star (*) alleles catalogued by the Human CYP450 Allele Nomenclature Database; however, CYP2D6 is challenging to interrogate due to highly homologous pseudogenes (CYP2D7 and CYP2D8).
Noncoding variants have recently been implicated in the regulation of CYP2D6 expression, and other important structural variants include CYP2D6 deletions and duplications as well as gene conversion alleles and tandem allele configurations.
CYP2D6 interrogation & allele discovery
The earliest CYP2D6 variant alleles were discovered by cloning and/or Sanger sequencing; however, the availability of long-range PCR subsequently enabled the use of multiplexed targeted genotyping across full-length CYP2D6 gene amplicons.
Short-read sequencing of CYP2D6 (including exome sequencing) is challenging due to nonspecific target enrichment and/or misalignment as a result of the homologous pseudogenes.
Long-read sequencing has recently been successfully applied to full-gene CYP2D6 amplicons, including long-range haplotyping, duplication-specific sequencing and novel allele characterization.
CYP2D6 phasing & star (*) allele haplotyping
Many CYP2D6 sequence variants are found in multiple star (*) allele haplotypes, underscoring the need for proper variant phasing, which recently has been enabled by automated software tools (Cypiripi, Astrolabe and Pharmacogenomics Clinical Annotation Tool).
CYP2D6 phenotype prediction & clinical pharmacogenetic testing
CYP2D6 metabolizer phenotype prediction infers four different categories (ultrarapid, normal, intermediate and poor metabolizer) based on the available genotype or sequencing data; however, the CYP2D6*1 haplotype is assigned by default in the absence of detected variant alleles, which can lead to inaccurate phenotype prediction.
An ‘activity score’ phenotype prediction system has been developed to facilitate more uniform CYP2D6 diplotype interpretations, and practice guidelines and resources are increasingly available for clinical laboratories and practitioners to enable CYP2D6 testing and pharmacogenetic-guided medical management.
Future perspective
To facilitate the clinical implementation of more thorough CYP2D6 sequencing strategies, it will be imperative to develop rapid functional assays that assess novel and rare CYP2D6 variants as they are identified.
Supplementary Material
Footnotes
Financial and competing interests disclosure
This work was supported in part by the National Institute of General Medical Sciences (NIGMS) of the NIH, through grant K23GM104401 (SA Scott). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Owen RP, Sangkuhl K, Klein TE, Altman RB. Cytochrome P450 2D6. Pharmacogenet. Genomics. 2009;19(7):559–562. doi: 10.1097/FPC.0b013e32832e0e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Zhou SF, Liu JP, Lai XS. Substrate specificity, inhibitors and regulation of human cytochrome P450 2D6 and implications in drug development. Curr. Med. Chem. 2009;16(21):2661–2805. doi: 10.2174/092986709788681985. [DOI] [PubMed] [Google Scholar]
- 4.US FDA. Table of pharmacogenomic biomarkers in drug labeling. https://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm
- 5.Alexanderson B, Evans DA, Sjoqvist F. Steady-state plasma levels of nortriptyline in twins: influence of genetic factors and drug therapy. Brit. Med. J. 1969;4(5686):764–768. doi: 10.1136/bmj.4.5686.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichelbaum M, Spannbrucker N, Steincke B, Dengler HJ. Defective N-oxidation of sparteine in man: a new pharmacogenetic defect. Eur. J. Clin. Pharmacol. 1979;16(3):183–187. doi: 10.1007/BF00562059. [DOI] [PubMed] [Google Scholar]
- 7.Mahgoub A, Idle JR, Dring LG, Lancaster R, Smith RL. Polymorphic hydroxylation of debrisoquine in man. Lancet. 1977;2(8038):584–586. doi: 10.1016/s0140-6736(77)91430-1. [DOI] [PubMed] [Google Scholar]
- 8.Bertilsson L, Dengler HJ, Eichelbaum M, Schulz HU. Pharmacogenetic covariation of defective N-oxidation of sparteine and 4-hydroxylation of debrisoquine. Eur. J. Clin. Pharmacol. 1980;17(2):153–155. doi: 10.1007/BF00562624. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez FJ, Skoda RC, Kimura S, et al. Characterization of the common genetic defect in humans deficient in debrisoquine metabolism. Nature. 1988;331(6155):442–446. doi: 10.1038/331442a0. [DOI] [PubMed] [Google Scholar]
- 10.Gough AC, Miles JS, Spurr NK, et al. Identification of the primary gene defect at the cytochrome P450 CYP2D locus. Nature. 1990;347(6295):773–776. doi: 10.1038/347773a0. [DOI] [PubMed] [Google Scholar]
- 11.Heim M, Meyer UA. Genotyping of poor metabolisers of debrisoquine by allele-specific PCR amplification. Lancet. 1990;336(8714):529–532. doi: 10.1016/0140-6736(90)92086-w. [DOI] [PubMed] [Google Scholar]
- 12.Kimura S, Umeno M, Skoda RC, Meyer UA, Gonzalez FJ. The human debrisoquine 4-hydroxylase (CYP2D) locus: sequence and identification of the polymorphic CYP2D6 gene, a related gene, and a pseudogene. Am. J. Hum. Genet. 1989;45(6):889–904. [PMC free article] [PubMed] [Google Scholar]
- 13.Eichelbaum M, Baur MP, Dengler HJ, et al. Chromosomal assignment of human cytochrome P-450 (debrisoquine/sparteine type) to chromosome 22. Br. J. Clin. Pharmacol. 1987;23(4):455–458. doi: 10.1111/j.1365-2125.1987.tb03075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS. Prediction of CYP2D6 phenotype from genotype across world populations. Genet. Med. 2017;19(1):69–76. doi: 10.1038/gim.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CYP2D6 allele nomenclature. http://www.cypalleles.ki.se/cyp2d6.htm
- 16.Sim SC, Ingelman-Sundberg M. Update on allele nomenclature for human cytochromes P450 and the human cytochrome P450 allele (CYP-allele) nomenclature database. Methods Mol. Biol. 2013;987:251–259. doi: 10.1007/978-1-62703-321-3_21. [DOI] [PubMed] [Google Scholar]
- 17.Caudle KE, Dunnenberger HM, Freimuth RR, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC) Genet. Med. 2016;19(2):215–223. doi: 10.1038/gim.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin. Pharmacol. Ther. 2008;83(2):234–242. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
- 19.Bell GC, Caudle KE, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin. Pharmacol. Ther. 2016 doi: 10.1002/cpt.598. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC(R)) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 Update. Clin. Pharmacol. Ther. 2016 doi: 10.1002/cpt.597. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake Inhibitors. Clin. Pharmacol. Ther. 2015;98(2):127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical pharmacogenetics implementation consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. 2014;95(4):376–382. doi: 10.1038/clpt.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swen JJ, Nijenhuis M, De Boer A, et al. Pharmacogenetics: from bench to byte-an update of guidelines. Clin. Pharmacol. Ther. 2011;89(5):662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 24.Capra JA, Williams AG, Pollard KS. ProteinHistorian: tools for the comparative analysis of eukaryote protein origin. PLoS Comput. Biol. 2012;8(6):e1002567. doi: 10.1371/journal.pcbi.1002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasukochi Y, Satta Y. Molecular evolution of the CYP2D subfamily in primates: purifying selection on substrate recognition sites without the frequent or long-tract gene conversion. Genome Biol. Evol. 2015;7(4):1053–1067. doi: 10.1093/gbe/evv056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.1000 Genomes Project Consortium. Auton A, Brooks LD. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundqvist E, Johansson I, Ingelman-Sundberg M. Genetic mechanisms for duplication and multiduplication of the human CYP2D6 gene and methods for detection of duplicated CYP2D6 genes. Gene. 1999;226(2):327–338. doi: 10.1016/s0378-1119(98)00567-8. [DOI] [PubMed] [Google Scholar]
- 29.Popitsch N, Consortium WGS, Schuh A, Taylor JC. ReliableGenome: annotation of genomic regions with high/low variant calling concordance. Bioinformatics. 2016;33(2):155–160. doi: 10.1093/bioinformatics/btw587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, Zhang B, Molony C, et al. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 2010;20(8):1020–1036. doi: 10.1101/gr.103341.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Poi MJ, Sun X, Gaedigk A, Leeder JS, Sadee W. Common CYP2D6 polymorphisms affecting alternative splicing and transcription: long-range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum. Mol. Genet. 2014;23(1):268–278. doi: 10.1093/hmg/ddt417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, Papp AC, Sun X. Functional characterization of CYP2D6 enhancer polymorphisms. Hum. Mol. Genet. 2015;24(6):1556–1562. doi: 10.1093/hmg/ddu566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 34.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10(8):551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaedigk A, Hernandez J, Garcia-Solaesa V, Sanchez S, Isidoro-Garcia M. Detection and characterization of the CYP2D6*9x2 gene duplication in two Spanish populations: resolution of AmpliChip CYP450 test no-calls. Pharmacogenomics. 2011;12(11):1617–1622. doi: 10.2217/pgs.11.107. [DOI] [PubMed] [Google Scholar]
- 36.Ramamoorthy A, Skaar TC. Gene copy number variations: it is important to determine which allele is affected. Pharmacogenomics. 2011;12(3):299–301. doi: 10.2217/pgs.11.5. [DOI] [PubMed] [Google Scholar]
- 37.Ramamoorthy A, Flockhart DA, Hosono N, Kubo M, Nakamura Y, Skaar TC. Differential quantification of CYP2D6 gene copy number by four different quantitative real-time PCR assays. Pharmacogenet. Genomics. 2010;20(7):451–454. doi: 10.1097/FPC.0b013e32833a1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaedigk A, Jaime LK, Bertino JS, et al. Identification of novel CYP2D7–2D6 hybrids: non-functional and functional variants. Front Pharmacol. 2010;1:121. doi: 10.3389/fphar.2010.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sim SC, Daly AK, Gaedigk A. CYP2D6 update: revised nomenclature for CYP2D7/2D6 hybrid genes. Pharmacogenet. Genomics. 2012;22(9):692–694. doi: 10.1097/FPC.0b013e3283546d3c. [DOI] [PubMed] [Google Scholar]
- 40.Gaedigk A, Bradford LD, Alander SW, Leeder JS. CYP2D6*36 gene arrangements within the CYP2D6 locus: association of CYP2D6*36 with poor metabolizer status. Drug Metab. Dispos. 2006;34(4):563–569. doi: 10.1124/dmd.105.008292. [DOI] [PubMed] [Google Scholar]
- 41.Black JL, Walker DL, O’kane DJ, Harmandayan M. Frequency of undetected CYP2D6 hybrid genes in clinical samples: impact on phenotype prediction. Drug Metab. Dispos. 2012;40(1):111–119. doi: 10.1124/dmd.111.040832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaedigk A, Bhathena A, Ndjountche L, et al. Identification and characterization of novel sequence variations in the cytochrome P4502D6 (CYP2D6) gene in African Americans. Pharmacogenomics J. 2005;5(3):173–182. doi: 10.1038/sj.tpj.6500305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marez D, Legrand M, Sabbagh N, et al. Polymorphism of the cytochrome P450 CYP2D6 gene in a European population: characterization of 48 mutations and 53 alleles, their frequencies and evolution. Pharmacogenetics. 1997;7(3):193–202. doi: 10.1097/00008571-199706000-00004. [DOI] [PubMed] [Google Scholar]
- 44.Gaedigk A. Complexities of CYP2D6 gene analysis and interpretation. Int. Rev. Psychiatry. 2013;25(5):534–553. doi: 10.3109/09540261.2013.825581. [DOI] [PubMed] [Google Scholar]
- 45.Qiao W, Yang Y, Sebra R, et al. Long-read single molecule real-time full gene sequencing of cytochrome P450–2D6 . Hum. Mutat. 2016;37(3):315–323. doi: 10.1002/humu.22936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wennerholm A, Dandara C, Sayi J, et al. The African-specific CYP2D617 allele encodes an enzyme with changed substrate specificity. Clin. Pharmacol. Ther. 2002;71(1):77–88. doi: 10.1067/mcp.2002.120239. [DOI] [PubMed] [Google Scholar]
- 47.Wennerholm A, Johansson I, Hidestrand M, Bertilsson L, Gustafsson LL, Ingelman-Sundberg M. Characterization of the CYP2D6*29 allele commonly present in a black Tanzanian population causing reduced catalytic activity. Pharmacogenetics. 2001;11(5):417–427. doi: 10.1097/00008571-200107000-00005. [DOI] [PubMed] [Google Scholar]
- 48.Lyon E, Gastier Foster J, Palomaki GE, et al. Laboratory testing of CYP2D6 alleles in relation to tamoxifen therapy. Genet. Med. 2012;14(12):990–1000. doi: 10.1038/gim.2012.108. [DOI] [PubMed] [Google Scholar]
- 49.Gaedigk A, Riffel AK, Leeder JS. CYP2D6 haplotype determination using long range allele-specific amplification: resolution of a complex genotype and a discordant genotype involving the CYP2D6*59 allele. J. Mol. Diagn. 2015;17(6):740–748. doi: 10.1016/j.jmoldx.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott SA, Tan Q, Baber U, et al. An allele-specific PCR system for rapid detection and discrimination of the CYP2C19 *4A, *4B, and *17 alleles: implications for clopidogrel response testing. J. Mol. Diagn. 2013;15(6):783–789. doi: 10.1016/j.jmoldx.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 51.Zook JM, Chapman B, Wang J, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nature Biotechnol. 2014;32(3):246–251. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
- 52.Twist GP, Gaedigk A, Miller NA, et al. Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole-genome sequences. NPJ Genom. Med. 2016;1:15007. doi: 10.1038/npjgenmed.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng W, Zhao S, Xue D, et al. Improving alignment accuracy on homopolymer regions for semiconductor-based sequencing technologies. BMC Genomics. 2016;17(Suppl. 7):521. doi: 10.1186/s12864-016-2894-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gordon AS, Fulton RS, Qin X, Mardis ER, Nickerson DA, Scherer S. PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet. Genomics. 2016 doi: 10.1097/FPC.0000000000000202. Epub ahead to print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ip CL, Loose M, Tyson JR, et al. MinION Analysis and Reference Consortium: Phase I data release and analysis. F1000Res. 2015;4:1075. doi: 10.12688/f1000research.7201.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.PacBio. New Chemistry Boosts Average Read Length to 10 kb – 15 kb for PacBio® RS II. http://www.pacb.com/blog/new-chemistry-boosts-average-read/
- 57.Ammar R, Paton TA, Torti D, Shlien A, Bader GD. Long read nanopore sequencing for detection of HLA and CYP2D6 variants and haplotypes. F1000Res. 2015;4:17. doi: 10.12688/f1000research.6037.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.GitHub. The amplicon long-read error correction. https://github.com/scottlab/ALECtext
- 59.Robarge JD, Li L, Desta Z, Nguyen A, Flockhart DA. The star-allele nomenclature: retooling for translational genomics. Clin. Pharmacol. Ther. 2007;82(3):244–248. doi: 10.1038/sj.clpt.6100284. [DOI] [PubMed] [Google Scholar]
- 60.PharmGKB. https://www.pharmgkb.org
- 61.Qiao W, Wang J, Pullman BS, Chen R, Yang Y, Scott SA. The CYP2D6 VCF Translator. Pharmacogenomics J. 2016 doi: 10.1038/tpj.2016.14. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Numanagic I, Malikic S, Pratt VM, Skaar TC, Flockhart DA, Sahinalp SC. Cypiripi: exact genotyping of CYP2D6 using high-throughput sequencing data. Bioinformatics. 2015;31(12):i27–i34. doi: 10.1093/bioinformatics/btv232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klein TE, Whirl-Carrillo M, Whaley RM, et al. The 66th Annual Meeting of The American Society of Human Genetics. Vancouver, Canada: 19 October 2016. Pharmacogenomics Clinical Annotation Tool (PharmCAT) Presented at. [Google Scholar]
- 64.GitHub. PharmGKB/PharmCAT. https://github.com/PharmGKB/PharmCAT
- 65.Hertz DL, Snavely AC, Mcleod HL, et al. In vivo assessment of the metabolic activity of CYP2D6 diplotypes and alleles. Brit. J. Clin. Pharmacol. 2015;80(5):1122–1130. doi: 10.1111/bcp.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hicks JK, Swen JJ, Gaedigk A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr. Drug Metab. 2014;15(2):218–232. doi: 10.2174/1389200215666140202215316. [DOI] [PubMed] [Google Scholar]
- 67.Wu AH. Genotype and phenotype concordance for pharmacogenetic tests through proficiency survey testing. Arch. Pathol. Lab. Med. 2013;137(9):1232–1236. doi: 10.5858/arpa.2012-0261-CP. [DOI] [PubMed] [Google Scholar]
- 68.Pratt VM, Everts RE, Aggarwal P, et al. Characterization of 137 genomic DNA reference materials for 28 pharmacogenetic genes: a GeT-RM collaborative project. J. Mol. Diagn. 2016;18(1):109–123. doi: 10.1016/j.jmoldx.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin. Pharmacol. Ther. 2014;96(4):482–489. doi: 10.1038/clpt.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.