Abstract
Homing endonucleases confer mobility to their host intervening sequence, either an intron or intein, by catalyzing a highly specific double-strand break in a cognate allele lacking the intervening sequence. These proteins are characterized by their ability to bind long DNA target sites (14–40 bp) and their tolerance of minor sequence changes in these sites. A wealth of biochemical and structural data has been generated for these enzymes over the past few years. Herein we review our current understanding of homing endonucleases, including their diversity and evolution, DNA-binding and catalytic mechanisms, and attempts to engineer them to bind novel DNA substrates.
INTRODUCTION
Homing is the lateral transfer of an intervening sequence (either an intron or intein) to a homologous allele that lacks the sequence (1). The process is catalyzed by an endonuclease that recognizes and cleaves the target allele. The ‘homing endonuclease’ itself is encoded by an open reading frame (ORF) embedded within the mobile intervening sequence. The mobile elements avoid disrupting host gene function by self-splicing at the RNA (introns) or protein (inteins) level.
Homing endonucleases are highly specific and have evolved to cleave target sequences within cognate alleles without being overly toxic to the organism. They tolerate some individual base variation at their homing site, which ensures their propagation despite evolutionary drift of their target sequence. Finally, homing endonucleases tend to be small proteins of <40 kDa, a property likely due to length limitations of the mobile sequences in which they reside.
This extra-Mendelian genetic phenomenon was first described for a group I intron of budding yeast. In the 1970s, the genetic marker ‘ω’ in Saccharomyces cerevisiae was found to transfer to strains lacking the marker when crossed to ω+ strains (2). This marker corresponded to a 1.1 kb group I intron found in the large ribosomal RNA (rRNA) gene of the mitochondrial genome. Subsequent analysis indicated that the gene duplication event required a double-strand break at the intron insertion site and the expression of an ORF within the intron itself (3,4). This ORF was further shown to encode a site-specific endonuclease capable of recognizing and cleaving the intronless allele, and thereby initiating the homing event (5–7). This protein, now called I-SceI based on current nomenclature (8), was the first of over 250 homing endonucleases since identified.
Homing appears to be widespread (reviewed in 8–13). Thirty percent of group I introns are estimated to contain internal ORFs, and a significant number of these appear to be mobile. In addition to group I introns, many group II introns, archaeal introns and inteins also engage in homing. Furthermore, intervening sequences capable of homing are found in all branches of life: eubacteria, archaea and eucaryota. Within eucaryotes, these elements are found within nuclear, mitochondrial and chloroplast genomes. Homing has evolved multiple times, as evidenced by the differing recognition/cleavage mechanisms between group I and group II introns, and by the existence of multiple distinct families of homing endonucleases.
In this review we will summarize our understanding of these enzymes. This will include the distribution of intervening sequences, group I and group II intron homing mechanisms, classification of group I homing endonuclease families, their evolution, their DNA recognition and cleavage mechanisms and the potential for engineering homing endonucleases with novel specificity.
DISTRIBUTION OF INTERVENING SEQUENCES
Intervening sequences are found in all branches of life and can be classified into multiple categories based on sequence homology and the mechanism by which the sequence is removed prior to host gene function (reviewed in 11,14–17). Common forms of intervening sequence include spliceosomal mRNA introns, group I introns, group II introns, archaeal introns and inteins.
Although ubiquitous in the nuclear genomes of higher eucaryotes, the spliceosomal mRNA introns are not known to be mobile or harbor homing endonucleases. Mobile sequences and their accompanying homing endonucleases, however, have been discovered in each of the other four categories. It is the mobility of these sequences that is hypothesized to have given rise to the large number of non-mobile introns commonly found in genomes. For example, structural similarities between group II introns and splicosomal mRNA introns have led to suggestions that some mRNA introns in the human genome might be relics of once-mobile group II introns (18,19). Further evidence for this theory is provided by recent experiments in which engineered group II introns invade ectopic chromosomal sites (20,21).
The remaining categories of intervening sequence are phylogenetically widespread. Self-splicing group I introns have been found in the mitochondrial DNA of fungi, but these elements have also been characterized in eubacteria and bacteriophages, the mitochondrial and chloroplast genomes of plants and algae, and the nuclei of ciliates, slime molds, algae and fungi (reviewed in 11,22,23). The self-splicing group II introns have been found in fungal and plant mitochondria, algal and plant chloroplasts, and eubacteria (24,25). They are the common form of intervening sequence in chloroplasts and mitochondria of higher plants, and are a minor form in fungal mtDNA. Archaeal introns, although rare, are located within many tRNA and rRNA genes (16). Finally, inteins, a distinct form of intervening sequence spliced out of the host gene at the protein level, are found mainly in archaea, but also in bacteria and a few eucaryotic organelles and nuclei (17).
GROUP I AND GROUP II INTRON HOMING MECHANISMS
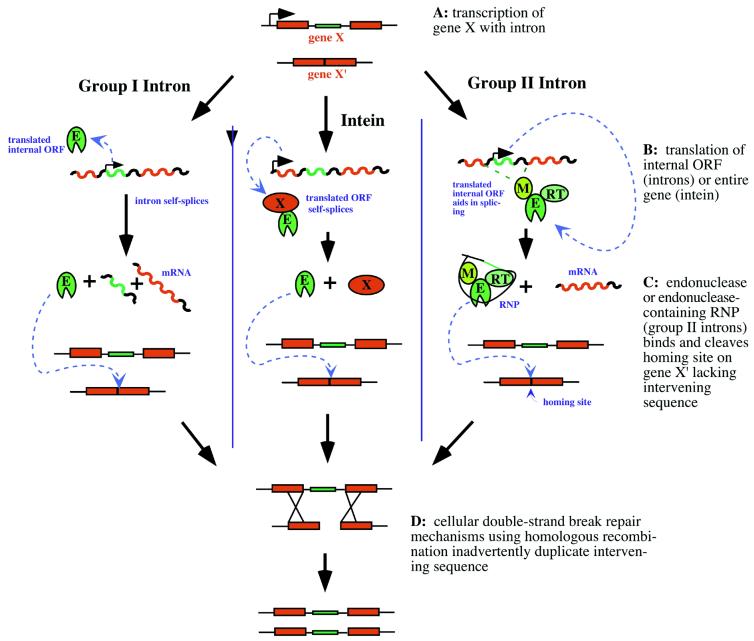
The homing mechanism of mobile group I introns requires the translation of the intron-encoded endonuclease (Fig. 1, left) (reviewed in 11,14–17). This protein product is highly specific for the intronless allele, binding to a homing site composed of the flanking exon sequences (into which the intervening sequence is copied). Once bound to the homing site, the endonuclease cleaves this site and cellular mechanisms relying on homologous recombination between alleles for proper double-strand break repair ensure the lateral transfer of the intervening sequence to the cleaved allele. Although less extensively studied, homing of archaeal introns and inteins (Fig. 1, center) is thought to proceed through an identical or similar mechanism.
Figure 1.
Homing mechanisms of group I introns (left), inteins (center) and group II introns (right) in which the intervening sequence of gene X is duplicated in its cognate allele, gene X′. Mobile ORFs and encoded products are green; host gene exons and products are red; other nucleic acid sequence is black. E, endonuclease; M, maturase; RT, reverse-transcriptase; RNP, ribonucleoprotein.
The homing mechanism employed by mobile group II introns (Fig. 1, right), termed ‘retrohoming,’ differs significantly from the mechanism used by group I and archaeal introns and inteins (26–33). As determined from group II introns found in yeast mitochondria and the bacterium Lactococcus lactis, this mechanism is more complex and requires the use of a ribonucleoprotein (RNP) consisting of the encoded protein endonuclease and the spliced intron RNA. The protein also functions as a maturase and reverse-transcriptase. Once the intron-encoded ORF has been translated, the protein product binds the RNA and aids in proper splicing. The spliced intron lariat/protein product (the RNP) recognizes the intronless allele via base-pairing between the protein-bound RNA and the DNA target site. The RNA lariat reverse splices into the target site, and the complementary DNA strand is usually cleaved by the protein’s endonuclease domain. Finally, the protein’s reverse-transcriptase domain synthesizes DNA using the invading RNA template. Cellular machinery completes the homing of the intervening sequence by replacing the invading RNA with DNA.
HOMING ENDONUCLEASE FAMILIES
The frequently observed mobility of these genetic elements has promoted their spread into all branches of life. As will be discussed more extensively later in this review, homing endonuclease genes are themselves thought to be mobile elements that invade self-splicing intervening sequences, thereby rendering mobility to the entire sequence (13). Self-splicing intervening sequences provide an ideal refuge within a host genome, as they allow propagation of the homing endonuclease without adversely affecting host gene function. Indeed, one group of homing endonucleases in particular, the LAGLIDADG family, has successfully exploited group I introns, archaeal introns and inteins. This family, along with the GIY-YIG, His-Cys box and HNH families, were first described as mobile sequences within group I introns (reviewed in 8). Since that time, members of both the GIY-YIG and HNH families, in addition to the LAGLIDADG family, have been described in systems other than group I introns. The majority of these ORFs, however, are still associated with the group I introns, and the four families are often collectively termed ‘group I homing endonuclease families’ in the literature.
The group I homing endonucleases have been more exhaustively studied biochemically and structurally and will be the focus of the remainder of this review. This discussion will also include other homing endonucleases closely related to group I homing endonucleases in structure and mechanism, namely the homing endonucleases from mobile inteins and archaeal introns.
LAGLIDADG family
This large protein family with >200 members has been variously termed ‘LAGLIDADG’, ‘DOD’, ‘dodecapeptide’, ‘dodecamer’ and ‘decapeptide’ (8,11,34). The LAGLIDADG endonucleases are the most phylogenetically diverse of the homing endonuclease families. This vast host distribution includes, for example, the genomes of plant and algal chloroplasts, fungal and protozoan mitochondria, bacteria and archaea. One reason for a wide distribution of LAGLIDADG ORFs appears to be their remarkable ability to invade unrelated types of intervening sequences, including group I introns, archaeal introns and inteins. Descendents of LAGLIDADG homing endonucleases are also found as freestanding endonuclease genes (35–37) and as maturases that assist in RNA splicing (38–40). Members of this family are defined by having either one or two copies of the conserved LAGLIDADG motif. Enzymes that contain a single copy of this motif, such as I-CreI (41) and I-CeuI (42) act as homodimers and recognize a nearly palindromic homing site (which, like a homodimeric protein, has inherent 2-fold symmetry). Enzymes that have two copies of this motif separated by 80–150 residues, such as I-DmoI (43) and PI-SceI (44) act as monomers. Unlike homodimers, monomers are not constrained to highly symmetrical DNA targets, and in fact their homing sites tend to be less palindromic. All LAGLIDADG endonucleases recognize long DNA sites (14–30 bp) and cleave the DNA to leave 4 nt 3′ overhangs. Like most nucleases, they require divalent cations for activity.
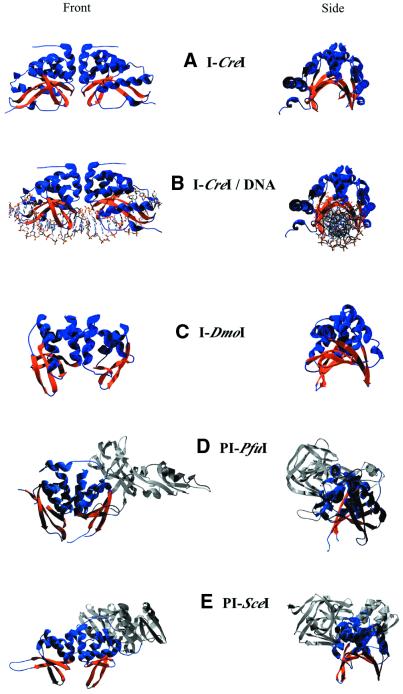
As shown in Figure 2, structural models using X-ray crystallography have been generated for the group I intron-encoded I-CreI (23S rRNA gene of Chlamydomonas reinhardtii chloroplast; 45), the archaeal intron-encoded I-DmoI (23S rRNA gene of Desulfurococcus mobilis; 46), the intein-encoded PI-SceI (ATP synthase gene of S.cerevisiae mitochondria; 47), and the archaeal intein-encoded PI-PfuI (RNR gene of Pyrococcus furiosus; 48). Structures of I-CreI bound to its DNA homing site have also been elucidated (49,50). The four enzyme structures reveal the functional significance of the LAGLIDADG motif, the nature of the DNA-binding interface, the location of the two active sites, and (in the case of the I-CreI–DNA complexes) details of the catalytic mechanism.
Figure 2.
Structures of LAGLIDADG family members. Endonuclease domains are blue with orange β-sheets showing DNA-binding saddle. Other intein domains are gray. (A) I-CreI homodimer. (B) I-CreI with DNA. (C) I-DmoI monomer. (D) PI-PfuI monomer. (E) PI-SceI monomer.
Despite little primary sequence homology outside of the LAGLIDADG motifs, the topology of I-CreI, I-DmoI and the endonuclease domains of PI-SceI and PI-PfuI are markedly similar. The core αββαββα fold of the I-CreI subunit is repeated twice in I-DmoI, PI-SceI and PI-PfuI and confers upon all three monomers a pseudo-dimeric structure.
The first α-helix of each domain or subunit contains the defining LAGLIDADG motif. The two LAGLIDADG helices of each protein form a tightly packed dimer or domain interface with direct van der Waals contacts between the protein backbones. The close packing of the helical backbone and the sharp turn provided by the highly conserved glycine residues at the bottom of the helices position conserved acidic (Asp or Glu) residues near the protein interface. These residues coordinate the divalent metal cations used by the two adjacent active sites. Because of the close packing of the LAGLIDADG helices, both active sites are accommodated within the width of the minor groove and are arranged appropriately to yield 4 nt 3′ overhangs. The precise catalytic mechanism of DNA cleavage is discussed later in this review.
The DNA-binding interface is formed by the four β-strands of each domain that fold into an antiparallel β-sheet in the shape of a saddle. As revealed by the I-CreI–DNA structure, the first two strands exhibit a curve complimentary to that of the major groove of the DNA-binding site. Within these strands, every second side chain makes a base-specific contact within the groove, while the basic residues of the flanking loops make contact to the phosphodiester backbone of the DNA. The β-sheets are stabilized by hydrophobic packing between the tops of the sheets and the next two α-helices of the topology.
The length of the DNA-binding saddles in I-CreI (75 Å) is longer than that of I-DmoI (50 Å), which corresponds to the differing length of their homing sites (22 versus 14 bp, respectively). The longer 30 bp homing site recognized by PI-SceI is due to an additional DNA-binding interface provided by a separate DNA recognition domain combined with the 60 Å endonuclease saddle (51,52). The PI-SceI structure suggests that the DNA homing site needs to be severely bent to accommodate the binding interface. This is corroborated by gel shift data that estimate a bend of ∼60° (53,54). Similarly, PI-PfuI induces a 73° bend in its DNA target site and extended DNA recognition is likely mediated by a unique stirrup domain (48,55).
PI-PfuI appears to be particularly interesting. It was isolated from a cosmid library of the archaeon P.furiosus in a screen for proteins with specific affinity for Holliday junction DNA (55). Although this activity was further characterized in vitro, its biological significance is unknown. One possible role for this activity, however, might be participation in a double-strand break repair pathway with a Holliday junction intermediate (55).
Many group I intron-encoded LAGLIDADG proteins also function as maturases and aid in the efficient splicing of introns. This activity was first described for the third cytochrome b gene in S.cerevisiae mitochondria (56,57). These proteins do not cleave the RNA; instead, it is thought that they simply aid in folding the RNA into a conformation that favors self-splicing. Many LAGLIDADG maturases, however, are also functional endonucleases, including I-AniI from Aspergillus nidulans (40,58) and I-ScaI from Saccharomyces capensis (39,59). Additional experiments further convey the close association between LAGLIDADG endonucleases and maturases. In the I-SceII homing endonuclease from S.cerevisiae, a single E→K mutation activates latent maturase activity (60). Conversely, a S.cerevisiae maturase (cyt b gene) closely related to I-SceI can be transformed into a homing endonuclease by two amino acid changes (59). These results suggest endonuclease and maturase activity to be closely coupled in both the function and evolution of LAGLIDADG proteins encoded within group I introns.
GIY-YIG family
This smaller family of endonucleases is characterized by the conserved GIY-(X10–11)-YIG motif (61). GIY-YIG endonucleases have been found in the T4 bacteriophage both as freestanding enzymes (F-TevI, F-TevII; 62) and within mobile group I introns (I-TevI, I-TevII; 63). GIY-YIG ORFs have also been reported in introns of fungal mitochondria (64–66), algal mitochondria (67,68) and algal chloroplasts (69,70).
Studies of the 28 kDa I-TevI and the 30 kDa I-TevII reveal these monomeric enzymes to recognize long homing sites (37 and 31 bp, respectively) and cleave their DNA many bases away from the intron insertion site (71–74). Both enzymes bind primarily across the minor groove and phosphate backbone and cleave their DNA substrates to leave 2 bp 3′ overhangs (74–76). Furthermore, I-TevI is extremely tolerant of base pair changes in its homing site. No specific bases are essential for activity and small insertions or deletions between the cleavage and insertion sites are permitted (72,76).
Limited proteolysis and footprinting experiments have shown I-TevI to be a bipartite enzyme with distinct catalytic and DNA-binding domains separated by a long flexible linker (76,77), similar to the type IIs restriction enzyme FokI (78). The C-terminal DNA-binding domain recognizes a 20 bp sequence that includes the intron insertion site. The cleavage site bound by the cleavage domain is ∼25 bp away (71,72). The GIY-YIG motif of I-TevI is located in the N-terminal catalytic domain. NMR studies reveal this domain to have a mixed α/β topology with the GIY-YIG residues located in the three-stranded β-sheet (61). Two other highly conserved residues (R27 and E75) reside in the α-helices and are required for catalytic activity (61). When independently expressed, the C-terminal binding domain binds with the same affinity as the wild-type enzyme suggesting its role as the primary structural region for DNA recognition (77). Crystallographic studies of this domain bound to its 20 bp DNA site demonstrate it to consist of three separate DNA-binding subdomains: a zinc finger, followed by an α-helix, and then a helix–turn–helix motif (V.Derbyshire, M.Belfort and P.Van Roey, personal communication). This structure reveals how multiple DNA-binding domains with low intrinsic specificity can be summed together and linked to a catalytic domain to yield an endonuclease with high overall specificity.
His-Cys box family
This small family of proteins is encoded within the only known mobile group I introns residing in nuclear genomes (79). All of these mobile introns are located within highly conserved regions of nuclear small and large subunit ribosomal DNA of slime molds, fungi and amoebae. The best-studied member of this family is I-PpoI from Physarum polycephalum (Fig. 3A) (80–84). This enzyme, along with I-DirI from Didymium iridis, has been experimentally shown to promote the homing of its intron in its natural hosts (80,85). Another closely related group of His-Cys box homing endonucleases from Naegleria, I-NjaI, I-NanI and I-NitI has recently been described (86,87), although experiments to verify their role in mobility of their intron have yet to be performed. As these enzymes are >85% identical and act as isoschizomers, they are collectively referred to as I-NxxI in this review. The His-Cys box endonucleases are characterized by a highly conserved series of histidines and cysteines over a central 100 residue region (79,81).
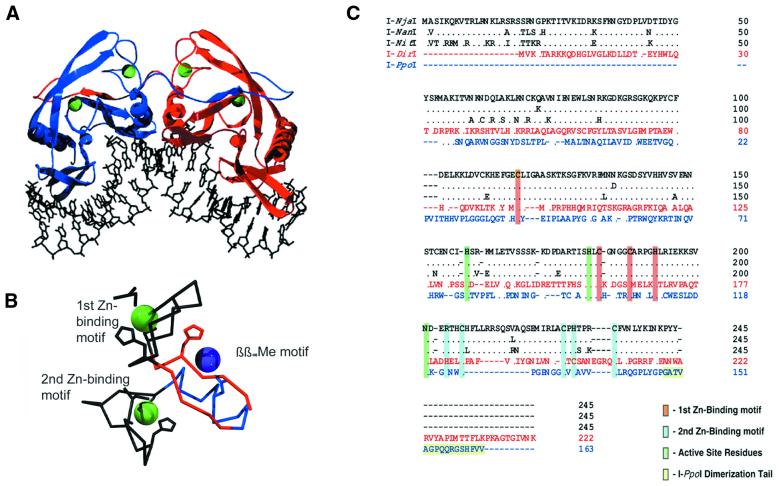
Figure 3.
His-Cys box family. (A) I-PpoI homodimer bound to its DNA target site. Note ‘domain-swapped’ C-terminal tails that form much of the dimer interface. Zinc atoms are green. (B) Core metal-binding motifs in I-PpoI. Zinc ions are green; magnesium ion is purple. β-strands and α-helix of the conserved ββαMe motif are red and blue, respectively. (C) Primary sequence alignment of known His-Cys box family members including I-NjaI, I-NanI, I-NitI, I-DirI and I-PpoI. The C-terminal dimerization tail of I-PpoI and the highly conserved zinc-binding and active sight residues are highlighted.
I-PpoI has been well-studied biochemically and structurally. This small homodimeric protein (18 kDa per monomer) recognizes a 14 bp pseudo-palindromic homing site that it bends severely and cleaves to yield 4 nt 3′ overhangs (81,82). Like all other homing endonucleases, it is dependent on divalent cations for cleavage activity. Several structures of I-PpoI have been solved by X-ray crystallography, including the apo-enzyme, the enzyme–product complex, the enzyme–substrate complex, and H98A and L116A mutant enzymes bound to DNA (88–90). These structures reveal the significance of the His-Cys box motif and other conserved residues and offer a detailed model of the cleavage mechanism.
An I-PpoI monomer has a mixed α/β topology consisting of two α-helices and ten β-strands folded into three separate β-sheets. Like I-CreI, I-PpoI uses β-sheets for recognition and binding to the major groove of its DNA homing site, and cleaves across the minor groove. Three structural features of I-PpoI are particularly interesting. First, I-PpoI lacks a tightly packed hydrophobic core found in most proteins. Instead, the highly conserved histidines and cysteines of the His-Cys box form novel zinc-binding folds that stabilize the structural core of the protein. The His-Cys box also provides two conserved histidines to each active site. Secondly, in contrast to the tightly packed active sites of the LAGLIDADG enzymes (10 Å separation), the active sites in I-PpoI are spaced 20 Å apart. To accommodate these active sites and still yield the same cleavage pattern as the LAGLIDADG endonucleases, I-PpoI severely bends and distorts its DNA substrate to widen the minor groove enough (20 Å, compared with 9–10 Å of B-form DNA) to place the scissile phosphates into its active sites. Finally, the central dimer interface of I-PpoI buries a relatively small surface area (only 700 Å2). Dimerization is stabilized by domain-swapped C-terminal tails that wrap around the opposing monomer to bury an additional 900 Å2 (Fig. 3A).
As only ∼10% of the residues of each I-PpoI, I-NxxI and I-DirI align well with either of the other two, these proteins are likely to have diverged long ago (Fig. 3C). Despite the limited overall similarity of the primary sequence, the amino acids that form the zinc-binding motifs and the active sites are highly conserved in both identity and position. The N-terminal zinc-binding motif in I-PpoI (C-X58-C-X4-C-X4-H) is easily identified in both I-NxxI and I-DirI (C-X65-C-X4-C-X4-H and C-X63-C-X5-C-X4-H, respectively). Similarly, the C-terminal zinc-binding motif of I-PpoI (R-X2-C-X6-C-X1-H-X3-C), which includes R122 that forms a hydrogen bond to the backbone oxygen of V136 to help stabilize the tight fold, is present in I-NxxI (R-X2-C-X19-C-X1-H-X3-C). Some divergence is seen in I-DirI (H-X2-C-X13-C-X1-C-X7-C), although the R→H and H→C substitutions are likely functional equivalents. While the precise active site mechanism of I-PpoI will be discussed later in this review, we can see from the alignment of these proteins that all active site residues are also strictly conserved. H78, H98 and N119 of I-PpoI align with H158, H180 and N201 of I-NxxI and H134, H156 and N178 of I-DirI.
Despite the excellent alignment of the zinc-binding motifs and the active site residues in I-PpoI, I-DirI and I-NxxI, striking structural differences must exist. Most importantly, I-NxxI cleaves to yield a novel 5 bp 3′ overhang, in marked contrast to the 4 bp 3′ overhang generated by I-PpoI (86,87). As already described, I-PpoI severely distorts its DNA target in order to accommodate the scissile phosphates within its active sites (separated by 20 Å). This distortion may not be required for I-NxxI. Unlike the 10 Å that separates I-PpoI scissile phosphates in B-form DNA, the scissile phosphates cleaved by I-NxxI are 15 Å apart in unbent DNA. Thus, if the active sites of I-PpoI and I-NxxI are in similar positions, the I-NxxI homing site might be much less distorted when bound to the protein. In support of this hypothesis, it is significant that I-NxxI lacks a homolog to I-PpoI L116. This residue is required for maximal bend in the DNA near the active sites and the formation of a well-ordered protein–DNA complex (90). Similarly, no obvious homolog to L116 exists in I-DirI. Although its homing site has not been characterized, this suggests I-DirI might not bend its DNA substrate severely and, therefore, might cleave with 5 bp overhangs analogous to I-NxxI.
I-NxxI lacks the C-terminal tail vital for I-PpoI dimerization (Fig. 3C). I-PpoI’s tail consists of the final 16 C-terminal residues; only three from I-NxxI overlap with these. It is likely that I-NxxI has evolved alternate means by which to stabilize a dimer interface. Other possibilities include simply burying more surface area at the interface near the active sites, or the use of its long N-terminal sequence—which is not conserved in I-PpoI—for formation of a novel dimer interface. I-DirI has both an extended N- and C-terminal tail, the significance of which is not known.
HNH family
Members of the HNH family are the least well characterized structurally and biochemically of all homing endonucleases. Interestingly, the HNH motif is also the least restricted to the group I homing endonuclease families. It has been identified in the non-specific endonucleases, such as the antibacterial colicins E7 and E9 (91,92), and in proteins encoded by mobile group II introns, including I-SceV, I-SceVI and I-LlaI (28,29,32). HNH proteins contain two pairs of conserved histidines surrounding a conserved asparagine within a 30–33 residue sequence (93,94). Members of the HNH family encoded within group I introns include I-HmuI and I-HmuII from the SPO1 and SP83 introns, respectively, of two closely related Bacillus subtilis bacteriophages (95–97) and I-TevIII from the nrdB intron of RB3 bacteriophage (73). This motif is also contained within I-CmoeI from the psbA gene of the Chlamydomonas moewusii chloroplast (98) and within a homologous, yet uncharacterized, ORF in the psbA gene of C.reinhardtii (70). Other ORFs containing HNH motifs, including inteins, have been reported but not studied (34,99,100).
The currently identified HNH enzymes share few biochemical properties outside of their defining motif. For example, I-HmuI and I-HmuII each cleave only one strand of their DNA substrate (97). I-TevIII is the only homing endonuclease known to generate 5′ overhangs (73), while I-CmoeI leaves 4 bp 3′ overhangs like the LAGLIDADG endonucleases (98). Furthermore, I-TevIII and I-CmoeI each appear to have additional domains found in other enzymes. I-TevIII contains two putative DNA-binding zinc finger domains (73); I-CmoeI contains a degenerate GIY-YIG sequence (98). Other than these readily identified primary sequence motifs, little detailed structural information is available for the HNH homing endonucleases.
Most current structural knowledge of the HNH proteins derives from biochemical and structural studies of the E7 (91) and E9 colicins (92). Colicins are a group of plasmid-encoded protein DNases from Escherichia coli that are synthesized and secreted to kill other E.coli and coliform bacteria (101). The structure of the colicin E9 shows residues in the conserved HNH motif fold into two β-strands and two α-helices wrapped around a bound phosphate molecule and a nickel ion. The conserved H103 coordinates the phosphate, while the invariant N118 forms a bridging and stabilizing hydrogen bond to the backbone of L105. The metal is coordinated by three histidines: the conserved H127, and the less well conserved H102 and H131. All of these residues appear to have homologs in I-HmuI, I-HmuII, I-TevIII and I-CmoeI.
Do the His-Cys box and HNH families constitute a ββαMe superfamily?
Although we treat members of the His-Cys box and HNH families as distinct groups in this review, recent structural evidence might suggest a closer relationship. One interesting aspect of the colicin structures is the significant degree to which a 22 residue stretch containing three of the four elements of secondary structure discussed above (two β-strands and an α-helix), including the active site, superimpose upon the core active site structure of I-PpoI (Fig. 3B; 102). In addition to this, others have noted a high level of structural homology between the active site of the non-specific Serratia nuclease (SN) and I-PpoI (103). Specifically, the metal ion is in identical positions in all three (magnesium in SN and I-PpoI and nickel in E9). N119, which coordinates the metal ion in I-PpoI, is N119 in SN and H127 in E9 (this substitution is thought to govern the identity of the metal ion; 102). The invariant H98 in I-PpoI superimposes with H89 in SN and H103 in E9. Similarly, the definitive N118 of the HNH motif in E9 that forms the stabilizing hydrogen bond to the protein backbone near the active site is homologous to N110 in SN. In I-PpoI, this residue superimposes with H110, which is part of the first structurally fundamental zinc-binding fold.
Given these remarkable similarities in active site structure, Kuhlmann et al. (102) have proposed a reclassification of the His-Cys box and HNH endonucleases into a larger family called the ββαMe family. Presumably, this family could encompass all His-Cys box and HNH homing endonucleases, the HNH non-specific endonucleases (colicins) and enzymes related to the SN (e.g. NucA). The recently reported structure of T4 endonuclease VII, a DNA junction resolvase, also shares this core fold (104).
It must be noted, however, that little or no structural homology exists outside of this central ββαMe core. For example, the defining features of the His-Cys box family, namely the two conserved zinc-binding folds that form the structural scaffold of I-PpoI, are not found in any other known protein. Furthermore, residues in similar positions within the ββαMe fold appear to have different functions in different families. While H154 of the HNH endonuclease I-CmoeI is required for binding its DNA substrate (presumably by coordinating the metal ion; 98), the homologous residue in I-PpoI, H98, functions as the general base that activates the nucleophilic water and is not required for the binding of substrate (89). These differences, as well as our lack of understanding of the HNH endonuclease catalytic mechanism, make it difficult to decipher the exact relationship between members of the HNH and His-Cys box families of homing endonucleases, and in general, other nucleases that share this fold.
EVOLUTION OF HOMING ENDONUCLEASES
At their most basic level, homing endonuclease genes are extremely efficient parasitic elements that take advantage of host DNA double-strand break repair mechanisms for propagation. They are closely associated with intervening sequences, and use these sequences as a refuge that enables the replication of their ORF without deleterious effects to a host gene. The existence of at least three—and possibly four—distinct families of homing endonucleases indicates multiple independent evolutionary origins for these elements. Furthermore, the distribution of homing endonucleases among intervening sequences suggests the origin of homing endonucleases and intervening sequences to also be independent (reviewed in 13). For example, homologous members of both the LAGLIDADG and HNH families are found in unrelated introns and inteins. Conversely, closely related introns, such as those from the T4 phage, harbor homing endonucleases from different families.
The relationship between homing endonuclease-containing mobile intervening sequences and their host genes is dynamic. The extra-Mendelian inheritance of mobile intervening sequences assures that they rapidly become fixed in a population. After this occurs, there is little selective pressure for the maintenance of a functional homing endonuclease, and this ORF is subsequently lost, followed eventually by the intron itself. Once the intron is lost, the site is ripe for re-invasion. This cycle has been illustrated in yeast by the ω element (105). A sampling of 20 species enabled the categorization of the ω element into functional, non-functional and absent states. Comparisons of the phylogenetic analyses of the intron, the ω endonuclease (I-SceI), and the host yeast species present evidence for multiple rounds of rapid invasion by horizontal transmission, slow degeneration and eventual loss of the intron with an invasion frequency of once about every two million years.
There has been speculation whether homing endonucleases and/or their mobile intervening sequences confer any benefit to their host gene or organism. We believe most sequences capable of homing to be nothing more than opportunistic selfish DNA. Evidence for this conjecture is provided by the cycle of invasion and elimination of the ω element as described above. If these invasive elements benefit their hosts, it is unlikely that they would be eliminated from the host genome at such a rapid rate. Conversely, it is possible that a small fraction of homing endonuclease genes have evolved a more host-beneficial role. In so doing, these enzymes may have generated selective pressure to be retained and, therefore, broken free of the invasion/elimination cycle of most homing sequences.
A few homing endonuclease-like proteins do seem to benefit their host organism. Maturases such as intron-encoded I-AniI (40,58), for example, aid in the splicing of their host intron. While the presence of the intron itself might not benefit the host per se, the efficient removal of the intervening sequence may have provided enough selective pressure to maintain a functional protein, which in turn aided in the survival of the intron and its homing endonuclease ORF. Furthermore, some homing endonucleases appear to have been adopted by the host as freestanding enzymes. One LAGLIDADG enzyme in particular, HO, is responsible for the mating switch in yeast (37,106). This protein is closely related to the endonuclease domain of the intein-encoded PI-SceI and may have arisen from a gene duplication event or from the remnants of a parallel invasion by the intervening sequence. Finally, I-HmuI and I-HmuII are atypical HNH intron-encoded endonucleases from closely related B.subtilis phage that cleave only one strand of their DNA substrate (97). These enzymes confer advantage to their host by specifically cleaving the DNA of the heterologous phage during mixed infections (97). Whether I-AniI, I-HmuI and/or I-HmuII have an invasion/elimination cycle analogous to ω has yet to be determined. Assuming their beneficial role, however, it is possible that these homing endonuclease genes with divergent functions have escaped this cycle.
Although the mechanism by which horizontal transfer of mobile intervening sequences between distantly related species is not well understood, a growing body of evidence suggests that it occurs frequently. One study of >300 diverse land plants reveals a recent massive invasion by a homing endonuclease-containing group I intron in the mitochondrial cox1 gene (107,108). Similarly, homologous group I introns and associated homing endonucleases also exist in identical positions of large subunit RNA genes in an algal chloroplast and an amoeboid protozoon mitochondria (109). The wide distribution of LAGLIDADG-containing inteins, as found in eucaryotes, bacteria and archaea, are also suggestive of horizontal transmission.
Two examples of relatively recent invasion of intervening sequence by a homing endonuclease gene offer further insight into the evolutionary relationship of the homing endonuclease gene and its host sequence. I-TevII, in particular, might suggest a mechanism by which intervening sequences are invaded to generate mobile introns. The intron sequences flanking the endonuclease ORF are remarkably similar to the exon sequences flanking the intron itself which, when fused in a cognate allele lacking the intervening sequence, constitutes the I-TevII homing site (110). This homology in flanking sequences indicates the homing endonuclease ORF invaded the intron through a cleavage event similar to that used to mobilize the intron.
I-PcII, a recently described GIY-YIG enzyme from a group I intron (cytb-i2) in Podospora curvicolla mitochondria, provides an alternate example of recent invasion and further emphasizes the evolutionary independence of homing endonuclease genes from intervening sequences (66). This ORF is unique in that it invaded an already mobile LAGLIDADG intron. The insertion of the GIY-YIG ORF, which includes a stop codon at its 3′ end, into the middle of the LAGLIDADG ORF destroys the activity of the resident LAGLIDADG homing endonuclease and the mobility of the intron host. The GIY-YIG endonuclease, however, confers mobility to its own ORF in a homing-like mechanism that specifically targets the LAGLIDADG-containing intron. This is the first known example of a homing endonuclease gene that directly invades and parasitizes another active homing endonuclease-containing mobile intron.
Finally, structural studies of the homing endonucleases have enabled us to infer some of the evolutionary history of individual families. Within the LAGLIDADG family, four widely divergent LAGLIDADG homing endonuclease structures have been solved (Fig. 2), including a homodimer from a chloroplast intron, I-CreI (45,49,50), a monomer from an archaeal intron, I-DmoI (46), a monomeric intein from fungal mitochondria, PI-SceI (47) and a monomeric intein from archaea, PI-PfuI (48). Despite limited sequence homology outside the LAGLIDADG motif(s), they all share a core topology that places the residues involved in DNA-binding and catalysis within the same domain. Furthermore, while the three monomers have a pseudo-dimeric structure that resembles the I-CreI homodimer, one half of each monomer is larger than the other. In I-DmoI, the N-terminal domain is larger and more closely related to an I-CreI subunit, while the opposite is true for both PI-SceI and PI-PfuI. This observation has led to speculation that the monomeric LAGLIDADG enzymes are descendents of multiple gene duplication events of an I-CreI-like ancestor (46).
The recent discovery of three I-CreI homologs also suggests a dynamic LAGLIDADG evolutionary history (111). Although these enzymes cleave the same DNA substrate, only seven of the 21 amino acids in I-CreI that contact the DNA (per subunit) are conserved. Other curious evolutionary events are also evident. I-MsoI, for example, shares only 35% identity with I-CreI in amino acid sequence but 52% in nucleotide sequence. At least five distinct sets of frameshift mutations account for this unusual observation.
Like the LAGLIDADG endonucleases, the His-Cys box proteins also contain DNA-binding and catalytic machinery within one domain (88). This family is much smaller than the LAGLIDADG family, and all known members are homodimers from nuclear group I introns in slime molds and amoebae. Their niche within nuclear genomes may have prevented them from spreading as widely as other homing endonuclease genes, such as those of the LAGLIDADG family. Interestingly, the His-Cys box enzymes also have a widely divergent primary sequence (Fig. 3C). The only homology readily identified at the amino acid level correlates to a core series of histidines and cysteines. The structure of I-PpoI reveals these conserved core residues to be located in the active site and the novel zinc-binding motifs that form the core internal structures of these proteins. The evolution of these enzymes is particularly interesting in that they have acquired unique cleavage patterns. While I-PpoI cleaves to yield 4 nt 3′ overhangs (81), the I-NxxI enzymes cleave to yield 5 nt 3′ overhangs (86,87). This difference might be the result of two different strategies to cleave across the minor groove. Similarly, the His-Cys box enzymes appear to have evolved different strategies for dimerization. I-PpoI uses an extended C-terminal tail (88), which is not conserved in the I-NxxI enzymes, to wrap around its sister subunit and stabilize the dimer interface.
Unlike the LAGLIDADG and His-Cys box enzymes that contain DNA-binding and catalytic functions within the same domain, the GIY-YIG and HNH motifs appear to be catalytic units that fuse with other DNA-binding domains to generate functional homing endonucleases. The structure of I-TevI reveals the GIY-YIG sequence to be part of a catalytic domain, and this is fused to a series of DNA-binding motifs, including a zinc finger, an α-helix and a helix–loop–helix, like beads on a string (V.Derbyshire, M.Belfort and P.Van Roey, personal communication). Similarly, the GIY-YIG enzyme I-TevII appears to utilize separate zinc finger domains for DNA recognition and binding (73). Finally, I-CmoeI contains both an HNH and a degenerate GIY-YIG motif (98), although the functional significance of the latter is unknown.
DNA-BINDING AND RECOGNITION BY HOMING ENDONUCLEASES
Two properties distinguish homing endonucleases from other site-specific endonucleases (Fig. 4). Most strikingly, homing endonucleases recognize and bind exceptionally long homing sites (14–40 bp) despite their relatively small size (<40 kDa). Furthermore, homing endonucleases tolerate subtle changes in homing site sequence, and often cleave mutant homing sites as efficiently as wild-type. Homing site length ensures high specificity and relatively low toxicity (associated with excessive cleavage of a host genome). Conversely, the ability to recognize and cleave variant homing sites ensures the continued propagation of the intervening sequences at ectopic sites within an evolving host and perhaps to similar sites within closely related species. These characteristics are in sharp contrast to the best-studied group of endonucleases, namely the large and diverse class II restriction enzymes (reviewed in 112).
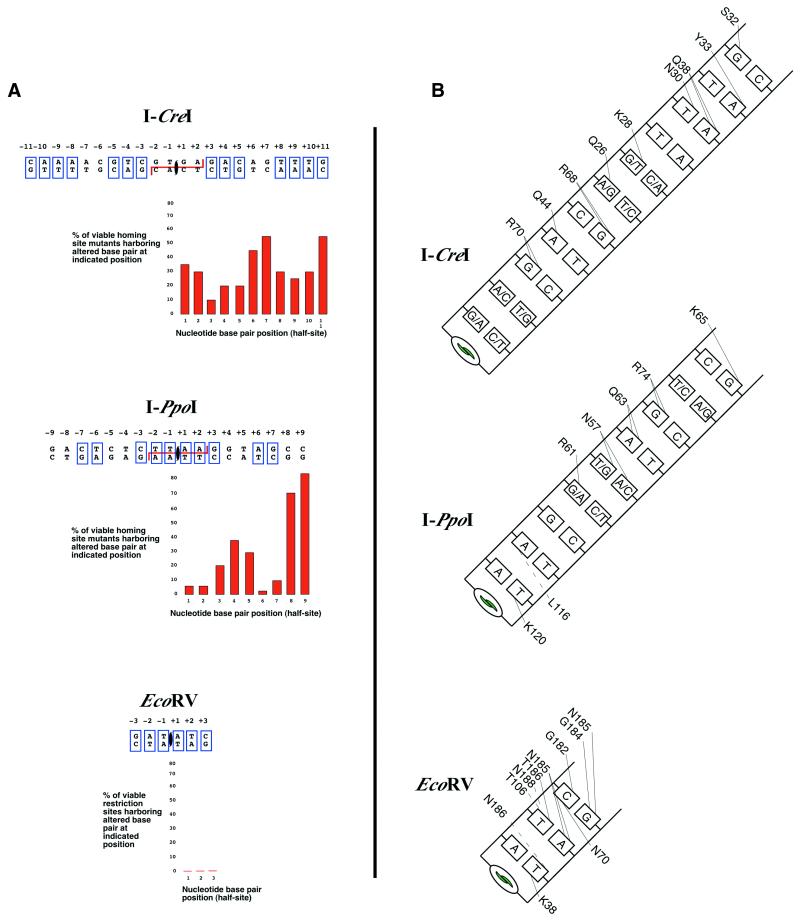
Figure 4.
DNA-binding by homing and restriction endonucleases. (A) Summary of actively cleaved target sites containing single base changes. I-CreI and I-PpoI recognize degenerate palindromes; EcoRV recognizes a strict palindrome. Palindromic bases are boxed. The homing endonucleases I-CreI and I-PpoI are tolerant of minor changes in their homing site as shown. The restriction endonuclease EcoRV is intolerant of any changes to its restriction site. Generally, the level of tolerance correlates inversely to number of specific protein–DNA contacts. Mutant homing site data taken from Argast et al. (114) and restriction site data from Pingoud and Jeltsch (112). (B) Base-specific contacts made by each endonuclease. Major groove contacts are drawn to the top of the base; minor groove contacts are drawn to the bottom of the base. Hydrogen bonds are represented as solid lines; other interactions are dashed lines.
The structures of the LAGLIDADG enzyme I-CreI and the His-Cys box enzyme I-PpoI bound to their DNA target sites provide much of our current understanding of the relationship between homing endonucleases and their homing sites (49,88). Specific recognition by both of these enzymes is provided by β-sheets that fold to complement the size and curvature of the DNA major groove. The spacing between alternating side chains extended from one side of a β-strand is nearly equal to that between every other DNA base pair. When two strands are staggered in a β-sheet, they can make contacts to consecutive base pairs within the major groove (113).
In I-CreI, a sheet and loop combination contacts nine consecutive base pairs per DNA half-site; in I-PpoI, a sheet contacts 5 bp in each half-site. Sequence specificity for DNA-binding is mediated by the unique pattern of hydrogen bond donors and acceptors presented by base pairs in the major groove. Each base pair type in a specific orientation presents a different combination of three or four hydrogen bond donors and acceptors. Protein contacts with two of these are usually sufficient to specify the preferred base pair at a position of the protein–DNA interface. A single contact is less specific, and often two different base pair identities can satisfy the hydrogen bond contact. This principle is reflected in the base contacts and specificities exhibited by both I-CreI and I-PpoI. Neither protein makes saturating contacts with all possible hydrogen bond donors and acceptors presented by the homing sequence throughout the DNA major groove. I-CreI makes two or more contacts per base pair at three positions in a recognition half-site. These positions are conserved between the two half-sites of the pseudo-palindromic DNA target and have been shown to be least tolerant to mutation (114). In addition to these contacts, I-CreI also makes single contacts to base pairs at five other positions within the half-site, some of which permit two different bases. For example, Gln26 acts as a hydrogen bond donor to the N7 of adenine in the A-T base pair at position +6. In the opposite half-site, the same residue in the other half of the dimer, Gln26′, donates a proton to the N7 of guanine in the G-C base pair at position –6. Neither a T-A nor a C-G base pair at that position would present a N7 hydrogen bond acceptor. In randomization studies of the recognition site these two identities (T-A or C-G) at this position are not recovered (114). Similar results are seen at the other positions with a single protein contact. The same is true for I-PpoI: base pairs that make two or more contacts to the protein are highly conserved, whereas positions with a single protein contact are found with two of the four possible base pair identities. This is illustrated in the structure at positions that either hold or break the palindrome of the recognition sequence, and by homing site mutation studies (114). One caveat in this analysis is the central 4 bp in I-PpoI. Despite no direct side chain contacts to these bases they are highly conserved. One likely explanation is the requirement of these bases for the severe bend of the DNA in this active site region as described above, and this might illustrate the functional importance of sequence-dependent DNA conformation and its role in sequence specificity.
Recently, Lucas et al. (111) identified three new LAGLIDADG homing endonucleases that reside within introns homologous to the one occupied by I-CreI. All are homodimers that share the same intron insertion site and cleave the same DNA substrates. Interestingly, many of the base-specific contacts described above are not conserved in these other proteins. For example, only four of the nine amino acids (per subunit) that I-CreI uses to make base-specific contacts are conserved in I-MsoI. A structure of any of these I-CreI homologs promises to suggest much about the flexibility of a protein–DNA interface of high specificity.
The structures of I-DmoI, PI-PfuI and PI-SceI (all determined in the absence of bound DNA) imply similar use of β-sheets for specific, yet tolerant, DNA recognition. In addition, the extra domains of the intein-encoded homing endonucleases appear to aid in DNA recognition. Footprinting and DNA cross-linking experiments of PI-SceI reveal the DNA recognition region to be required for complete site-specific DNA-binding (51–54,115,116). Substrate docking models also suggest it likely that the unique stirrup domain of PI-PfuI contacts part of its homing site to provide a larger DNA–protein interface than the endonuclease domain alone (48). Protein–substrate co-crystal structures of either PI-SceI or PI-PfuI will provide greater insight into the precise functional role of these additional domains.
The GIY-YIG enzyme I-TevI is distinct from other homing endonucleases in both its extreme tolerance to base pair changes within its homing site and its composition of many small and relatively non-specific DNA-binding subdomains (61,72,76,77). It lacks multiple hydrogen bond contacts directly to the bases of its target site and instead appears to derive high specificity from numerous minor contacts over a long 37 bp protein–DNA interface. The relationship between inherent or induced conformation of the DNA and recognition by I-TevI, or other homing endonucleases, has yet to be determined.
The structures of many type II restriction endonucleases bound to DNA emphasize structural and functional differences between these enzymes and homing endonucleases. Not only do the larger restriction endonucleases bind a much shorter DNA sequence, but they are also intolerant of variation in their binding site. Unlike most homing endonucleases that make limited hydrogen bond contacts within the major groove, restriction endonucleases saturate nearly all possible hydrogen bonds in both the major and minor grooves of their DNA target site. For example, EcoRV makes 22 hydrogen bond contacts in the major and minor grooves over its 6 bp restriction site (Fig. 4) (117,118), or ∼3.5 contacts per base pair. In contrast, I-CreI makes 24 specific contacts to a 24 bp homing site, or one contact per base pair (49). The high number of sequence-specific contacts ensures precise recognition of a DNA site; restriction sequence variants are cleaved several orders of magnitude less efficiently.
The differences between homing and restriction endonucleases correlate with their distinct functional roles. Both groups require sequence-specific recognition, though they differ in their level of specificity and their tolerance to changes in their target site. Homing endonucleases have evolved to recognize and cleave one site in a genome, and to be able to recognize this DNA target despite small evolutionary changes, or mutations, in the homing site. As such, they must be highly specific but capable of recognizing a slightly deviant homing site. Conversely, restriction endonucleases have evolved to identify and eliminate short foreign DNA sequences invading their host cells. They need to be less specific (by recognizing shorter restriction sites that are usually <6 bp) to increase their chances of finding a useful target site in the foreign sequences. At the same time, however, they must be inflexible in their target site recognition to prevent random cleavage of their own host genome. The precision with which restriction endonucleases recognize their restriction site is further underscored by the cell’s ability to protect its own DNA from restriction endonuclease-based cleavage via the addition of one or more methyl groups to the restriction sequence as part of the restriction–modification system.
HOMING ENDONUCLEASE CLEAVAGE MECHANISMS: STRUCTURAL INSIGHTS
Homing endonucleases, like restriction endonucleases, break the covalent bond between the phosphate and the oxygen at the 3′ position of the deoxyribose sugar in what appears to be a classic SN2 reaction (Fig. 5A). Cleavage of this bond requires a general base to activate and position a nucleophile—usually a water molecule—for inline attack of the electrophilic 5′ phosphate. A Lewis acid is necessary to provide positive charge and stabilize the pentacoordinate intermediate, as is a general acid to donate a proton to the 3′ oxygen leaving group. Most nuclease catalysts employ one or two divalent metal ions for the reaction. These metal ions are usually thought to alter the pKa of a bound water molecule for easy deprotonation and to position it for nucleophilic attack. The positive charge of metal ions can also stabilize the increased negative charge of the phosphoanion transition state and/or the leaving group.
Figure 5.
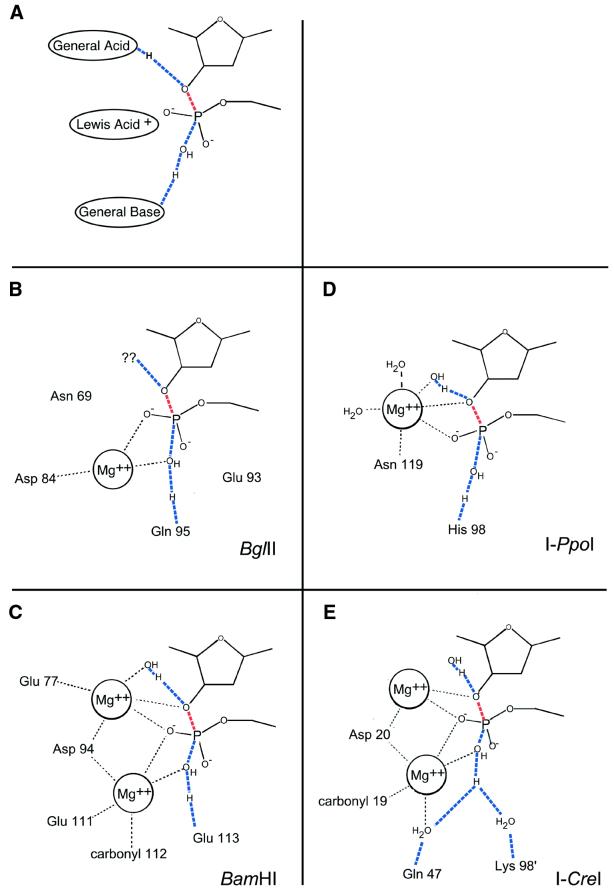
Endonuclease cleavage transition states. In all figures, the blue lines represent bonds in transition. Red lines represent the breaking bond of the DNA backbone. (A) Phosphodiester bond cleavage requires three chemical entities: a general base to activate the nucleophile, a Lewis acid to stabilize the pentacoordinate phosphoanion transition state and a general acid to protonate the 3′ leaving group. Water molecules usually function as nucleophiles and often as general acids. Divalent metal cations function as Lewis acids, can help lower the pKa of associated waters and position the nucleophile. Other residues act as general bases and/or to position waters and metal ions. (B) BglII uses a single metal in its mechanism. The magnesium functions to lower pKa of nucleophile and stabilize the transition state. The general base appears to be Gln95; the general acid has not been conclusively identified. (C) In BamHI, two magnesium ions aid in catalysis by positioning and lowering the pKa of the nucleophile and proton donor and stabilizing the phosphoanion transition state. Glu113 is the general base and water is the general acid. (D) I-PpoI, like BglII, uses a single-metal mechanism. In I-PpoI, the metal stabilizes the transition state and activates the general acid, an associated water molecule. It does not appear to participate in positioning and/or activating the nucleophile. The general base is His98. (E) I-CreI uses a two-metal mechanism, the central metal being shared by two separate active sites. The metals position and lower the pKa of the nucleophile, stabilize the transition state and stabilize the 3′ oxygen leaving group. The general base and acid appear to be within a well-ordered solvent network (positioned by peripheral residues and the first metal ion); for clarity, only three of these water molecules—other than the nucleophile—are drawn.
Studies of the cleavage mechanisms used by members of the type II restriction endonucleases provide insight into the chemistry of nuclease active sites. These enzymes reveal nuclease active sites to be divergent; however, they consistently emphasize the chemical requirements listed above. For example, BamHI uses a two metal mechanism (Fig. 5C) (119). A glutamate functions as the general base that activates the nucleophilic water, which the first metal positions for attack. The second metal positions and activates water that, as a general acid, donates a proton to the 3′ leaving group. Both metals act as Lewis acids and stabilize the phosphoanion transition state.
Although its mechanism is less well understood, BglII only uses one metal in its active site (Fig. 5B) (120). A glutamine appears to be in position to be a general base and activate the nucleophilic water. This water is coordinated by the metal ion, which also stabilizes the transition state. The identity of the general acid is unclear.
Like restriction endonucleases, homing endonucleases have widely divergent active sites and utilize different catalytic mechanisms. Despite identical 3′ 4 bp cleavage chemistry, for example, I-CreI and I-PpoI have evolved completely different catalytic mechanisms. Indeed, even the means by which they place the scissile phosphates into their active sites differs. While the LAGLIDADG proteins have tightly packed adjacent active sites that fit inside the minor groove of nearly B-form DNA (49), I-PpoI severely distorts its DNA substrate to widen the minor groove a full 20 Å to fit the scissile phosphates into its active sites (88). Furthermore, these differences in scissile phosphate placement, and the relative positions of the active sites in general, are integral to the active site architectures and catalytic mechanisms.
DNA cleavage by the LAGLIDADG endonucleases
Superposition of the active sites in the four known LAGLIDADG structures, I-CreI, I-DmoI, PI-SceI and PI-PfuI reveals surprisingly high divergence in both the identity and position of active site residues (Fig. 6). These differences, along with a low-resolution I-CreI–DNA structure (49) that prevented all active site components from being visualized has impeded our understanding of the LAGLIDADG catalytic mechanism. Two recently published structures, however, offer high-resolution images of I-CreI bound to both an uncleaved and a cleaved DNA substrate and reveal a mechanism with many novel properties (Fig. 5E) (50).
Figure 6.
Structural alignment of LAGLIDADG active sites. View is top-down through protein body to active site residues and DNA below. In all three diagrams, subunits of the homodimer I-CreI are blue and purple and are labeled as I-CreI and I-CreI′. The DNA backbone (gray) and the three divalent ions (black) from I-CreI–DNA co-crystal structures are shown for reference. Note the enormous divergence in both amino acid identity and position at the periphery of the active sites. (A) I-DmoI over I-CreI. (B) PI-PfuI over I-CreI. (C) PI-SceI over I-CreI. Alignments generated by superposition of LAGLIDADG α-helices.
Most surprisingly, these structures of I-CreI reveal that this enzyme uses three metal ions within its two closely positioned active sites (Figs 6 and 7). The two outer metals are used by each active site individually; the third is located precisely in the middle at the protein subunit interface. Essentially, this enzyme employs a two-metal mechanism analogous to BamHI described above, except that the central metal is the ‘second’ for each active site and is shared between the two.
Figure 7.
Diagram of the I-CreI active site (A) in the presence of calcium (non-cleaved DNA substrate) and (B) in the presence of magnesium (cleaved DNA substrate). DNA is red, metal ions are green, protein side chains are black. Waters are light blue; distances between waters are shown in Å by the associated blue lines representing the hydrogen-bonding network. The nucleophilic water is purple. Note that the protein makes no direct contact to the nucleophile, the scissile phosphate (red outlined in black), or the 3′ leaving group and that the solvent network extends from the nucleophile to the leaving group.
Furthermore, I-CreI makes no direct protein contact to any of the components of the chemical reaction including the nucleophilic water, the scissile phosphate and the 3′ leaving group (Fig. 7). Instead, these are all contacted by metal ions and a large number of well-ordered water molecules. These solvent molecules, which appear to be positioned by several side chain residues, form a hydrogen-bonding network that extends around the reaction center from the nucleophile to the leaving group.
The nucleophile is positioned by the outer metal, and is probably activated for attack by one of two peripheral waters that act as the general base (Fig. 7A). The phosphoanion transition state is thought to be stabilized by the two metal ions that making direct contact to the scissile phosphate, and the 3′ oxygen leaving group by the central metal. A nearby water molecule is a likely proton donor to this leaving group.
Many lines of evidence suggest this mechanism is conserved across the entire LAGLIDADG family. Homologs to the I-CreI metal-binding residue, D20, are essential for activity in all enzymes tested, including PI-SceI (121), PI-PfuI (55), I-DmoI (122) and I-CeuI (123). Indeed, this is the only residue that is strictly conserved in all known LAGLIDADG active sites. In the monomer PI-SceI, changing each of two aspartates to cysteines, which preferentially binds manganese ions, allows for cleavage only in the presence of this metal and not magnesium. These results confirm D218 and D326, which are D20 and D20′ in the I-CreI homodimer, to be the primary metal-binding residues of PI-SceI (124). Furthermore, biochemical experiments show that the monomers PI-SceI and PI-PfuI, like I-CreI, each contain two distinct active sites (48,125). Metal-mapping experiments in PI-SceI (125) and I-DmoI (122) reveal metals to be in similar positions as in I-CreI. Finally, the only other common feature noticeable in the LAGLIDADG superposition is what they lack: residues that extend into and fill the pocket occupied by the solvent network in I-CreI. Thus, the pocket containing the solvent network in each active site of the I-CreI structures is conserved across the LAGLIDADG family.
Based on the wide divergence in both position and identity of residues in the periphery of the LAGLIDADG active sites (i.e. the ones lining the pockets in I-CreI), one might predict them to be of limited importance for cleavage activity. Experimental results demonstrate the opposite to be true. Changes to Q47, R51 and K98 abolish activity in I-CreI (126), while mutations to D229, K301, H343, T341 and K403 all reduce or eliminate activity in PI-SceI (52,124,125,127). In I-CreI, these peripheral residues coordinate waters of a solvent network that, in turn, bonds to the nucleophile. These results suggest that not only is a pocket required for solvent, but that these water molecules must be positioned in such a way as to promote activation of the nucleophile. The divergence in LAGLIDADG active sites further suggests there to be many possible architectures by which the waters can be ordered into an active assembly. Any changes to the peripheral, water-coordinating, residues in a LAGLIDADG active site, however, would likely disrupt the actively arranged network of water molecules and reduce or inhibit the cleavage reaction.
DNA cleavage by the His-Cys box endonucleases
Like the LAGLIDADG enzymes, multiple high-resolution structures of I-PpoI complexed with both uncleaved and cleaved DNA substrate provide a detailed understanding of a novel, single-metal catalytic mechanism. I-PpoI has two separate active sites, each of which contains the strictly conserved H98 and N119 residues, a single magnesium ion, the scissile phosphate and three to four distinct water molecules (depending on substrate status). Experiments that trapped enzyme–substrate (ES) intermediates just prior to bond cleavage, in addition to the previously published enzyme–product complex, reveal the precise role of each member of the active site.
Three different uncleaved ES complexes have been visualized using crystallography: one with a non-bridging oxygen on the scissile phosphate replaced by a sulfur atom (in order to inhibit metal binding), a second in which the divalent magnesium is replaced by a monovalent sodium and a third in which H98 is mutated to an alanine (88–90). N119 positions the magnesium ion in the active site. This metal is required to function as a Lewis acid for stabilization of the pentacoordinate transition state and to induce a bonded water molecule to donate a proton to the 3′ leaving group. H98 is the general base that activates the attacking water molecule.
These specific roles are revealed by the various I-PpoI–DNA structures. In the H98A mutant structure, the water molecule is visible, but unable to attack the phosphate backbone that remains uncleaved. When magnesium is substituted by sodium, the charge density of the bound metal ion is reduced, leading to inadequate stabilization of the phosphoanion transition state. Furthermore, the 6-fold coordination of the metal is strained in all three ES complexes. This is relieved in the product complex, and implies the metal has an additional role in destabilizing the ES complex prior to cleavage. After cleavage, a fourth water molecule moves in to complete the octahedral coordination of the metal. Independent biochemical studies corroborate many of these mechanistic conclusions (128).
Substrate bending and cleavage are closely related for I-PpoI (90). The enzyme displays a 5° rigid-body rotation of individual subunits upon binding its homing site. The homing site is not bent in the absence of protein, but severely bent in the presence of I-PpoI. Together, these observations suggest an induced fit model of binding and catalysis. Mutational analyses demonstrate L116, which faces the DNA at the most severe bend point, to be required for both binding and catalysis. In the structure of the L116A mutant–DNA complex, the protein subunits are not rotated and the uncleaved DNA not as severely bent. Therefore, L116 is important for forming a well-ordered protein–DNA complex at the cleavage site and appears to stabilize a maximally bent DNA by desolvating the partially unstacked nucleotide bases. As described above, the other His-Cys box enzymes probably need not tweak their DNA substrate as much, and lack obvious homologs to L116.
It is significant to note that I-PpoI can be activated by numerous divalent cations, including calcium (83). Most other endonucleases, including restriction endonucleases and the LAGLIDADG enzymes, are limited in their use of metals. Promiscuous use of metal ions by an endonuclease might indicate that the bound cation is predominantly for charge stabilization of the transition state. In contrast, greater sensitivity of cleavage rates with respect to metal species might imply a metal ion is also used to precisely position and activate a nucleophile.
Structural studies of the SN reveal an identical active site architecture to I-PpoI (129,130), and this enzyme is hypothesized to share the same catalytic mechanism. The HNH family member colicin E9 also has a similar active site, except the conserved asparagine is replaced by a histidine (92). This difference is thought to govern the identity of the metal ion, which is a nickel in the E9 structure. It is possible that E9 also uses a mechanism similar to I-PpoI, and this may be representative of the entire HNH family. Therefore, while novel, I-PpoI’s use of a histidine as a general base and the metal as a Lewis acid and specific acid activator, is not unique and may be definitive of a larger family of endonuclease active sites.
ENGINEERING ENDONUCLEASES: GENERATION OF NOVEL ENZYMES WITH HIGH SPECIFICITY
Ever since researchers began to address challenges involving very large pieces of DNA, such as genomic mapping and gene therapy applications, there has been considerable interest in the identification of new enzymes with high specificity. Although class II restriction enzymes are common reagents in most laboratories and have revolutionized molecular biology, they evolved to recognize and eliminate relatively short DNA sequences invading their host cell. As such, their natural restriction sites are usually ≤6 bp, which corresponds to roughly one cleavage site in every 4000 bp of a random DNA target. Homing endonucleases offer an alternative reagent with higher specificity, though the few commercially available, such as I-CreI, PI-SceI and I-PpoI, have limited target site repertoires in comparison to the large variety of restriction endonucleases offered. The demand for novel rare cutting enzymes has led to great interest in the possibility of engineering enzymes capable of recognizing and cleaving novel target sites. Current research toward this goal is focused on the engineering of at least four different protein substrates: restriction enzymes, group I homing endonucleases, recognition/cleavage domain fusions and group II homing endonucleases.
Many engineering attempts have centered on modifying restriction endonucleases. Unfortunately, the precision of these enzymes, provided by many redundant contacts to the bases in their restriction sites, has made this venture extremely challenging. Despite a wealth of structural information on, for example, EcoRV, well-designed experiments to engineer this enzyme to recognize novel sites have been only moderately successful. Some progress has been made, however, including the generation of an EcoRV mutant that prefer deoxyuridine over thymidine and another that prefers a methylphosphonate substitution in one position of the restriction sequence (131,132). Other EcoRV mutants have a nearly 100-fold preference for distinct base pairs flanking the 6 bp restriction site (133).
In contrast to restriction endonucleases, homing endonucleases do not present redundant contacts to bases in their target site. This implies that single amino acid changes at the DNA–protein interface should have a higher probability for successfully altering base preferences within a homing site. Furthermore, the structures of I-PpoI and I-CreI reveal most of the base-specific contacts to be mediated through alternating amino acids within a few β-strands. One significant characteristic of this β-sheet DNA-binding motif is the close proximity within the primary sequence of many amino acids responsible for specificity of DNA recognition. This presents the opportunity of replacing one or two short stretches of DNA within the ORF with randomized sequences, and thereby generating combinatorial libraries in which all or most of the amino acids making specific contacts to the bases are varied. The wide variety of homing endonucleases found in nature, as exemplified by the large LAGLIDADG family and their unique homing sites, further suggests these enzymes to be malleable and should offer a strong foundation for engineering novel DNA-binding proteins. Although these enzymes appear to be promising substrates for engineering, few results have yet been published.
An alternative strategy with marked success is the joining of non-specific endonuclease domains, such as the cleavage domain of FokI, to DNA-binding domains via a flexible linker. This strategy has successfully created novel endonucleases, including a FokI fusion to the Gal4 transcription factor, capable of binding and cleaving the Gal4 recognition site (134). Similarly, multiple FokI–zinc finger fusions have also generated novel enzymes (135,136). This strategy has considerable promise due to the engineering flexibility offered by zinc fingers (reviewed in 137). Series of linked modular zinc fingers have been generated to target a wide variety of specific target sequences with high affinity (138–142), and engineered zinc fingers influence expression of target genes in vivo (143).
A final strategy takes advantage of the RNP complex of group II homing endonucleases (20). As recognition occurs through base pairing interactions between the DNA target site and the RNA of the RNP, modification of the RNA sequence directly leads to novel substrate-binding. Although the RNP complex is likely to be too unstable for common in vitro uses as laboratory reagents, initial results in which redesigned group II introns/homing endonucleases inserted into targeted sites (HIV-1 proviral DNA and the human CCR5 gene) within human cells suggests this strategy might offer great promise for genetic engineering, functional genomics and gene therapy applications (144).
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Marlene Belfort, Victoria Derbyshire, Patrick Van Roey and Morten Elde for sharing data and results prior to publication. We also thank Eric Galburt and Ray Monnat for their critical reading of this review prior to submission. B.L.S. is funded for this work by the NIH (GM49857); B.S.C. is funded through a NIH Interdisciplinary Training Grant (5T32CA80416).
References
- 1.Dujon B., Belfort,M., Butow,R.A., Jacq,C., Lemieux,C., Perlman,P.S. and Vogt,V.M. (1989) Mobile introns: definition of terms and recommended nomenclature. Gene, 82, 115–118. [DOI] [PubMed] [Google Scholar]
- 2.Coen D., Deutsch,J., Netter,P., Petrochilo,E. and Slonimski,P.P. (1970) Mitochondrial genetics. I. Methodology and phenomenology. Symp. Soc. Exp. Biol., 23, 449–496. [PubMed] [Google Scholar]
- 3.Dujon B. (1980) Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell, 20, 185–197. [DOI] [PubMed] [Google Scholar]
- 4.Bos J.L., Heyting,C., Borst,P., Arnberg,A.C. and Van Bruggen,E.F. (1978) An insert in the single gene for the large ribosomal RNA in yeast mitochondrial DNA. Nature, 275, 336–338. [DOI] [PubMed] [Google Scholar]
- 5.Colleaux L., d’Auriol,L., Betermier,M., Cottarel,G., Jacquier,A., Galibert,F. and Dujon,B. (1986) Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E.coli as a specific double strand endonuclease. Cell, 44, 521–533. [DOI] [PubMed] [Google Scholar]
- 6.Macreadie I.G., Scott,R.M., Zinn,A.R. and Butow,R.A. (1985) Transposition of an intron in yeast mitochondria requires a protein encoded by that intron. Cell, 41, 395–402. [DOI] [PubMed] [Google Scholar]
- 7.Jacquier A. and Dujon,B. (1985) An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell, 41, 383–394. [DOI] [PubMed] [Google Scholar]
- 8.Belfort M. and Roberts,R.J. (1997) Homing endonucleases: keeping the house in order. Nucleic Acids Res., 25, 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dujon B. (1989) Group I introns as mobile genetic elements: facts and mechanistic speculations. Gene, 82, 91–114. [DOI] [PubMed] [Google Scholar]
- 10.Belfort M. and Perlman,P.S. (1995) Mechanisms of intron mobility. J. Biol. Chem., 270, 30237–30240. [DOI] [PubMed] [Google Scholar]
- 11.Lambowitz A.M. and Belfort,M. (1993) Introns as mobile genetic elements. Annu. Rev. Biochem., 62, 587–622. [DOI] [PubMed] [Google Scholar]
- 12.Jurica M.S. and Stoddard,B.L. (1999) Homing endonucleases: structure, function and evolution. Cell. Mol. Life Sci., 55, 1304–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimble F.S. (2000) Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol. Lett., 185, 99–107. [DOI] [PubMed] [Google Scholar]
- 14.Cech T.R. (1990) Self-splicing of group I introns. Annu. Rev. Biochem., 59, 543–568. [DOI] [PubMed] [Google Scholar]
- 15.Saldanha R., Mohr,G., Belfort,M. and Lambowitz,A.M. (1993) Group I and group II introns. FASEB J., 7, 15–24. [DOI] [PubMed] [Google Scholar]
- 16.Lykke-Andersen J., Aagaard,C., Semionenkov,M. and Garrett,R.A. (1997) Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem. Sci., 22, 326–331. [DOI] [PubMed] [Google Scholar]
- 17.Perler F.B., Olsen,G.J. and Adam,E. (1997) Compilation and analysis of intein sequences. Nucleic Acids Res., 25, 1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiner A.M. (1993) mRNA splicing and autocatalytic introns: distant cousins or the products of chemical determinism? Cell, 72, 161–164. [DOI] [PubMed] [Google Scholar]
- 19.Sharp P.A. (1994) Split genes and RNA splicing. Cell, 77, 805–815. [DOI] [PubMed] [Google Scholar]
- 20.Mohr G., Smith,D., Belfort,M. and Lambowitz,A.M. (2000) Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev., 14, 559–573. [PMC free article] [PubMed] [Google Scholar]
- 21.Cousineau B., Lawrence,S., Smith,D. and Belfort,M. (2000) Retrotransposition of a bacterial group II intron. Nature, 404, 1018–1021. [DOI] [PubMed] [Google Scholar]
- 22.Cech T.R. (1988) Conserved sequences and structures of group I introns: building an active site for RNA catalysis. Gene, 73, 259–271. [DOI] [PubMed] [Google Scholar]
- 23.Michel F. and Westhof,E. (1990) Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol., 216, 585–610. [DOI] [PubMed] [Google Scholar]
- 24.Michel F., Umesono,K. and Ozeki,H. (1989) Comparative and functional anatomy of group II catalytic introns. Gene, 82, 5–30. [DOI] [PubMed] [Google Scholar]
- 25.Michel F. and Ferat,J.L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- 26.Eskes R., Yang,J., Lambowitz,A.M. and Perlman,P.S. (1997) Mobility of yeast mitochondrial group II introns: engineering a new site specificity and retrohoming via full reverse splicing. Cell, 88, 865–874. [DOI] [PubMed] [Google Scholar]
- 27.Yang J., Zimmerly,S., Perlman,P.S. and Lambowitz,A.M. (1996) Efficient integration of an intron RNA into double-stranded DNA by reverse splicing. Nature, 381, 332–335. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerly S., Guo,H., Perlman,P.S. and Lambowitz,A.M. (1995) Group II intron mobility occurs by target DNA-primed reverse transcription. Cell, 82, 545–554. [DOI] [PubMed] [Google Scholar]
- 29.Zimmerly S., Guo,H., Eskes,R., Yang,J., Perlman,P.S. and Lambowitz,A.M. (1995) A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell, 83, 529–538. [DOI] [PubMed] [Google Scholar]
- 30.Mills D.A., McKay,L.L. and Dunny,G.M. (1996) Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol ., 178, 3531–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shearman C., Godon,J.J. and Gasson,M. (1996) Splicing of a group II intron in a functional transfer gene of Lactococcus lactis. Mol. Microbiol., 21, 45–53. [DOI] [PubMed] [Google Scholar]
- 32.Matsuura M., Saldanha,R., Ma,H., Wank,H., Yang,J., Mohr,G., Cavanagh,S., Dunny,G.M., Belfort,M. and Lambowitz,A.M. (1997) A bacterial group II intron encoding reverse transcriptase, maturase and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev., 11, 2910–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cousineau B., Smith,D., Lawrence-Cavanagh,S., Mueller,J.E., Yang,J., Mills,D., Manias,D., Dunny,G., Lambowitz,A.M. and Belfort,M. (1998) Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell, 94, 451–462. [DOI] [PubMed] [Google Scholar]
- 34.Dalgaard J.Z., Klar,A.J., Moser,M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res., 25, 4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watabe H., Shibata,T. and Ando,T. (1981) Site-specific endo-deoxyribonucleases in eukaryotes: endonucleases of yeasts, Saccharomyces and Pichia. J. Biochem. (Tokyo), 90, 1623–1632. [DOI] [PubMed] [Google Scholar]
- 36.Watabe H., Iino,T., Kaneko,T., Shibata,T. and Ando,T. (1983) A new class of site-specific endodeoxyribonucleases. Endo.Sce I isolated from a eukaryote, Saccharomyces cerevisiae. J. Biol. Chem., 258, 4663–4665. [PubMed] [Google Scholar]
- 37.Kostriken R., Strathern,J.N., Klar,A.J., Hicks,J.B. and Heffron,F. (1983) A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell, 35, 167–174. [DOI] [PubMed] [Google Scholar]
- 38.Schafer B., Wilde,B., Massardo,D.R., Manna,F., Del Giudice,L. and Wolf,K. (1994) A mitochondrial group-I intron in fission yeast encodes a maturase and is mobile in crosses. Curr. Genet., 25, 336–341. [DOI] [PubMed] [Google Scholar]
- 39.Monteilhet C., Dziadkowiec,D., Szczepanek,T. and Lazowska,J. (2000) Purification and characterization of the DNA cleavage and recognition site of I-ScaI mitochondrial group I intron encoded endonuclease produced in Escherichia coli. Nucleic Acids Res., 28, 1245–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho Y., Kim,S.J. and Waring,R.B. (1997) A protein encoded by a group I intron in Aspergillus nidulans directly assists RNA splicing and is a DNA endonuclease. Proc. Natl Acad. Sci. USA, 94, 8994–8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson A.J., Yuan,X., Kudlicki,W. and Herrin,D.L. (1992) Cleavage and recognition pattern of a double-strand-specific endonuclease (I-CreI) encoded by the chloroplast 23S rRNA intron of Chlamydomonas reinhardtii. Gene, 119, 247–251. [DOI] [PubMed] [Google Scholar]
- 42.Marshall P. and Lemieux,C. (1992) The I-CreI endonuclease recognizes a sequence of 19 base pairs and preferentially cleaves the coding strand of the Chlamydomonas moewusii chloroplast large subunit rRNA gene. Nucleic Acids Res., 20, 6401–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dalgaard J.Z., Garrett,R.A. and Belfort,M. (1993) A site-specific endonuclease encoded by a typical archaeal intron. Proc. Natl Acad. Sci. USA, 90, 5414–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gimble F.S. and Thorner,J. (1992) Homing of a DNA endonuclease gene by meiotic gene conversion in Saccharomyces cerevisiae. Nature, 357, 301–306. [DOI] [PubMed] [Google Scholar]
- 45.Heath P.J., Stephens,K.M., Monnat,R.J.,Jr and Stoddard,B.L. (1997) The structure of I-Crel, a group I intron-encoded homing endonuclease. Nature Struct. Biol., 4, 468–476. [DOI] [PubMed] [Google Scholar]
- 46.Silva G.H., Dalgaard,J.Z., Belfort,M. and Van Roey,P. (1999) Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI. J. Mol. Biol., 286, 1123–1136. [DOI] [PubMed] [Google Scholar]
- 47.Duan X., Gimble,F.S. and Quiocho,F.A. (1997) Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity. Cell, 89, 555–564. [DOI] [PubMed] [Google Scholar]
- 48.Ichiyanagi K., Ishino,Y., Ariyoshi,M., Komori,K. and Morikawa,K. (2000) Crystal structure of an archaeal intein-encoded homing endonuclease PI-PfuI. J. Mol. Biol., 300, 889–901. [DOI] [PubMed] [Google Scholar]
- 49.Jurica M. S., Monnat,R.J.,Jr and Stoddard,B.L. (1998) DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol. Cell ., 2, 469–476. [DOI] [PubMed] [Google Scholar]
- 50.Chevalier B.S., Monnat,R.J.,Jr and Stoddard,B.L. (2001) The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nature Struct. Biol., 8, 312–316. [DOI] [PubMed] [Google Scholar]
- 51.Grindl W., Wende,W., Pingoud,V. and Pingoud,A. (1998) The protein splicing domain of the homing endonuclease PI-SceI is responsible for specific DNA binding. Nucleic Acids Res., 26, 1857–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He Z., Crist,M., Yen,H., Duan,X., Quiocho,F.A. and Gimble,F.S. (1998) Amino acid residues in both the protein splicing and endonuclease domains of the PI-SceI intein mediate DNA binding. J. Biol. Chem., 273, 4607–4615. [DOI] [PubMed] [Google Scholar]
- 53.Gimble F.S. and Wang,J. (1996) Substrate recognition and induced DNA distortion by the PI-SceI endonuclease, an enzyme generated by protein splicing. J. Mol. Biol., 263, 163–180. [DOI] [PubMed] [Google Scholar]
- 54.Wende W., Grindl,W., Christ,F., Pingoud,A. and Pingoud,V. (1996) Binding, bending and cleavage of DNA substrates by the homing endonuclease Pl-SceI. Nucleic Acids Res., 24, 4123–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komori K., Ichiyanagi,K., Morikawa,K. and Ishino,Y. (1999) PI-PfuI and PI-PfuII, intein-coded homing endonucleases from Pyrococcus furiosus. II. Characterization of the binding and cleavage abilities by site-directed mutagenesis. Nucleic Acids Res., 27, 4175–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Ommen G.J., Boer,P.H., Groot,G.S., De Haan,M., Roosendaal,E., Grivell,L.A., Haid,A. and Schweyen,R.J. (1980) Mutations affecting RNA splicing and the interaction of gene expression of the yeast mitochondrial loci cob and oxi-3. Cell, 20, 173–183. [DOI] [PubMed] [Google Scholar]
- 57.Lazowska J., Jacq,C. and Slonimski,P.P. (1980) Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell, 22, 333–348. [DOI] [PubMed] [Google Scholar]
- 58.Ho Y. and Waring,R.B. (1999) The maturase encoded by a group I intron from Aspergillus nidulans stabilizes RNA tertiary structure and promotes rapid splicing. J. Mol. Biol., 292, 987–1001. [DOI] [PubMed] [Google Scholar]
- 59.Szczepanek T. and Lazowska,J. (1996) Replacement of two non-adjacent amino acids in the S.cerevisiae bi2 intron-encoded RNA maturase is sufficient to gain a homing-endonuclease activity. EMBO J., 15, 3758–3767. [PMC free article] [PubMed] [Google Scholar]
- 60.Dujardin G., Jacq,C. and Slonimski,P.P. (1982) Single base substitution in an intron of oxidase gene compensates splicing defects of the cytochrome b gene. Nature, 298, 628–632. [DOI] [PubMed] [Google Scholar]
- 61.Kowalski J.C., Belfort,M., Stapleton,M.A., Holpert,M., Dansereau,J.T., Pietrokovski,S., Baxter,S.M. and Derbyshire,V. (1999) Configuration of the catalytic GIY-YIG domain of intron endonuclease I-TevI: coincidence of computational and molecular findings. Nucleic Acids Res., 27, 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharma M., Ellis,R.L. and Hinton,D.M. (1992) Identification of a family of bacteriophage T4 genes encoding proteins similar to those present in group I introns of fungi and phage. Proc. Natl Acad. Sci. USA, 89, 6658–6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bell-Pedersen D., Quirk,S., Clyman,J. and Belfort,M. (1990) Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: mechanistic and evolutionary implications. Nucleic Acids Res., 18, 3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian G.L., Michel,F., Macadre,C., Slonimski,P.P. and Lazowska,J. (1991) Incipient mitochondrial evolution in yeasts. II. The complete sequence of the gene coding for cytochrome b in Saccharomyces douglasii reveals the presence of both new and conserved introns and discloses major differences in the fixation of mutations in evolution. J. Mol. Biol., 218, 747–760. [DOI] [PubMed] [Google Scholar]
- 65.Paquin B., Laforest,M.J. and Lang,B.F. (1994) Interspecific transfer of mitochondrial genes in fungi and creation of a homologous hybrid gene. Proc. Natl Acad. Sci. USA, 91, 11807–11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saguez C., Lecellier,G. and Koll,F. (2000) Intronic GIY-YIG endonuclease gene in the mitochondrial genome of Podospora curvicolla: evidence for mobility. Nucleic Acids Res., 28, 1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kroymann J. and Zetsche,K. (1997) The apocytochrome-b gene in Chlorogonium elongatum (Chlamydomonadaceae): an intronic GIY-YIG ORF in green algal mitochondria. Curr. Genet., 31, 414–418. [DOI] [PubMed] [Google Scholar]
- 68.Denovan-Wright E.M., Nedelcu,A.M. and Lee,R.W. (1998) Complete sequence of the mitochondrial DNA of Chlamydomonas eugametos. Plant Mol. Biol., 36, 285–295. [DOI] [PubMed] [Google Scholar]
- 69.Paquin B., O’Kelly,C.J. and Lang,B.F. (1995) Intron-encoded open reading frame of the GIY-YIG subclass in a plastid gene. Curr. Genet., 28, 97–99. [DOI] [PubMed] [Google Scholar]
- 70.Holloway S.P., Deshpande,N.N. and Herrin,D.L. (1999) The catalytic group-I introns of the psbA gene of Chlamydomonas reinhardtii: core structures, ORFs and evolutionary implications. Curr. Genet., 36, 69–78. [DOI] [PubMed] [Google Scholar]
- 71.Mueller J.E., Smith,D., Bryk,M. and Belfort,M. (1995) Intron-encoded endonuclease I-TevI binds as a monomer to effect sequential cleavage via conformational changes in the td homing site. EMBO J., 14, 5724–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bryk M., Belisle,M., Mueller,J.E. and Belfort,M. (1995) Selection of a remote cleavage site by I-TevI, the td intron-encoded endonuclease. J. Mol. Biol., 247, 197–210. [DOI] [PubMed] [Google Scholar]
- 73.Eddy S.R. and Gold,L. (1991) The phage T4 nrdB intron: a deletion mutant of a version found in the wild. Genes Dev., 5, 1032–1041. [DOI] [PubMed] [Google Scholar]
- 74.Loizos N., Silva,G.H. and Belfort,M. (1996) Intron-encoded endonuclease I-TevII binds across the minor groove and induces two distinct conformational changes in its DNA substrate. J. Mol. Biol., 255, 412–424. [DOI] [PubMed] [Google Scholar]
- 75.Bell-Pedersen D., Quirk,S.M., Bryk,M. and Belfort,M. (1991) I-TevI, the endonuclease encoded by the mobile td intron, recognizes binding and cleavage domains on its DNA target. Proc. Natl Acad. Sci. USA, 88, 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bryk M., Quirk,S.M., Mueller,J.E., Loizos,N., Lawrence,C. and Belfort,M. (1993) The td intron endonuclease I-TevI makes extensive sequence-tolerant contacts across the minor groove of its DNA target. EMBO J., 12, 2141–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Derbyshire V., Kowalski,J.C., Dansereau,J.T., Hauer,C.R. and Belfort,M. (1997) Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J. Mol. Biol., 265, 494–506. [DOI] [PubMed] [Google Scholar]
- 78.Wah D.A., Hirsch,J.A., Dorner,L.F., Schildkraut,I. and Aggarwal,A.K. (1997) Structure of the multimodular endonuclease FokI bound to DNA. Nature, 388, 97–100. [DOI] [PubMed] [Google Scholar]
- 79.Johansen S., Embley,T.M. and Willassen,N.P. (1993) A family of nuclear homing endonucleases. Nucleic Acids Res., 21, 4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muscarella D.E. and Vogt,V.M. (1989) A mobile group I intron in the nuclear rDNA of c. Cell, 56, 443–454. [DOI] [PubMed] [Google Scholar]
- 81.Muscarella D.E., Ellison,E.L., Ruoff,B.M. and Vogt,V.M. (1990) Characterization of I-Ppo, an intron-encoded endonuclease that mediates homing of a group I intron in the ribosomal DNA of Physarum polycephalum. Mol. Cell. Biol., 10, 3386–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ellison E.L. and Vogt,V.M. (1993) Interaction of the intron-encoded mobility endonuclease I-PpoI with its target site. Mol. Cell. Biol., 13, 7531–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wittmayer P.K. and Raines,R.T. (1996) Substrate binding and turnover by the highly specific I-PpoI endonuclease. Biochemistry, 35, 1076–1083. [DOI] [PubMed] [Google Scholar]
- 84.Wittmayer P.K., McKenzie,J.L. and Raines,R.T. (1998) Degenerate DNA recognition by I-PpoI endonuclease. Gene, 206, 11–21. [DOI] [PubMed] [Google Scholar]
- 85.Johansen S., Elde,M., Vader,A., Haugen,P., Haugli,K. and Haugli,F. (1997) In vivo mobility of a group I twintron in nuclear ribosomal DNA of the myxomycete Didymium iridis. Mol. Microbiol., 24, 737–745. [DOI] [PubMed] [Google Scholar]
- 86.Elde M., Haugen,P., Willassen,N.P. and Johansen,S. (1999) I-NjaI, a nuclear intron-encoded homing endonuclease from Naegleria, generates a pentanucleotide 3′ cleavage-overhang within a 19 base-pair partially symmetric DNA recognition site. Eur. J. Biochem., 259, 281–288. [DOI] [PubMed] [Google Scholar]
- 87.Elde M., Willassen,N.P. and Johansen, S. (2000) Functional characterization of isoschizomeric His-Cys box homing endonucleases from Naegleria. Eur. J. Biochem., 267, 7257–7266. [DOI] [PubMed] [Google Scholar]
- 88.Flick K.E., Jurica,M.S., Monnat,R.J.,Jr and Stoddard,B.L. (1998) DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature, 394, 96–101. [DOI] [PubMed] [Google Scholar]
- 89.Galburt E.A., Chevalier,B., Tang,W., Jurica,M.S., Flick,K.E., Monnat,R.J.,Jr and Stoddard,B.L. (1999) A novel endonuclease mechanism directly visualized for I-PpoI. Nature Struct. Biol., 6, 1096–1099. [DOI] [PubMed] [Google Scholar]
- 90.Galburt E.A., Chadsey,M.S., Jurica,M.S., Chevalier,B.S., Erho,D., Tang,W., Monnat,R.J.,Jr and Stoddard,B.L. (2000) Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site. J. Mol. Biol., 300, 877–887. [DOI] [PubMed] [Google Scholar]
- 91.Ko T.P., Liao,C.C., Ku,W.Y., Chak,K.F. and Yuan,H.S. (1999) The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Struct. Fold. Des., 7, 91–102. [DOI] [PubMed] [Google Scholar]
- 92.Kleanthous C., Kuhlmann,U.C., Pommer,A.J., Ferguson,N., Radford,S.E., Moore,G.R., James,R. and Hemmings,A.M. (1999) Structural and mechanistic basis of immunity toward endonuclease colicins. Nature Struct. Biol., 6, 243–252. [DOI] [PubMed] [Google Scholar]
- 93.Shub D.A., Goodrich-Blair,H. and Eddy,S.R. (1994) Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci., 19, 402–404. [DOI] [PubMed] [Google Scholar]
- 94.Gorbalenya A.E. (1994) Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci., 3, 1117–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goodrich-Blair H., Scarlato,V., Gott,J.M., Xu,M.Q. and Shub,D.A. (1990) A self-splicing group I intron in the DNA polymerase gene of Bacillus subtilis bacteriophage SPO1. Cell, 63, 417–424. [DOI] [PubMed] [Google Scholar]
- 96.Goodrich-Blair H. and Shub,D.A. (1994) The DNA polymerase genes of several HMU-bacteriophages have similar group I introns with highly divergent open reading frames. Nucleic Acids Res., 22, 3715–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goodrich-Blair H. and Shub,D.A. (1996) Beyond homing: competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell, 84, 211–221. [DOI] [PubMed] [Google Scholar]
- 98.Drouin M., Lucas,P., Otis,C., Lemieux,C. and Turmel,M. (2000) Biochemical characterization of I-CmoeI reveals that this H-N-H homing endonuclease shares functional similarities with H-N-H colicins. Nucleic Acids Res., 28, 4566–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gorbalenya A.E. (1998) Non-canonical inteins. Nucleic Acids Res., 26, 1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pietrokovski S. (1998) Modular organization of inteins and C-terminal autocatalytic domains. Protein Sci., 7, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Braun V., Pilsl,H. and Gross,P. (1994) Colicins: structures, modes of action, transfer through membranes and evolution. Arch. Microbiol., 161, 199–206. [DOI] [PubMed] [Google Scholar]
- 102.Kuhlmann U.C., Moore,G.R., James,R., Kleanthous,C. and Hemmings,A.M. (1999) Structural parsimony in endonuclease active sites: should the number of homing endonuclease families be redefined? FEBS Lett., 463, 1–2. [DOI] [PubMed] [Google Scholar]
- 103.Friedhoff P., Franke,I., Meiss,G., Wende,W., Krause,K.L. and Pingoud,A. (1999) A similar active site for non-specific and specific endonucleases. Nature Struct. Biol., 6, 112–113. [DOI] [PubMed] [Google Scholar]
- 104.Raaijmakers H., Vix,O., Toro,I., Golz,S., Kemper,B. and Suck,D. (1999) X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J., 18, 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goddard M.R. and Burt,A. (1999) Recurrent invasion and extinction of a selfish gene. Proc. Natl Acad. Sci. USA, 96, 13880–13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Raveh D., Hughes,S.H., Shafer,B.K. and Strathern,J.N. (1989) Analysis of the HO-cleaved MAT DNA intermediate generated during the mating type switch in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet., 220, 33–42. [PubMed] [Google Scholar]
- 107.Cho Y., Qiu,Y.L., Kuhlman,P. and Palmer,J.D. (1998) Explosive invasion of plant mitochondria by a group I intron. Proc. Natl Acad. Sci. USA, 95, 14244–14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gray M.W. (1998) Mass migration of a group I intron: promiscuity on a grand scale. Proc. Natl Acad. Sci. USA, 95, 14003–14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Turmel M., Cote,V., Otis,C., Mercier,J.P., Gray,M.W., Lonergan,K.M. and Lemieux,C. (1995) Evolutionary transfer of ORF-containing group I introns between different subcellular compartments (chloroplast and mitochondrion). Mol. Biol. Evol., 12, 533–545. [DOI] [PubMed] [Google Scholar]
- 110.Loizos N., Tillier,E.R. and Belfort,M. (1994) Evolution of mobile group I introns: recognition of intron sequences by an intron-encoded endonuclease. Proc. Natl Acad. Sci. USA, 91, 11983–11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lucas P., Otis,C., Mercier,J.P., Turmel,M. and Lemieux,C. (2001) Rapid evolution of the DNA-binding site in LAGLIDADG homing endonucleases. Nucleic Acids Res., 29, 960–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pingoud A. and Jeltsch,A. (1997) Recognition and cleavage of DNA by type-II restriction endonucleases. Eur. J. Biochem., 246, 1–22. [DOI] [PubMed] [Google Scholar]
- 113.Phillips S.E. (1994) The beta-ribbon DNA recognition motif. Annu. Rev. Biophys. Biomol. Struct., 23, 671–701. [DOI] [PubMed] [Google Scholar]
- 114.Argast G.M., Stephens,K.M., Emond,M.J. and Monnat,R.J.,Jr (1998) I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J. Mol. Biol., 280, 345–353. [DOI] [PubMed] [Google Scholar]
- 115.Hu D., Crist,M., Duan,X., Quiocho,F.A. and Gimble,F.S. (2000) Probing the structure of the PI-SceI–DNA complex by affinity cleavage and affinity photocross-linking. J. Biol. Chem., 275, 2705–2712. [DOI] [PubMed] [Google Scholar]
- 116.Christ F., Steuer,S., Thole,H., Wende,W., Pingoud,A. and Pingoud,V. (2000) A Model for the PI-SceI–DNA complex based on multiple base and phosphate backbone-specific photocross-links. J. Mol. Biol., 300, 841–849. [DOI] [PubMed] [Google Scholar]
- 117.Winkler F.K., Banner,D.W., Oefner,C., Tsernoglou,D., Brown,R.S., Heathman,S.P., Bryan,R.K., Martin,P.D., Petratos,K. and Wilson,K.S. (1993) The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J., 12, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kostrewa D. and Winkler,F.K. (1995) Mg2+ binding to the active site of EcoRV endonuclease: a crystallographic study of complexes with substrate and product DNA at 2 Å resolution. Biochemistry, 34, 683–696. [DOI] [PubMed] [Google Scholar]
- 119.Viadiu H. and Aggarwal,A.K. (1998) The role of metals in catalysis by the restriction endonuclease BamHI. Nature Struct. Biol., 5, 910–916. [DOI] [PubMed] [Google Scholar]
- 120.Lukacs C.M., Kucera,R., Schildkraut,I. and Aggarwal,A.K. (2000) Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution. Nature Struct. Biol., 7, 134–140. [DOI] [PubMed] [Google Scholar]
- 121.Gimble F.S. and Stephens,B.W. (1995) Substitutions in conserved dodecapeptide motifs that uncouple the DNA binding and DNA cleavage activities of PI-SceI endonuclease. J. Biol. Chem., 270, 5849–5856. [DOI] [PubMed] [Google Scholar]
- 122.Lykke-Andersen J., Garrett,R.A. and Kjems,J. (1997) Mapping metal ions at the catalytic centres of two intron-encoded endonucleases. EMBO J., 16, 3272–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Turmel M., Otis,C., Cote,V. and Lemieux,C. (1997) Evolutionarily conserved and functionally important residues in the I-CeuI homing endonuclease. Nucleic Acids Res., 25, 2610–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schottler S., Wende,W., Pingoud,V. and Pingoud,A. (2000) Identification of Asp218 and Asp326 as the principal Mg2+ binding ligands of the homing endonuclease PI-SceI. Biochemistry, 39, 15895–15900. [DOI] [PubMed] [Google Scholar]
- 125.Christ F., Schoettler,S., Wende,W., Steuer,S., Pingoud,A. and Pingoud,V. (1999) The monomeric homing endonuclease PI-SceI has two catalytic centres for cleavage of the two strands of its DNA substrate. EMBO J., 18, 6908–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Seligman L.M., Stephens,K.M., Savage,J.H. and Monnat,R.J.,Jr (1997) Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli. Genetics, 147, 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gimble F.S., Duan,X., Hu,D. and Quiocho,F.A. (1998) Identification of Lys-403 in the PI-SceI homing endonuclease as part of a symmetric catalytic center. J. Biol. Chem., 273, 30524–30529. [DOI] [PubMed] [Google Scholar]
- 128.Mannino S.J., Jenkins,C.L. and Raines,R.T. (1999) Chemical mechanism of DNA cleavage by the homing endonuclease I-PpoI. Biochemistry, 38, 16178–16186. [DOI] [PubMed] [Google Scholar]
- 129.Miller M.D., Cai,J. and Krause,K.L. (1999) The active site of Serratia endonuclease contains a conserved magnesium-water cluster. J. Mol. Biol., 288, 975–987. [DOI] [PubMed] [Google Scholar]
- 130.Friedhoff P., Franke,I., Krause,K.L. and Pingoud,A. (1999) Cleavage experiments with deoxythymidine 3′,5′-bis-(p-nitrophenyl phosphate) suggest that the homing endonuclease I-PpoI follows the same mechanism of phosphodiester bond hydrolysis as the non-specific Serratia nuclease. FEBS Lett., 443, 209–214. [DOI] [PubMed] [Google Scholar]
- 131.Lanio T., Selent,U., Wenz,C., Wende,W., Schulz,A., Adiraj,M., Katti,S.B. and Pingoud,A. (1996) EcoRV-T94V: a mutant restriction endonuclease with an altered substrate specificity towards modified oligodeoxynucleotides. Protein Eng., 9, 1005–1010. [DOI] [PubMed] [Google Scholar]
- 132.Wenz C., Selent,U., Wende,W., Jeltsch,A., Wolfes,H. and Pingoud,A. (1994) Protein engineering of the restriction endonuclease EcoRV: replacement of an amino acid residue in the DNA binding site leads to an altered selectivity towards unmodified and modified substrates. Biochim. Biophys. Acta, 1219, 73–80. [DOI] [PubMed] [Google Scholar]
- 133.Lanio T., Jeltsch,A. and Pingoud,A. (1998) Towards the design of rare cutting restriction endonucleases: using directed evolution to generate variants of EcoRV differing in their substrate specificity by two orders of magnitude. J. Mol. Biol., 283, 59–69. [DOI] [PubMed] [Google Scholar]
- 134.Kim Y.G., Smith,J., Durgesha,M. and Chandrasegaran,S. (1998) Chimeric restriction enzyme: Gal4 fusion to FokI cleavage domain. Biol. Chem., 379, 489–495. [DOI] [PubMed] [Google Scholar]
- 135.Kim Y.G., Cha,J. and Chandrasegaran,S. (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA, 93, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim Y.G., Shi,Y., Berg,J.M. and Chandrasegaran,S. (1997) Site-specific cleavage of DNA-RNA hybrids by zinc finger/FokI cleavage domain fusions. Gene, 203, 43–49. [DOI] [PubMed] [Google Scholar]
- 137.Klug A. (1999) Zinc finger peptides for the regulation of gene expression. J. Mol. Biol., 293, 215–218. [DOI] [PubMed] [Google Scholar]
- 138.Desjarlais J.R. and Berg,J.M. (1993) Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc. Natl Acad. Sci. USA, 90, 2256–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rebar E.J. and Pabo,C.O. (1994) Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science, 263, 671–673. [DOI] [PubMed] [Google Scholar]
- 140.Greisman H.A. and Pabo,C.O. (1997) A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science, 275, 657–661. [DOI] [PubMed] [Google Scholar]
- 141.Moore M., Klug,A. and Choo,Y. (2001) Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc. Natl Acad. Sci. USA, 98, 1437–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Moore M., Choo,Y. and Klug,A. (2001) Design of polyzinc finger peptides with structured linkers. Proc. Natl Acad. Sci. USA, 98, 1432–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Choo Y., Sanchez-Garcia,I. and Klug,A. (1994) In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature, 372, 642–645. [DOI] [PubMed] [Google Scholar]
- 144.Guo H., Karberg,M., Long,M., Jones,J.P.,III, Sullenger,B. and Lambowitz,A.M. (2000) Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science, 289, 452–457. [DOI] [PubMed] [Google Scholar]