Abstract
Background
The Campylobacter species usually causes infection between humans and livestock interaction via livestock breeding. The studies of the Campylobacter species thus far in all clinical isolates were to show the many kinds of antibiotic phenomenon that were produced. Their integrons cause the induction of antibiotic resistance between bacterial species in the Campylobacter species.
Results
The bacterial strains from the diarrhea of pediatric patient which isolated by China Medical University Hospital storage bank. These isolates were identified by MALDI-TOF mass spectrometry. The anti-microbial susceptibility test showed that Campylobacter species resistant to cefepime, streptomycin, tobramycin and trimethoprim/sulfamethoxazole (all C. jejuni and C. coli isolates), ampicillin (89% of C. jejuni; 75% of C. coli), cefotaxime (78% of C. jejuni; 100% of C. coli), nalidixic acid (78% of C. jejuni; 100% of C. coli), tetracycline (89% of C. jejuni; 25% C. coli), ciprofloxacin (67% of C. jejuni; 50% C. coli), kanamycin (33% of C. jejuni; 75% C. coli) and the C. fetus isolate resisted to ampicillin, cefotaxime, nalidixic acid, tetracycline, ciprofloxacin, kanamycin by disc-diffusion method. The effect for ciprofloxacin and tetracycline of the Campylobacter species was tested using an E-test. The tet, erm, and integron genes were detected by PCR assay. According to the sequencing analysis (type I: dfr12-gcuF-aadA2 genes and type II: dfrA7 gene), the cassette type was identified. The most common gene cassette type (type I: 9 C. jejuni and 2 C. coli isolates; type II: 1 C. coli isolates) was found in 12 class I integrase-positive isolates.
Conclusions
Our results suggested an important information in the latency of Campylobacter species with resistance genes, and irrational antimicrobial use should be concerned.
Electronic supplementary material
The online version of this article (doi:10.1186/s13099-017-0199-4) contains supplementary material, which is available to authorized users.
Keywords: Campylobacter species, Pediatric patient, Integrons, Antibiotic resistance gene
Background
The Campylobacter species, a bacillus, causes diseases in animals and humans [1]. Most species of animal including cattle, chicken, turkey, dog, cat, mink, ferret, pig, and primate is susceptible to infection [2]. Animals can be exposed to the Campylobacter species bacteria by direct contact with infected animals, or through contaminated feed or water. Raw or undercooked meat fed to pets may also contain the Campylobacter species [3]. In the 1970s, the Campylobacter species was first identified as a human pathogen [4, 5]. The Campylobacter species is a zoonotic pathogen, and it infects humans through contaminated food, water, or milk [6]. Infections caused by the Campylobacter species has emerged as a leading cause of acute gastroenteritis worldwide [7, 8]. The clinical characteristics of infection can create diarrhea, abdominal pain, fever, and vomiting [9]. The major pathogens of the Campylobacter species in humans is Campylobacter jejuni, Campylobacter coli, Campylobacter lari, and Campylobacter fetus [10]. The group with the highest incidence rate of Campylobacter species infection is infants younger than 2 years of age [11]. Campylobacter species’ infection causes moderate to severe diarrhea in infants and young children [11].
Most campylobacteriosis need not be therapy, but patient requires antibiotic treatment if the symptoms is severe. Fluoroquinolones, erythromycin, and tetracycline are the first-line antibiotic agents for treatment of Campylobacter species’ infections [12]. The inappropriate use of anti-microbial agents in animal husbandry has led to the development of antibiotic resistance in Campylobacter treatment [12]. Recently, the Centers for Disease Control and Prevention (CDC) has recorded drug-resistant Campylobacter as a thoughtful threat in the United States. Resistance of Campylobacter to antibiotics agents is interceded by multiple mechanisms [13]. The resistance mechanisms of the Campylobacter species bacteria include horizontal gene transfer (HGT) or multidrug efflux pump [13]. Integron is considered to be vectors for rapid HGT that causes antibiotic resistance between bacterial species [14, 15]. In Gram-negative bacteria, integrons are possibly a major factor in distribution of multidrug resistance [16]. Integron encoding the anti-microbial resistance gene (s) may act an important role for the dissemination of resistance in Campylobacter isolates. In addition, the class 1 integrons associated with the aadA9 gene (aminoglycoside-resistance gene) located on an R-plasmid have been reported in Campylobacter isolates [12].
The aim of this study was to identify the major pathogens and to assess the molecular basis of resistance to antibiotic agents in Campylobacter species isolated from the diarrhea of pediatric patient in Taiwan.
Methods
Isolation and identification of bacterial strains
The 14 Campylobacter strains used in this study were isolated from feces sample of pediatric diarrhea patient in China Medical University Hospital storage bank from January 2014 to February 2017. The Campylobacter species was cultured on Campy-BAP plates (Becton–Dickinson and Company, USA) containing five anti-microbial agents (amphotericin B, polymyxin B, cephalothin, trimethoprim, and vancomycin) and 10% sheep blood overnight at 42 °C and in an air condition of 5% O2, 10% CO2, and 85% N2. Subsequently, the strains were all re-cultivated under microaerophilic conditions. The isolated strains were then placed in storage in skim milk medium containing 20% glycerol stock at a temperature of −80 °C [17, 18].
MALDI-TOF mass spectrometry
The cultured bacteria were suspended in 300 μl of bi-distilled water and mixed with 900 ml of ethanol (Carl Roth GmbH). The measurement of the sample was then conducted following procedures previously described by El-Ashker et al. [19]. Measurement was performed with an Ultraflex III TOF/TOF mass spectrometer (Bruker Daltonics) equipped with a 200-Hz smartbeam 1 laser. The parameter setting was as follows: delay, 80 ns; ion source, 1 V, 25 kV; ion source, 2 V, 23.4 kV; lens voltage, 6 kV; and mass range 0–20 137 kDa. The raw spectra was analyzed by MALDI BIOTYPER 2.0 software (Bruker Daltonics, Billerica, MA, USA) using the default settings. The procedure of the MALDI-TOF mass spectrometry measurement was performed automatically without any user intervention. The software generated a list of peaks up to 100. The peaks with a mass-to-charge ratio difference of <250 ppm were considered to be identical. The peak list that was generated was used for matches against the reference library by directly using the integrated pattern-matching algorithms of the software. All parameters were the same regardless of the presumptive bacterial species analyzed. The BIOTYPER 2.0 database was composed using only Campylobacter and related species [20].
Antibiotics resistance screening in Campylobacter species by Kirby–Bauer disc diffusion test and Epsilometer test
The Kirby–Bauer disc diffusion test and Epsilometer test (Etest) were used to determine the anti-microbial susceptibility of the Campylobacter spp. isolates [21–23]. The susceptibility of each isolate to each antibiotic was determined according to the Neo-Sensitabs™ “user’s guide” for susceptibility testing and the latest guidelines of the Clinical and Laboratory Standards Institute (CLSI). To conduct the Kirby–Bauer disc diffusion test, sixteen antibiotic discs were chosen, including amikacin (30 μg/disc), ampicillin (30 μg/disc), cefepime (30 μg/disc), cefotaxime (30 μg/disc), chloramphenicol (30 μg/disc), ciprofloxacin (5 μg/disc), erythromycin (15 μg/disc), gentamicin (10 μg/disc), imipenem (10 μg/disc), kanamycin (5 μg/disc), nalidixic acid (30 μg/disc), streptomycin (30 μg/disc), sulfamethoxazole/trimethoprim (1.25/23.75 μg/disc), tetracycline (5 μg/disc) and tobramycin (10 μg/disc). All of which were purchased from Mast Group Ltd. (Merseyside, UK). The Campy-BAP plates were prepared and inoculated with the Campylobacter spp. isolates according to the procedures recommended by the NCCLS. The plates were incubated at 37 °C under the same microaerophilic conditions already described in the “Isolation and identification of bacterial strains” section above for 48 h.
For the Epsilometer test (Etest), five antibiotic Epsilometer strips were used, including strips containing amikacin, ciprofloxacin, imipenem, piperacillin/tazobactam, and tetracycline. For each tested organism, a Campy-BAP plate was inoculated by swabbing the plate evenly in three directions with a 1.0 McFarland standard of the test organism. The Etest strips were then applied to the surface of the plate and were incubated under the same conditions as indicated above for the disc diffusion test [22].
Detection of tetracycline- and erythromycin-resistant genes
The plasmid DNA was extracted using a Plasmid Miniprep Purification kit (GeneMark, GMbiolab, Taichung, Taiwan) and the genomic DNA extract was prepared with the PureLink Genomic DNA Mini kit (Carlsbad, CA, USA). The polymerase chain reaction (PCR) assay was applied to detect tetracycline-resistant genes reported by Abdi-Hachesoo et al. [24, 25]. The 9 pairs of primers were used to detect tet (A), tet (B), tet (K), tet (L), tet (M), tet (O), tet (Q), tet (S), and tet (W) gene in plasmid and genomic DNA of Campylobacter isolates, respectively. The specific genes, applicons, primers, and annealing temperatures are listed in Additional file 1: Table S1. Briefly, we performed the PCR with O’In 1 DNA polymerase (Yeastern Biotech, Taipei, Taiwan) with 10 pmol of forward and reverse primers (Mission Biotech Co., Ltd., Taipei, Taiwan) and 60 ng of the extracted plasmid or chromosomal DNA. The Campylobacter jejunii isolate 3 was positive for tet (A), tet (L), tet (M), tet (O), and tet (Q), we further identified the PCR product by DNA sequencing. We used the Campylobacter jejunii-isolate 3 as positive control for these genes. The erythromycin-resistant (erm) genes were detected, and the PCR-amplification of primers for erm (A), erm (B), erm (C), and erm (F), respectively, were used to detect any Erm-R genes in the individual Campylobacter isolates. Each gene of the PCR conditions, primers, and size that was listed in Additional file 1: Table S1.
The class 1 integrase and integron detection
The gene cassettes of integron were harbored in all the integrase-positive isolates. In order to screen these isolates for the presence of the class 1 integrase gene, the specific primers IntI1F and IntI1R were used [26]. The specific oligonucleotide PCR primer used in this study is listed in Additional file 1: Table S1. Following the manufacturer’s instructions, template DNA from all the isolates was prepared using Plasmid Miniprep Purification kits (GeneMark). PCR was then performed in a total volume of 25 μl consisting of 1 μl of target DNA, 17 μl of distilled water, 1 μl of each primer (10 μM), and 5 μl of 5× Master Mix (PCR Master Mix Kit, GeneMark), which in turn consisted of 0.75 U Taq Plus DNA polymerase, 250 μM dNTP, 2 mM MgCl2, and PCR buffer. More specifically, the amplicons that were used for the gene cassette of class I integron analysis were determined by the specific primers Cassette F and Cassette R [27], while the specific oligonucleotide PCR primer used is listed in Additional file 1: Table S1. The PCR procedures were executed under the same conditions as indicated above for the integrase gene detection (except that the annealing temperature was set at 55 °C for 30 s for detecting the intI1 gene while it was set at 57 °C for 45 s for detecting the gene cassette). PCR products of integron gene cassette were then sequenced from both sides. The same company (i.e., Mission Biotech Co., Ltd.) also synthesized the oligonucleotide primers used for the DNA sequencing. The nucleotide sequences were analyzed using the Blast program available on the National Center for Biotechnology Information (NCBI) website (http://blast.ncbi.nlm.nih.gov/Blast.cgi) in order to search for the closest sequences in the GenBank database (NCBI, US National Library of Medicine).
Molecular typing
The genotyping of the 14 Campylobacter species strains was performed according to a standard operating procedure which was recommended by the Centers for Disease Control and Prevention (CDC) for PulseNet PFGE of Campylobacter spp. (http://www.cdc.gov/pulsenet). The DNA of agarose plugs was performed with SmaI restriction enzyme. The electrophoresis conditions consisted of an initial switch time of 6.8 s and a final switch time of 35.4 s and a gradient of 6 V/cm for 19 h on CHEF-DR III System (Bio-Rad, Hercules, CA, USA). The PFGE patterns were analyzed using Gel Compare II version 6.5 software (Applied Maths, Sint-Matenslatem, Belgium). The DNA size marker was used by standard Salmonella serovar Branderup H9812 strain (digested with XbaI restriction enzyme). The pair comparisons of types and cluster analyses were performed by the Dice correlation coefficient and UPGMA (unweighted pair group method with arithmetic averages) clustering algorithm.
Results
The C. jejuni, C. coli, and C. fetus strains are the major Campylobacter species in the diarrhea of pediatric patient
According to the MALDI-TOF mass spectrometry results, the detection ratio of the clinical isolates for all the patient treated from January 2015 to December 2015 was 5%. The bacterial species of clinical isolates were identified by the adequate level (score ≥2.0; the addition of biochemical tests for score ≤2.0). In total samples, ten C. jejuni strains, three C. coli strains, and one C. fetus strain were isolated. Overall, the respective population rates for the C. jejuni, C. coli, and C. fetus strains were 71% (10/14), 21% (3/14), and 8% (1/14). These results indicate that C. jejuni, C. coli, and C. fetus strains were the major dominant Campylobacter species found in the samples from the diarrhea of pediatric patient.
Anti-microbial susceptibility patterns of Campylobacter isolates
The Kriby–Bauer disc diffusion test was used in this experiment. More specifically, a variation of the standard disc diffusion method that has been approved by the CLSI and EUCAST as an acceptable means of screening for the susceptibility of Campylobacter isolates so as to be treated with fifteen antibiotics, and the result is summarized in Table 1. It should be noted that because the breakpoints for amikacin, cefepime, streptomycin, and tobramycin have not yet been defined by the CLSI, the suggested E. coli breakpoint was used for those particular antibiotics instead. All the Campylobacter isolates exhibited full resistance to cefepime, streptomycin, and trimethoprim/sulfamethoxazole, as well as potential resistance to ampicillin, ciprofloxacin, cefotaxime, kanamycin, nalidixic acid, tobramycin, and tetracycline. Conversely, all the isolates were susceptible to amikacin, chloramphenicol, erythromycin, gentamicin, and imipenem.
Table 1.
The disc diffusion test of Campylobacter specie isolates from China Medical University Hospital stored bank
| Antibiotics | Resistance | Breakpoints (mm) |
|---|---|---|
| Amikacin (AK) | 4 (29%) | 14–17 |
| Ampicillin (AP) | 12 (86%) | 23–28 |
| Cefepime (CFM) | 14 (100%) | 18–25 |
| Cefotaxime (CTX) | 12 (86%) | 23–28 |
| Chloramphenicol (C) | 5 (36%) | 23–28 |
| Ciprofloxacin (CIP) | 9 (64%) | 20–24 |
| Erythromycin (E) | 5 (36%) | 12–16 |
| Gentamicin (GM) | 3 (21%) | 12–15 |
| Imipenem (IPM) | 5 (36%) | 23–28 |
| Kanamycin (K) | 7 (50%) | 13–18 |
| Nalidixic acid (NA) | 12 (86%) | 13–19 |
| Streptomycin (S) | 14 (100%) | 4–32 |
| Tobramycin (TM) | 13 (96%) | 18–26 |
| Trimethoprim/sulfamethoxazole (SXT) | 14 (100%) | 23–28 |
| Tetracycline (TE) | 10 (71%) | 22–26 |
All the isolates were tested by Epsilometer test (Etest) for their susceptibility to four antibiotics (amikacin, ciprofloxacin, imipenem and tetracycline), and the result is summarized in Additional file 1: Table S2. Because the breakpoints for amikacin and imipenem have not yet been defined by the CLSI, the suggested E. coli breakpoint from the same reference paper was used instead. All the isolates showed potential resistance to ciprofloxacin and tetracycline, while only five isolates exhibited resistance to amikacin, and only two isolates exhibited resistance to imipenem. Our prediction is that the will be isolates susceptible to amikacin and imipenem.
Tetracycline and erythromycin-resistant genes in plasmid
All Tet-R genes were found in plasmid, but not in genomic DNA, of tested Campylobacter spp. isolates. According the results of the screening for tetracycline-resistant genes, 79% (11/14) of the Campylobacter spp. isolates tested positive for tet (A) and tet (O) total, with tet (A) being seen in 89% (8/9) of the C. jejuni isolates and 75% (3/4) of the C. coli isolates. The tet (O) was found in 89% (8/9) of the C. jejuni isolates, 50% of the C. coli isolates, and 100% of the C. fetus isolates. Ten of the Campylobacter spp. isolates (71%) total tested positive for tet (M): it was detected in 78% of C. jejuni isolates (7/9) and 75% of the C. coli isolates (3/4). The tet (Q) resistant genes were seen in 36% of these Campylobacter spp. isolates total, being found in 33% of the C. jejuni isolates (3/9) and 50% of the C. coli isolates (2/4). The tet (L) (Fig. 1), which had the lowest prevalence rate, was found in only three of the Campylobacter spp. isolates total (21%), being detected in 22% of the C. jejuni isolates (2/9) and 25% of C. coli isolates (1/4). The Campylobacter isolates were screened to determine which harbored any of nine tetracycline-resistant genes, and the result is summarized in Additional file 1: Table S3. The tet (B), tet (K), tet (S) and tet (W) resistant genes were not identified in any of the tested Campylobacter spp. isolates.
Fig. 1.
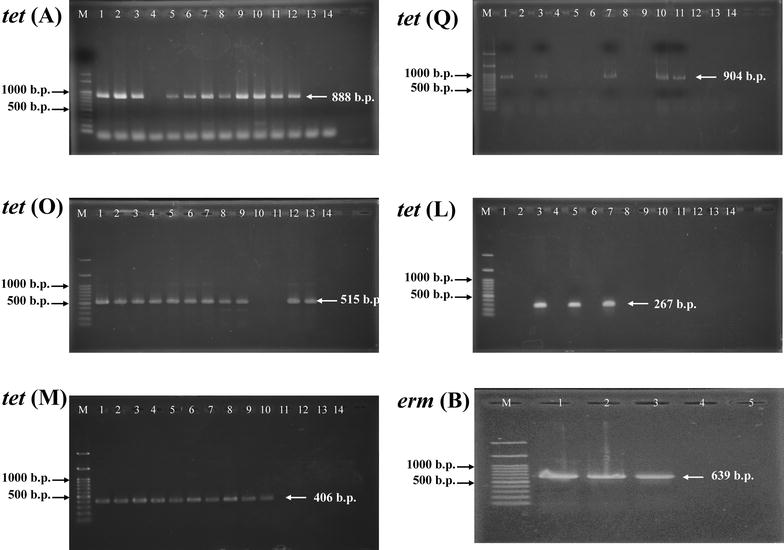
Polymerase chain reaction amplification of the tet (A) gene, tet (O), tet (M), tet (Q), tet (L) and erm (B) gene. (M: 100 b.p. DNA marker; Campylobacter jejuni: lines 1–6, 9, 11, and 12; Campylobacter coli: lines 7, 8, 10, and 13; Campylobacter fetus: line 14)
The result of the PCR detect Erm-R genes in 36% (5/14) erythromycin-resistant isolates showed that only the erm (B) (Fig. 1) (2 C. jejuni isolates, 22%; 1 C. coli isolate, 25%) was found (Additional file 1: Table S4). The erm (A), erm (C), and erm (F) resistant genes were not identified in any of the tested Campylobacter spp. isolates.
Class 1 integron detection in Campylobacter species
In Additional file 1: Table S5, we show the characteristics of various gene cassette types in class 1 integron of Campylobacter species isolates. The integrase gene (intI1) was detected in 86% of the fourteen Campylobacter isolates (12/14) identified in this study. Among the intI1-positive strains, twelve isolates (86%) harbored class 1 integron containing various sizes of gene cassettes ranging from 750 to 1907 b.p. in length.
The results of the PCR testing to detect Class I integron cassette genes indicated that 86% (12/14) of the Campylobacter spp. isolates tested positive for Class I integron cassette genes (Fig. 2). According to the sequencing analysis, two gene cassette types were identified in this study (namely, type I: dfr12-gcuF-aadA2 genes and type II: dfrA7 gene). Each cassette type had a unique inserted gene cassette pattern harboring various antibiotic-resistant genes. The most common gene cassette type (type I: dfr12-gcuF-aadA2 genes) was found in 92% of the 12 intI1-positive strains (9 C. jejuni strains and 3 C. coli strains) and was responsible for trimethoprim, streptomycin, and spectinomycin resistance in those strains. The other cassette type (type II: dfrA7 gene) was found in only 8% of the intI1-positive strains (i.e., one C. coli strain) and was responsible for trimethoprim resistance in that strain.
Fig. 2.

Polymerase chain reaction amplification of the class I integron gene cassette. (M: 1000 b.p. DNA marker; Campylobacter jejuni: lines 1–6, 9, 11, and 12; Campylobacter coli: lines 7, 8, 10, and 13; Campylobacter fetus: line 14)
Pulsed-field gel electrophoresis of Campylobacter isolates
In Fig. 3, among 14 Campylobacter isolates were typeable by PFGE, that the guidelines of PFGE patterns were applied by Tenover [28]. PFGE analysis was performed in order to determine the genetic diversity of these isolates and the relationship between the isolates retrieved from different patients. Analysis of the PFGE profiles of the Campylobacter isolates suggested that the isolates possessed diverse genotypes, when comparing profiles of isolates belonging to different species. The similarity of the isolates was from 50 to 100%. These 14 isolates had 13 different PFGE genotypes, only one PFGE type was recovered between two C. jejuni isolates. Furthermore, by using a cut-off similarity value of 75%, profiles of the C. jejuni were classified to 6 clusters and C. coli were classified to 2 clusters.
Fig. 3.
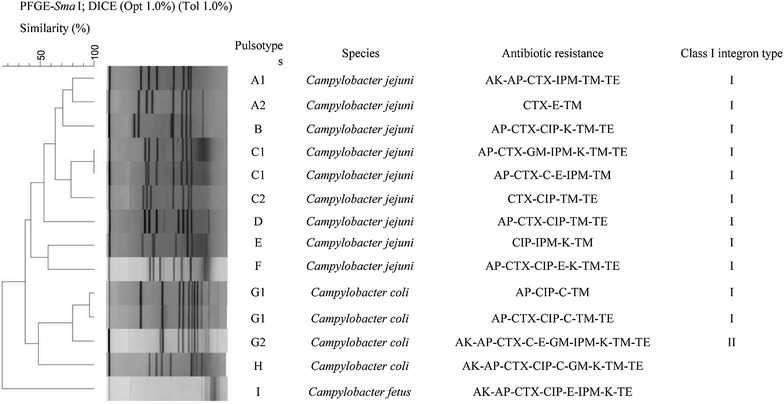
The dendrogram of SmaI digested PFGE profile of 14 Campylobacter strains. AK amikacin, AP ampicilin, CFM cefepime, CTX cefotaxime, C chloramphenicol, CIP ciprofloxacin, E erythromycin, GM gentamicin, IPM imipenem, K kanamycin, NA nalidixic acid, S streptomycin, TM tobramycin, T tetracyclin, SXT trimethoprim/sulfamethoxazole
Discussion
The Campylobacter species is some of the many bacterial foodborne pathogens worldwide. After Campylobacter infection, chronic sequelae can be very serious, causing lifelong morbidity. Campylobacter species infection is also associated with various chronic sequelae. In addition, C. jejuni in particular is among the most frequent in acute enteritis [1]. The major Campylobacter species in humans is C. jejuni, C. coli, C. lari, and C. fetus [10]. In Taiwan, Wang et al. demonstrated that in 104 enteric campylobacteriosis patient, C. coli was found in 24 patient (23.1%), while C. jejuni was found in 80 patient (76.9%) [9]. In our study, we used the MALDI-TOF–MS analysis method to determine the major Campylobacter species in the diarrhea of pediatric patient, with our results showing that C. jejuni, C. coli, and C. fetus were the major Campylobacter species found in our samples. Thus, our study is in agreement with previous findings.
Campylobacter species infection leads to campylobacteriosis, so fluid supplementation and antibiotic treatments are the most important therapies. The first-line antibiotic agent used in the treatment of Campylobacter infections is such as erythromycin and ciprofloxacin. Tetracycline can be an alternative choice in the treatment of clinical campylobacteriosis [29]. Antibiotic resistance in Campylobacter species therapy has now become a major public health concern worldwide. In this study, the disc diffusion test and the Etest were used to detect the antibiotic-resistant effect in Campylobacter species. Our results demonstrated that all the isolates presented resistance to cefepime, streptomycin, and trimethoprim/sulfamethoxazole, ampicillin, ciprofloxacin, cefotaxime, kanamycin, nalidixic acid, tobramycin, and tetracycline (Additional file 1: Tables S1, S2). In addition, the PCR results indicate that all the Tet-R genes and Erm-R genes were found in plasmid (Additional file 1: Tables S3, S4; Fig. 1).
Resistance to tetracycline is converted by the gene in Campylobacter species. The Tet (O) and tet (S) genes are transferred as plasmid-encoded genes and are not self-mobile in the chromosome. The tet (A) and tet (B), efflux genes, code for an approximately 46-kDa membrane bound efflux protein for membrane-associated proteins that export tetracycline from the cell. Our results demonstrate that 79% (11/14) of the Campylobacter species isolates tested positive for tet (A) and tet (O) (Fig. 1), and tet (A) was seen in 80% of the C. jejuni isolates and 100% of the C. coli isolates. The tet (O) gene was found in 80% of the C. jejuni isolates, 67% of the C. coli isolates, and 100% of the C. fetus isolates. (Additional file 1: Table S3). It has been reported that the () gene was also found in 33–76% of tetracycline-resistant C. jejuni isolates lacking plasmids in Canada and Australia, respectively [30].
Erythromycin is the first macrolide antibiotics agent. Erythromycin inhibits bacterial RNA-dependent protein synthesis in ribosomal 50S subunit. There are two mechanisms of macrolide resistance, one is 23S rRNA and the mutations of ribosomal proteins L4 and L22, and the other is antibiotic efflux by the multidrug efflux pump CmeABC [31]. It has been reported that the rRNA methylase gene erm (B) mediates resistance to erythromycin in one C. coli isolate of farm-animal origin [31, 32]. Our results demonstrate that only the erm (B) was seen in C. jejuni and C. coli (Additional file 1: Table S4; Fig. 1). Thus, our study is in agreement with previous studies.
Integron is a major genetic element in multidrug-resistant Gram-negative bacteria. The integron contains an integrase gene and a site-specific integration site where the integrase can link antibiotic-resistant gene cassettes. There are nine classes of integron and over 60 distinct antibiotic-resistant gene cassettes have been characterized within integrons. Our results demonstrate that the integrase gene (intI1) was detected in 86% of the fourteen Campylobacter isolates (12/14). 71% (10/14) of the Campylobacter spp. isolates tested positive for Class I integron gene cassette by PCR (Fig. 2). The sequencing analysis and PCR demonstrated that two gene cassette types were identified, including type I: dfr12-gcuF-aadA2 genes; and type II: dfrA7 gene. It has already been reported that the class 1 integron gene is associated with antibiotic resistance in Campylobacter jejuni isolated from the broiler chicken house environment [33].
Conclusions
This is the first extensive study of as well as new findings in the differential categorization of antibiotic-resistant genes and integron gene cassettes in clinical Campylobacter species. To our knowledge, C. jejuni isolate harbored erm (B) in plasmid was only found in strains from animal, and we first reported that patients infected with erm (B)-positive C. jejuni, two types of class I integron gene cassette pattern were different from previous studies. Therefore, an important information in the latence of Campylobacter species with antibiotic-resistant genes and the irrational using antimicrobial agents in therapy are what concerned us.
Authors’ contributions
Conceived the design of study, reviewed the literature performed necessary interventions including laboratory investigations YCC and NT. Case identification, entry and data analysis JSY, CCL, FJT, TJH and IKW. Prepared the manuscript with the help of JSY, CCL and FJT. All authors read and approved the final manuscript.
Acknowledgements
We would like to express our thanks to Dr. Shen-Min Huang (Show Chwan Memorial Hospital, Changhua, Taiwan) for providing two Campylobacter species isolates.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable as the work has been done using China Medical University Hospital storage bank archived strains.
Funding
This study was supported by a Grant from Taiwan Ministry of Health and Welfare Clinical Trial Center (MOHW106-TDU-B-212-113004) and by the Ministry of Science and Technology (Taiwan) (grant MOST 103-2815-C-039-040-B). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13099-017-0199-4) contains supplementary material, which is available to authorized users.
Contributor Information
Yi-Chih Chang, Phone: +886-422053366, Email: yichih@mail.cmu.edu.tw.
Ni Tien, Email: t6719@mail.cmuh.org.tw.
Jai-Sing Yang, Email: jaisingyang@gmail.com.
Chi-Cheng Lu, Email: a722353@gmail.com.
Fuu-Jen Tsai, Email: d0704@mail.cmuh.org.tw.
Tsurng-Juhn Huang, Email: stevenhcj@gmail.com.
I-Kuan Wang, Email: ikwang@mail.cmuh.org.tw.
References
- 1.O’Brien SJ. The consequences of Campylobacter infection. Curr Opin Gastroenterol. 2017;33(1):14–20. doi: 10.1097/MOG.0000000000000329. [DOI] [PubMed] [Google Scholar]
- 2.Sahin O, Yaeger M, Wu Z, Zhang Q. Campylobacter-Associated Diseases in Animals. Annu Rev Anim Biosci 2017;5(21-42. [DOI] [PubMed]
- 3.Skarp CP, Hanninen ML, Rautelin HI. Campylobacteriosis: the role of poultry meat. Clin Microbiol Infect. 2016;22(2):103–109. doi: 10.1016/j.cmi.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Said B, Wright F, Nichols GL, Reacher M, Rutter M. Outbreaks of infectious disease associated with private drinking water supplies in England and Wales 1970–2000. Epidemiol Infect. 2003;130(3):469–479. [PMC free article] [PubMed] [Google Scholar]
- 5.Figueras MJ, Levican A, Collado L. Updated 16S rRNA-RFLP method for the identification of all currently characterised Arcobacter spp. BMC Microbiol. 2012;12:292. doi: 10.1186/1471-2180-12-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Dong Y, Deng F, Liu D, Yao H, Zhang Q, Shen J, Liu Z, Gao Y, Wu C, Shen Z. Species shift and multidrug resistance of Campylobacter from chicken and swine, China, 2008–2014. J Antimicrob Chemother. 2016;71(3):666–669. doi: 10.1093/jac/dkv382. [DOI] [PubMed] [Google Scholar]
- 7.Fitzgerald C. Campylobacter. Clin Lab Med. 2015;35(2):289–298. doi: 10.1016/j.cll.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Oosterom J. Campylobacter jejuni: an important causative agent of food infection in man. An overview. Tijdschr Diergeneeskd. 1984;109(11):446–455. [PubMed] [Google Scholar]
- 9.Wang SC, Chang LY, Hsueh PR, Lu CY, Lee PI, Shao PL, Hsieh YC, Yen FP, Lee CY, Huang LM. Campylobacter enteritis in children in northern Taiwan—a 7-year experience. J Microbiol Immunol Infect. 2008;41(5):408–413. [PubMed] [Google Scholar]
- 10.Sheppard SK, Maiden MC. The evolution of Campylobacter jejuni and Campylobacter coli. Cold Spring Harb Perspect Biol. 2015;7(8):a018119. doi: 10.1101/cshperspect.a018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan H, Ge Y, Xu H, Zhang J, Kuang D, Yang X, Su X, Huang Z, Shi X, Xu X, Meng J. Molecular characterization, antimicrobial resistance and Caco-2 cell invasion potential of Campylobacter jejuni/coli from young children with diarrhea. Pediatr Infect Dis J. 2016;35(3):330–334. doi: 10.1097/INF.0000000000001016. [DOI] [PubMed] [Google Scholar]
- 12.Ekkapobyotin C, Padungtod P, Chuanchuen R. Antimicrobial resistance of Campylobacter coli isolates from swine. Int J Food Microbiol. 2008;128(2):325–328. doi: 10.1016/j.ijfoodmicro.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Su CC, Yin L, Kumar N, Dai L, Radhakrishnan A, Bolla JR, Lei HT, Chou TH, Delmar JA, Rajashankar KR, Zhang Q, Shin YK, Yu EW. Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump. Nat Commun. 2017;8(1):171. doi: 10.1038/s41467-017-00217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meinersmann RJ, Phillips RW, Ladely SR. Inter- and intra-genomic heterogeneity of the intervening sequence in the 23S ribosomal RNA gene of Campylobacter jejuni and Campylobacter coli. Syst Appl Microbiol. 2009;32(2):91–100. doi: 10.1016/j.syapm.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Angulo FJ, Baker NL, Olsen SJ, Anderson A, Barrett TJ. Antimicrobial use in agriculture: controlling the transfer of antimicrobial resistance to humans. Semin Pediatr Infect Dis. 2004;15(2):78–85. doi: 10.1053/j.spid.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Bakhshi B, Kalantar M, Rastegar-Lari A, Fallah F. PFGE genotyping and molecular characterization of Campylobacter spp. isolated from chicken meat. Iranian J Vet Res. 2016;17(3):177–183. [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Ma L, Li Y, Jia H, Wei J, Shao D, Liu K, Shi Y, Qiu Y, Ma Z. Antimicrobial resistance of Campylobacter species isolated from broilers in live bird markets in Shanghai, China. Foodborne Pathog Dis. 2017;14(2):96–102. doi: 10.1089/fpd.2016.2186. [DOI] [PubMed] [Google Scholar]
- 18.Hiett KL. Campylobacter jejuni isolation/enumeration from environmental samples. Methods Mol Biol. 2017;1512:1–8. doi: 10.1007/978-1-4939-6536-6_1. [DOI] [PubMed] [Google Scholar]
- 19.El-Ashker M, Gwida M, Tomaso H, Monecke S, Ehricht R, El-Gohary F, Hotzel H. Staphylococci in cattle and buffaloes with mastitis in Dakahlia Governorate, Egypt. J Dairy Sci. 2015;98(11):7450–7459. doi: 10.3168/jds.2015-9432. [DOI] [PubMed] [Google Scholar]
- 20.Bessede E, Solecki O, Sifre E, Labadi L, Megraud F. Identification of Campylobacter species and related organisms by matrix assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry. Clin Microbiol Infect. 2011;17(11):1735–1739. doi: 10.1111/j.1469-0691.2011.03468.x. [DOI] [PubMed] [Google Scholar]
- 21.Kilonzo-Nthenge A, Nahashon SN, Chen F, Adefope N. Prevalence and antimicrobial resistance of pathogenic bacteria in chicken and guinea fowl. Poult Sci. 2008;87(9):1841–1848. doi: 10.3382/ps.2007-00156. [DOI] [PubMed] [Google Scholar]
- 22.Stone D, Davis M, Baker K, Besser T, Roopnarine R, Sharma R. MLST genotypes and antibiotic resistance of Campylobacter spp. isolated from poultry in Grenada. Biomed Res Int. 2013;2013:794643. doi: 10.1155/2013/794643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge B, Wang F, Sjolund-Karlsson M, McDermott PF. Antimicrobial resistance in Campylobacter: susceptibility testing methods and resistance trends. J Microbiol Methods. 2013;95(1):57–67. doi: 10.1016/j.mimet.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 24.Abdi-Hachesoo B, Khoshbakht R, Sharifiyazdi H, Tabatabaei M, Hosseinzadeh S, Asasi K. Tetracycline resistance genes in Campylobacter jejuni and C. coli isolated from poultry carcasses. Jundishapur J Microbiol. 2014;7(9):e12129. doi: 10.5812/jjm.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mottola C, Matias CS, Mendes JJ, Melo-Cristino J, Tavares L, Cavaco-Silva P, Oliveira M. Susceptibility patterns of Staphylococcus aureus biofilms in diabetic foot infections. BMC Microbiol. 2016;16(1):119. doi: 10.1186/s12866-016-0737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindstedt BA, Heir E, Nygard I, Kapperud G. Characterization of class I integrons in clinical strains of Salmonella enterica subsp. enterica serovars Typhimurium and Enteritidis from Norwegian hospitals. J Med Microbiol. 2003;52(Pt 2):141–149. doi: 10.1099/jmm.0.04958-0. [DOI] [PubMed] [Google Scholar]
- 27.Ng LK, Mulvey MR, Martin I, Peters GA, Johnson W. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob Agents Chemother. 1999;43(12):3018–3021. doi: 10.1128/aac.43.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wieczorek K, Osek J. Antimicrobial resistance mechanisms among Campylobacter. Biomed Res Int. 2013;2013:340605. doi: 10.1155/2013/340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibreel A, Tracz DM, Nonaka L, Ngo TM, Connell SR, Taylor DE. Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet(O)-mediated tetracycline resistance. Antimicrob Agents Chemother. 2004;48(9):3442–3450. doi: 10.1128/AAC.48.9.3442-3450.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin S, Wang Y, Zhang Q, Zhang M, Deng F, Shen Z, Wu C, Wang S, Zhang J, Shen J. Report of ribosomal RNA methylase gene erm(B) in multidrug-resistant Campylobacter coli. J Antimicrob Chemother. 2014;69(4):964–968. doi: 10.1093/jac/dkt492. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Zhang M, Deng F, Shen Z, Wu C, Zhang J, Zhang Q, Shen J. Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob Agents Chemother. 2014;58(9):5405–5412. doi: 10.1128/AAC.03039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee MD, Sanchez S, Zimmer M, Idris U, Berrang ME, McDermott PF. Class 1 integron-associated tobramycin-gentamicin resistance in Campylobacter jejuni isolated from the broiler chicken house environment. Antimicrob Agents Chemother. 2002;46(11):3660–3664. doi: 10.1128/AAC.46.11.3660-3664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


