Abstract
Background
Small islands serve as potential malaria reservoirs through which new infections might come to the mainland and may be important targets in malaria elimination efforts. This study investigated malaria vector species diversity, blood-meal hosts, Plasmodium infection rates, and long-lasting insecticidal net (LLIN) coverage on Mageta, Magare and Ngodhe Islands of Lake Victoria in western Kenya, a region where extensive vector control is implemented on the mainland.
Results
From trapping for six consecutive nights per month (November 2012 to March 2015) using CDC light traps, pyrethrum spray catches and backpack aspiration, 1868 Anopheles mosquitoes were collected. Based on their cytochrome oxidase I (COI) and intergenic spacer region PCR and sequencing, Anopheles gambiae s.l. (68.52%), Anopheles coustani (19.81%) and Anopheles funestus s.l. (11.67%) mosquitoes were differentiated. The mean abundance of Anopheles mosquitoes per building per trap was significantly higher (p < 0.001) in Mageta than in Magare and Ngodhe. Mageta was also the most populated island (n = 6487) with low LLIN coverage of 62.35% compared to Ngodhe (n = 484; 88.31%) and Magare (n = 250; 98.59%). Overall, 416 (22.27%) engorged Anopheles mosquitoes were analysed, of which 41 tested positive for Plasmodium falciparum infection by high-resolution melting (HRM) analysis of 18S rRNA and cytochrome b PCR products. Plasmodium falciparum infection rates were 10.00, 11.76, 0, and 18.75% among blood-fed An. gambiae s.s. (n = 320), Anopheles arabiensis (n = 51), An. funestus s.s. (n = 29), and An. coustani (n = 16), respectively. Based on HRM analysis of vertebrate cytochrome b, 16S rRNA and COI PCR products, humans (72.36%) were the prominent blood-meal hosts of malaria vectors, but 20.91% of blood-meals were from non-human vertebrate hosts.
Conclusions
These findings demonstrate high Plasmodium infection rates among the primary malaria vectors An. gambiae s.s. and An. arabiensis, as well as in An. coustani for the first time in the region, and that non-human blood-meal sources play an important role in their ecology. Further, the higher Anopheles mosquito abundances on the only low LLIN coverage island of Mageta suggests that high LLIN coverage has been effective in reducing malaria vector populations on Magare and Ngodhe Islands.
Keywords: Malaria vector, Blood-meal, Malaria parasite, Plasmodium falciparum, Malaria transmission
Background
Small island communities are harder to reach than mainland populations and do not have the same access to malaria control interventions and are less researched. Malaria continues to be a major public health problem and a key impediment to socio-economic development in areas around Lake Victoria, western Kenya where recent studies have focused on the mainland [1–4] and large islands, such as Rusinga Island [5, 6]. Despite higher control efforts in these regions, traffic from these small islands might sustain transmission if not equally targeted. The Lake Victoria island region provides suitable habitats for abundant and diverse anophelines that support efficient endemic malaria transmission all year round [7, 8], with various health facilities reporting more than 40% prevalence rates based on rapid diagnostic tests (RDTs) at public hospitals [9]. The main anophelines present in this region are Anopheles gambiae sensu stricto (s.s.) and Anopheles arabiensis, with Anopheles funestus bridging transmission during dry seasons and Anopheles coustani as a secondary vector [7, 10, 11].
The current front-line strategies for malaria control in this region are malaria patient management [12], following the WHO strategy of diagnostic testing, treatment and malaria surveillance [13], and protecting people from receiving infectious bites through long-lasting insecticidal nets (LLINs) [14]. LLINs have been used extensively to reduce mosquito density and biting activity, causing a decline in malaria transmission in the Lake Victoria region of western Kenya [14–16]. Despite the decline, malaria transmission remains, undermining the current drive to eliminate malaria [17].
Local transmission can be maintained by importation of infection through movement of infected people and/or mosquitoes within an endemic area [18, 19], creating malaria hotspots that facilitate widespread transmission [20]. Therefore, monitoring localized transmission dynamics is important in determining actual effectiveness of control strategies deployed, and can guide appropriate adjustments. Malaria control interventions can be more effective if applied based on knowledge of geographically localized transmission dynamics as influenced by vector species and their blood-meal sources. This study was undertaken to investigate LLIN coverage and compare the distribution of malaria vectors, blood-meal sources, and Plasmodium infection rates in three small islands with neglected communities in Lake Victoria, western Kenya.
Methods
Study location
The study was conducted in Mageta, Magare and Ngodhe Islands of Lake Victoria in western Kenya (Fig. 1). Mageta and Magare Islands are in Siaya County, while Ngodhe Island is in Homa Bay County. The sizes of Mageta, Magare and Ngodhe are approximately 7.02, 0.20 and 0.90 sq km, respectively, while the distance between Mageta and Magare is about 0.30 km, and Ngodhe is about 28.50 km from Mageta. Mageta and Magare are about 7.5 km from the mainland, while Ngodhe is about 3 km from a relatively larger island (Rusinga). The islands are only accessible by ferry services or boats. In addition, all three islands have a rocky terrain. Therefore, the islands are difficult to reach when compared with the mainland. The main economic activities in the islands are fishing, fishmongering and small-scale agriculture.
Fig. 1.
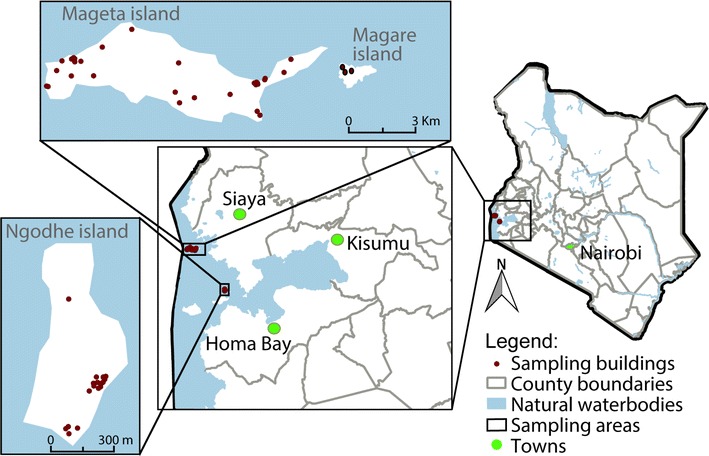
Map showing study locations. The three study islands are located in Lake Victoria, western Kenya. Mageta and Magare are located in Siaya County, while Ngodhe is in Homa Bay County. Areas of Mageta, Magare and Ngodhe are 7.02, 0.20 and 0.90 sq km, respectively. The distance between Mageta and Magare Islands is about 0.30, which are about 28.5 km north-west of Ngodhe Island
Household information
All buildings from which mosquitoes were samples on the three islands, including unoccupied mills that were part of residential complexes, were identified and geo-referenced using a hand-held geographical positioning system (eTrex, Vista, Garmin, USA). The roofing and wall types were recorded and the nature of eaves scored as either opened or closed. With the household head/adult member (i.e., aged over 18 years) of the household as the respondent, a survey was carried out to determine island populations and LLIN coverage (the percentage of households having at least one LLIN).
Adult mosquito trapping
Adult mosquitoes were trapped indoors or outdoors from 16 geo-referenced residential buildings on each island for six consecutive nights each month between November 2012 to September 2014 (23 months; 2208 trap nights) in Ngodhe, and June 2013 to March 2015 (22 months; 2112 trap nights) in Magare and Mageta. Residential buildings were defined as housing units comprising one or two people sharing a living space. When members of a family lived in the same compound with several buildings, each building was regarded as a separate residential building if at least one member of the family spends the night in the housing unit. One randomized trapping method (unbaited indoor or outdoor CDC light traps, indoor pyrethrum spray catches (PSC) or indoor backpack aspiration (ASP) was used per geo-referenced residential building in a sampling month. In Magare and Ngodhe, buildings were resampled across months (with different methods) due to the limited availability of residential houses on these islands. The trapping nights were selected to coincide with the period before full moon [21, 22]. Overall, 96 trappings were done per month on each island, with Ngodhe having an additional trapping month of 96 collections. Host-seeking vectors were trapped using unbaited CDC light traps set indoors or outdoors in the evening at 18:00 and removed the following morning at 06:00. The CDC indoors light traps were set on the foot side of a person’s bed (about 1.5 m from the ground), irrespective of whether they were sleeping under a bed net or not. Outdoors, CDC light traps were set about 20 m from a sampling building and a present cattle shed. Indoor-resting vectors were collected using PSC and ASP between 06:00 and 08:00.
After collection, mosquitoes were anaesthetized with chloroform [23], identified using morphological keys and sexed [24]. All the females belonging to An. gambiae, An. funestus and An. coustani species complexes were counted and classified on the basis of their abdominal status as blood-fed (engorged), gravid, half-gravid, or unfed (not engorged) [25]. Further, the females were preserved individually in barcoded vials containing isopropanol, stored at room temperature in the field and at −20 °C in the laboratory for further analysis. The labelling indicated details of collection method, building identification, site identification, morphological identification, sex and collection date.
Nucleic acid extraction
The engorged abdomens of field-collected and laboratory-reared adult anophelines were separated from the rest of the body (head, thorax, legs) using sterile forceps and dissection pins, and transferred into individual sterile 1.5-mL microtubes. Genomic DNA from the engorged abdomens, remaining body parts (head, thorax, legs), and known vertebrate whole blood samples were extracted separately using DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) following the manufacturer’s extraction protocol with some modifications. Briefly, 200 µL of PBS (phosphate-buffered saline, pH 7.4) was added to the sample in a 1.5-mL microcentrifuge tube containing 20 µL proteinase K and the sample mixed by vortexing for 15 s. Then, 200 µL lysis buffer was added, vortexed and the mixture incubated at 56 °C for 2 h. Subsequently, 200 µL of absolute ethanol was added. The solution was mixed by vortexing and homogenate transferred into a mini spin column placed in a 2-mL collection tube before centrifugation for 1 min at 6000×g. The flow-through and collection tubes were discarded and the spin column transferred to a new 2-mL collection tube, 500 µL of buffer AW1 was added and centrifuged for 3 min at 20,000×g. The flow-through and the collection tubes were discarded and the spin column transferred to a new 1.5-mL micro-centrifuge tube. The DNA was finally eluted by adding 30 µL buffer AE to the centre of the spin column membrane, incubated for 3 min at room temperature (25 °C) and centrifuged for 1 min at 6000×g, and stored at −20 °C.
Molecular identification of anopheline mosquitoes
Molecular identification of engorged An. coustani, An. funestus sensu lato (s.l.) and An. gambiae mosquitoes involved polymerase chain reaction (PCR) amplification and sequencing of the cytochrome oxidase subunit 1 (COI) region [26], polymorphic ITS2 region of ribosomal DNA [27, 28] and analysing melt curve differences of 165 base pairs (bp) intergenic spacer region (IGS) gene amplicons obtained using IGS gene primers [29, 30]. Colony-reared, sugar-fed An. gambiae s.s. Mbita strain (established in 2001) and An. arabiensis Mwea strain (established in 2004) from the International Centre of Insect Physiology and Ecology (icipe) in Nairobi, Kenya, served as standard reference positive controls for An. gambiae s.l. sibling species identification and negative controls for blood-meal analysis.
Ten microlitre PCR reactions were prepared with 0.5 μM final concentrations for each primer, 2 μL of 5× Hot Firepol Evagreen HRM Mix (Solis BioDyne, Tartu, Estonia) and 1 μL of DNA template. Thermal cycling conditions for COI and ITS2 were as follows: initial denaturation at 95 °C for 15 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s and extension at 72 °C for 1 min 30 s, and a final extension at 72 °C for 7 min. For IGS amplification, the conditions were as follows: initial denaturation at 95 °C for 15 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 57 °C for 30 s, and extension at 72 °C for 45 s and a final extension at 72 °C for 7 min. PCR reactions for COI and ITS2 were conducted on Veriti thermocycler (Applied Biosystems), and for IGS, a high-resolution melting (HRM) capable Rotor-Gene Q real time PCR thermocycler (QIAGEN, Hilden, Germany) was used. Following PCR, HRM analysis of amplicons was conducted by gradually increasing the temperature by 0.1 °C after every 2 s from 75 to 92 °C, resulting in a plot of the change in fluorescence with time (dF/dT). PCR-HRM protocols were validated for accuracy and sensitivity using standard reference controls. ExoSAP-IT (USB Corporation, Cleveland, OH, USA) was used to remove unincorporated dNTPs and PCR primers before sequencing. Sequences were edited in Geneious 7.0.5 (http://www.geneious.com) [31] and used to query GenBank [32].
Blood-meal source detection
High-resolution melting profiles obtained from PCR products of vertebrate cytochrome b (cyt b) [33–36], 16S ribosomal (r)RNA [33] and COI gene primers were used to distinguish different vertebrate hosts in anopheline mosquito blood-meals. Using DNA extracted from known vertebrate whole blood as positive controls and sugar-fed colony-reared mosquito DNA extracts as negative controls, PCRs were carried out in final volumes of 10 μL, containing 6 µL of PCR water, 0.5 μM concentrations of each primer, 2 µL of 5× Hot Firepol Evagreen HRM Mix (Solis BioDyne, Tartu, Estonia) and 1 µL of DNA template. Thermal cycling conditions for cyt b and 16S rRNA primers [33] were used for all engorged anophelines. The thermal cycling conditions used for COI primers were as follows: initial denaturation for 15 min at 95 °C, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s and extension at 72 °C for 60 s followed by a final extension at 72 °C for 7 min. Following PCR, HRM profile analysis of amplicons was conducted as previously stated with normalization regions between 75.0–78.0 and 88.00–95.0 °C. Known vertebrate whole blood from cow (Bos tarsus), pig (Sus scrofa), goat (Capra hircus), chicken (Gallus gallus), dog (Canis familiaris), and human (Homo sapiens) DNA collected in a previous study [33], as well as Swiss mouse (Mus musculus) and rabbit (Oryctolagus cuniculus) whole blood samples sourced from icipe’s animal rearing unit, served as standard reference positive controls for blood-meal analysis. Whole blood from livestock samples was obtained from a local abattoir. Blood-meal sources were identified by comparison of HRM melt curves to those of the reference control species. Amplicons with unique cyt b, 16S rRNA or COI HRM melt curves were purified for sequencing as previously stated.
Detection of malaria parasites
Malaria parasites in salivary glands of engorged mosquitoes were detected by analysing species-specific HRM profiles generated from PCR products of 18S rRNA [37] and 183-bp cyt b gene [38] primers. PCRs were carried out in final volumes of 10 μL, containing 6 µL of PCR water, 0.5 μM concentrations of each primer, 2 µL of 5× Hot Firepol Evagreen HRM Mix (Solis BioDyne, Tartu, Estonia) and 1 µL of DNA template. nPCR-HRM was used for 18S rRNA, the touchdown thermal cycling conditions [37] were used on all field-collected engorged anopheline mosquitoes. The thermal cycling conditions used for cyt b primers were as follows: initial denaturation for 15 min at 95 °C followed by 40 cycles of denaturation at 95 °C for 20 s, annealing at 45 °C for 60 secs and extension at 72 °C for 45 s followed by a final extension at 72 °C for 10 min. Following PCR, HRM profile analysis of amplicons was conducted as previously stated with normalization regions between 64.0–66.0 and 86.00–92.0 °C. Plasmodium falciparum infection was detected by comparison of melt curves to those of a standard reference positive control, P. falciparum DNA acquired from the National Institute for Biological Standards and Control (NIBSC; Hertfordshire, UK). Representative positive samples were purified for sequencing as previously stated. The sequences were edited in Geneious 7.0.5 software [31] and queried in GenBank using BLAST [32].
Statistical analysis
Field entomological data were analysed using R version 3.3.0 [39]. Anopheline mosquito abundance was estimated as the mean number of Anopheles mosquitoes collected per building per trap. Chi square tests were used to compare these mean abundances of anophelines among the three study islands. Species composition was estimated in terms of relative abundances, the mean percentages of specific anopheline mosquito species collected per trap per building. Differences in P. falciparum infection rates among engorged vector species were compared in a Bayesian fashion using the Bayesian First Aid package [40]. The Bayesian approach was adopted because these data were sparse, thus rendering the classical Chi square approach for comparing proportions unreliable. Differences were considered significant if the 95% credibility interval (Bayesian equivalent of classical 95% confidence interval) did not include zero.
Results
Demographic information and LLIN coverage
A total of 2671 residential buildings were geo-referenced within the three islands with Mageta, Magare, and Ngodhe having 2446, 71, and 154 residential buildings, respectively (Table 1). The biggest island, Mageta, had 6487 inhabitants, while Magare and Ngodhe had 250 and 484 inhabitants, respectively. Mageta had LLIN coverage of 62% (n = 1525), which is below the universal WHO-recommended target of 80%, while Magare had 99% (n = 70) and Ngodhe had 88% (n = 136) LLIN coverage (Table 1). Most houses were made of mud walls with iron-sheet roofing and open eaves [Mageta 95% (n = 2331); Magare 100% (n = 71), Ngodhe 96.75% (n = 149)] between the wall and the roof (Table 1).
Table 1.
Demographic information of Mageta, Magare and Ngodhe, including long-lasting insecticidal net coverage
| Study island | Residential buildings | Open eaves | Population | LLIN coverage |
|---|---|---|---|---|
| Mageta | 2446 | 2331 | 6487 | 1525 (62.35%) |
| Magare | 71 | 71 | 250 | 70 (98.59%) |
| Ngodhe | 154 | 149 | 484 | 136 (88.31%) |
LLINs: long-lasting insecticide nets
Anopheline population dynamics
During the study period, between November 2012 and March 2015, a total of 7350 mosquitoes were collected in pools from CDC light trap, PSC and ASP collections. Of the mosquitoes collected 2752 (37.44%) were Aedes aegypti, 2266 (30.83%) were Mansonia species and 2332 (31.73%) were anopheline species (An. gambiae s.l., An. funestus and An. coustani). A confirmatory An. coustani COI sequence was deposited into GenBank (accession MF782553). The anopheline mosquitoes collected were comprised of 464 (19.90%) males and 1868 (80.10%) females. Of the female anopheline mosquitoes sampled, 1452 (77.73%) were collected indoors and 416 (22.27%) were collected outdoors (Table 2). Of these vectors collected, 1280 (68.52%) were An. gambiae s.l., 370 (19.81%) were An. coustani, and 218 (11.67%) were An. funestus s.l. The mean abundance of female anophelines was significantly higher in the largest island, Mageta, compared to Magare and Ngodhe (χ2 = 193.26, df = 2, p < 0.001) and similar in Magare and Ngodhe (χ2 = 0.56, df = 1, p < 0.453) (Fig. 2a). In Mageta and Ngodhe, the predominant species in terms of relative abundance was An. gambiae s.l., while in Magare, which had the highest LLIN coverage, An. coustani was predominant (Table 2; Fig. 2b).
Table 2.
Distribution of anophelines
| Study area | Species | n (%) | Indoor | Outdoor | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASP | PSC | CDC | CDC | |||||||||||||||
| BF | UF | G | HG | BF | UF | G | HG | BF | UF | G | HG | BF | UF | G | HG | |||
| Mageta | An. gambiae s.l. | 1067 (70.38) | 63 | 35 | 14 | 41 | 384 | 53 | 19 | 81 | 29 | 267 | 31 | 7 | 5 | 35 | 2 | 1 |
| An. funestus | 134 (8.84) | 11 | 1 | 5 | 11 | 27 | 0 | 1 | 8 | 8 | 42 | 7 | 2 | 0 | 10 | 1 | 0 | |
| An. coustani | 315 (20.78) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 5 | 0 | 0 | 35 | 266 | 4 | 0 | |
| Total (%) | 1516 | 182 (12.01) | 573 (37.80) | 402 (26.52) | 359 (23.68) | |||||||||||||
| Magare | An. gambiae s.l. | 20 (28.57) | 0 | 0 | 1 | 0 | 4 | 2 | 0 | 1 | 0 | 11 | 0 | 0 | 0 | 1 | 0 | 0 |
| An. funestus | 22 (31.43) | 0 | 0 | 0 | 0 | 8 | 3 | 1 | 0 | 1 | 8 | 0 | 0 | 0 | 0 | 1 | 0 | |
| An. coustani | 28 (40) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 26 | 0 | 0 | |
| Total (%) | 70 | 1 (1.43) | 19 (27.14) | 20 (28.57) | 30 (42.86) | |||||||||||||
| Ngodhe | An. gambiae s.l. | 193 (68.44) | 11 | 18 | 6 | 3 | 9 | 11 | 12 | 2 | 14 | 60 | 37 | 4 | 2 | 2 | 2 | 0 |
| An. funestus | 62 (21.99) | 1 | 1 | 3 | 3 | 7 | 2 | 1 | 2 | 4 | 18 | 15 | 4 | 0 | 0 | 1 | 0 | |
| An. coustani | 27 (9.57) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 6 | 0 | 0 | 2 | 16 | 2 | 0 | |
| Total (%) | 282 | 46 (16.31) | 46 (16.31) | 163 (57.80) | 27 (9.57) | |||||||||||||
n: number of anophelines; ASP: aspirator; PSC: pyrethrum spray collector; CDC: CDC light trap; BF: blood-fed; UF: unfed; G: gravid; HG: half gravid
Fig. 2.
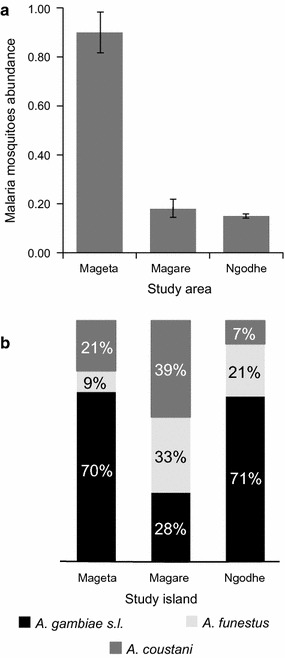
Anopheline abundance and species composition. a Mean abundances per trap per building of anophelines in each of the three study islands. b Relative abundances of malaria vector species by islands
Blood-meal sources of engorged field-collected anopheline mosquitoes
The majority of 632 blood-fed (engorged) anophelines were collected indoors (92.72%, n = 586), while 7.28% (n = 46) were collected outdoors (Table 2). Of the analysed 416 (22.27%) malaria vectors, An. gambiae s.s. (76.92%, n = 320) was the most frequent blood-fed species, followed by An. arabiensis (12.26%, n = 51), An. funestus s.s. (6.97%, n = 29), and An. coustani (3.85%, n = 16) (Table 3).
Table 3.
Number of blood-meal sources of engorged anopheline species
| Study area | Species | n | Vertebrate host | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Chicken | Sheep | Cow | Goat | Pig | Frog | Rat | Dog | Bird | UN | |||
| Mageta | An. gambiae s.s. | 310 | 236.5a | 9a | 1 | 25.5a | 4a | 2 | 1 | 5 | 6 | 1 | 19 |
| An. arabiensis | 40 | 26a | 1 | 4 | 7a | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |
| An. funestus s.s. | 16 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| An. coustani | 8 | 1 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total (%) | 374 | 278.5a (74.47%) | 10a (2.67%) | 5 (1.34%) | 39.5a (10.56%) | 4a (1.07%) | 2 (0.53%) | 1 (0.27%) | 5 (1.34%) | 6 (1.60%) | 1 (0.27%) | 22 (5.88%) | |
| Magare | An. gambiae s.s. | 7 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 |
| An. arabiensis | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| An. funestus s.s. | 8 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |
| An. coustani | 8 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Total (%) | 25 | 10 (40%) | 0 | 0 | 8 (32%) | 0 | 0 | 0 | 1 (4%) | 0 | 0 | 6 (24%) | |
| Ngodhe | An. gambiae s.s. | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| An. arabiensis | 9 | 7.5a | 0 | 0 | 1.5a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| An. funestus s.s. | 5 | 2a | 0 | 0 | 3a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total (%) | 17 | 12.5a (73.53%) | 0 | 0 | 4.5a (26.47%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total | 416 | 297 | 10 | 5 | 53 | 4 | 2 | 2 | 6 | 6 | 1 | 27 | |
n: numbers of engorged anophelines analysed; UN: numbers of blood-meals whose sources identification was not successful
aInclude mixed anophelines blood-meals
Blood-meal sources were identified from 389 Anopheles mosquitoes, representing 93.51% of all analysed engorged anophelines (n = 416). Overall, 10 blood-meal hosts, including humans, goats (Capra hircus), cows (Bos taurus), sheep (Ovis aries), dogs (Canis lupus), domesticated birds (chicken, Gallus gallus), pigs (Sus scrofa), rats (Rattus rattus), grass frogs (Ptychadena nilotica), and a wild bird (Dendrocincla turdina), were identified (Fig. 3; Table 3).
Fig. 3.
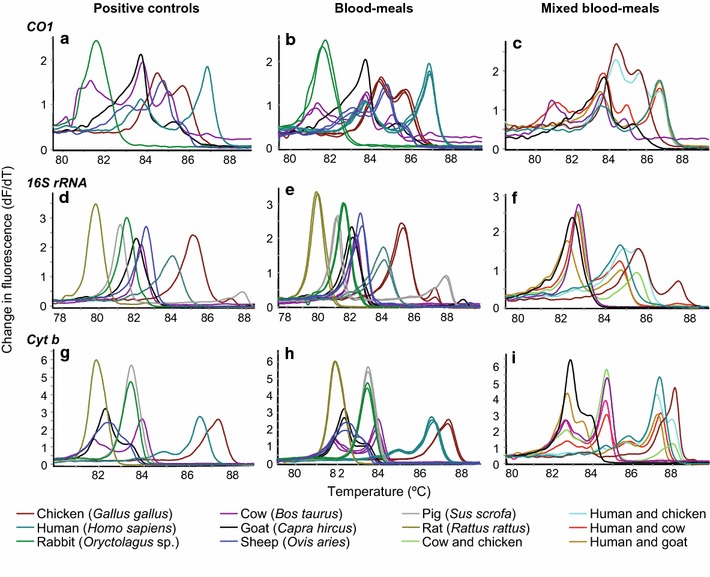
Representative melt rate profiles of anopheline blood-meal sources melt rate profiles of COI a positive controls, b single blood-meals, c mixed blood-meals, 16S rRNA d positive controls, e single blood-meals, f mixed blood-meal and cyt b g positive controls, h single blood-meals; and, i mixed blood-meals
A total of 3.61% of all analysed blood-meals (n = 15) were from mixed blood-meals. In addition to humans, An. gambiae s.s., An. arabiensis and An. funestus had fed on cow, goat or chicken, respectively. Anopheles gambiae s.s. mosquitoes also fed on goat. Mixed blood-meal melt curves showed double peaks with melting temperatures similar to those of more than one positive controls (Fig. 3c, f, i). In Mageta, blood-meal sources were 74.46% human and 19.65% non-human. In Magare, 40% of blood-meal sources were from humans and 36% from other vertebrate species. In Ngodhe, 73.53% of blood-meal sources were from humans and 26.47% from non-humans. Conversely, An. coustani, which were predominantly collected outdoors, showed greater tendency of blood-feeding on cow (Table 3). The range of host species varied in all study areas (Fig. 4).
Fig. 4.
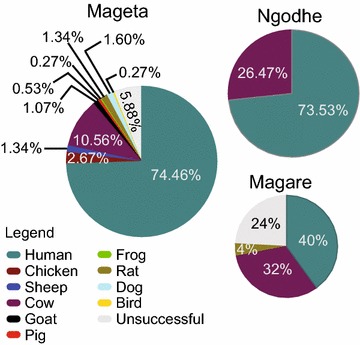
Proportion of blood-meal sources of anophelines. The pie charts show proportions of blood-meal sources among captured anophelines. The range of host species varied in all study areas
Malaria parasite infection in field-collected mosquitoes
The 416 engorged anopheline mosquitoes (320 An. gambiae s.s., 51 An. arabiensis, 29 An. funestus s.s., 16 An. coustani) were further tested for presence of malaria parasites using 18S rRNA [37], and cyt b [38] (Fig. 5). Overall, P. falciparum infection rate was 9.86%. Anopheles gambiae s.s. had an individual P. falciparum infection rate of 10.00%, while the infection rate in An. arabiensis was 11.76%. A small number of engorged An. coustani analysed had a P. falciparum infection rate of 18.75% (Table 4). However, none of the engorged An. funestus samples tested positive for P. falciparum infection. There were no significant differences in P. falciparum infection between the primary malaria vectors An. gambiae s.s., An. arabiensis and An. funestus s.s. or between human fed and non-human fed mosquitoes.
Fig. 5.
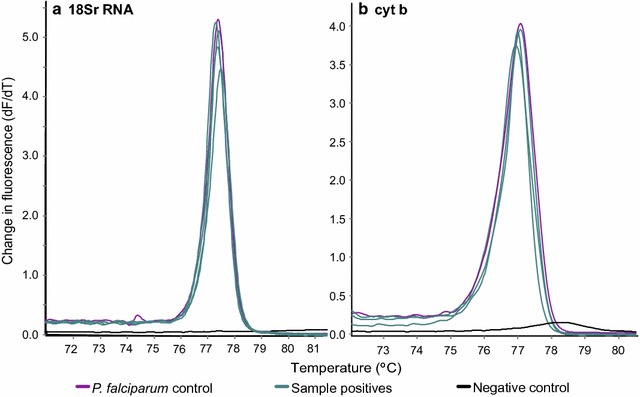
Representative melt rate profiles of Plasmodium falciparum control and field-collected sample positives. Melt rate profiles of a 18S rRNA and b cyt b PCR products. The y-axes indicate change in fluorescence units with increasing temperatures (dF/dT), with temperature shown in the x-axes. The peak melt rates represent empirical melting temperatures (Tm) P. falciparum PCR products
Table 4.
Plasmodium falciparum infection among engorged field collected anophelines
| Study area | Mosquito species | n | HF | NHF | MF | UF | nPfal (%) | ERF (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Mageta | An. gambiae s.s. | 310 | 23 | 4 | 1 | 2 | 30 (9.68) | 0.10 (0.067, 0.13) |
| An. arabiensis | 40 | 3 | 0 | 0 | 1 | 4 (10.00) | 0.11 (0.032, 0.21) | |
| An. funestus s.s. | 16 | 0 | 0 | 0 | 0 | 0 (0.00) | 0.04 (3.9e−06, 0.16) | |
| An. coustani | 8 | 1 | 0 | 0 | 1 | 2 (25.00) | 0.29 (0.055, 0.57) | |
| Total | 374 | 27 | 4 | 1 | 4 | 36 (9.63) | ||
| Magare | An. gambiae s.s. | 7 | 1 | 0 | 0 | 1 | 2 (28.57) | 0.32 (0.059, 0.62) |
| An. arabiensis | 2 | 0 | 0 | 0 | 1 | 1 (50.00) | 0.50 (0.085, 0.89) | |
| An. funestus s.s. | 8 | 0 | 0 | 0 | 0 | 0 (0.00) | 0.07 (5.4e−07, 0.28) | |
| An. coustani | 8 | 0 | 1 | 0 | 0 | 1 (12.50) | 0.18 (0.011, 0.44) | |
| Total | 25 | 1 | 1 | 0 | 2 | 4 (16.00) | ||
| Ngodhe | An. gambiae s.s. | 3 | 0 | 0 | 0 | 0 | 0 (0.00) | 0.16 (5.4e−07, 0.53) |
| An. arabiensis | 9 | 1 | 0 | 0 | 0 | 1 (11.11) | 0.16 (0.0068, 0.39) | |
| An. funestus s.s. | 5 | 0 | 0 | 0 | 0 | 0 (0.00) | 0.11 (3.4e−06, 0.40) | |
| Total | 17 | 1 | 0 | 0 | 0 | 1 (5.88) | ||
| Total (%) | 416 | 29 (6.97) | 6 (1.44) | 1 (0.24) | 5 (1.20) | 41 (9.86) |
n: number of engorged anophelines analysed; nPfal: overall number of engorged anophelines with P. falciparum infection confirmed by 18S and cyt b markers; HF: human-fed engorged anophelines with P. falciparum infection; NHF: non-human fed engorged anophelines with P. falciparum infection; MF: mixed-fed (human and cow) engorged anophelines with P. falciparum infection; UF: unsuccessfully identified blood-meal sources of engorged anophelines with P. falciparum infection; ERF: estimated relative frequency of success; CI: Credibility interval
Discussion
Consistent with previous studies in other parts of the region [10, 11], this study identified four anopheline species, namely An. gambiae s.s., An. arabiensis, An. funestus s.s., and An. coustani. Overall, anophelines were most abundant in Mageta compared to Magare and Ngodhe, likely due to Mageta’s relatively low LLIN coverage. In all islands, the majority of engorged An. gambiae s.s., An. arabiensis and An. funestus s.s. were collected indoors by CDC light, revealing their tendency of feeding indoors, while An. coustani mosquitoes were collected predominantly outdoors. Further, the blood-meals from the species collected indoors were fresh, showing that they rested indoors after feeding [41], predominantly on cattle. Unlike a previous study in East Africa, which showed that An. funestus largely completes its developmental cycle indoors [42], there were fewer gravid anophelines, including An. funestus, collected with the resting traps (ASP and PSC) compared to the blood-fed vectors, indicating that gonotrophic development was completed outdoors.
Mosquitoes feed on potentially diverse hosts [2, 3, 33, 43]. Therefore, it is necessary to establish mosquito blood-feeding patterns to understand malaria transmission dynamics and provide strategies for optimal vector control. Vector control strategies depend largely on LLINs. However, their use is threatened by changes in mosquito behaviour and human behaviour/activities such as night fishing that alter disease transmission dynamics. Therefore, adequate knowledge of vector species and feeding patterns can inform the efficacy of and allow appropriate deployment of other vector control strategies, including LLINs.
Ten blood-meal hosts were identified by matching HRM profiles obtained using COI, 16S rRNA and cyt b genes from engorged abdomens of field-collected mosquitoes to those obtained from standard reference positive controls and verified by sequencing. Although HRM analysis of the three distinct genetic markers could resolve a broad diversity of vertebrate hosts, this study did not detect host DNA from 7.21% of engorged abdomens and there is uncertainty why there were these amplification failures, despite the analysis involving the use of a shorter COI fragment (130-bp) that is suitable for degraded DNA samples from digested blood-meals [44]. The amplification failures could be potentially attributed to much older, degraded blood-meals. Nonetheless, this study demonstrates that anopheline mosquitoes in these small islands have a wide host range, able to sustain mosquito populations, and decrease transmission intensity. However, in the largest island, Mageta, LLIN coverage was low, allowing for increased human feeding indoors by An. gambiae s.s., An. arabiensis, and An. funestus s.s, and contributing to transmission.
Overall, the data also show that An. coustani mosquitoes fed predominantly on cattle outdoors. In An. gambiae s.s, humans were the most common source of blood-meals; however, blood-meal sources also included diverse non-human hosts. Blood-meals from both An. arabiensis and An. funestus collected indoors were more substantially from humans, unlike in a study in Mwea which found significantly higher bovine feeding among indoor-collected An. arabiensis. Indoor-collected An. funestus also preferentially fed on humans [45]. Although a human blood-meal was identified in one An. coustani mosquito, which was also P. falciparum positive, this species preferred feeding outdoors on cows as previously shown [46].
Anopheline feeding patterns depend on the density and diversity of host species [47], which by their availability form readily accessible blood-meal sources. While PSC and ASP traps enabled us to understand anophelines’ endophily, by placing CDC traps indoors and outdoors that target host-seeking anophelines, this study aimed to maximize the recovery of data on feeding patterns (exophagy versus endophagy) to enhance understanding of localized anopheline feeding dynamics. Although the majority of trapping was done indoors, this study found that the range of host species can vary depending on study areas. Mageta had the highest numbers of engorged anophelines and broadest range of host species. In contrast, Ngodhe had the fewest engorged anopheline and narrowest range of host species as determined by PCR-HRM. The range of host species did not differ significantly in Magare compared to Ngodhe, perhaps reflecting lower vertebrate species diversity on these small islands compared to Mageta.
The blood-meal identification approach in this study allowed for high sensitivity identification of mixed blood-meals from individual mosquitoes. Mixed blood-meal sources could have been a result of blood-feeding anophelines resuming blood-feeding on a different host in an effort to complete an unsuccessful blood-meal, a characteristic that is common with anophelines infected with sporozoite-stage malaria parasites [48, 49]. The finding that mixed feeding on vertebrate hosts included blood-meals from cow, chicken and goat further confirmed malaria vectors feeding on readily available blood-meal sources [47], underlining the economic activities of the study population, which provide other blood-meal sources important for vector survival. However, the analysis was not able to differentiate multiple hosts of the same species within individual mosquito, blood-meals such as in The Gambia where one malaria vector was suspected to have taken blood-meals from children sharing rooms on one night [50]; this would require more extensive genotyping of blood-meals.
Overall, 9.86% of engorged anophelines harboured malaria parasites. There was no significant difference among engorged anopheline species in terms of P. falciparum infection rates among all four species or among the three islands. Further, neither island sizes nor blood-meal sources influenced P. falciparum infection rates in the vector. However, the vector species play an important role in malaria transmission by harbouring malaria parasites. These findings suggest that human-mosquito contact is still very frequent in the islands and has not been well controlled, therefore presenting a risk of malaria transmission.
Conclusions
This study shows that on the small remote islands of Mageta, Magare and Ngodhe in Kenya’s Lake Victoria, anopheline mosquitoes are maintained by humans and, in part, by other blood-meal hosts. Among these islands, only Mageta had below WHO-recommended LLIN coverage, and significantly higher Anopheles mosquito abundances. The availability of alternative blood-meal sources not affected by LLINs can lead to opportunistic feeding events that present a challenge to malaria control efforts. This study indicates that there is an urgent need to target indoor-feeding mosquito populations by achieving full LLIN coverage. In addition, this is the first report of An. coustani with sporozoite infection in the area, highlighting the need to target secondary vectors with a more exophilic behaviour.
Authors’ contributions
EO, JV, JM, and DKM conceived and designed the study; EO, DM, DO, JM, and DKM supervised the field activities and conducted the research. EO, JV, BO, and DKM analysed the data; EO, JV, JM, VO, and DKM wrote and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We gratefully acknowledge the technical contribution of Daniel Ouso, Geoffrey Jagero, Sharon Towett, Enock Mararo and Lilian Mbaisi of icipe’s ML-EID laboratory, and Collins Omogo and Simon Ngao of Molecular Biology and Bioinformatics Unit. We are grateful to logistical support of Lillian Igweta, Lisa Omondi, and Margaret Ochanda of the Capacity Building Unit at icipe and Esther Waweru of Molecular Biology and Bioinformatics Unit.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data and materials are available upon request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The present study was approved by the Kenya Medical Research Institute (KEMRI) Ethics Review Committee (Non-SSC Protocol #388 and Non-SSC Protocol #310). Informed consent was obtained from village elders on the study activities and from household heads before inclusion of their households in the study.
Funding
This work was financially supported by Training Health Researchers into Vocational Excellence (THRiVE) in East Africa (Grant Number 087540) funded by Wellcome Trust; the UK’s Department for International Development (DFID); the Swedish International Development Cooperation Agency (Sida); the Swiss Agency for Development and Cooperation (SDC); the Kenyan Government; USAID-PEER (AID-OAA-A-11-00012) National Academy of Sciences sub-grant award (2000006204) and Wellcome Trust Fellowship (Grant Number 101166/Z/13/Z). The funding bodies did not play a role in the design of this study, the collection, analyses and interpretation of data, the writing of the manuscript, or decision to submit the manuscript for publication.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CI
credibility interval
- ERF
estimated relative frequency of success
- HRM
high-resolution melting
- KEMRI
Kenya Medical Research Institute
- LLINs
long-lasting insecticides nets
- nPCR-HRM
nested polymerase chain reaction-high-resolution melting
- PCR
polymerase chain reaction
- PSC
pyrethrum spray catches
- WHO
World Health Organization
Contributor Information
Edwin Ogola, Email: edwinogola@gmail.com.
Jandouwe Villinger, Email: jandouwe@icipe.org.
Danspaid Mabuka, Email: mabukadan@gmail.com.
David Omondi, Email: omondiouma08@gmail.com.
Benedict Orindi, Email: benedict.orindi@gmail.com.
James Mutunga, Email: mutungajames@gmail.com.
Vincent Owino, Email: vowino@gmail.com.
Daniel K Masiga, Email: dmasiga@icipe.org.
References
- 1.Cooke MK, Kahindi SC, Oriango RM, Owaga C, Ayoma E, Mabuka D, et al. “A bite before bed”: exposure to malaria vectors outside the times of net use in the highlands of western Kenya. Malar J. 2015;14:259. doi: 10.1186/s12936-015-0766-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ototo EN, Mbugi JP, Wanjala CL, Zhou G, Githeko AK, Yan G. Surveillance of malaria vector population density and biting behaviour in western Kenya. Malar J. 2015;14:244. doi: 10.1186/s12936-015-0763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ndenga BA, Mulaya NL, Musaki SK, Shiroko JN, Dongus S, Fillinger U. Malaria vectors and their blood-meal sources in an area of high bed net ownership in the western Kenya highlands. Malar J. 2016;15:76. doi: 10.1186/s12936-016-1115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Njoroge MM, Tirados I, Lindsay SW, Vale GA, Torr SJ, Fillinger U. Exploring the potential of using cattle for malaria vector surveillance and control: a pilot study in western Kenya. Parasit Vectors. 2017;10:18. doi: 10.1186/s13071-016-1957-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olanga EA, Okombo L, Irungu LW, Mukabana WR. Parasites and vectors of malaria on Rusinga Island, western Kenya. Parasit Vectors. 2015;8:250. doi: 10.1186/s13071-015-0860-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Homan T, Hiscox A, Mweresa CK, Masiga D, Mukabana WR, Oria P, et al. The effect of mass mosquito trapping on malaria transmission and disease burden (SolarMal): a stepped-wedge cluster-randomised trial. Lancet. 2016;388:1193–1201. doi: 10.1016/S0140-6736(16)30445-7. [DOI] [PubMed] [Google Scholar]
- 7.Minakawa N, Sonye G, Dida GO, Futami K, Kaneko S. Recent reduction in the water level of Lake Victoria has created more habitats for Anopheles funestus. Malar J. 2008;7:119. doi: 10.1186/1475-2875-7-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ndenga BA, Simbauni JA, Mbugi JP, Githeko AK, Fillinger U. Productivity of malaria vectors from different habitat types in the western Kenya highlands. PLoS ONE. 2011;6:e19473. doi: 10.1371/journal.pone.0019473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Githinji S, Noor AM, Malinga J, Macharia PM, Kiptui R, Omar A, et al. A national health facility survey of malaria infection among febrile patients in Kenya, 2014. Malar J. 2016;15:591. doi: 10.1186/s12936-016-1638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minakawa N, Dida GO, Sonye GO, Futami K, Njenga SM. Malaria vectors in Lake Victoria and adjacent habitats in western Kenya. PLoS ONE. 2012;7:e32725. doi: 10.1371/journal.pone.0032725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCann RS, Messina JP, MacFarlane DW, Bayoh MN, Vulule JM, Gimnig JE, et al. Modeling larval malaria vector habitat locations using landscape features and cumulative precipitation measures. Int J Health Geogr. 2014;13:17. doi: 10.1186/1476-072X-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zurovac D, Githinji S, Memusi D, Kigen S, Machini B, Muturi A, et al. Major improvements in the quality of malaria case-management under the “test and treat” policy in Kenya. PLoS ONE. 2014;9:e92782. doi: 10.1371/journal.pone.0092782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO. T3: Test. Treat. Track initiative. Geneva: World Health Organization; 2016. http://www.who.int/malaria/areas/test_treat_track/en/.
- 14.Ter Kuile FO, Terlouw DJ, Phillips-Howard PA, Hawley WA, Friedman JF, Kolczak MS, et al. Impact of permethrin-treated bed nets on malaria and all-cause morbidity in young children in an area of intense perennial malaria transmission in western Kenya: cross-sectional survey. Am J Trop Med Hyg. 2003;68:100–107. [PubMed] [Google Scholar]
- 15.Phillips-Howard PA, Nahlen BL, Kolczak MS, Hightower AW, Ter Kuile FO, Alaii JA. Efficacy of permetrin-treated bed nets in the prevention of mortality in young children in an area of high perennial malaria transmission in western Kenya. Am J Trop Med Hyg. 2003;68:23–29. [PubMed] [Google Scholar]
- 16.Lim SS, Fullman N, Stokes A, Ravishankar N, Masiye F, Murray CJL, et al. Net benefits: a multicountry analysis of observational data examining associations between insecticide-treated mosquito nets and health outcomes. PLoS Med. 2011;8:e1001091. doi: 10.1371/journal.pmed.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO . Eliminating malaria. Geneva: World Health Organization; 2016. [Google Scholar]
- 18.Crowell V, Hardy D, Briët O, Chitnis N, Maire N, Smith T. Can we depend on case management to prevent re-establishment of P. falciparum malaria, after local interruption of transmission? Epidemics. 2012;4:1–8. doi: 10.1016/j.epidem.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Smith DL, Cohen JM, Chiyaka C, Johnston G, Gething PW, Gosling R, et al. A sticky situation: the unexpected stability of malaria elimination. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120145. doi: 10.1098/rstb.2012.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bousema T, Stresman G, Baidjoe AY, Bradley J, Odongo W, Shagari S, et al. The impact of hotspot-targeted interventions on malaria transmission in Rachuonyo south district in the western Kenyan highlands: a cluster-randomized controlled trial. PLoS Med. 2016;13:e1001993. doi: 10.1371/journal.pmed.1001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowden J, Morris MG. The influence of moonlight on catches of insects in light-traps in Africa. III. The effective radius of a mercury-vapour light-trap and the analysis of catches using effective radius. Bull Entomol Res. 1975;65:303–348. doi: 10.1017/S000748530000599X. [DOI] [Google Scholar]
- 22.Moon phase calendar for any location and year. http://www.timeanddate.com/moon/phases/. Accessed 29 Feb 2016.
- 23.O’Guinn ML, Turell MJ. Effect of triethylamine on the recovery of selected south american alphaviruses, flaviviruses, and bunyaviruses from mosquito (Diptera: Culicidae) pools. J Med Entomol. 2002;39:806–808. doi: 10.1603/0022-2585-39.5.806. [DOI] [PubMed] [Google Scholar]
- 24.Gillies M, Coetzee M. A supplement to the anophelinae of Africa South of the Sahara. Publ South African Inst Med Res. 1987;55:63. [Google Scholar]
- 25.WHO Division of Malaria and Other Parasitic Diseases . Manual on pratical entomology in malaria. Geneva: World Health Organization; 1975. [Google Scholar]
- 26.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 27.Koekemoer LL, Kamau L, Hunt RH, Coetzee M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am J Trop Med Hyg. 2002;66:804–811. doi: 10.4269/ajtmh.2002.66.804. [DOI] [PubMed] [Google Scholar]
- 28.Cohuet A, Simard F, Toto JC, Kengne P, Coetzee M, Fontenille D. Species identification within the Anopheles funestus group of malaria vectors in Cameroon and evidence for a new species. Am J Trop Med Hyg. 2003;69:200–205. [PubMed] [Google Scholar]
- 29.Zianni MR, Nikbakhtzadeh MR, Jackson BT, Panescu J, Foster WA. Rapid discrimination between Anopheles gambiae s.s. and Anopheles arabiensis by high-resolution melt (HRM) analysis. J Biomol Tech. 2013;24:1–7. doi: 10.7171/jbt.13-2401-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ajamma YU, Mararo E, Omondi D, Onchuru T, Muigai AWT, Masiga D, et al. Rapid and high throughput molecular identification of diverse mosquito species by high resolution melting analysis. F1000Research. 2016;5:1949. doi: 10.12688/f1000research.9224.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Omondi D, Masiga D, Ajamma Y, Fielding B, Njoroge L, Villinger J. Unraveling host-vector-arbovirus interactions by two-gene high resolution melting mosquito bloodmeal analysis in a Kenyan wildlife-livestock interface. PLoS ONE. 2015;10:e0134375. doi: 10.1371/journal.pone.0134375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boakye DA, Tang J, Truc P, Merriweather A, Unnasch TR. Identification of bloodmeals in haematophagous Diptera by cytochrome B heteroduplex analysis. Med Vet Entomol. 1999;13:282–287. doi: 10.1046/j.1365-2915.1999.00193.x. [DOI] [PubMed] [Google Scholar]
- 35.Peña VH, Fernández GJ, Gómez-Palacio AM, Mejía-Jaramillo AM, Cantillo O, Triana-Chávez O. High-resolution melting (HRM) of the cytochrome B gene: a powerful approach to identify blood-meal sources in Chagas disease vectors. PLoS Negl Trop Dis. 2012;6:e1530. doi: 10.1371/journal.pntd.0001530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lutomiah J, Omondi D, Masiga D, Mutai C, Mireji PO, Ongus J. Blood meal analysis and virus detection in blood-fed mosquitoes collected during the 2006–2007 Rift Valley fever outbreak in Kenya. Vector-Borne Zoonotic Dis. 2014;14:656–664. doi: 10.1089/vbz.2013.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kipanga PN, Omondi D, Mireji PO, Sawa P, Masiga DK, Villinger J. High-resolution melting analysis reveals low Plasmodium parasitaemia infections among microscopically negative febrile patients in western Kenya. Malar J. 2014;13:429. doi: 10.1186/1475-2875-13-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fornadel CM, Norris LC, Franco V, Norris DE. Unexpected anthropophily in the potential secondary malaria vectors Anopheles coustani s.l. and Anopheles squamosus in Macha, Zambia. Vector-Borne Zoonotic Dis. 2011;11:1173–1179. doi: 10.1089/vbz.2010.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Development Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2010. http://www.R-project.org. p. 38.
- 40.Bååth R. Bayesian first aid: a package that implements bayesian alternatives to the classical test functions in R. Proc UseR. 2014;33:2. [Google Scholar]
- 41.Faye O, Konate L, Mouchet J, Fontenille D, Sy N, Hebrard G, Herve JP. Indoor resting by outdoor biting females of Anopheles gambiae complex (Diptera:Culicidae) in the Sahel of northern Senegal. J Med Entomol. 1997;34:285–289. doi: 10.1093/jmedent/34.3.285. [DOI] [PubMed] [Google Scholar]
- 42.Gillies MT. Studies in house leaving and outside resting of Anopheles gambiae Giles and Anopheles funestus Giles in East Africa. II. The exodus from houses and the house resting population. Bull Entomol Res. 1954;45:361–373. doi: 10.1017/S0007485300027188. [DOI] [Google Scholar]
- 43.Nanda N, Joshi H, Subbarao SK, Yadav RS, Shukla RP, Dua VK, et al. Anopheles fluviatilis complex: host feeding patterns of species S, T, and U. J Am Mosq Control Assoc. 1996;12:147–149. [PubMed] [Google Scholar]
- 44.Meusnier I, Singer GA, Landry J-F, Hickey DA, Hebert PD, Hajibabaei M. A universal DNA mini-barcode for biodiversity analysis. BMC Genom. 2008;9:214. doi: 10.1186/1471-2164-9-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muriu SM, Muturi EJ, Shililu JI, Mbogo CM, Mwangangi JM, Jacob BG, et al. Host choice and multiple blood feeding behaviour of malaria vectors and other anophelines in Mwea rice scheme, Kenya. Malar J. 2008;7:43. doi: 10.1186/1475-2875-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mwangangi JM, Muturi EJ, Muriu SM, Nzovu J, Midega JT, Mbogo C. The role of Anopheles arabiensis and Anopheles coustani in indoor and outdoor malaria transmission in Taveta District, Kenya. Parasit Vectors. 2013;6:114. doi: 10.1186/1756-3305-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lardeux F, Loayza P, Bouchité B, Chavez T. Host choice and human blood index of Anopheles pseudopunctipennis in a village of the Andean valleys of Bolivia. Malar J. 2007;6:8. doi: 10.1186/1475-2875-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koella JC. Stage-specific manipulation of a mosquito’s host-seeking behavior by the malaria parasite Plasmodium gallinaceum. Behav Ecol. 2002;13:816–880. doi: 10.1093/beheco/13.6.816. [DOI] [Google Scholar]
- 49.Cator LJ, Lynch PA, Read AF, Thomas MB. Do malaria parasites manipulate mosquitoes? Trends Parasitol. 2012;28:467–470. doi: 10.1016/j.pt.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindsay S. Malaria in a peri-urban area of The Gambia. Ann Trop Med Parasitol. 1990;84:553–562. doi: 10.1080/00034983.1990.11812510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials are available upon request.


