Abstract
Genome editing using the CRISPR/Cas9 RNA guided endonuclease system has rapidly become a driving force for discovery in modern biomedical research. This simple yet elegant system has been widely used to generate both loss-of-function alleles and precision knock-in mutations using single stranded donor oligonucleotides. Our CRISPRtools platform supports both of these applications in order to facilitate the use of CRISPR/Cas9. While there are several tools that facilitate CRISPR/Cas9 design and screen for potential off-target sites, the process is typically performed sequentially on single genes, limiting scalability for large-scale programs. Here, the design principle underlying gene ablation is based upon using paired guides flanking a critical region/exon of interest to create deletions. Guide pairs are rank ordered based upon published efficiency scores and off-target analyses, and reported in a concise format for downstream implementation. The exon deletion strategy simplifies characterization of founder animals and is the strategy employed for the majority of knockouts in the mouse. In proof-of-principle experiments, the effectiveness of this approach is demonstrated using microinjection and electroporation to introduce CRISPR/Cas9 components into mouse zygotes to delete critical exons.
Keywords: CRISPR, gene editing, mouse knockout, IMPC
Introduction
The adaption of the type II CRISPR/Cas system from Streptococcus pyogenes (S. py) to perform genome editing has opened up new avenues of research and increased the rate of animal model production (Cong et al. 2013; Jinek et al. 2012; Mali et al. 2013; Wang et al. 2013). This two component RNA guided endonuclease (RGN) system relies on a single guide RNA (sgRNA) of 18–20 nucleotides in length that can complex with Cas9 protein to target specific regions of the genome and induce double strand breaks(Cong et al. 2013; Jinek et al. 2012). These breaks are repaired imprecisely by the non-homologous end joining (NHEJ) complex resulting in small insertion or deletion events (indels) that can lead to mutagenic lesions impairing gene function. In addition to these applications, CRISPR/Cas9 has also been shown to enhance homology directed gene targeting in zygotes when Cas9 mRNA is co-injected with sgRNA and a donor DNA repair template (Chu et al. 2016; Richardson et al. 2016; Yang et al. 2013). A potential limitation of the CRISPR/Cas9 system is that regions targeted by the sgRNA must be adjacent to a protospacer adjacent motif (PAM) sequence in the form of NGG, thereby limiting the regions of the genome that can be targeted by S. py Cas9. However, despite this limitation, Cas9 has proven to be a versatile tool spanning a wide-range of applications from modulating gene expression to enhanced cell imaging (Chen et al. 2013; Cheng et al. 2013; Gilbert et al. 2013; Qi et al. 2013).
These advancements in CRISPR/Cas9 technology have been accompanied by the development of a number of computational tools that enable guide prediction, scoring for off-target sites and calculation of efficiency scores (Bae et al. 2014; Cradick et al. 2014; Doench et al. 2016; Doench et al. 2014; Heigwer et al. 2014; Hsu et al. 2013; Labun et al. 2016; Moreno-Mateos et al. 2015; Oliveros et al. 2016). Several different metrics have been introduced to weigh off-target predictions (Bae et al. 2014; Cradick et al. 2014; Hsu et al. 2013) and guide efficiencies have been evaluated both in vitro and in vivo (Doench et al. 2016; Doench et al. 2014; Moreno-Mateos et al. 2015). A limitation of the current tool set is that most focus on the prediction of single guides to impair gene function via the production of frame-shifting indels. However, these mutations are often difficult and expensive to maintain due to the challenges associated with genotyping small indels. In order to address these challenges, we set out to design a tool that would support a comprehensive analysis of putative guides and provide final a report of recommended guide combinations to create deletions using CRISPR/Cas9.
CRISPRtools provides a generalized and simplified approach to generate null alleles in mouse using CRISPR/Cas9 by designing deletions that remove internal coding regions or critical exons. This design approach follows the same rationale underlying traditional conditional allele design such that data from animal models generated using this approach can be directly compared to previously made alleles. Additionally, the deletions are a cost-effective way to generate new animal models that can be maintained using conventional genotyping approaches. The CRISPRtools platform supports alternative sgRNA scoring options and can be easily extended to other organisms. In addition to exon deletion alleles, we provide a guide ranking system for performing nucleotide substitution studies using CRISPR/Cas9. The feasibility of this approach is demonstrated by generating whole exon deletions using both microinjection and electroporation to perform genome editing in zygotes obtained from inbred mice.
Materials and Methods
Critical exon identification and genome annotation
For multi-exon genes, critical exons were determined by manual curation to be the most 5′ exon common to all isoforms that when removed will result in a frameshift. Ensembl exon identifiers or genomic coordinates are used to indicate the critical exon or region to be deleted. Gene annotations are obtained from Ensembl and locally stored in a SQLite database. The current version of CRISPRtools supports mouse genome build GRCm38/mm10.
Guide identification, ranking and deletion design
Guide sequences are identified by searching the defined sequence region for all 20 base pair (bp) sequences adjacent to a PAM, NGG. Each guide is then queried against a locally running instance of ots_server, which is provided as part of the Sanger CRISPR-Analyser tool (https://github.com/htgt/CRISPR-Analyser) to perform off-target analysis. Next, we calculate efficiency scores for each guide as described (Doench et al. 2014; Moreno-Mateos et al. 2015). For whole exon deletions, guide pairs are then identified within the upstream and downstream region flanking the exon by first filtering on the mismatch score with the criteria of [0:1, 2:0, 1:0]. This condition selects for uniquely mapping guides and eliminates guides that map to alternative loci with one or two mismatches, but allows guides that have three or more mismatches to potential off-target sites. Guides passing this filter can then be analyzed to determine if there are any linked off-target hits defined by a 10 megabase (Mb) interval centered on the on-target guide. This search can be limited to linked off-targets within exons. The remaining guides are then ranked by a user selected efficiency score and analyzed for overlap with each other and neighboring exons. Guides that overlap a nearby exon are also removed. The final deletion design is determined by selecting the top two non-overlapping guides within the upstream and downstream region. For internal exon deletions, the same criteria are applied as described above with the exception that the distance between guides meeting the mismatch criteria is maximized.
sgRNA guide synthesis
Individual guides were synthesized as Ultramers (IDT) with the guide sequence embedded between the T7 promoter and portion of stem loop as described (Bassett et al. 2013). DNA templates were generated via overlapping PCR by pooling all forward oligos with a universal reverse primer as described in (Bassett et al. 2013) (Table S1). DNA templates were column purified (Qiagen) prior to in vitro transcription reaction. sgRNA pools were column purified (Zymogen) and quantified via Nanodrop prior to microinjection.
Zygote microinjection and electroporation
All procedures performed in studies involving animals were in accordance with the ethical standards of The Jackson Laboratory Animal Care and Use Committee (ACUC protocol 99066). One-cell embryos were obtained from superovulated C57BL/6NJ (B6NJ; JAX stock number 5304) female donors crossed to B6NJ males. For microinjection, reagents were injected into the cytoplasm at the following concentrations (100 ng/μl Cas9 mRNA (Trilink); 200 ng/μl sgRNA pool) or (250 ng/μl Cas9 protein (ThermoFisher); 200 ng/μl sgRNA pool). For electroporation, guides were diluted into microinjection buffer at a concentration of 2400 ng/ul and mixed with Cas9 mRNA (1200 ng/ul). The final prep was diluted with an equal volume of opti-MEM (Thermofisher) prior to electroporation. Embryos were processed in batches of twenty and electroporated as previously described (Wang et al. 2016). Ribonucleoprotein complexes were generated by mixing Cas9 protein with sgRNAs and incubating for 15 minutes at 37°C prior to electroporation or microinjection.
Genotyping
DNA was obtained from tail tip biopsies extracted in 25 mM NaOH/0.2 mM EDTA at 95°C for 10 minutes and neutralized with equal volume of 40 mM Tris-HCl (pH5.5). PCR primers were designed to be at least 150 bp away from the guide cut sites. Gene specific PCR amplicons were generated using standard conditions and confirmed using Sanger sequencing. Primers used in this study are as listed with expected product sizes for wild-type alleles indicated in parentheses: Rab6b-F AGGGTGAAGAAAGGGTAGGC; Rab6b-R GCCCCTTGACATAACCAAACT (614 bp); Neil2-F GCTCATCATTAATCCATCTCCTG; Neil2-R GCCTTTCTTCCGAGAGAGC (847 bp); Pcnx2-F TCTCGTAACGTCCTCAGTAACG; Pcnx2-R CACACATTGGCCTGGAACT (805 bp).
Results
Automated workflow for generating exon deletions
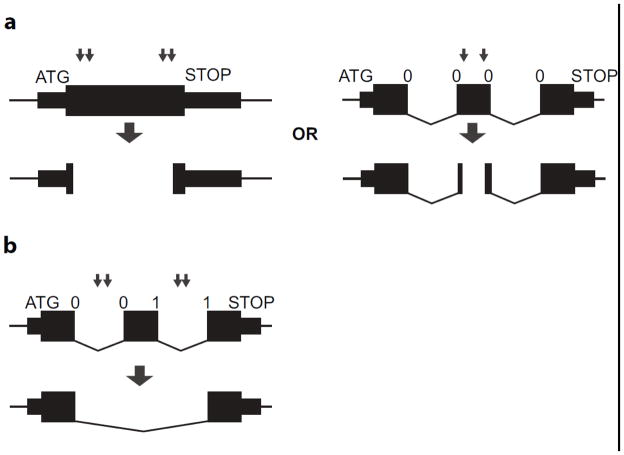
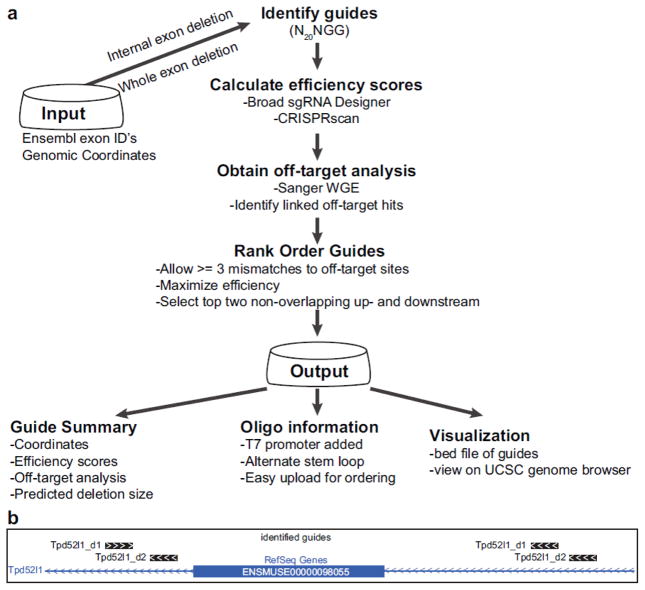
CRISPRtools supports two general design strategies depending on the gene structure: (1) internal exon deletions; and (2) whole exon deletions (Fig. 1). Internal exon deletions provide access to difficult gene models such as single exon genes or gene models where all exons are in the same phase. Whole exon deletions are designed to remove a critical exon resulting in a frameshift followed by degradation via the nonsense mediated decay (NMD) pathway. The whole exon deletion strategy most closely mimics conditional allele designs that flank the critical exon with recombination recognition sites (e.g. loxP or frt). The minimal deletion region is set by default to be greater than 50 bp in order to facilitate ease of genotyping using standard PCR followed by gel electrophoresis. These design principles are implemented as an automated pipeline to enable batch designs that takes as input a list of Ensembl mouse exon identifiers or genomic sequence coordinates and returns all of the required components for performing a downstream CRISPR/Cas9 experiment (http://crispr-tools.jax.org) (Fig. 2a,b and Online Resource 1).
Figure 1.
Hypothetical gene models illustrating deletion strategy for producing null alleles using paired CRISPR guides flanking the region of interest. (a) Two examples of internal exon deletions are shown to indicate removal of large portion of coding sequence for single exon gene. The other example shows an internal deletion within an exon for a multi-exon gene that has all exons in the same phase. (b). Paired guides flank a critical exon within a multi-exon gene that when deleted will result in a frameshift mutation. Exon phases (0,1,2) are indicated above the gene model for multi-exon genes and guide cut sites are shown as arrows.
Figure 2.
CRISPRtools workflow for designing null alleles. (a) Critical exon identifiers or genomic coordinates are used as input and rank ordered guides are provided as output with a series of summary files containing guide positions, scores, expected deletion sizes, oligo information, and a bed file for visualization on genome browser. (b) Stylized representation of UCSC genome browser view illustrating exon deletion strategy using four guides.
Internal exon deletions are suitable for removing a large portion of the coding region from single exon genes or small genes that have the majority of coding sequence contained within a single exon. In this mode, the user can specify the amount of overlap allowed with the neighboring UTR or intron to maximize the removal of the majority of the coding regions since single exon genes are not targeted by the NMD pathway (Maquat and Li 2001). In addition to single exon genes, internal exon deletions are suitable for genes that have all exons in the same phase since removal of an exon will not result in frameshift. Here, the strategy is to generate a deletion within a downstream exon that will remove critical coding sequences while at the same time preserve normal splicing. Whole exon deletions are designed by selecting pairs of guides flanking a critical exon within a user defined distance upstream and downstream. Our program supports single exon deletions as well as paired exon deletions to remove two closely spaced exons.
In both cases, all possible guide sequences adjacent to a PAM motif (NGG) are identified and subsequently filtered using specificity and efficiency scores. Guide sequences are first evaluated for their potential off-target cutting using the mismatch scores determined by the Sanger CRISPR-Analyser (Hodgkins et al. 2015). Only uniquely mapping guides with a mismatch tolerance set to allow guides that have three or more mismatches to off-target sites are considered as possible candidates. To minimize potential off-target effects, additional filtering options are provided to remove guides that are linked within 5 Mb on either side of the on-target guide and overlap an exon. This separation distance helps to reduce the risk that an off-target hit will segregate with the desired mutation in subsequent generations.
Next, guide efficiency scores are assigned to all guides using two alternative scoring matrices (Doench et al. 2014; Moreno-Mateos et al. 2015). A final set of rank ordered guides is then generated by selecting the top two non-overlapping guides upstream and downstream of the target region that meet the mismatch criteria and are ordered by their efficiency scores as determined by user selected scoring algorithm. Overlapping guides are reported when no other suitable guides are found within the search region. By default, the resulting KO designs all contain four guides consisting of a pair of upstream and downstream guides to increase the likelihood of generating the desired deletion. However, this restriction can be relaxed in order to allow designs that have fewer than two guides on either side to pass filtering.
The final output contains a results file that includes the selected guide sequences and their associated mismatch scores, efficiency scores, genomic position, expected deletion sizes and region of interest. Additionally, guides selected in the final design are provided as a bed file for easy viewing on a genome browser. Each guide is also reported as an oligo sequence with a prepended T7 sequence and sgRNA stem loop appended for use with a universal reverse primer to generate a DNA template via overlapping PCR that can be used to make the sgRNA (Bassett et al. 2013). A message log is included to report exons that failed to meet the filtering criteria and a list of linked off-targets. Lastly, the CRISPRtools also supports the use of an alternative stem loop previously reported to increase sgRNA stability (Chen et al. 2013).
Guide identification and ranking for nucleotide substitution studies
CRISPRtools supports guide prioritization and ranking for guides closest to a genomic position of interest. This functionality assists in choosing guides when performing a knock-in experiment such as introducing a single-nucleotide polymorphism (SNP) using CRISPR/Cas9. Guides within +/− 50 bp of the point of interest are ordered and classified according to their position relative to the desired insertion site and predicted efficiency scores are provided. Entries to the program are made in the form of Sirt2,chr7:28776872, T>C, where the target gene, location and desired substitution are provided by the user.
Efficient production of deletions in the mouse
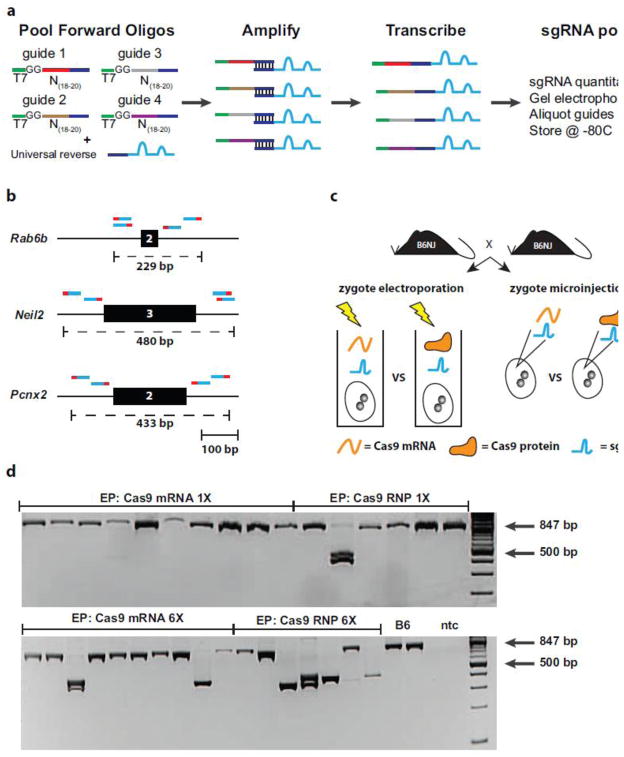
To test our design algorithm, we generated whole exon deletions within three target genes selected for production in the Knockout Mouse Phenotyping Program (KOMP2) (Bradley et al. 2012; Skarnes et al. 2011). For each target gene, we generated sgRNA pools by parallel amplification of all four DNA templates in a single reaction followed by in vitro transcription (Fig. 3a; Table S1). This method significantly reduces reagent costs and enables scalable production of sgRNA pools suitable for large-scale projects. The range of exon deletion sizes ranged from ~250–500 bp (Fig. 3b and Table S2; Online Resources 2–4).
Figure 3.
Strategy for generating exon deletions in mouse zygotes. (a) Individual oligos containing T7 promoter followed by the guide sequence are pooled with a universal reverse primer for parallel amplification via PCR. The DNA template pool is subsequently purified and used as input for in vitro transcription reaction. (b) Schematic showing guide positions relative to critical exon (black box) for selected target genes. Paired guides flanking the critical exon are shown above with PAM colored as red to indicate orientation. The expected maximal deletion is shown below as a dashed line. (c) Comparison of delivery method and Cas9 biological form into C57BL/6NJ zygotes. (d) Representative PCR genotyping results for Neil2 showing increased deletion frequency with increased pulse number. Two B6 control DNA samples and no template control (ntc) are included.
We performed a series of side-by-side comparisons using our guide pools to determine the impact of Cas9 (protein versus mRNA) as well as the method for introducing the reagents-- microinjection as compared to zygote electroporation (Fig. 3c; Table 1) (Qin et al. 2015; Wang et al. 2016). For electroporation, two different protocols were also tested that varied the number of times the program was run such that 1X delivered 2 pulses (Qin et al. 2015) and 6X delivered 12 pulses (Wang et al. 2016). Consistently, increasing the pulse number increased the level of mutations with Cas9 protein showing higher levels of activity when compared to mRNA for 2/3 target genes (Fig. 3d). These findings are in agreement with a recent report demonstrating increased efficiency with protein and higher pulse numbers (Wang et al. 2016). The only gene that was refractory to deletions using electroporation was Pcnx2 although there were founders carrying indels. Microinjection into the cytoplasm of zygotes produced the highest mutation rate and successfully generated exon deletions for all three target genes with the highest deletion rate observed for Rab6b. There were several founders identified for Rab6b that carried bi-allelic deletions of varying size with most variability occurring at the boundaries; however, there were also cases where the guides flanking the exon cut individually creating small indels that did not result in removal of the critical exon (Online Resource 5). Interestingly, there was no clear difference when comparing Cas9 protein versus mRNA when introduced via microinjection. In summary, these data show that our automated design workflow, combined with our pooled sgRNA production scheme, can efficiently produce exon deletion alleles in a scalable framework that can support the high-throughput needs large scale genome modification efforts, like the KOMP2 program.
Table 1.
Summary of exon deletions generated by Cas9 RGN using electroporation and microinjection
| Target | Condition1 | Cas9 | Embryos | Live Born | NHEJ2 | Deletions |
|---|---|---|---|---|---|---|
| Pcnx2 (433 bp del) | EP-1X | mRNA | 80 | 34 | 2 (6%) | 0 (0%) |
| EP-6X | mRNA | 60 | 21 | 4 (19%) | 0 (0%) | |
| EP-1X | Protein | 80 | 31 | 1 (3%) | 0 (0%) | |
| EP-6X | Protein | 60 | 12 | 0 (0%) | 0 (0%) | |
| MIJ | mRNA | 75 | 18 | 8 (44%) | 7 (39%) | |
| MIJ | Protein | 70 | 21 | 8 (38%) | 7 (33%) | |
| Neil2 (481 bp del) | EP-1X | mRNA | 60 | 27 | 8 (30%) | 3 (11%) |
| EP-6X | mRNA | 57 | 14 | 8 (57%) | 2 (14%) | |
| EP-1X | Protein | 60 | 12 | 4 (33%) | 1 (8%) | |
| EP-6X | Protein | 54 | 6 | 1 (17%) | 4 (67%) | |
| MIJ | mRNA | 50 | 10 | 0 (0%) | 6 (60%) | |
| MIJ | Protein | 60 | 10 | 1 (10%) | 7 (70%) | |
| Rab6b (232 bp del) | EP-1X | mRNA | 44 | 8 | 1 (13%) | 3 (38%) |
| EP-6X | mRNA | 60 | 12 | 3 (25%) | 6 (50%) | |
| EP-1X | Protein | 51 | 8 | 3 (38%) | 3 (38%) | |
| EP-6X | Protein | 60 | 10 | 3 (30%) | 6 (60%) | |
| MIJ | mRNA | 56 | 18 | 1 (6%) | 15 (83%) | |
| MIJ | Protein | 54 | 12 | 1 (8%) | 10 (83%) |
Conditions tested were electroporation (EP) with program repeated one or six times (1X, 6X) versus microinjection (MIJ).
Percentages are based upon number of non-homologous (NHEJ) events or deletions obtained over total number of live born.
Discussion
The increasing use of CRISPR/Cas9 to perform highly effective, rapid genome editing in a wide variety of organisms has quickly made it one of the most widely applied laboratory techniques for generating loss-of-function alleles and other more complex alleles. The CRISPRtools platform focuses on applying CRISPR/Cas9 to create exon deletion alleles that build upon the long history of targeted mutagenesis in the mouse. In proof-of-principle studies, this guide design strategy effectively generated exon deletions for all three target genes when tested directly in vivo. Moreover, exon-sized deletions were achieved using both electroporation and microinjection approaches in an inbred strain of mouse when using four guides per target gene. While exon deletions were obtained for all targets, the size of the deletions varied from founder to founder due in part to the imprecise repair via the NHEJ pathway and the different deletion outcomes possible when using four guides. One potential option for limiting this complexity is to reduce the number of guides, which has worked well for internal deletions (data not shown); however, this does not overcome the stochastic nature of NHEJ repair process, and could reduce the overall success rate if one guide’s efficiency is suboptimal. Given these limitations, it is critical to sequence confirm the N1 generation from individual founder animals to isolate a specific deletion allele of interest. Despite these potential drawbacks, these findings hold promise for implementing this strategy at a genome-wide scale.
Currently, the platform supports the identification of guides adjacent to NGG PAM of S. pyogenes but can easily be extended to include additional PAM sequences as other systems become more widely used such as CRISPR from Prevotella and Francisella 1 (Cpf1) enzyme identified in Acidaminococcus (AsCpf1) and Lachnospiraceae (LbCpf1) (Zetsche et al. 2015). Additionally, this approach is not limited to mouse but can easily be extended to other organisms for which complete genomic sequence is available. The deletion strategy can also be generalized beyond exons to include the removal of specific genomic regions such as enhancers (Li et al. 2014; Zhang et al. 2016). The generic framework of CRISPRtools enables future improvements such as support for additional efficiency guide scoring algorithms as well as alternate approaches for accounting for off-target matches. The ability to stitch together several independent analyses simplifies the workflow for the user and provides flexibility for growth in the midst of an ever-expanding genome editing toolbox.
Supplementary Material
Acknowledgments
We would like to thank Charles Vejnar, Miguel Moreo-Mateos and A. Giraldez at Yale University for kindly providing the CRISPRscan code; Susan Kales and Rachel Urban for technical support; Haoyi Wang and Wen-bo Wang for providing insight to electroporation experiments; and the Genetic Engineering Technologies and Transgenic Genotyping Services at the Jackson Laboratory. This work was supported by the Office Of The Director, National Institutes Of Health under Award Number OD011185 and UM1OD023222 (to S.A.M.), and OD010972 (to L.G.R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu J-L. Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System Cell Reports. 2013;4 doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley A, et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V, et al. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnology. 2016;16:4. doi: 10.1186/s12896-016-0234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, NY) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites Molecular therapy. Nucleic acids. 2014:3. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nature Biotechnology. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- Hodgkins A, Farne A, Perera S, Grego T, Parry-Smith DJ, Skarnes WC, Iyer V. WGE: a CRISPR database for genome engineering. Bioinformatics (Oxford, England) 2015;31:3078–3080. doi: 10.1093/bioinformatics/btv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, NY) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic acids research. 2016;44:6. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 2014;9:e114485. doi: 10.1371/journal.pone.0114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Li X. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA. 2001;7:445–456. doi: 10.1017/s1355838201002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin J-DD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nature methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros JC, Franch M, Tabas-Madrid D, San-Leon D, Montoliu L, Cubas P, Pazos F. Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res. 2016;44:W267–271. doi: 10.1093/nar/gkw407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, et al. Efficient CRISPR/Cas9-Mediated Genome Editing in Mice by Zygote Electroporation of Nuclease. Genetics. 2015;200:423–430. doi: 10.1534/genetics.115.176594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor. DNA Nature biotechnology. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, et al. Delivery of Cas9 Protein into Mouse Zygotes through a Series of Electroporation Dramatically Increases the Efficiency of Model Creation. J Genet Genomics. 2016;43:319–327. doi: 10.1016/j.jgg.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, Meyerson M. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2016;48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.