Abstract
Drosophila imaginal discs, the larval precursors of adult structures such as the wing and leg, are capable of regenerating after damage. During the course of regeneration, discs can sometimes generate structures that are appropriate for a different type of disc, a phenomenon termed transdetermination. Until recently, these phenomena were studied by physically fragmenting discs and then transplanting them into the abdomens of adult female flies. This field has experienced a renaissance following the development of genetic ablation systems that can damage precisely defined regions of the disc without the need for surgery. Together with more traditional approaches, these newer methods have generated many novel insights into wound healing, the mechanisms that drive regenerative growth, plasticity during regeneration and systemic effects of tissue damage and regeneration.
Introduction
Regeneration is the process by which tissues, organs or organisms restore missing or damaged parts. Regeneration is widespread in nature and observed in diverse taxa (reviewed by [1, 2]) including the cnidarian Hydra, flatworms, urodele amphibians (e.g. salamanders), and more recently, zebrafish. Approaches mostly derived from experimental embryology, such as amputation of limbs and transplantation of tissues, have been applied in each of these organisms and have provided important insights into cellular aspects of regeneration. While it is now possible, at least in principle, to modify genes in each of these organisms using genome-editing technologies, it is still difficult to carry out large-scale forward genetic screens.
In contrast, Drosophila melanogaster is an organism that has been studied by geneticists for over a hundred years. As a result, a number of sophisticated genetic tools are available to study biological processes. These include tools to manipulate gene expression in a tissue-specific manner as well as the efficient generation of genetic mosaics. Additionally, it is possible to conduct genetic screens in a variety of ways (e.g. chemical mutagenesis, RNAi screens) that can be used to deconstruct a complex process such as regeneration that occurs in a living organism.
In this review we focus on studies of regeneration in Drosophila imaginal discs which are the larval primordia of adult structures such as the wing and eye. Experiments on imaginal disc regeneration were pioneered by the group of Ernst Hadorn ([3–6] and reviewed in [7, 8]). Although these studies revealed many fascinating aspects of regeneration including the propensity of tissues to change fate during regeneration (transdetermination) [9], the technical difficulties associated with these experiments curtailed the development of this field, and until recently, relatively few laboratories studied regeneration in imaginal discs. The development of genetic approaches to ablate tissues in a predictable and spatially defined way without needing surgery [10, 11] has resulted in a renaissance of the study of imaginal disc regeneration. In this article, we first summarize the classical literature on regeneration and then discuss recent work derived from genetic approaches.
Discovery of the regenerative capacity of Drosophila imaginal discs
In pioneering studies that began in the 1940s, imaginal discs were cut into fragments that were subsequently implanted and cultured in adult female abdomens where regeneration occurred [3–6]. In the absence of molecular techniques, the only way true regeneration could be distinguished from other forms of growth was to demonstrate that the new tissue acquired the developmental potential of portions of the disc that had been ablated. This was assessed by re-implanting the cultured fragments into a larva where they differentiated during metamorphosis. Morphological markers were used to assess the extent of regeneration. Implanted fragments generated additional tissue by localized cell proliferation, reminiscent of a regeneration blastema observed following limb amputation in salamanders. Additionally, the isolated blastemas alone were shown capable of regenerating the lost structures [12], and confrontation of blastemas from distant parts induced regrowth of missing structures between those parts (intercalary growth) [13].
These studies also provided two unexpected results. First, certain types of disc fragments, instead of regenerating the missing portion, generated mirror-image duplications [6, 14]. Second, after long periods of culture, structures that normally derive from other discs were sometimes derived from the proliferating blastema, a phenomenon known as transdetermination [15, 16].
Development of genetic ablation systems
In addition to the studies that demonstrated regeneration after physical fragmentation, diffuse damage to discs, such as with X-ray irradiation, was found to elicit additional cell divisions from the surviving cells [17]. Some authors use the term “compensatory proliferation” to distinguish this type of response to injury from the more localized proliferation that results from physical damage. Both physical fragmentation and X-ray irradiation were, however, not capable of deleting defined portions of the disc with much precision. This problem was overcome by the development of genetic approaches to tissue ablation in imaginal discs [10, 11] that took advantage of the Gal4/UAS system to target a pro-apoptotic gene (e.g. eiger, reaper) to a defined region of the imaginal disc and the temperature-sensitive version of the Gal80 repressor (Gal80ts) to restrict the ablation to a specific time window of imaginal disc development. This offers the opportunity to compare the regenerative properties of imaginal discs of different maturity. Genetic ablation offers two advantages over the traditional fragmentation approach. First, since the discs are ablated in situ, they are able to generate the appropriate adult structures and thus the extent of regeneration can be assessed in a live fly; a transplanted disc develops within the abdomen of the host and needs to be excised from the abdomen of the recipient to be studied. Second, because it is far less laborious, genetic manipulations such as screens can be conducted in these flies thus enabling the discovery of novel regulators of regeneration. Other pro-apoptotic transgenes such as debcl (the pro-apoptotic Drosophila Bcl-2 ortholog)[18] and hid [19] have subsequently been used with this system. An analogous system was developed independently to study regeneration in the adult midgut [20]. Patches of tissue can also be ablated, albeit at random locations, by generating clones of tissue that are mutant for a temperature-sensitive cell-lethal mutation such as sec5ts [21].
As with fragmentation, genetic ablation usually results in localized regenerative growth characterized by an increase in the rate of proliferation of adjacent surviving cells [10, 11]. However, clear differences are apparent in the response to ablation with different pro-apototic genes. These differences likely reflect the signaling pathways that are activated and the rate of cell killing by each pro-apoptotic gene leading to differences in the type of cell extrusion (apical versus basal), the extent to which proliferation is localized, and whether regeneration occurs concurrently with tissue loss [19].
Wound healing and early responses to tissue damage
The cut edges of fragmented discs heal very efficiently. Imaginal discs are epithelial sacs that consist of two layers of cells, the columnar epithelium (the “disc proper”) and the squamous peripodial epithelium. Transient heterotypic contacts between cut edges of the two layers appear during the first day of in vivo culture and involves contact mediated by filopodia and closure facilitated by an actin-rich cable [22–24]. Within 48 hours in culture, the heterotypic interactions are resolved and continuity is re-established of both the disc proper and the peripodial eptihelium. Wound healing has been studied in other tissues in Drosophila including the epidermis of the embryo [25, 26], the larva [27], the pupa [28] and the adult [29]; there are similarities and differences between those processes and wound healing in discs.
T.H. Morgan proposed that regeneration can occur either by local stimulation of cell proliferation (epimorphosis) or by re-patterning of existing tissue (morphollaxis) [1]. Most studies of imaginal disc regeneration demonstrate DNA synthesis or mitoses near the wound i.e. a regeneration blastema [30–35] although a recent study suggests that morphollaxis could also occur to some extent [36]. Importantly, the first signs of cell proliferation precede the completion of wound healing [12, 32, 34, 35, 37]. This argues strongly against the notion that cell proliferation is triggered by the juxtaposition of tissues with disparate positional identities and favor a mechanism where tissue damage directly stimulates cell proliferation. Fragmented discs respond to wounding by activation of the Jun N-terminal kinase (JNK), concentrated in regions near the edges of the wound. JNK is required for wound healing and also for cell proliferation in imaginal disc blastemas [24, 35, 37–39]. In addition to JNK, the p38 stress-activated protein kinase is also activated upon tissue damage and is required for regeneration [40]. Reactive oxygen species (ROS) are produced rapidly after damage and act as chemoattractants for macrophages. This has been demonstrated in both the imaginal disc [41] and the embryo [42] and ROS are required for activation of JNK and p38 [40]. In embryonic wounds, the NADPH oxidase DUOX, which acts as a source of H2O2, is activated by calcium, and Ca2+ flashes have been found upon damage [43]. Intercellular Ca2+ waves, propagated via gap junctions are triggered by injury to imaginal discs [44, 45].
Functional screens for genes that regulate the early stages of wound healing in Drosophila imaginal discs are only beginning, and one such screen demonstrated a role for Plexin A in the repair of wounds in imaginal discs [46]. Since plexins and semaphorins, which have known functions in axonal pathfinding, predate the evolution of the nervous system, it is possible that the ancestral function of this pathway is to repair damaged epithelia.
Regenerative growth
The pathways that drive proliferation following tissue damage seem to be the same as those that regulate growth during normal development. Physical damage to discs results in the upregulation the WNT protein Wingless (Wg) [10, 47–49]. As has been shown for disc growth during development [50], wg functions in regenerative growth by increasing Myc levels via a double-repression mechanism involving Notch [10]. The JAK/STAT pathway [40, 49, 51–53] and the Hippo pathway effector Yorkie (Yki) [18, 54–56] also drive regenerative growth. There are ways in which these regulators are utilized during regenerative growth that differs from normal growth. First, rather than by developmentally regulated signals, these pathways are activated by damage responsive signals. The expression of wg is activated by a damage-responsive enhancer [57, 58]. Expression of Unpaired, the ligand upstream of the JAK/STAT pathway is activated by both JNK [40, 49, 51] and the p38 kinase [40]. Following tissue damage, the Hippo pathway is inhibited by the LIM-domain protein Ajuba, likely in response to changes in cell tension, resulting in increased Yki activity [54, 55]. Cross-talk between these pathways is likely; for example, Yki can activate Myc expression [59].
In addition, the rate of cell proliferation is higher during regenerative growth than during normal growth [21, 60]. Thus hypomorphic mutations in genes necessary for cell proliferation might not impair normal growth but can curtail regenerative growth. Finally, in contrast to the preferred orientation of mitotic cell divisions during normal growth (proximodistal in the wing pouch), cells re-orient their axes of division during regeneration to facilitate the efficient replacement of lost cells [56].
As for tissues in many organisms, the capacity for discs to regenerate diminishes as they mature [10]. This results, in significant part from muted upregulation of genes that appear necessary for regenerative growth (e.g. wg, Mmp1). In the wg locus, the inability to upregulate wg correlates with a localized increase in H3K27 trimethylation at the damage-responsive enhancer as discs mature [58]. This localized silencing is mediated by a silencing element that is adjacent to, and separate from, the damage-responsive module.
Patterning and regeneration
During normal development, patterning occurs concurrently with growth and the patterns of expression of morphogens such as Wg and Dpp have been well characterized. Following ablation, these same proteins are expressed in non-physiological patterns and their normal patterns of expression are often not re-established until regenerative growth is complete [10]. During regeneration, cell fates (such as vein and intervein fates in the wing disc) can be re-specified to intercalate missing pattern elements [56]. Similarly, cells from the hinge of the wing disc appear capable of generating cells that contribute to the pouch [10, 19, 52]. A normal feature of imaginal discs is the presence of compartments which are composed of lineage-restricted cells that remain separate from each other [61]. Compartment boundaries are re-established soon after tissue ablation [10, 11]. However, cells near the compartment boundary appear capable of changing fates and adopting new compartmental identities [62]. Indeed, previous studies with disc fragmentation demonstrated changes in compartment identity during regeneration [47, 63]. The activation of JNK, which has been shown to reduce the activity of Polycomb-dependent silencing [38] facilitates fate changes. The chromatin regulator Taranis has recently been shown to stabilize compartmental identities during regeneration [64].
The phenomenon of transdetermination is another example of enhanced plasticity during regeneration (reviewed in [8, 9]). Hadorn’s group was able to maintain imaginal discs in long-term culture by repeatedly fragmenting discs and culturing fragments in the abdomens of female adult flies until regeneration was complete (330 transfers over 12 years!). During these studies they observed that portions of these discs adopted the fates of other discs, as assessed by the adult structures they generated when implanted into larvae that were allowed to proceed through metamorphosis. Transdetermination appears to occur as a collective fate change of a small number of cells and not a single cell [65]. Moreover, there are parts of discs termed “weak points” that are more prone to transdetermination and coincide with locations where the expression of the two morphogens Dpp and Wingless coincide [6, 66–68]. Indeed ectopic expression of wg in the foreleg disc can promote leg to wing transdetermination [66, 68]. Transdetermination involves JNK-mediated downregulation of Polycomb-group proteins [38]. Investigations of transdetermination are being conducted at multiple levels; changes in gene expression have been cataloged [69] and genetic changes that modulate it have been documented [48]. However, our understanding of transdetermination is still at a rudimentary level.
Systemic effects of regeneration
Early studies pointed to mechanisms in Drosophila larvae that delay pupariation in response to X-irradiation [70] or damage to imaginal discs [71]. Retinoids mediate part of this delay although the mechanism by which they do so is still not known [72]. An exciting development was the discovery of a protein of the insulin/relaxin family, Dilp8, which is released by damaged imaginal discs [73, 74]. Expression of Dilp8 is activated by JNK [49, 73], Yki [75] and, indirectly, by the chromatin modifying enzyme Trithorax [76]. Dilp8 binds to a G-protein-coupled receptor, Lgr3 [77–80], that is expressed on a subset of neurons. The Lgr3-expressing neurons synapse with PTTH-secreting neurons which innervate the prothoracic gland (PG) and promote the synthesis of ecdysone, the hormone that promotes metamorphosis. This neural pathway likely mediates the developmental delay. In addition, Lgr3 is also expressed in the PG itself, where it activates nitric oxide synthase. NOS activity is necessary to slow the growth of undamaged discs while damaged tissue is being repaired [80, 81].
Concluding remarks
Studies of regeneration in Drosophila imaginal discs have identified novel components of the network of signals that regulated wound healing, revealed epigenetic mechanisms that regulate regenerative capacity and plasticity during regeneration, and uncovered systemic responses that facilitate regeneration. These findings are likely to promote the search for similar mechanisms in other organisms such as mammals.
Figure 1. Regeneration in wing-imaginal discs.
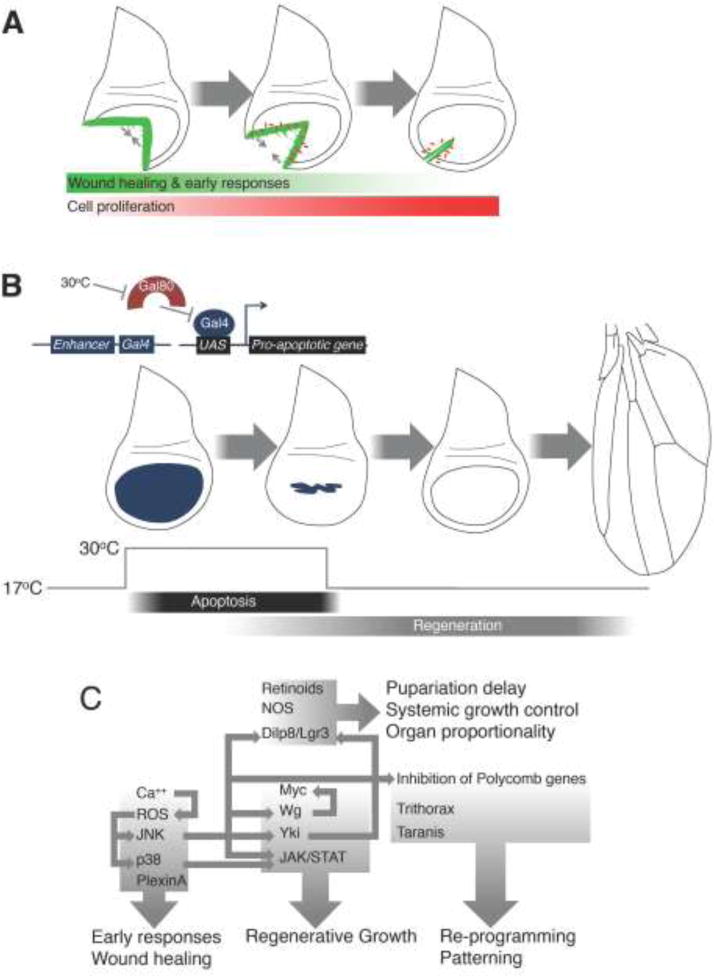
(A) Regeneration following physical fragmentation of a disc that has been implanted in an adult abdomen. Wound healing (gray arrows) is facilitated by actin cables and filopodia in regions of JNK activation (green). Damaged-induced proliferation (red) occurs prior to the completion of wound healing.
(B) Genetic ablation of tissue in the wing disc. Expression of a pro-apoptotic gene (blue) during the ablation phase (30°C) eliminates most of the pouch. Regenerative growth restores the pouch that can differentiate into a wing of normal size and shape.
(C) Summary of regulators implicated in imaginal disc regeneration.
Highlights.
Drosophila imaginal discs are capable of regeneration after damage.
Genetic tools enable specific regions of imaginal discs to be ablated in situ.
Reactive oxygen species, protein kinases and cytokines function in would healing.
Regenerative growth uses many of the same genes that function in normal growth.
Chromatin states impact regenerative capacity and plasticity during regeneration.
Acknowledgments
We apologize to investigators whose work we could not discuss due to space constraints. We thank Linda Setiawan, Melanie Worley, and Paula Santabárbara-Ruiz for comments on the manuscript. IKH is funded by the NIH (grants GM061672 and GM085576) and a Research Professor Award from the American Cancer Society (RP-16-238-06-COUN). FS is supported by the Spanish Ministerio de Economía y Competitividad grant BFU2015-67623-P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morgan TH. Regeneration. New York: Macmillan; 1901. [Google Scholar]
- 2.Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21(1):172–85. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadorn E, Bertani G, Gallera J. Regulative capacity and field organization of male genital discs in Drosophila melanogaster. Roux’s Arch Dev Biol. 1949;144:31–70. doi: 10.1007/BF00575293. [DOI] [PubMed] [Google Scholar]
- 4.Ursprung H. Fragmentation and radiation experiments to determine the determination and fate map of the Drosophila genital disc. Roux’s Arch Dev Biol. 1959;151:501–558. [Google Scholar]
- 5.Hadorn E, Buck D. On the differentiation of transplanted wing imaginal disc fragments of Drosophila melanogaster. Rev Suisse Zool. 1962;69:302–310. [Google Scholar]
- 6.Schubiger G. Regeneration, duplication and transdetermination in fragments of the leg disc of Drosophila melanogaster. Dev Biol. 1971;26(2):277–95. doi: 10.1016/0012-1606(71)90127-8. [DOI] [PubMed] [Google Scholar]
- 7.Bergantinos C, Vilana X, Corominas M, Serras F. Imaginal discs: Renaissance of a model for regenerative biology. Bioessays. 2010;32(3):207–17. doi: 10.1002/bies.200900105. [DOI] [PubMed] [Google Scholar]
- 8.Worley MI, Setiawan L, Hariharan IK. Regeneration and transdetermination in Drosophila imaginal discs. Annu Rev Genet. 2012;46:289–310. doi: 10.1146/annurev-genet-110711-155637. [DOI] [PubMed] [Google Scholar]
- 9.Hadorn E. Transdetermination. In: Ashburner M, Wright TRF, editors. The Genetics and Biology of Drosophila. Academic Press; London: 1978. pp. 556–617. [Google Scholar]
- 10.Smith-Bolton RK, Worley MI, Kanda H, Hariharan IK. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev Cell. 2009;16(6):797–809. doi: 10.1016/j.devcel.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergantinos C, Corominas M, Serras F. Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development. 2010;137(7):1169–79. doi: 10.1242/dev.045559. [DOI] [PubMed] [Google Scholar]
- 12.Karpen GH, Schubiger G. Extensive regulatory capabilities of a Drosophila imaginal disk blastema. Nature. 1981;294(5843):744–7. doi: 10.1038/294744a0. [DOI] [PubMed] [Google Scholar]
- 13.Haynie JL, Bryant PJ. Intercalary regeneration in imaginal wing disk of Drosophila melanogaster. Nature. 1976;259(5545):659–62. doi: 10.1038/259659b0. [DOI] [PubMed] [Google Scholar]
- 14.Bryant PJ. Regeneration and duplication following operations in situ on the imaginal discs of Drosophila melanogaster. Dev Biol. 1971;26(4):637–51. doi: 10.1016/0012-1606(71)90146-1. [DOI] [PubMed] [Google Scholar]
- 15.Hadorn E. Problems of determination and transdetermination. Brookhaven Symp Biol. 1965;18:148–161. [Google Scholar]
- 16.Schubiger G, Hadorn E. Auto- and allotypic differentiation in vivo cultivated foreleg blastemas of Drosophila melanogaster. Dev Biol. 1968;17(5):584–602. doi: 10.1016/0012-1606(68)90007-9. [DOI] [PubMed] [Google Scholar]
- 17.Haynie JL, Bryant PJ. The effects of X-rays on the proliferation dynamics of cells in the imaginal disc of Drosophila melanogaster. Roux’s Arch Dev Biol. 1977;183:85–100. doi: 10.1007/BF00848779. [DOI] [PubMed] [Google Scholar]
- 18.Grusche FA, Degoutin JL, Richardson HE, Harvey KF. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Dev Biol. 2011;350(2):255–66. doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 19.Herrera SC, Martin R, Morata G. Tissue homeostasis in the wing disc of Drosophila melanogaster: immediate response to massive damage during development. PLoS Genet. 2013;9(4):e1003446. doi: 10.1371/journal.pgen.1003446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137(7):1343–55. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerhold AR, Richter DJ, Yu AS, Hariharan IK. Identification and characterization of genes required for compensatory growth in Drosophila. Genetics. 2011;189(4):1309–26. doi: 10.1534/genetics.111.132993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinhardt CA, Hodgkin NM, Bryant PJ. Wound healing in the imaginal discs of Drosophila. I. Scanning electron microscopy of normal and healing wing discs. Dev Biol. 1977;60(1):238–57. doi: 10.1016/0012-1606(77)90122-1. [DOI] [PubMed] [Google Scholar]
- 23.Reinhardt CA, Bryant PJ. Wound healing in the imaginal discs of Drosophila. II. Transmission electron microscopy of normal and healing wing discs. J Exp Zool. 1981;216(1):45–61. doi: 10.1002/jez.1402160107. [DOI] [PubMed] [Google Scholar]
- 24.Bosch M, Serras F, Martin-Blanco E, Baguna J. JNK signaling pathway required for wound healing in regenerating Drosophila wing imaginal discs. Dev Biol. 2005;280(1):73–86. doi: 10.1016/j.ydbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Rämet M, Lanot R, Zachary D, Manfruelli P. JNK signaling pathway is required for efficient wound healing in Drosophila. Dev Biol. 2002;241(1):145–56. doi: 10.1006/dbio.2001.0502. [DOI] [PubMed] [Google Scholar]
- 26.Wood W, Jacinto A, Grose R, Woolner S, Gale J, Wilson C, Martin P. Wound healing recapitulates morphogenesis in Drosophila embryos. Nat Cell Biol. 2002;4(11):907–12. doi: 10.1038/ncb875. [DOI] [PubMed] [Google Scholar]
- 27.Galko MJ, Krasnow MA. Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol. 2004;2(8):E239. doi: 10.1371/journal.pbio.0020239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antunes M, Pereira T, Cordeiro JV, Almeida L, Jacinto A. Coordinated waves of actomyosin flow and apical cell constriction immediately after wounding. J Cell Biol. 2013;202(2):365–79. doi: 10.1083/jcb.201211039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Losick VP, Fox DT, Spradling AC. Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr Biol. 2013;23(22):2224–32. doi: 10.1016/j.cub.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dale L, Bownes M. Is regeneration in Drosophila the result of epimorphic regulation? Wilhelm Roux’s Arch Dev Biol. 1980;189:91–96. doi: 10.1007/BF00848497. [DOI] [PubMed] [Google Scholar]
- 31.Adler PN, MacQueen M. Cell proliferation and DNA replication in the imaginal wing disc of Drosophila melanogaster. Dev Biol. 1984;103(1):28–37. doi: 10.1016/0012-1606(84)90004-6. [DOI] [PubMed] [Google Scholar]
- 32.Kiehle CP, Schubiger G. Cell proliferation changes during pattern regulation in imaginal leg discs of Drosophila melanogaster. Dev Biol. 1985;109(2):336–46. doi: 10.1016/0012-1606(85)90460-9. [DOI] [PubMed] [Google Scholar]
- 33.O’Brochta DA, Bryant PJ. Distribution of S-phase cells during the regeneration of Drosophila imaginal wing discs. Dev Biol. 1987;119(1):137–42. doi: 10.1016/0012-1606(87)90215-6. [DOI] [PubMed] [Google Scholar]
- 34.Bryant PJ, Fraser SE. Wound healing, cell communication, and DNA synthesis during imaginal disc regeneration in Drosophila. Dev Biol. 1988;127(1):197–208. doi: 10.1016/0012-1606(88)90201-1. [DOI] [PubMed] [Google Scholar]
- 35.Bosch M, Baguna J, Serras F. Origin and proliferation of blastema cells during regeneration of Drosophila wing imaginal discs. Int J Dev Biol. 2008;52(8):1043–50. doi: 10.1387/ijdb.082608mb. [DOI] [PubMed] [Google Scholar]
- 36.Diaz-Garcia S, Baonza A. Pattern reorganization occurs independently of cell division during Drosophila wing disc regeneration in situ. Proc Natl Acad Sci U S A. 2013;110(32):13032–7. doi: 10.1073/pnas.1220543110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattila J, Omelyanchuk L, Kyttala S, Turunen H, Nokkala S. Role of Jun N-terminal Kinase (JNK) signaling in the wound healing and regeneration of a Drosophila melanogaster wing imaginal disc. Int J Dev Biol. 2005;49(4):391–9. doi: 10.1387/ijdb.052006jm. [DOI] [PubMed] [Google Scholar]
- 38.Lee N, Maurange C, Ringrose L, Paro R. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature. 2005;438:234–237. doi: 10.1038/nature04120. [DOI] [PubMed] [Google Scholar]
- 39.Blanco E, Ruiz-Romero M, Beltran S, Bosch M, Punset A, Serras F, Corominas M. Gene expression following induction of regeneration in Drosophila wing imaginal discs. Expression profile of regenerating wing discs. BMC Dev Biol. 2010;10:94. doi: 10.1186/1471-213X-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santabarbara-Ruiz P, Lopez-Santillan M, Martinez-Rodriguez I, Binagui-Casas A, Perez L, Milan M, Corominas M, Serras F. ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Drosophila Regeneration. PLoS Genet. 2015;11(10):e1005595. doi: 10.1371/journal.pgen.1005595. ROS are generated after apoptosis or after physical injury to discs. They function upstream of JNK and p38 to induce the cytokine Unpaired to promote regeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fogarty CE, Diwanji N, Lindblad JL, Tare M, Amcheslavsky A, Makhijani K, Bruckner K, Fan Y, Bergmann A. Extracellular Reactive Oxygen Species Drive Apoptosis-Induced Proliferation via Drosophila Macrophages. Curr Biol. 2016;26(5):575–84. doi: 10.1016/j.cub.2015.12.064. Cells undergoing apoptosis generate extracellular ROS which act on macrophages to induce production of the TNF ortholog Eiger. Eiger binds to receptors on epithelial cells to promote JNK activation which promotes apoptosis in some cells and compensatory proliferation in others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreira S, Stramer B, Evans I, Wood W, Martin P. Prioritization of competing damage and developmental signals by migrating macrophages in the Drosophila embryo. Curr Biol. 2010;20(5):464–70. doi: 10.1016/j.cub.2010.01.047. [DOI] [PubMed] [Google Scholar]
- 43.Razzell W, Evans IR, Martin P, Wood W. Calcium flashes orchestrate the wound inflammatory response through DUOX activation and hydrogen peroxide release. Curr Biol. 2013;23(5):424–9. doi: 10.1016/j.cub.2013.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narciso C, Wu Q, Brodskiy P, Garston G, Baker R, Fletcher A, Zartman J. Patterning of wound-induced intercellular Ca(2+) flashes in a developing epithelium. Phys Biol. 2015;12(5):056005. doi: 10.1088/1478-3975/12/5/056005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Restrepo S, Basler K. Drosophila wing imaginal discs respond to mechanical injury via slow InsP3R-mediated intercellular calcium waves. Nat Commun. 2016;7:12450. doi: 10.1038/ncomms12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoo SK, Pascoe HG, Pereira T, Kondo S, Jacinto A, Zhang X, Hariharan IK. Plexins function in epithelial repair in both Drosophila and zebrafish. Nat Commun. 2016;7:12282. doi: 10.1038/ncomms12282. A screen for genes that function in wound healing in imaginal discs identified PlexinA. A role for plexins in epithelial homeostasis may be evolutionarily more ancient than their well-known function in axonal pathfinding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibson MC, Schubiger G. Hedgehog is required for activation of engrailed during regeneration of fragmented Drosophila imaginal discs. Development. 1999;126(8):1591–9. doi: 10.1242/dev.126.8.1591. [DOI] [PubMed] [Google Scholar]
- 48.McClure KD, Sustar A, Schubiger G. Three genes control the timing, the site and the size of blastema formation in Drosophila. Dev Biol. 2008;319(1):68–77. doi: 10.1016/j.ydbio.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katsuyama T, Comoglio F, Seimiya M, Cabuy E, Paro R. During Drosophila disc regeneration, JAK/STAT coordinates cell proliferation with Dilp8-mediated developmental delay. Proc Natl Acad Sci U S A. 2015;112(18):E2327–36. doi: 10.1073/pnas.1423074112. Analysis of the transcriptome of regenerating discs showed that the cytokine Unpaired promotes regenerative proliferation together with Wingless and that JAK/STAT signaling also causes production of the insulin/relaxin family member Dilp8 which delays pupariation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herranz H, Perez L, Martin FA, Milan M. A Wingless and Notch double-repression mechanism regulates G1-S transition in the Drosophila wing. Embo J. 2008;27(11):1633–45. doi: 10.1038/emboj.2008.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pastor-Pareja JC, Wu M, Xu T. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis Model Mech. 2008;1(2–3):144–54. doi: 10.1242/dmm.000950. discussion 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verghese S, Su TT. Drosophila Wnt and STAT Define Apoptosis-Resistant Epithelial Cells for Tissue Regeneration after Irradiation. PLoS Biol. 2016;14(9):e1002536. doi: 10.1371/journal.pbio.1002536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.La Fortezza M, Schenk M, Cosolo A, Kolybaba A, Grass I, Classen AK. JAK/STAT signalling mediates cell survival in response to tissue stress. Development. 2016;143(16):2907–19. doi: 10.1242/dev.132340. [DOI] [PubMed] [Google Scholar]
- 54.Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev Biol. 2011;350(1):139–51. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meserve JH, Duronio RJ. Scalloped and Yorkie are required for cell cycle re-entry of quiescent cells after tissue damage. Development. 2015;142(16):2740–51. doi: 10.1242/dev.119339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Repiso A, Bergantinos C, Serras F. Cell fate respecification and cell division orientation drive intercalary regeneration in Drosophila wing discs. Development. 2013;140(17):3541–51. doi: 10.1242/dev.095760. [DOI] [PubMed] [Google Scholar]
- 57.Schubiger M, Sustar A, Schubiger G. Regeneration and transdetermination: the role of wingless and its regulation. Dev Biol. 2010;347(2):315–24. doi: 10.1016/j.ydbio.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris RE, Setiawan L, Saul J, Hariharan IK. Localized epigenetic silencing of a damage-activated WNT enhancer limits regeneration in mature Drosophila imaginal discs. Elife. 2016;5:e11588. doi: 10.7554/eLife.11588. The loss of regenerative capacity as imaginal discs mature correlates with an inability to upregulate the expression of genes necessary for regeneration such as wingless. A damage-responsive enhancer in the wingless locus is epigenetically silenced in a localized manner in mature discs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neto-Silva RM, de Beco S, Johnston LA. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell. 2010;19(4):507–20. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wells BS, Yoshida E, Johnston LA. Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr Biol. 2006;16(16):1606–15. doi: 10.1016/j.cub2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Bellido A, Ripoll P, Morata G. Developmental compartmentalisation of the wing disk of Drosophila. Nat New Biol. 1973;245(147):251–3. doi: 10.1038/newbio245251a0. [DOI] [PubMed] [Google Scholar]
- 62.Herrera SC, Morata G. Transgressions of compartment boundaries and cell reprogramming during regeneration in Drosophila. Elife. 2014;3:e01831. doi: 10.7554/eLife.01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szabad J, Simpson P, Nothiger R. Regeneration and compartments in Drosophila. J Embryol Exp Morphol. 1979;49:229–41. [PubMed] [Google Scholar]
- 64.Schuster KJ, Smith-Bolton RK. Taranis Protects Regenerating Tissue from Fate Changes Induced by the Wound Response in Drosophila. Dev Cell. 2015;34(1):119–28. doi: 10.1016/j.devcel.2015.04.017. Following damage to wing imaginal discs, the chromatin regulator Taranis is necessary to maintain expression of Engrailed at normal levels in posterior cells and preventing their transformation into anterior cells. [DOI] [PubMed] [Google Scholar]
- 65.Gehring W. Clonal analysis of determination dynamics in cultures of imaginal disks in Drosophila melanogaster. Dev Biol. 1967;16(5):438–56. doi: 10.1016/0012-1606(67)90058-9. [DOI] [PubMed] [Google Scholar]
- 66.Maves L, Schubiger G. Wingless induces transdetermination in developing Drosophila imaginal discs. Development. 1995;121(5):1263–72. doi: 10.1242/dev.121.5.1263. [DOI] [PubMed] [Google Scholar]
- 67.Johnston LA, Schubiger G. Ectopic expression of wingless in imaginal discs interferes with decapentaplegic expression and alters cell determination. Development. 1996;122(11):3519–29. doi: 10.1242/dev.122.11.3519. [DOI] [PubMed] [Google Scholar]
- 68.Maves L, Schubiger G. A molecular basis for transdetermination in Drosophila imaginal discs: interactions between wingless and decapentaplegic signaling. Development. 1998;125(1):115–24. doi: 10.1242/dev.125.1.115. [DOI] [PubMed] [Google Scholar]
- 69.Klebes A, Sustar A, Kechris K, Li H, Schubiger G, Kornberg TB. Regulation of cellular plasticity in Drosophila imaginal disc cells by the Polycomb group, trithorax group and lama genes. Development. 2005;132(16):3753–65. doi: 10.1242/dev.01927. [DOI] [PubMed] [Google Scholar]
- 70.Hussey RG, Thompson WR, Calhoun ET. The influence of X-rays on the development of Drosophila larvae. Science. 1927;66(1698):65–66. doi: 10.1126/science.66.1698.65. [DOI] [PubMed] [Google Scholar]
- 71.Simpson P, Berreur P, Berreur-Bonnenfant J. The initiation of pupariation in Drosophila: dependence on growth of the imaginal discs. J Embryol Exp Morphol. 1980;57:155–65. [PubMed] [Google Scholar]
- 72.Halme A, Cheng M, Hariharan IK. Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr Biol. 2010;20(5):458–63. doi: 10.1016/j.cub.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Colombani J, Andersen DS, Leopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. 2012;336(6081):582–5. doi: 10.1126/science.1216689. [DOI] [PubMed] [Google Scholar]
- 74.Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science. 2012;336(6081):579–82. doi: 10.1126/science.1216735. [DOI] [PubMed] [Google Scholar]
- 75.Boone E, Colombani J, Andersen DS, Leopold P. The Hippo signalling pathway coordinates organ growth and limits developmental variability by controlling dilp8 expression. Nat Commun. 2016;7:13505. doi: 10.1038/ncomms13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skinner A, Khan SJ, Smith-Bolton RK. Trithorax regulates systemic signaling during Drosophila imaginal disc regeneration. Development. 2015;142(20):3500–11. doi: 10.1242/dev.122564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vallejo DM, Juarez-Carreno S, Bolivar J, Morante J, Dominguez M. A brain circuit that synchronizes growth and maturation revealed through Dilp8 binding to Lgr3. Science. 2015;350(6262):aac6767. doi: 10.1126/science.aac6767. [DOI] [PubMed] [Google Scholar]
- 78.Colombani J, Andersen DS, Boulan L, Boone E, Romero N, Virolle V, Texada M, Leopold P. Drosophila Lgr3 Couples Organ Growth with Maturation and Ensures Developmental Stability. Curr Biol. 2015;25(20):2723–9. doi: 10.1016/j.cub.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 79.Garelli A, Heredia F, Casimiro AP, Macedo A, Nunes C, Garcez M, Dias AR, Volonte YA, Uhlmann T, Caparros E, Koyama T, Gontijo AM. Dilp8 requires the neuronal relaxin receptor Lgr3 to couple growth to developmental timing. Nat Commun. 2015;6:8732. doi: 10.1038/ncomms9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jaszczak JS, Wolpe JB, Bhandari R, Jaszczak RG, Halme A. Growth Coordination During Drosophila melanogaster Imaginal Disc Regeneration Is Mediated by Signaling Through the Relaxin Receptor Lgr3 in the Prothoracic Gland. Genetics. 2016;204(2):703–709. doi: 10.1534/genetics.116.193706. References 77–80 describe the indentification of Lgr3 as the receptor for Dilp8. Lgr3 is expressed on neurons and in the prothoracic gland. It mediates the pupariation delay and the slowing of growth caused by the production of Dilp8 by regenerating or overgrown tissue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jaszczak JS, Wolpe JB, Dao AQ, Halme A. Nitric Oxide Synthase Regulates Growth Coordination During Drosophila melanogaster Imaginal Disc Regeneration. Genetics. 2015;200(4):1219–28. doi: 10.1534/genetics.115.178053. Growth coordination between regenerating and undamaged imaginal discs is dependent upon Dilp8 activation of nitric oxide synthase. [DOI] [PMC free article] [PubMed] [Google Scholar]


