Abstract
Background and Objective
Short-term fasting can alter drug exposure but it is unknown whether this is an effect of altered oral bioavailability and/or systemic clearance. Therefore, the aim of our study was to assess the effect of short-term fasting on oral bioavailability and systemic clearance of different drugs.
Methods
In a randomized, controlled, crossover trial, 12 healthy subjects received a single administration of a cytochrome P450 (CYP) probe cocktail, consisting of caffeine (CYP1A2), metoprolol (CYP2D6), midazolam (CYP3A4), omeprazole (CYP2C19) and warfarin (CYP2C9), on four occasions: an oral (1) and intravenous (2) administration after an overnight fast (control) and an oral (3) and intravenous (4) administration after 36 h of fasting. Pharmacokinetic parameters of the probe drugs were analyzed using the nonlinear mixed-effects modeling software NONMEM.
Results
Short-term fasting increased systemic caffeine clearance by 17% (p = 0.04) and metoprolol clearance by 13% (p < 0.01), whereas S-warfarin clearance decreased by 19% (p < 0.01). Fasting did not affect bioavailability.
Conclusion
The study demonstrates that short-term fasting alters CYP-mediated drug metabolism in a non-uniform pattern without affecting oral bioavailability.
Key Points
| Short-term fasting influences systemic drug metabolism mediated by cytochrome P450 (CYP) 1A2, CYP2C9 and CYP2D6 but did not affect oral bioavailability. |
| The effect of fasting is enzyme specific since short-term fasting affected systemic clearance in a non-uniform pattern. |
| Additional research is warranted to determine if dose adjustments of drugs metabolized by CYP are necessary to improve drug treatment in patients with fasting-related consequences, such as malnutrition, or in combination with diets based on therapeutic fasting. |
Introduction
The ultimate goal of personalized medicine is to predict the best treatment strategy for the individual patient. To achieve this, it is necessary to understand the factors that contribute to variability within and between patients, which remains a challenge [1]. There is considerable variability in drug metabolism, which may result in treatment failure or, conversely, in untoward side effects. Cytochrome P450 (CYP) enzymes play an important role in drug metabolism since this enzyme family catalyzes the oxidative phase I biotransformation of most drugs [2]. Whereas monogenic polymorphisms explain an important part of the variability for a few CYP enzymes, most enzymes are multifactorially controlled by genetic, physiologic, pharmacologic, environmental, and nutritional factors such as fasting [3].
Short-term fasting can modulate the activity of some CYP enzymes in preclinical studies and in humans [4–8]. In a previous study, we have demonstrated that short-term fasting increased clearance of caffeine by 20% but decreased clearance of S-warfarin by 25%, when administered in an oral cocktail of five different drugs [8]. This cocktail consisted of the following CYP probes: caffeine (CYP1A2), metoprolol (CYP2D6), midazolam (CYP3A4), omeprazole (CYP2C19), and warfarin (CYP2C9) [9]. Together, these enzymes account for more than 70% of all phase I-dependent metabolism of drugs, nutraceuticals, and herbal remedies [3].
CYP enzymes not only reside in the liver but also in the gastrointestinal tract. CYP3A4 is abundantly expressed in the small intestine and, to a lesser extent, CYP1A2, CYP2C9, CYP2C19 and CYP2D6 [10]. The intestinal metabolism by CYP3A substrates is often similar to, or even exceeds, hepatic metabolism even though the total content of, for example, CYP3A in the entire human small intestine is only 1% of that in the liver [11]. In our previous study, the drug cocktail was administered orally. It is unknown whether the effects of fasting on drug metabolism were caused by altered oral bioavailability and/or altered systemic clearance. Therefore, the aim of our current study was to assess the effect of short-term fasting on oral bioavailability and systemic clearance by using the cocktail approach in healthy volunteers.
Materials and Methods
Subjects
Twelve healthy male subjects were recruited to participate in the trial. Inclusion criteria were as follows: (1) age 18 years or older; and (2), healthy, as determined by an experienced physician, and with normal renal and liver function. Exclusion criteria were (1) major illness in the past 3 months; (2) gastrointestinal disease that may influence drug absorption; (3) abnormal values of the following laboratory parameters: alanine aminotransferase, alkaline phosphatase, aspartate aminotransferase, bilirubin, γ-glutamyl transferase, and creatinine; (4) excessive alcohol intake (more than three units of alcohol per day) or use of alcohol for at least 2 days prior to each study day; (5) drugs of abuse; (6) smokers; (7) strenuous exercise at least 3 days prior to each study day, defined as more than 1 h of exercise per day; (8) use of prescription or nonprescription drugs; (9) consumption of caffeine-containing foods or beverages within 1 day prior to the study; and (10) consumption of grapefruit and grapefruit-containing products or starfruit for at least 2 days prior to each study day [8].
Study Design
We performed an open-label, randomly assigned, crossover intervention study in healthy male subjects. After approval of the protocol (Amendment 2, ABRnr: NL40834.018.12) by the Institutional Ethics Review Board, this study was performed at the Academic Medical Center, University of Amsterdam, The Netherlands. Each subject received a single oral or intravenous administration of a drug cocktail on four occasions, with washout periods of 4 weeks: an oral (1) or intravenous (2) administration after an overnight fast (control), and an oral (3) or intravenous (4) administration after 36 h of fasting. Subjects were randomly assigned for the order in which they received the drug cocktail. On all occasions, the drug cocktail was administered at 8:00 a.m. In order to minimize the effect of food intake in the morning on the bioavailability of the drug cocktail, subjects fasted from 10:00 p.m. the preceding evening while participating in the control interventions [occasions (1) and (2)]. In the fasting interventions [occasions (3) and (4)], subjects fasted from 8:00 p.m. starting two evenings prior to administration of the cocktail. This ensures a period of 36 h of fasting at the time of administration of the cocktail. On each of the four occasions, subjects had a standard fluid meal (Nutridrink Compact; Nutricia, Zoetermeer, The Netherlands) at noon. The meal was standardized to prevent differences in caloric intake between the interventions to affect the pharmacokinetics of the drug cocktail. After 4:00 p.m. subjects were allowed to consume their habitual diet [8].
Subjects kept a diary containing dietary instructions to standardize their diet in the 3 days preceding each of the four occasions. Furthermore, the following biomarkers were measured at baseline on each occasion in order to check adherence to the fasting protocol: glucose, β-hydroxybutyrate, free fatty acids, and acetoacetate [12].
Cytochrome P450 (CYP) Probe Cocktail
Subjects received a CYP probe drug cocktail that had previously been validated by Turpault et al. and consisted of caffeine (CYP1A2), metoprolol (CYP2D6), midazolam (CYP3A4), omeprazole (CYP2C19), and S-warfarin (CYP2C9) [9]. The cocktail administered orally consisted of caffeine 100 mg (10 mg/mL, 1 mL ampoules; VU University Medical Center [VUMC], Amsterdam, The Netherlands), racemic warfarin 5 mg (5 mg tablet; Crescent Pharma Ltd, Hampshire, UK), omeprazole 20 mg (20 mg capsule; Teva Pharmachemie, Haarlem, The Netherlands), metoprolol 100 mg (100 mg tablet; Teva Pharmachemie), and midazolam 0.03 mg kg−1 (1 mg/mL oral solution; University Medical Centre Groningen, Groningen, The Netherlands) [8]. The intravenous administration of the cocktail consisted of caffeine 50 mg (10 mg/mL, 1 mL ampoules; VUMC), racemic warfarin 5 mg (5 mg/mL, 3 mL ampoules; Radboud University Medical Center, Nijmegen, The Netherlands), omeprazole 20 mg (40 mg powder for solution for infusion; AstraZeneca BV, Zoetermeer, The Netherlands), metoprolol 20 mg (1 mg/mL, 5 mL ampoules; AstraZeneca BV), and midazolam 0.015 mg kg−1 (5 mg/mL, 1 mL ampoules; Roche Nederland BV, Woerden, The Netherlands).
Blood Sampling and Bioanalysis of the CYP Probe Drugs
For the estimation of pharmacokinetic parameters, blood samples were collected pre-dose and at 1, 2, 3, 4, 5, 6, 7, 8 and 10 h after oral administration of the drug cocktail. For the intravenous treatment arms, samples were taken pre-dose and at 2, 11.5, 15, 29, 41.5, 60, 90, 135, 173, 180, 195 min and 3.5, 4, 5, 7 and 9 h after intravenous administration of the drug cocktail blood. Furthermore, pharmacokinetic samples were obtained at days 2, 3, 8 and 15, of which the latter two were due to the long elimination half-life of warfarin [13]. Plasma was separated by centrifugation and stored at −80 °C until analysis.
The plasma concentrations of the drugs in the cocktail were simultaneously determined using a validated liquid chromatography/tandem mass spectrometry (LC-MS/MS) method as previously described [14]. The lower and upper limits of quantification (LLOQ and ULOQ) were 50–5000 ng mL−1 for caffeine, 1–200 ng mL−1 for metoprolol, 0.5–100 ng mL−1 for midazolam, 2–500 ng mL−1 for omeprazole, and 4–1000 ng mL−1 for S-warfarin. Linearity was R 2 ≥ 0.995 for all components. For all analytes, the mean process efficiency was >95% and the mean ionization efficiency was >97%. Furthermore, for all analytes the accuracy was between 94.9 and 108%, and the within- and between-run imprecision was <11.7% for the LLOQ and <12.6% for the middle level and ULOQ [14].
Pharmacogenetic Analysis
Genomic DNA was isolated from whole blood using a total nucleic acid extraction kit on a MagnaPure LC (Roche Diagnostics GmbH, Penzberg, Germany). Genotyping was performed using predesigned DME Taqman allelic discrimination assays on the Life Technologies Taqman 7500 system. Each assay consisted of two allele-specific minor groove binding (MGB) probes, labeled with the fluorescent dyes VIC and FAM. Polymerase chain reactions (PCRs) were performed in a reaction volume of 10 µL, containing assay-specific primers, allele-specific Taqman MGB probes, Abgene Absolute QPCR Rox Mix, and genomic DNA (20 ng). The thermal profile consisted of 40 cycles of denaturation at 95 °C for 20 s and annealing at 92 °C for 3 s, as well as extension at 60 °C for 30 s. Genotypes were scored by measuring allele-specific fluorescence using the 7500 software v2.3 for allelic discrimination (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA): CYP1A2 -3860G>A (*1C allele), -163C>A (*1F and *1K alleles), -729C>T (*1K allele); for CYP2C9 430C>T (*2) and 1075A>C (*3); for CYP3A4 -392A>G (*1B), g.20230G>A (*1G), 664T>C (*2), 1334T>C (*3), 352A>G (*4), 653G>C (*5), 520G>C (*10), 1117C>T (*12), 566T>C (*17), 878T>C (*18) and g.15389C>T (*22). CYP2C19 and CYP2D6 were analyzed on INFINITY Plus (Autogenomics, San Diego, CA, USA) according to the manufacturer’s instructions. For CYP2C19, variants analyzed were 681G>A (*2), 636G>A (*3), 1A>G (*4), 1297C>T (*5), 395G>A (*6), g.19294T>A (*7), 358T>C (*8), 431G>A (*9), 680C>T (*10) and -806C>T (*17); for CYP2D6, 2-1584C>G (*2), 2549delA (*3), 1846G>A (*4), gene deletion (*5), 1707delT (*6), 2935A>C (*7), 1758G>T (*8), 2615_2617delAAG (*9), 100C>T (*4, *10), 124G>A (*14), 1023C>T (*17), 1659G>A (*29), 2988G>A (*41) and gene duplication. The absence of investigated single nucleotide polymorphisms (SNPs) gave the default allele assignment “*1”.
Pharmacokinetic Analysis
Data were analyzed using the first-order conditional estimation with interaction (FOCE-I) method in the nonlinear mixed-effects modeling software NONMEM version 7.2 (Globomax LLC, Hanover, MD, USA). Nonlinear mixed-effects compartmental modeling was preferred instead of noncompartmental analysis because of the ability to accurately study the time-dependent effects of fasting on the pharmacokinetics of the probe drugs [8]. Furthermore, NONMEM allows to study only the variability between both interventions (i.e. the effect of fasting versus the control intervention) without incorporating other factors that may bias this variability, such as time-based interoccasion variability [15].
Structural Model
The concentration data were log-transformed for all compounds; one-, two-, and three-compartment models were fitted to the data. The population models were built in a stepwise manner. The following parameters were quantified: clearance (CL), intercompartment clearance (Q), and volume of distribution of the central (V 1) and peripheral compartment (V 2). For caffeine, midazolam and S-warfarin the absorption rate constant (K a) could not be estimated and was fixed to 6 h−1. In order to account for the delay between administration of omeprazole and absorption from the gut, also known as transit time, transit compartments were incorporated in the omeprazole pharmacokinetic model [16]. The mean transit time (MTT) between the gut and systemic circulation was estimated by dividing the ratio of the number of transit compartments (n) by the transition rate constant (K tr) between the compartments (MTT = n/K tr) [16].
For all parameter estimates, inter- and intraindividual variability were assessed assuming a log-normal distribution and an exponential error model [8, 15]. Residual variability was estimated with an additional error model.
Software such as R version 64 3.0.1 (The R Foundation for Statistical Computing, Vienna, Austria) and Xpose version 4 (Uppsala University, Dept. of Pharmaceutical Biosciences, Uppsala, Sweden) were used to visualize and evaluate the models [17]. Pirana software (Pirana Software & Consulting BV, Denekamp, The Netherlands) was used as an interface between NONMEM, R and Xpose [18].
The log-likelihood ratio test was used to discriminate between different structural and statistical models. A reduction in the objective function value (OFV) of ≥3.9 points was considered statistically significant (p < 0.05 for one degree of freedom) [15]. In addition, goodness-of-fit plots (population or individual predictions versus observations of measured drug concentrations, and conditional weighted residuals (CWRES) versus time and population predictions) and η and ε shrinkage were assessed [19]. Furthermore, the confidence interval (CI) of the parameter estimates, the correlation matrix, and visual improvement of the individual plots were used to evaluate the model. Ill-conditioning was assessed by the ratio between the largest and smallest eigenvalue of the covariance matrix of the estimate from the NONMEM output. A ratio of >1000 indicates ill-conditioning of the model and is often due to overparameterization [20].
Covariate Analysis
The effect of fasting on pharmacokinetic parameters, systemic clearance (CL), bioavailability (F), and volume of distribution (V) was evaluated by stepwise inclusion in the models [8, 21].
In order to study a possible time dependency of fasting on the pharmacokinetics of the drugs in the cocktail, a time cut-point covariate model was used in which the pharmacokinetic parameter was increased or decreased due to fasting before the time cut-point (θ cut) and comparable with the control intervention after θ cut [8]. The effect of fasting was tested for one pharmacokinetic parameter at a time and statistically tested by the likelihood ratio test. When fasting significantly affected more than one parameter, the model with the largest decrease in the OFV was chosen as the basis to sequentially explore the influence of additional parameters. The final model containing the effect of fasting was further evaluated as discussed in the structural model section.
Model Validation
To evaluate validity and robustness of the final models, simulation-based diagnostics (visual predictive checks [VPCs]) and bootstrap diagnostics were used [22, 23]. The bootstrap analysis was performed using the Perl modules Pearl-speaks-NONMEM. The model-building dataset was resampled 1000 times to create new datasets similar in size [22, 24]. Parameter estimates obtained by the bootstrap analysis (median values and the 2.5th and 97.5th percentiles of parameter distribution) were compared with the parameter estimates of the final pharmacokinetic models. VPC plots were used to compare the 10th and 90th percentiles of simulated concentration–time profiles (1000 replicates) with the observed concentrations [23].
Statistical Analysis
A paired t test (normally distributed data) and a Wilcoxon signed-rank test (not normally distributed data) were used to test differences in biochemical parameters between the occasions, and the Shapiro–Wilk test was used to assess the normality of data distribution. A p-value ≤0.05 was considered significant. Statistical analysis was performed using IBM SPSS Statistics version 23.0 (IBM Corporation, Armonk, NY, USA).
Results
Healthy Subjects and Study Design
Twelve healthy male subjects (mean age 23.6 years) were recruited to participate in the trial. Eight subjects completed all four interventions. This study was based on an amendment of our previously published study in which nine subjects received the cocktail orally [8]. Of these nine subjects, six also received the cocktail intravenously. In addition, the data of the other three subjects who received the drug cocktail on the two occasions after oral administration, and the data of one other subject who completed the two intravenous interventions plus one oral intervention, were included to further optimize the models.
No adverse events were reported, and baseline characteristics are shown in Table 1.
Table 1.
Baseline characteristics
| Patient characteristics | Median (range) |
|---|---|
| Age, years | 23 (19–33) |
| Height, cm | 186 (175–194) |
| Body weight, kg | 80 (68–103) |
| Ethnicity [n = 12] | Caucasian |
| Genotype [n = 9] | n (%) |
|---|---|
| CYP1A2 | |
| *1A/*1F (EM) | 5 (56) |
| *1F/*1F (EM) | 4 (44) |
| CYP2C9 | |
| *1/*1 (EM) | 1 (11) |
| *1/*2 (IM) | 5 (56) |
| *1/*3 (IM) | 3 (33) |
| CYP2C19 | |
| *1/*1 (EM) | 2 (22) |
| *1/*2 (IM) | 4 (44) |
| *1/*4 (IM) | 1 (11) |
| *1/*8 (IM) | 1 (11) |
| *1/*17 (EM) | 1 (11) |
| CYP2D6 | |
| *1/*1 (EM) | 1 (11) |
| *1/*1 (xN) (UM) | 1 (11) |
| *1/*2 (EM) | 1 (11) |
| *1/*3 (IM) | 1 (11) |
| *1/*4 (IM) | 4 (44) |
| *4/*4 (PM) | 1 (11) |
| CYP3A4 | |
| *1A/*1A (EM) | 9 (100) |
EM extensive metabolizer, IM intermediate metabolizer, PM poor metabolizer, UM ultra-rapid metabolizer, xN allele duplication
DNA for the analysis of CYP1A2, CYP2C9, CYP2C19, CYP2D6, and CYP3A4 polymorphisms was available in nine subjects. The distribution of genotypes are shown in Table 1. Subjects were characterized as either extensive metabolizers (EMs, normal CYP enzyme activity) and/or intermediate metabolizers (IMs, slightly reduced CYP enzyme activity compared with EMs) for CYP1A2, CYP2C9, CYP2C19 and CYP3A4. For CYP2D6, one subject was characterized genotypically as a poor metabolizer (PM, little or no CYP2D6 enzyme activity) and another subject was characterized as an ultra-rapid metabolizer (UM, multiple copies of the CYP2D6 gene and therefore increased CYP2D6 enzyme activity) (Table 1).
After 36 h of fasting, the biomarkers for fasting (glucose, γ-hydroxybutyrate, free fatty acids, and acetoacetate) were all significantly altered in comparison with the control condition, which indicates compliance to the fasting protocol (Table 2).
Table 2.
Effect of 36 h of fasting on biochemical parameters in healthy subjects
| Value | Oral administration [n = 11] | Intravenous administration [n = 9] | ||||
|---|---|---|---|---|---|---|
| Control | Fasting (36 h) | p-Value | Control | Fasting (36 h) | p-Value | |
| Glucose, mmol/l | 5.07 (4.60–5.90) | 4.05 (3.60–5.10) | 0.011 | 4.97 (4.40–5.40) | 4.40 (3.60–5.00) | 0.024 |
| β-Hydroxybutyrate, mmol/l | 0.11 (0.00–0.40) | 0.69 (0.30–1.60) | 0.005 | 0.10 (0.00–0.20) | 0.83 (0.30–2.70) | 0.011 |
| Acetoacetate, mmol/l | 0.05 (0.01–0.15) | 0.19 (0.08–0.33) | 0.005 | 0.05 (0.01–0.09) | 0.25 (0.06–0.80) | 0.012 |
| Free fatty acids, mmol/l | 0.49 (0.15–0.91) | 1.03 (0.60–1.55) | 0.018 | 0.48 (0.19–0.73) | 1.29 (0.63–2.57) | 0.012 |
Data are expressed as median (range)
Pharmacokinetics of CYP Probe Drugs
The pharmacokinetics of the five probe drugs after both oral and intravenous administration were characterized by nonlinear mixed-effects modeling (NONMEM). The data of all 12 subjects included in the trial were used to develop pharmacokinetic models. Since not all subjects received the four administrations, this may introduce an unbalanced design. However, one of the advantages of NONMEM over noncompartmental analysis is the effective way of incorporating an unbalanced design [25]. Therefore, this does not preclude accurate analysis of the effect of fasting within subjects. The plasma concentration versus time profiles were described using a one-compartment model for caffeine, a two-compartment model for metoprolol and omeprazole, and a three-compartment model for metoprolol and S-warfarin (Table 3).
Table 3.
Pharmacokinetic parameters of the CYP probe drugs in 12 healthy male subjects, and median parameter values (2.5–97.5%) of nonparametric bootstrap replicates of the final pharmacokinetic models
| Parameters | Caffeine (CYP1A2) | Metoprolol (CYP2D6) | Midazolam (CYP3A4) | Omeprazole (CYP2C19) | S-Warfarin (CYP2C9) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Estimates [typical value (95% CI)] | Bootstrap [median (2.5–97.5%)] | Estimates [typical value (95% CI)] | Bootstrap [median (2.5–97.5%)] | Estimates [typical value (95% CI)] | Bootstrap [median (2.5–97.5%)] | Estimates [typical value (95% CI)] | Bootstrap [median (2.5–97.5%)] | Estimates [typical value (95% CI)] | Bootstrap [median (2.5–97.5%)] | |
| CL, L h−1 | 6.67 (5.32–8.02) | 6.65 (5.45–8.22) | 65.8 (57.4–74.3) | 65.8 (57.8–73.9) | 24.1 (22.5–25.7) | 23.9 (20.1–25.6) | 14.3 (12.1–16.6) | 14.3 (12.1–16.7) | 0.19 (0.16–0.22) | 0.19 (016–0.22) |
| θ CL Fasting | 1.17 (1.06–1.28) | 1.16 (1.05–1.28) | 1.13 (1.06–1.20) | 1.13 (1.06–1.21) | – | – | – | – | 0.81 (0.67–0.96) | 0.81 (0.68–0.99) |
| θ CL,cutp, h | – | – | – | – | – | – | – | – | 13.9 (12.1–15.7) | 14.0 (2.7–24.4) |
| θ CL Slow metab | – | – | 0.44 (fixed) | 0.44 (fixed) | – | – | – | – | – | – |
| θ CL UR metab | – | – | 2.01 (fixed) | 2.01 (fixed) | – | – | – | – | – | – |
| V 1, L | 50.5 (44.2–56.8) | 50.3 (44.7–57.1) | 187 (147–227) | 185 (146–232) | 12.5 (8.8–16.2) | 12.4 (9.2–16.3) | 9.2 (6.9–11.5) | 9.3 (6.9–12.0) | 10.3 (9.2–11.4) | 10.3 (9.1–11.3) |
| θ V1,Fasting | 0.91 (0.83–0.99) | 0.91 (0.83–0.99) | – | – | – | – | – | – | 0.79 (0.75–0.84) | 0.79 (0.75–0.84) |
| θ V1,cutp, h | – | – | – | – | – | – | – | – | 25.6 (20.3–30.9) | 26.1 (11.6–31.7) |
| Ka, fixed | 6 | 6 | 1.19 (0.92–1.46) | 1.21 (0.95–1.57) | 6 | 6 | – | – | 6 | 6 |
| F | 0.97 (0.87–1.07) | 0.97 (0.87–1.07) | 0.45 (0.39–0.50) | 0.45 (0.39–0.51) | 0.35 (0.29–0.41) | 0.35 (0.30–0.41) | 0.44 (0.37–0.52) | 0.44 (0.37–0.52) | 1 (fixed) | 1 (fixed) |
| θ FSlow metab | – | – | 2.28 (fixed) | 2.28 (fixed) | – | – | – | – | – | |
| θ FUR metab | – | – | 0.43 (fixed) | 0.43 (fixed) | – | – | – | – | – | |
| F predose | 0.04 (0.01–0.06) | 0.04 (0.02–0.08) | – | – | – | – | – | – | – | – |
| MTT, h | – | – | – | – | – | – | 1.60 (1.42–1.78) | 1.60 (1.42–1.77) | – | – |
| Q, L h−1 | – | – | 74.5 (40.2–108) | 74.0 (44.0–129) | 13.3 (11.0–15.6) | 13.3 (11.3–16.9) | 5.2 (2.3–8.1) | 5.1 (2.6–9.7) | 1.41 (1.05–1.78) | 1.42 (1.09–1.84) |
| V 2, L | – | – | 97.6 (71.3–124) | 98.3 (66.9–126) | 52.1 (35.8–68.5) | 52.9 (40.7–125) | 3.7 (2.7–4.7) | 3.7 (2.6–4.8) | 6.0 (5.3–6.8) | 5.93 (5.06–6.74) |
| Q 2, L h−1 | – | – | – | – | 139 (84–194) | 138 (94–195) | – | – | 0.11 (0.08–0.15) | 0.11 (0.09–0.16) |
| V 3, L | – | – | – | – | 30.2 (25.6–34.8) | 30.3 (26.6–34.8) | – | – | 27.2 (19.1–35.3) | 26.8 (18.4–38.6) |
| Interindividual variability | ||||||||||
| CL, % | 34.6 (22.1–44.2) | 32.7 (20.6–42.7) | 26.3 (18.0–32.5) | 25.1 (16.9–32.4) | 13.4 (7.9–17.3) | 12.9 (7.7–17.9) | 20.7 (9.9–27.7) | 19.4 (8.7–27.2) | 24.1 (10.5–32.8) | 22.9 (11.2–31.2) |
| V 1, % | – | – | – | – | – | – | – | – | 20.7 (10.1–27.7) | 19.4 (10.8–26.8) |
| Q 2, % | – | – | – | – | – | – | – | – | – | – |
| Intraindividual variability | ||||||||||
| CL, % | 17.5 (10.0–22.6) | 16.8 (11.3–23.0) | 11.1 (5.5–14.7) | 10.3 (6.7–15.7) | 11.2 (5.4–14.9) | 11.0 (6.6–15.8) | 16.7 (11.9 –20.5) | 16.5 (12.7–22.0) | – | |
| F predose, % | 1250 (670–2300) | 1147 (628–1890) | – | – | – | – | – | – | – | – |
| MTT, % | – | – | – | – | – | – | 22.8 (13.5–29.5) | 22.3 (14.3–30.0) | – | – |
| Residual variability | ||||||||||
| Prop. error | 0.13 (0.11–0.16) | 0.13 (0.10–0.16) | 0.20 (0.15–0.25) | 0.20 (0.14–0.24) | 0.16 (0.15–0.18) | 0.16 (0.14–0.17) | 0.35 (0.27–0.42) | 0.34 (0.26–0.41) | 0.17 (0.15–0.20) | 0.17 (0.14–0.19) |
CL clearance, CYP cytochrome P450, F bioavailability, Ka absorption rate constant, MTT mean transit time, Q inter-compartment clearance, V 1 volume of distribution of the central compartment, V 2 volume of distribution of the peripheral compartment, θ difference in PK parameter, θ cut time cut-point
Model Validation
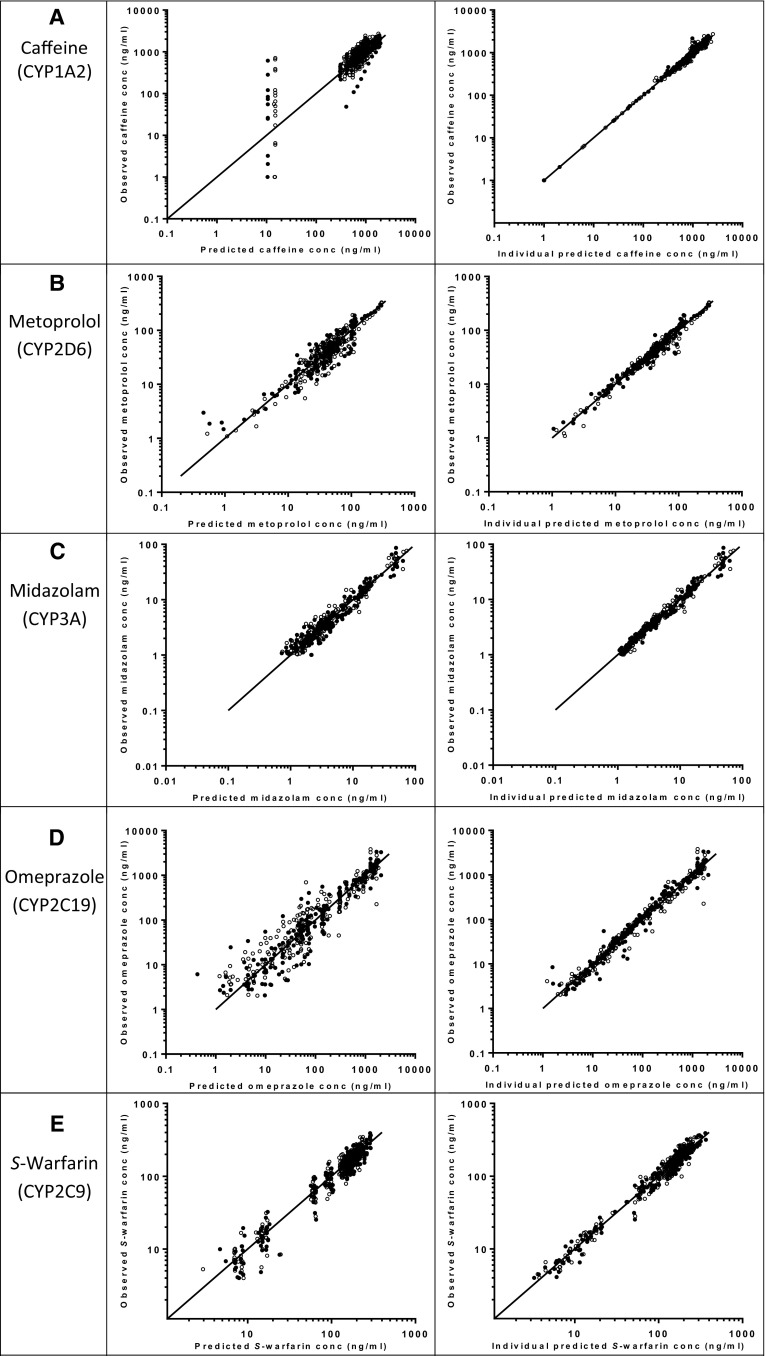
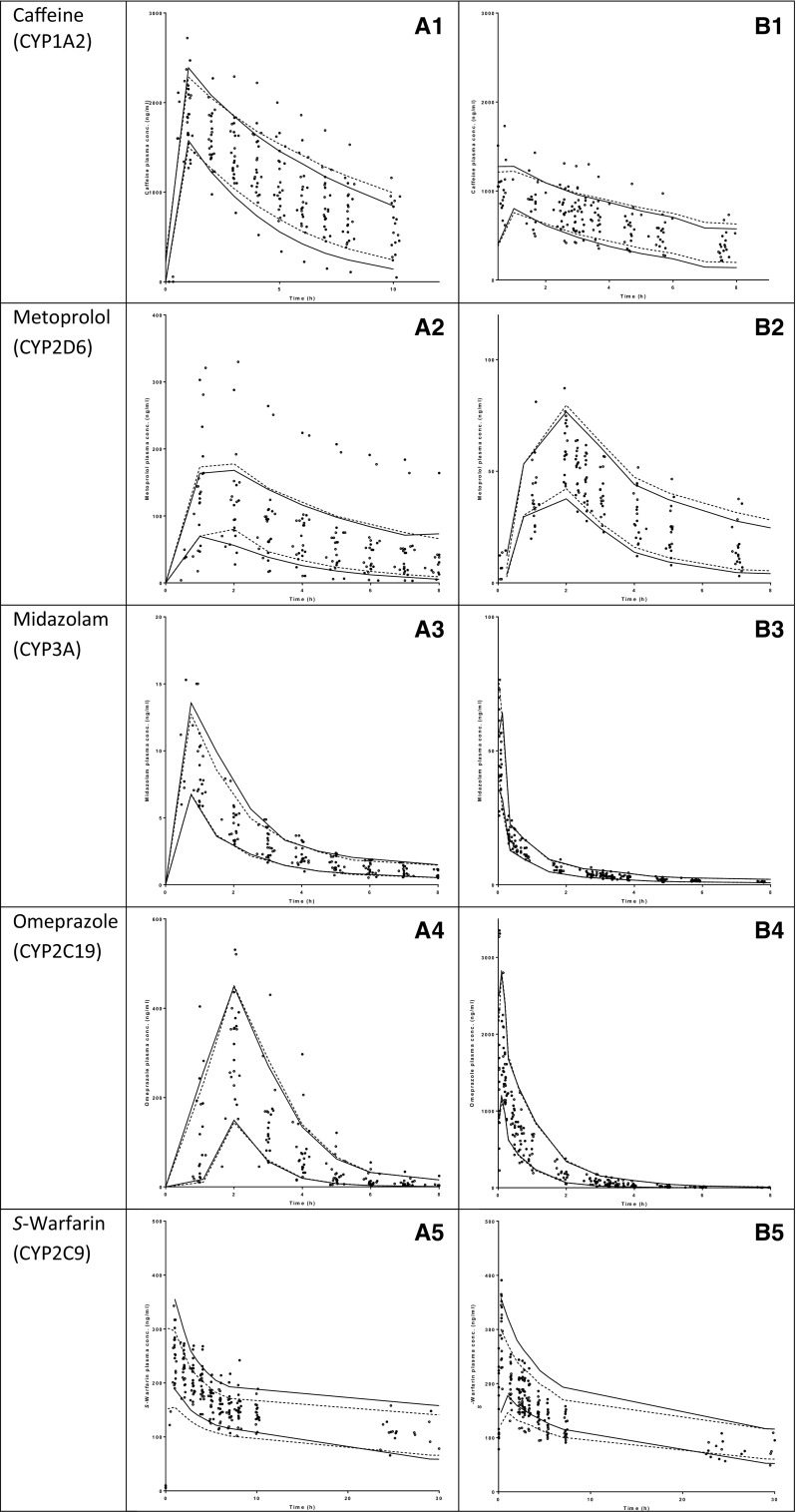
The observed data were described well by the developed models, as demonstrated by the goodness-of-fit plots (Fig. 1). Furthermore, no trends were observed in the plots of CWRES versus time or model-predicted concentrations (plots not shown). The η and ε shrinkage of the pharmacokinetic parameters and residual variability were <20%. Table 3 gives an overview of the parameter estimates of the final models and the nonparametric bootstraps (n = 1000 replicates per model). As the latter were in agreement with those of the final pharmacokinetic models, the parameter estimates of the final models are considered reliable. VPC plots further demonstrate the validity of the models since the central tendency and variability of the simulated data is comparable with the observed data (Fig. 2).
Fig. 1.
Goodness-of-fit plots of the five CYP probe drugs. Observed concentrations versus population-predicted (left panel) and individual-predicted (right panel) concentrations: a caffeine (CYP1A2); b metoprolol (CYP2D6); c midazolam (CYP3A4); d omeprazole (CYP2C19); e S-warfarin (CYP2C9). The closed circles represent the 36 h of fasting intervention and the open circles represent the control intervention. The solid line is the line of identity. CYP cytochrome P450, conc concentration
Fig. 2.
Visual predictive check plots of the five CYP probe drugs after oral [left panel (1)] and intravenous [right panel (2)] administration: a caffeine (CYP1A2); b metoprolol (CYP2D6); c midazolam (CYP3A4); d omeprazole (CYP2C19); e S-warfarin (CYP2C9). The closed circles represent the observed data points after 36 h of fasting and the open circles represent the control observations. The solid (36 h fasting) and dashed (control) lines represent the 10th and 90th percentiles of the simulated data. CYP cytochrome P450, conc concentration
Effect of Fasting on Oral Bioavailability and Systemic Clearance
Caffeine (CYP1A2)
Although restricted by the study protocol, preadministration plasma concentrations of caffeine were observed (range 0–709 mg/L). To account for this variable pre-intake of caffeine, we incorporated a fictive caffeine dose of 100 mg orally or 50 mg intravenously, with variable bioavailability in the model that was administered 12 h before administration of the cocktail. The typical bioavailability and its interoccasion variability of this pre-intake were estimated in the NONMEM analysis. The mean pre-intake of caffeine was low since the typical bioavailability was 4.0%, whereas the variability was high (1250%) due to three subjects with observed caffeine plasma concentrations at baseline.
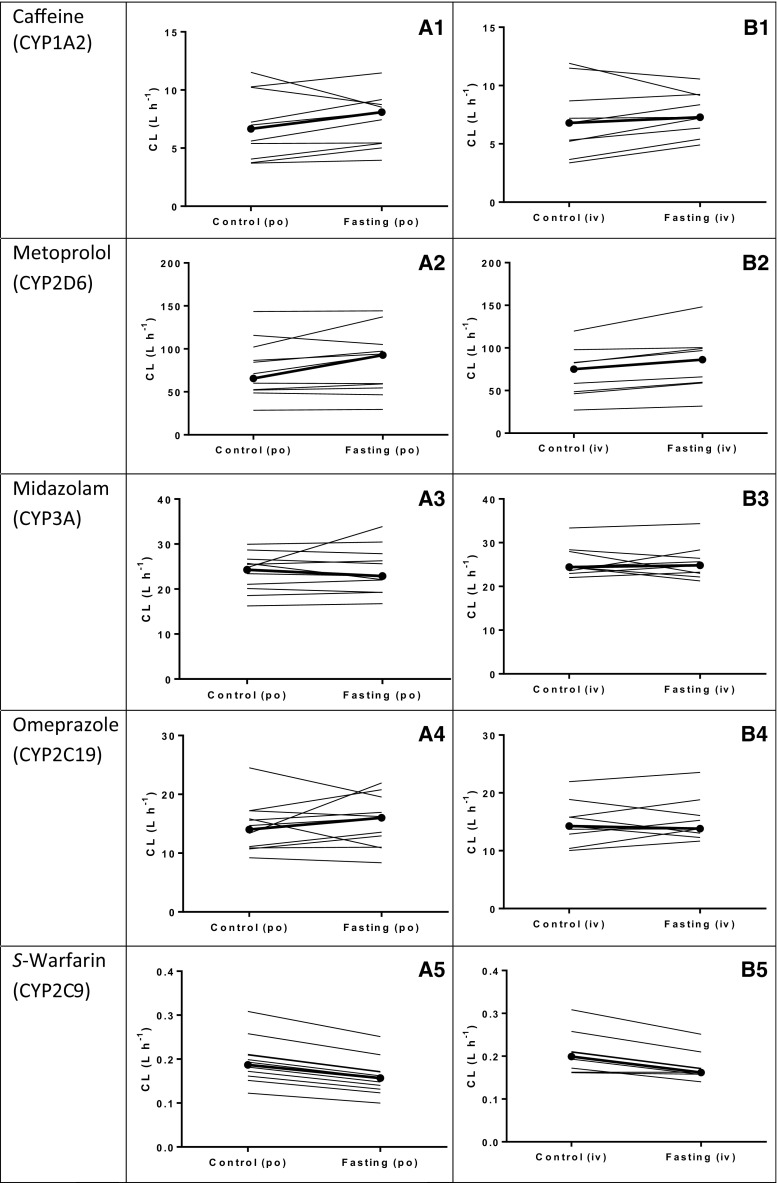
The typical subject had a systemic caffeine clearance (CLcaffeine) of 6.67 L/h. Fasting increased CLcaffeine by 17% (θ CL,fasting = 1.17, 95% CI 1.06–1.28, p = 0.04) (Table 3). The accompanying VPC plot also illustrates this effect (Fig. 2A1, B1). After post hoc analysis, 36 h of fasting increased the median caffeine clearance after oral administration (CLPO-caffeine,posthoc) from 6.67 L/h (range 3.71–11.52) in the control group to 8.09 L/h (range 3.95–17.47). After intravenous administration, fasting increased the median post hoc caffeine clearance (CLIV-caffeine,posthoc) from 6.80 L/h (range 3.37–11.91) in the control group to 7.29 L/h (range 4.91–10.56) (Fig. 3A1, B1). Furthermore, 36 h of fasting decreased the central volume of distribution (V 1) by 9% (V 1,caffeine = 0.91, 95% CI 0.83–0.99, p = 0.01). Fasting did not affect the oral bioavailability of caffeine (F caffeine) (Table 3).
Fig. 3.
Effect of fasting on post hoc individual clearance rates of the five CYP probe drugs after oral (left panel) and intravenous (right panel) administration of the cocktail: a caffeine (CYP1A2); b metoprolol (CYP2D6); c midazolam (CYP3A4); d omeprazole (CYP2C19); e S-warfarin (CYP2C9). The closed circles represent the median values. CYP cytochrome P450, po oral, iv intravenous
Metoprolol (CYP2D6)
For two subjects, the exposure of metoprolol clearly deviated from the other subjects based on the plasma concentration–time curves. Both subjects were also characterized genotypically as a CYP2D6 PM (CYP2D6 *4/*4) and UM (CYP2D6 *1/*xN1), respectively (Table 1). Systemic CLmetoprolol was 65.8 L/h for the typical subject, but 56% lower for the PM. Furthermore, typical bioavailability of metoprolol was 45% and was more than twofold higher in this subject. For the UM, CLmetoprolol was doubled and bioavailability (F UR) was lower, with a value of 19%. Estimation of the difference in bioavailability and clearance of the PM and UM significantly improved the final model of metoprolol based on OFV (ΔOFV = −87.6), but also led to ill-conditioning. Therefore, bioavailability and clearance of the PM and UM were determined using a similar NONMEM model that only included the data of the control intervention without taking the effect of fasting into account, and both parameters were then FIXED in the final model.
Fasting increased systemic CLmetoprolol by 13% (θ CL,fasting = 1.13, 95% CI 1.06–1.20, p < 0.01), but did not affect oral bioavailability of metoprolol (F metoprolol) (Table 3; Fig. 2A2, B2). Following oral administration of metoprolol, short-term fasting increased the median post hoc estimates for systemic clearance from 65.7 L/h (range 28.6–143.4) after the control intervention to 92.7 L/h (range 29.5–144.2) after 36 h of fasting. After intravenous administration, short-term fasting increased the median metoprolol clearance (CLIV-metoprolol,posthoc) from 75.2 L/h (range 27.1–119.8) to 86.2 L/h (range 31.8–148.2) (Fig. 3A2, B2).
Midazolam (CYP3A4)
The systemic clearance (CLmidazolam) or oral bioavailability (F midazolam) of midazolam was not affected by fasting (Table 3; Fig. 2A3, B3). Median post hoc estimates for systemic midazolam clearance after oral administration were 24.3 L/h (range 16.3–30.0) after the control intervention and 22.87 L/h (range 16.75–33.89) after 36 h of fasting. Following intravenous administration, the median clearance of midazolam (CLIV-midazolam,posthoc) after the control intervention was 24.43 L/h (range 23.56–33.34) and 24.29 L/h (range 16.27–29.96) after 36 h of fasting (Fig. 3A3, B3).
Omeprazole (CYP2C19)
Since omeprazole is known to show a delay (transit time) between administration and absorption from the gut, we incorporated 10 transit compartments in the model [16]. The MTT was 1.6 h, with an intraindividual variability of 23% (Table 3).
Omeprazole systemic clearance (CLomeprazole) or oral bioavailability (F omeprazole) were not affected by fasting (Table 3; Fig. 2A4, B4). Median post hoc estimates for clearance following oral administration were 14.02 L/h (range 9.20–24.50) and 16.00 L/h (range 8.36–19.54) after the control intervention and 36 h of fasting, respectively. After intravenous administration of omeprazole, the median clearance was 14.27 L/h (range 10.03–21.93) after the control intervention and 13.80 L/h (range 11.65–23.54 L/h) after 36 h of fasting (Fig. 3A4, B4).
S-Warfarin (CYP2C9)
Estimation of oral bioavailability (F S-warfarin) resulted in an approximate value of F S-warfarin ≅ 1, indicating that bioavailability after oral administration is circa 100%, which is also described in the literature [13]. Since estimation of bioavailability did not improve the model, this parameter was fixed to F S-warfarin = 1.
Until 14 h after cocktail administration, fasting decreased S-warfarin systemic clearance by 19% compared with the control group (θ CL,fasting = 0.81, 95% CI 0.67–0.96, p < 0.01). Fasting also decreased the central volume of distribution by 21% (θ V1,fasting = 0.79, 95% CI 0.75–0.84, p < 0.001); the corresponding time cut-point was 25 h (Table 3; Fig. 2A5, B5). As both CL and V 1 decreased at approximately the same amount, an effect of fasting on bioavailability may also explain the result after oral administration of the cocktail. However, similar results were found after intravenous administration of the cocktail, which indicates that bioavailability does not play a role. After post hoc analysis, short-term fasting decreased the median systemic S-warfarin clearance following oral administration from 0.19 L/h (range 0.12–0.31) after the control intervention to 0.16 L/h (range 0.12–0.25) after 36 h of fasting. After intravenous administration of warfarin, fasting decreased the median clearance from 0.20 L/h (range 0.16–0.31) after the control intervention to 0.17 L/h (range 0.14–0.26) after 36 h of fasting (Fig. 3A5, B5).
Discussion
In this crossover intervention study, we determined the effects of short-term fasting on oral bioavailability and systemic clearance related to CYP-mediated drug metabolism in healthy subjects, and found that short-term fasting increased systemic clearance of caffeine and metoprolol. This indicates that fasting increased the activity of CYP1A2 and CYP2D6, considering that caffeine and metoprolol are probes for the activity of these enzymes, respectively. Furthermore, short-term fasting decreased systemic S-warfarin clearance, which indicates decreased activity of CYP2C9, considering that S-warfarin is a probe of CYP2C9 activity. Although short-term fasting affected systemic clearance mediated by several CYP enzymes, fasting did not affect oral bioavailability of the five CYP probe drugs. The drug cocktail used has previously been validated by Turpault et al. [9]. The absence of a pharmacokinetic interaction between the probe drugs makes this cocktail useful for the in vivo evaluation of metabolism-based interactions [9].
The effects of fasting on systemic clearance of caffeine and S-warfarin are in line with our previous findings that short-term fasting alters oral clearance of both drugs in a non-uniform pattern [8]. We can now confirm that fasting affects systemic clearance rather than an effect on oral bioavailability. In contrast to our previous study, short-term fasting also altered metoprolol clearance, which is likely due to increased statistical power because of a larger sample size.
Estimation of the difference in bioavailability between the control and the fasting intervention with NONMEM did not improve the models. However, we were able to estimate bioavailability of the probe drugs for the typical subject, and the results are all in agreement with the product characteristics of the drugs: F caffeine ≈ 0.97, F metoprolol ≈ 0.45, F midazolam ≈ 0.35, F omeprazole ≈ 0.44, and F s-warfarin ≈ 1.0 [13]. This strengthens our conclusion that no effect of short-term fasting on bioavailability was found.
Despite the study restrictions, pre-intake plasma concentrations of caffeine were observed at baseline, which may question the compliance to the protocol regarding the other fasting-related restrictions. Compliance to the fasting protocol was tested by measuring biomarkers for fasting (glucose, β-hydroxybutyrate, free fatty acids, and acetoacetate) before administration of the cocktail at each intervention. For all subjects, short-term fasting readily changed the plasma concentrations of these biomarkers, which indicates compliance to the fasting protocol.
Subjects were genotyped for the CYP1A2, CYP2C9, CYP2C19, CYP2D6, and CYP3A4 polymorphisms (Table 1). For metoprolol only, the genotype improved the model. However, the randomized crossover design in which each subject serves as his own control enabled us to only study the effect of fasting, instead of a mixture of other factors that may also influence drug metabolism, such as genotype, and physiologic and environmental factors.
Although the data acquisition in this randomized, controlled, crossover intervention study was sufficiently dense and individuals served as their own controls, NONMEM was preferred over noncompartmental analysis because of its ability to accurately study time-dependent effects of fasting on the pharmacokinetics of the probe drugs. This was based on recently reported findings of a variable time-dependent effect of fasting on S-warfarin clearance that diminished when returned to the nonfasting state [8]. The observation of the caffeine pre-intake at baseline further strengthens our choice for NONMEM since we were able to account for this variable pre-intake of caffeine, thereby excluding this type of bias (this would not have been possible when using noncompartmental analysis).
In addition to the potential effects on oral bioavailability, drug clearance can also be affected by differences in protein binding. The observed differences in clearance of caffeine, metoprolol, and S-warfarin may therefore not only be an effect of fasting on intrinsic clearance but also on protein binding, as described by the well-stirred model of drug metabolism (Eqs. 1 and 2) [26]:
| 1 |
| 2 |
where Q H indicates hepatic blood flow, f u indicates fraction of unbound drug in plasma, CLint indicates intrinsic clearance of unbound drug, and E H indicates the hepatic extraction ratio.
For drugs with a low extraction ratio (E H < 0.3), such as caffeine (f u ≅ 0.65) and S-warfarin (f u ≅ 0.01), the hepatic clearance can mainly be influenced by changes in protein binding and intrinsic clearance [27]. We have shown that fasting increased caffeine clearance but decreased S-warfarin clearance in a non-uniform pattern. If fasting affects protein binding, we would have expected a similar pattern for both drugs. Therefore, it is more likely that the observed effect of fasting on caffeine and S-warfarin clearance is based on intrinsic clearance caused by differences in CYP enzyme activity. In addition, this non-uniform pattern in drug clearance is in line with the expression of hepatic messenger RNA (mRNA) of the corresponding CYP enzymes in rat [8].
Following intravenous administration of drugs with an intermediate extraction ratio (E H = 0.3–0.7), such as metoprolol (E H = 0.67), midazolam (E H = 0.31), and omeprazole (E H = 0.35), drug clearance can be affected by changes in a combination of three determinants: Q H, CLint, and f u [13, 27]. We have shown that fasting increases metoprolol clearance but it is unlikely that this is caused by an effect of fasting on Q H or f u instead of CLint. The fraction of unbound metoprolol is already high (90–95%) and therefore is not likely to be significantly affected by fasting [13]. Furthermore, it is unlikely that fasting would have increased hepatic blood flow to retrieve the observed effect since the opposite has been described in literature [28, 29]. Additional research needs to be performed to study the effect of fasting on protein binding.
Our findings imply that dosage adjustments of drugs metabolized by CYP enzymes could be necessary to improve drug treatment in patients with malnutrition. Recently, studies have shown that short-term fasting can have a positive effect on the treatment of cancer [30–32]. By short-term fasting, the susceptibility to chemotherapy can differ between healthy somatic and cancer cells, a phenomenon called differential stress resistance [30]. Because it is likely that the effects of short-term fasting will be enhanced if the period of fasting is prolonged, clinical trials are now being performed in the field of oncology, with low protein fasting mimicking diets to ease the burden of prolonged fasting [31, 33]. In order to optimize drug treatment, it would be of interest to study not only the effect of these diets on differential stress resistance but also to study the effect of the fasting-based diet on the metabolism of the drugs that are administered during this diet, especially for drugs with a small therapeutic range and/or metabolized by the specific CYP enzymes that were affected by fasting (CYP1A2, CYP2D6, and CYP2C9), e.g. bendamustine, cyclophosphamide, flutamide, gefitinib, idarubicin, or tamoxifen [13, 34].
Although more preclinical than clinical research has been performed to study the pharmacological mechanism behind our findings, the exact mechanism of action of fasting still remains unsolved. Nuclear receptors such as the pregnane X receptor (PXR) and constitutive androstane receptor (CAR) regulate the activity of many of the drug-metabolizing CYP enzymes [35]. The fact that fasting differentially affects CYP enzymes showing either increased (CYP1A2, CYP2D6) or decreased (CYP2C9) activity indicates a complex interplay of different factors that regulate CYP enzyme activity.
Conclusion
Our study provides evidence that short-term fasting alters systemic clearance of CYP-mediated drug metabolism but does not affect changes in oral bioavailability of drugs. Additional research is warranted to study the clinical implications of our findings.
Acknowledgements
The authors would like to thank M. Pistorius and M. Van der Meer for their specific contribution to the laboratory analysis of this study, and L. Kuijsten for her technical assistance with the clinical trial experiments.
Compliance with Ethical Standards
Funding
No funding was received.
Conflicts of Interest
Laureen A. Lammers, Roos Achterbergh, Ron H. N. van Schaik, Johannes A. Romijn, and Ron A. A. Mathôt have no conflicts of interest to declare.
Ethical Approval
The study was conducted in accordance with the ethical principles for medical research involving human subjects (Helsinki Declaration of 1975, as revised in 2013).
Informed Consent
All subjects provided written informed consent prior to participation in this study.
References
- 1.Lin JH. Pharmacokinetic and pharmacodynamic variability: a daunting challenge in drug therapy. Curr Drug Metab. 2007;8(2):109–136. doi: 10.2174/138920007779816002. [DOI] [PubMed] [Google Scholar]
- 2.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Ingelman-Sundberg M. Pharmacogenetics: an opportunity for a safer and more efficient pharmacotherapy. J Intern Med. 2001;250(3):186–200. doi: 10.1046/j.1365-2796.2001.00879.x. [DOI] [PubMed] [Google Scholar]
- 4.Murray M. Altered CYP expression and function in response to dietary factors: potential roles in disease pathogenesis. Curr Drug Metab. 2006;7(1):67–81. doi: 10.2174/138920006774832569. [DOI] [PubMed] [Google Scholar]
- 5.Merrell MD, Cherrington NJ. Drug metabolism alterations in nonalcoholic fatty liver disease. Drug Metab Rev. 2011;43(3):317–334. doi: 10.3109/03602532.2011.577781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longo V, Ingelman-Sundberg M, Amato G, Salvetti A, Gervasi PG. Effect of starvation and chlormethiazole on cytochrome P450s of rat nasal mucosa. Biochem Pharmacol. 2000;59(11):1425–1432. doi: 10.1016/S0006-2952(00)00266-5. [DOI] [PubMed] [Google Scholar]
- 7.Qu W, Rippe RA, Ma J, Scarborough P, Biagini C, Fiedorek FT, et al. Nutritional status modulates rat liver cytochrome P450 arachidonic acid metabolism. Mol Pharmacol. 1998;54(3):504–513. doi: 10.1124/mol.54.3.504. [DOI] [PubMed] [Google Scholar]
- 8.Lammers LA, Achterbergh R, de Vries EM, van Nierop FS, Klumpen HJ, Soeters MR, et al. Short-term fasting alters cytochrome P450-mediated drug metabolism in humans. Drug Metab Dispos. 2015;43(6):819–828. doi: 10.1124/dmd.114.062299. [DOI] [PubMed] [Google Scholar]
- 9.Turpault S, Brian W, Van Horn R, Santoni A, Poitiers F, Donazzolo Y, et al. Pharmacokinetic assessment of a five-probe cocktail for CYPs 1A2, 2C9, 2C19, 2D6 and 3A. Br J Clin Pharmacol. 2009;68(6):928–935. doi: 10.1111/j.1365-2125.2009.03548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 11.Thummel KE, O’Shea D, Paine MF, Shen DD, Kunze KL, Perkins JD, et al. Oral first-pass elimination of midazolam involves both gastrointestinal and hepatic CYP3A-mediated metabolism. Clin Pharmacol Ther. 1996;59(5):491–502. doi: 10.1016/S0009-9236(96)90177-0. [DOI] [PubMed] [Google Scholar]
- 12.Foster KJ, Alberti KG, Hinks L, Lloyd B, Postle A, Smythe P, et al. Blood intermediary metabolite and insulin concentrations after an overnight fast: reference ranges for adults, and interrelations. Clin Chem. 1978;24(9):1568–1572. [PubMed] [Google Scholar]
- 13.Micromedex . Micromedex solutions—drug monographs. 2014. Greenwood Village: Truven Health Analytics Inc; 2014. [Google Scholar]
- 14.Lammers LA, Achterbergh R, Pistorius MC, Bijleveld Y, de Vries EM, Boelen A, et al. Quantitative method for simultaneous analysis of a 5-probe cocktail for cytochrome P450 enzymes. Ther Drug Monit. 2016;38(6):761–768. doi: 10.1097/FTD.0000000000000338. [DOI] [PubMed] [Google Scholar]
- 15.Mould DR, Upton RN. Basic concepts in population modeling, simulation, and model-based drug development-part 2: introduction to pharmacokinetic modeling methods. CPT Pharmacomet Syst Pharmacol. 2013;2:e38. doi: 10.1038/psp.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savic RM, Jonker DM, Kerbusch T, Karlsson MO. Implementation of a transit compartment model for describing drug absorption in pharmacokinetic studies. J Pharmacokinet Pharmacodyn. 2007;34(5):711–726. doi: 10.1007/s10928-007-9066-0. [DOI] [PubMed] [Google Scholar]
- 17.Jonsson EN, Karlsson MO. Xpose: an S-PLUS based population pharmacokinetic/pharmacodynamic model building aid for NONMEM. Comput Methods Programs Biomed. 1999;58(1):51–64. doi: 10.1016/S0169-2607(98)00067-4. [DOI] [PubMed] [Google Scholar]
- 18.Keizer RJ, van Benten M, Beijnen JH, Schellens JH, Huitema AD. Pirana and PCluster: a modeling environment and cluster infrastructure for NONMEM. Comput Methods Programs Biomed. 2011;101(1):72–79. doi: 10.1016/j.cmpb.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Karlsson MO, Savic RM. Diagnosing model diagnostics. Clin Pharmacol Ther. 2007;82(1):17–20. doi: 10.1038/sj.clpt.6100241. [DOI] [PubMed] [Google Scholar]
- 20.Bijleveld YA, de Haan TR, van der Lee HJ, Groenendaal F, Dijk PH, van Heijst A, et al. Altered gentamicin pharmacokinetics in term neonates undergoing controlled hypothermia. Br J Clin Pharmacol. 2016;81(6):1067–1077. doi: 10.1111/bcp.12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandema JW, Verotta D, Sheiner LB. Building population pharmacokinetic–pharmacodynamic models. I. Models for covariate effects. J Pharmacokinet Biopharm. 1992;20(5):511–528. doi: 10.1007/BF01061469. [DOI] [PubMed] [Google Scholar]
- 22.Ette EI. Stability and performance of a population pharmacokinetic model. J Clin Pharmacol. 1997;37(6):486–495. doi: 10.1002/j.1552-4604.1997.tb04326.x. [DOI] [PubMed] [Google Scholar]
- 23.Bergstrand M, Hooker AC, Wallin JE, Karlsson MO. Prediction-corrected visual predictive checks for diagnosing nonlinear mixed-effects models. AAPS J. 2011;13(2):143–151. doi: 10.1208/s12248-011-9255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindbom L, Pihlgren P, Jonsson EN. PsN-Toolkit: a collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Comput Methods Programs Biomed. 2005;79(3):241–257. doi: 10.1016/j.cmpb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Sun X, Li J. PKreport: report generation for checking population pharmacokinetic model assumptions. BMC Med Inform Decis Mak. 2011;11:31. doi: 10.1186/1472-6947-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benet LZ, Hoener BA. Changes in plasma protein binding have little clinical relevance. Clin Pharmacol Ther. 2002;71(3):115–121. doi: 10.1067/mcp.2002.121829. [DOI] [PubMed] [Google Scholar]
- 27.Verbeeck RK. Pharmacokinetics and dosage adjustment in patients with hepatic dysfunction. Eur J Clin Pharmacol. 2008;64(12):1147–1161. doi: 10.1007/s00228-008-0553-z. [DOI] [PubMed] [Google Scholar]
- 28.Lomax MA, Baird GD. Blood flow and nutrient exchange across the liver and gut of the dairy cow. Effects of lactation and fasting. Br J Nutr. 1983;49(3):481–496. doi: 10.1079/BJN19830057. [DOI] [PubMed] [Google Scholar]
- 29.Eipel C, Abshagen K, Vollmar B. Regulation of hepatic blood flow: the hepatic arterial buffer response revisited. World J Gastroenterol. 2010;16(48):6046–6057. doi: 10.3748/wjg.v16.i48.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raffaghello L, Lee C, Safdie FM, Wei M, Madia F, Bianchi G, et al. Starvation-dependent differential stress resistance protects normal but not cancer cells against high-dose chemotherapy. Proc Natl Acad Sci USA. 2008;105(24):8215–8220. doi: 10.1073/pnas.0708100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Groot S, Vreeswijk MP, Welters MJ, Gravesteijn G, Boei JJ, Jochems A, et al. The effects of short-term fasting on tolerance to (neo) adjuvant chemotherapy in HER2-negative breast cancer patients: a randomized pilot study. BMC Cancer. 2015;15:652. doi: 10.1186/s12885-015-1663-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huisman SA, Bijman-Lagcher W, Ijzermans JN, Smits R, de Bruin RW. Fasting protects against the side effects of irinotecan but preserves its anti-tumor effect in Apc15lox mutant mice. Cell Cycle. 2015;14(14):2333–2339. doi: 10.1080/15384101.2015.1044170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandhorst S, Choi IY, Wei M, Cheng CW, Sedrakyan S, Navarrete G, et al. A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab. 2015;22(1):86–99. doi: 10.1016/j.cmet.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 pharmacogenetics and cancer. Oncogene. 2006;25(11):1679–1691. doi: 10.1038/sj.onc.1209377. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez JP, Mota LC, Baldwin WS. Activation of CAR and PXR by dietary, environmental and occupational chemicals alters drug metabolism, intermediary metabolism, and cell proliferation. Curr Pharmacogenom Pers Med. 2009;7(2):81–105. doi: 10.2174/187569209788654005. [DOI] [PMC free article] [PubMed] [Google Scholar]