Abstract
The human gut is colonized by a variety of large amounts of microbes that are collectively called intestinal microbiota. Most of these microbial residents will grow within the mucus layer that overlies the gut epithelium and will act as the first line of defense against both commensal and invading microbes. This mucus is essentially formed by mucins, a family of highly glycosylated protein that are secreted by specialize cells in the gut. In this Review, we examine how commensal members of the microbiota and pathogenic bacteria use mucus to their advantage to promote their growth, develop biofilms and colonize the intestine. We also discuss how mucus-derived components act as nutrient and chemical cues for adaptation and pathogenesis of bacteria and how bacteria can influence the composition of the mucus layer.
Keywords: mucus, commensals, pathogens, biofilm, microbiota, microflora, goblet cells
Introduction
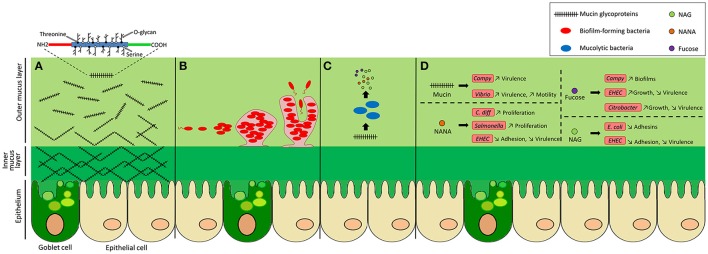
The gastrointestinal tract harbors a complex bacterial community called the intestinal microbiota that, in healthy conditions, maintains a commensal relationship with our body. Various mechanisms are used by the host to keep intestinal homeostasis and to prevent aberrant immune responses directed against the microbiota. One of these is the production of a mucus layer that covers the epithelial cells of the gut. This mucus is synthesized and secreted by host goblet cells and form an integral structural component of the mammal intestine. Its major function is to protect the intestinal epithelium from damage caused by food and digestive secretions (Deplancke and Gaskins, 2001). The mucus layer provides a niche for bacterial colonization because it contains attachment sites and is also a carbon source (Harel et al., 1993). Effectively, the mucus is a direct source of carbohydrates that are released in the lumen. Therefore, several bacterial species of the microbiota can use mucus glycan as a carbon source (Ouwerkerk et al., 2013). An alteration in glycan availability modifies the composition of the microbiota (Martens et al., 2008). The mucus layer also prevents pathogens from reaching and persisting on the intestinal epithelial surfaces and thereby is a major component of innate immunity. It is constantly renewed and acts as a trap for commensal residents, but also for pathogens, preventing their access to the epithelia (Johansson et al., 2008; Bertin et al., 2013). Although its composition and thickness vary along the gut, the mucus layer is mainly formed of glycoproteins containing different glycans; nonspecific antimicrobial molecules, such as antimicrobial peptides (AMP); secreted antibodies targeting specific microbial antigens; and other intestinal proteins (McGuckin et al., 2011; Antoni et al., 2014). Interaction with the mucus layer is important for the colonization of gut commensals as well as some pathogens that have evolved to adhere to mucus and exploit it (Juge, 2012). Some pathogens also use mucus components as a cue to modulate the expression of virulence genes and thereby adapt to the host environment. In this Review, we describe the interactions between bacteria and components of the human mucus layer: their use as carbon sources, adhesion sites and their genetic adaptation (Figure 1).
Figure 1.
Bacterial activities in the colonic mucus layer environment. The colonic epithelium is covered by a mucus gel layer formed of glycoproteins called mucins. (A) Mucins consist of a protein core and a high number of O-linked glycans. They are secreted by goblet cells and are assembled into a net-like structure that forms a dense inner layer, firmly attached to cells, that does not allow bacteria to penetrate. Further from the epithelium, the outer layer becomes loose and permissive, providing a niche for intestinal bacteria. (B) Mucus oligosaccharides can act as adhesion sites for bacteria, facilitating their colonization. Some bacteria are able to form microcolonies and biofilms. (C) Bacteria with mucolytic activity can release monosaccharides from mucin O-glycans and metabolize them. These sugars can also be utilized by nearby bacteria. (D) Mucus components can influence the behavior of pathogenic bacteria by increasing or decreasing their virulence expression, adhesion, motility, proliferation, or growth.
The gastrointestinal mucus
Mucus composition
The intestinal mucus is composed mainly of mucins that are complex agglomerates of structural glycoproteins with specific O-linked glycans (O-glycans) produced by specialized cells of the host called goblet cells (Forstner, 1995). Mucins can either be secreted and form a gel, or be produced as membrane-bound glycoproteins that are part of the epithelial glycocalyx (Johansson et al., 2008, 2011; Jonckheere et al., 2013; Nilsson et al., 2014). These glycoproteins share a common structure made of tandem repeated amino acids rich in proline, threonine and serine and are call PTS domains. These sequences of amino acid provide sites for the covalent attachment of the polysaccharides and are widely O-glycosylated (Moran et al., 2011). Four different types of polysaccharide core structures are commonly found in mucin glycoproteins. These cores are formed by a combination of three polysaccharides, galactose, N-acetyl-galactosamine and N-acetyl-glucosamine (Larsson et al., 2009; Juge, 2012). Different chains of glycan will be attached to the core. The terminal monosaccharide is usually a fucose or a sialic acid (Larsson et al., 2009; Juge, 2012). Oligosaccharide chains are also sulfated, especially in colonic regions (Rho et al., 2005). The mucin proteins MUC1, MUC5AC, and MUC6 mainly form the mucus layer in the stomach, whereas MUC2 is the most abundant mucin in the small intestine and the colon (Johansson et al., 2009; Moran et al., 2011). The thickness of the mucus layer varies through the gut. The colon, which harbors the highest density of microorganisms, is covered by the thickest mucus layer (Gum et al., 1994). It is composed of an inner layer that is dense and firmly attached to the epithelium and an outer loose layer that is exposed to bacterial proteolytic activity. The numerous O-glycans of the outer layer can serve as adhesion sites and as nutrients for bacteria while the inner layer is less permissive to bacterial penetration in healthy individuals (Johansson et al., 2008, 2011). Most bacterial residents are present in the outer mucus layer and the competition for survival in this niche shapes the composition of the microbiota. The differential resource utilization of bacterial species participates to the establishment of distinct communities that includes non-mucolytic bacteria (Li et al., 2015).
Role of the mucus layer
The mucus barrier has an important role in regulating the severity of infectious diseases. It provides protection against many intestinal pathogens, including Yersinia enterocolitica, Shigella flexneri, Salmonella, and Citrobacter rodentium (Mantle and Rombough, 1993; Bergstrom et al., 2010; Arike and Hansson, 2016). MUC2 (Mouse, Muc2) plays a crucial role during infection. Using Muc2-deficient mice, it was shown that the glycoprotein is critical in controlling Salmonella infection (Zarepour et al., 2013). Moreover, Muc2−/− mice revealed higher susceptibility to attaching and effacing (A/E) Citrobacter rodentium infections (Bergstrom et al., 2010).
An alteration of mucosal integrity is generally associated with health problems, such as inflammatory bowel diseases, including ulcerative colitis and Crohn's disease (Trabucchi et al., 1986; Hanski et al., 1999). During ulcerative colitis, alteration of mucus integrity results in a thinner mucus layer due to goblet cell depletion (Pullan et al., 1994) and a reduced O-glycosylation and sulfation of mucins (Raouf et al., 1992; Larsson et al., 2011). During Crohn's disease, the mucus layer is essentially continuous and comparable to healthy mucosa (Strugala et al., 2008) although there is evidence of abnormal expression and glycosylation of the mucin (Buisine et al., 2001; Moehle et al., 2006; Dorofeyev et al., 2013). These changes in the mucosal environment could also be linked to dysbiosis, an abnormal change in the composition of the intestinal microbiota due to Crohn's disease. Once impaired, the mucus barrier becomes permeable to bacteria that are able to access the epithelium and therefore cause inflammation (Antoni et al., 2014; Johansson et al., 2014), which is why the integrity of the mucus layer is critical for the upkeep of a homeostatic relationship between the intestinal microbiota and its host.
Mucin as a growth substrate
Mucin proteins are highly glycosylated and therefore constitute a carbon and energy source for intestinal microbiota. A key nutritional aspect of the mucus layer for gut bacteria is its high polysaccharide content with up to 80% of the mucin biomass being composed of mostly O-linked glycans (Johansson et al., 2009, 2011; Marcobal et al., 2013).
Mucolytic bacteria
A distinct subset of intestinal bacteria possesses the enzymatic activity, such as glycosidases, necessary for the degradation of mucin oligosaccharides, which can be further metabolized by resident microbiota (Koropatkin et al., 2012; Ouwerkerk et al., 2013). Indeed, various anaerobic bacteria species of gut microbiota, such as Akkermansia muciniphila (Derrien et al., 2004; Png et al., 2010), Bacteroides thetaiotaomicron (Xu et al., 2003; Sonnenburg et al., 2005), Bifidobacterium bifidum (Crociani et al., 1994; Png et al., 2010; Garrido et al., 2011), Bacteroides fragilis (Macfarlane and Gibson, 1991; Swidsinski et al., 2005a; Huang et al., 2011), Ruminococcus gnavus (Png et al., 2010; Crost et al., 2013), and Ruminococcus torques (Hoskins et al., 1985; Png et al., 2010) are now known as mucin-degrading specialists. These bacteria will use their specific enzymatic activities to release monosaccharides attached to the mucin glycoproteins. Some mucolytic bacteria, such as B. thetaiotaomicron, that possess an important variety of glycosidases, are better suited for the utilization of a wide range of glycans (Xu et al., 2003; Marcobal et al., 2013). To complete the degradation of mucins, a combination of enzymatic activity of several mucolytic bacteria is needed (Derrien et al., 2010; Marcobal et al., 2013). Therefore, MUC2 glycans act as nutritional sources for bacteria that can utilize the mucus-derived sugars, but lack the enzymes necessary for cleaving sugar linkages (Johansson et al., 2015; Arike and Hansson, 2016). Commonly, several bacteria collaborate in a community and it has been shown that the sulfatase activity of some commensal bacteria on sulfomucin allows glycosidases to access and act on mucins (Rho et al., 2005). Released saccharides, such as N-acetyl-D-glucosamine (GlcNAc also called NAG), N-acetylgalactosamine (GalNAc), galactose, fucose and sialic acid (N-acetylneuraminic acid also called NANA) can then be used by the degrader itself or by other resident bacteria (Bjursell et al., 2006; Martens et al., 2008; Sonnenburg et al., 2010). As example, commensal E. coli that are limited to growth on mono- or disaccharides, are unable to degrade the complex polysaccharides that constitute mucin (Hoskins et al., 1985) and therefore use such carbohydrate sources (Chang et al., 2004; Png et al., 2010; Bertin et al., 2013). Another example is vancomycin-resistant Enterococcus that can grow on mucin pre-digested with extracts from human stools, but not on purified mucin. This suggests that Enterococcus can benefit of the microbiota activity on mucin and uses released mucus-derived products (Pultz et al., 2006). In this way, mucolytic bacteria make mucus O-glycan derived products also available for other bacterial residents.
Use of mucus-derived nutrients by pathogens
Intestinal pathogens have developed strategies to compete with commensal microflora for nutrients, such as carbohydrates and these strategies have been reviewed in Conway and Cohen (2015), Vogt et al. (2015), and Baumler and Sperandio (2016). Pathogenic and commensal E. coli strains displayed considerable catabolic diversity when colonizing streptomycin-treated mice, indicating that nutrient availability can influence their colonization success and their niche adaptation (Maltby et al., 2013). For example, pathogenic E. coli such as enterohemorrhagic E. coli (EHEC) strain EDL933 efficiently utilizes some mucus-derived monosaccharides. This can provide competitive growth compared to that of commensal E. coli (Fabich et al., 2008). Moreover, the metabolic flexibility of some pathogenic strains to use both glycolytic and gluconeogenic nutrients may be advantageous (Bertin et al., 2013). The pathogen Vibrio cholerae's preferential use of mucus-derived monosaccharides, such as GlcNAc and sialic acid confers an advantage in the infant mouse model of infection (Almagro-Moreno et al., 2015). C. jejuni also possess the ability to metabolize fucose. Its growth is enhanced in culture medium supplemented with it (Alemka et al., 2012). In addition, antibiotic treatment also perturbs the microbiota and therefore affects the availability of mucin carbohydrates. The concentration of free fucose and sialic acid reaching high levels during antibiotic treatment facilitates expansion of pathogens such as Salmonella enterica serotype Typhimurium and Clostridium difficile (Ng et al., 2013). In addition, Salmonella serotype Typhimurium is known both to bind glycoprotein containing sialic acids (Vimal et al., 2000) and to have the ability to release the carbohydrate using its sialidase (Hoyer et al., 1992). Thereby, to colonize specific niches, many pathogens have evolved in a way to use mucus-derived sugars as a carbon source.
Bacterial adhesion to mucins
Mucins proteins are highly glycosylated. Their O-glycans are used as ligands for bacterial adhesins (Juge, 2012). It can be speculated that adhesion to mucins may initiate colonization of the intestine. The carbohydrate structures on mucins can provide initial attachment site to bacteria including specialized pathogens and could facilitate the invasion of epithelial cells (Derrien et al., 2010). As example, pathogenic microorganisms, such as Campylobacter and enterotoxinogenic E. coli (ETEC) are known to adhere to the glycoprotein MUC1 that is present in human breast milk. This interferes with colonization of these pathogens in the infant GI tract (Martin-Sosa et al., 2002; Ruiz-Palacios et al., 2003). Although no specific mucus-adherent microflora was identified (van der Waaij et al., 2005), there are evidence that bacteria can bind directly to mucins by expressing specific proteins, pili, fimbriae and flagella (Table 1).
Table 1.
Bacterial adhesion to mucin components.
| Bacteria | Adhesin | Mucin glycoprotein | Mucin component | References | |
|---|---|---|---|---|---|
| COMMENSAL BACTERIA | |||||
| Bacteroides fragilis | Huang et al., 2011 | ||||
| Bifidobacterium bifidum | Extracellular transaldolase | Marcobal et al., 2013 | |||
| Bifidobacterium longum subsp. infantis | Family 1 of solute binding proteins | Mucin oligosaccharides | Garrido et al., 2011 | ||
| Escherichia coli Nissle 1917 | Flagellum | Troge et al., 2012 | |||
| Lactic acid bacteria | MUB | Boekhorst et al., 2006 | |||
| Pili | Kankainen et al., 2009; Le et al., 2013 | ||||
| PATHOGENS | |||||
| Campylobacter jejuni | Carbohydrate-lectin, FlaA, MOMP | MUC2 | Tu et al., 2008; Naughton et al., 2013; Mahdavi et al., 2014 | ||
| Clostridium difficile | FliC | Cecal mucus | Tasteyre et al., 2001 | ||
| FliD | |||||
| Escherichia coli | UPEC CFT073 | F9 fimbriae | Galβ1-3GlcNAc structures | Wurpel et al., 2014 | |
| EPEC E2348/69 | H6 flagella | MUC2 | Mucin-type core 2 O-glycan | Erdem et al., 2007; Ye et al., 2015 | |
| EHEC EDL933 | H7 flagella | MUC2 | Mucin-type core 2 O-glycan | Erdem et al., 2007; Ye et al., 2015 | |
| Listeria monocytogenes | LPXTG-internalin proteins (MucBP) LmiA | Bierne et al., 2007; Mariscotti et al., 2014 | |||
| Salmonellae enterica serotype Typhimurium | Fimbrial adhesin (std operon) | Alpha1-2 fucosylated receptor(s) | Chessa et al., 2009 | ||
| Vibrio cholerae | Vibrio polysaccharide (VPS) | Liu et al., 2015 | |||
| Chitin-binding protein (GbpA) | N-acetyl D-glucosamine | Bhowmick et al., 2008 | |||
Interactions between mucin and surface proteins
To adhere to mucus, commensal and pathogenic bacteria use different strategies. First, they can produce proteins that specifically bind the mucus. Mucus-binding proteins (MUB) are cell-surface proteins mainly described in lactic acid bacteria (LAB) (Boekhorst et al., 2006), especially in Lactobacillus reuteri (Roos and Jonsson, 2002; MacKenzie et al., 2009). MUB contain domains that are similar to the model mucin-binding protein (MucBP) from the Pfam database (Boekhorst et al., 2006). The MucBP domain is found in a variety of bacterial proteins that are known for their capacity to adhere to mucus (Juge, 2012). MUB also share structural and functional homology with pathogenic Gram-positive adhesins that have specificity to sialylated mucin glycans (Etzold et al., 2014). For example, some surface proteins of Listeria monocytogenes contain a MucBP domain similar to those found in Lactobacillus, allowing them to adhere to mucin (Bierne et al., 2007; Mariscotti et al., 2014). The causative agent of cholera, V. cholerae, can also bind to mucin using surface protein called GbpA (chitin-binding protein) that binds specifically to N-acetyl D-glucosamine residues of intestinal mucins (Bhowmick et al., 2008). In addition, C. jejuni is well-known for its ability to interact with different human histoblood group antigens (HBGAs) expressed in mucosa (Naughton et al., 2013). The major outer membrane protein (MOMP) of C. jejuni is involved in these interactions (Mahdavi et al., 2014). This way, C. jejuni can interact with intestinal mucin MUC2 in the intestine (Tu et al., 2008). Furthermore, Bifidobacterium spp. is also known for its specific adhesion to mucus. For example, in a B. bifidum mucin-binding assay, the expression of an extracellular transaldolase correlated with a positive mucin-binding phenotype (Gonzalez-Rodriguez et al., 2012). B. longum subsp. infantis is another species that binds specifically to mucin using family-1 solute binding proteins (Kankainen et al., 2009). Interestingly, a study using gnotobiotic mice colonized by B. fragilis and E. coli revealed that the commensal bacterium B. fragilis was found in the mucus layer while E. coli was only found in the lumen. Further analysis showed that B. fragilis specifically binds to highly purified mucins. This indicated that a direct bond with intestinal mucus could be a mechanism used by B. fragilis for gut colonization (Huang et al., 2011).
Interactions between mucin and pili/fimbriae
In addition to produce specific mucus binding proteins, some bacteria can also use cell-surface appendix, such as pili or fimbriae to bind the mucus. For example, production of pili by LAB was shown to be implicated in mucus-binding activity (Douillard et al., 2013) and moreover, the SpaC pilus protein of L. rhamnosus GG was shown to strongly binds the human mucins (Kankainen et al., 2009). An in vitro study using mucus-secreting HT29-MTX intestinal epithelial cell model showed that the adhesion of Salmonellae enterica serotype Typhimurium to mucus-secreting intestinal epithelial cells was higher than in non- and low-mucus producing cells (Gagnon et al., 2013). Moreover, virulent strains seem to bind more efficiently to mucus than avirulent strains and the binding that preferentially targets the neutral mucin is mannose-dependant (Vimal et al., 2000). As with some uropathogenic E. coli (Wurpel et al., 2014), the adhesion of S. enterica serotype Typhimurium could be the result of interaction between fimbrial adhesin and mucin glycans, more specifically terminal fucose residues (Chessa et al., 2009). The E. coli K88 (F4) fimbriae is also able to bind mucus from the small intestines of 35-day-old piglets with a specificity to the glycolipid galactosylceramide (Blomberg et al., 1993). Hence, pili and fimbriae are involved in specific adhesion to mucus.
Interactions between mucin and flagella
Many enteric bacteria also produce flagellum. In addition to their role in motility, flagella are also involved in adhesion. As example, the E. coli probiotic strain Nissle 1917 was shown to be able to interact, via its flagella, with human and porcine mucus but not with murine mucus. Furthermore, the mucus component gluconate has been identified as one receptor for the adhesion of these flagella (Troge et al., 2012). Other studies have revealed the role of the flagella for the binding of mucin glycoproteins by C. difficile (Tasteyre et al., 2001) and pathogenic E. coli (Erdem et al., 2007). Indeed, a mutation of the flagellum element fliC prevents the adhesion of EPEC and EHEC to mucins (Erdem et al., 2007). More recently, the flagella of EPEC (O127:H6) and EHEC (O157:H7) were shown to adhere to mucin-type core 2 O-glycan in MUC2. C. jejuni is another pathogen that uses its flagella to bind mucin. It was showed that the major flagella subunit protein (FlaA) is also involved in the adhesion to HBGA in the mucus. Therefore, flagella can be used in attachment strategies by gut residents.
Bacterial biofilm and mucus
There are more mucus-associated bacteria in the proximal region of the colon than in distal colonic sites. Among the complex microbial communities within the gut, some are believed to form mucosal biofilm, that is a complex and self-produced polymeric matrix where microorganisms can attach to each other and be attached to the mucosal surface (de Vos, 2015). The rapid growth of the intestinal mucus and the lack of effective preservation techniques complicated the study investigating biofilms in healthy individuals (Bollinger et al., 2007; de Vos, 2015). However, biofilms were observed in artificial mucin gels that simulate the proximal and distal colon (Macfarlane et al., 2005), and also by electron microscopy in uninflamed proximal large bowel of mice (Swidsinski et al., 2005a), rat, baboon, and humans (Palestrant et al., 2004). Some evidence, such as the rates of plasmids transfer and the expression of colonization factors by gut bacteria, plead for the presence of biofilms in the gut (Macfarlane et al., 1997; Licht et al., 1999; Hooper and Gordon, 2001). In addition, components of the mucus layer, such as secretory IgA (SIgA) and mucins are likely to play a role in biofilm formation as they have been shown to modulate biofilm production in vitro (Bollinger et al., 2003, 2006; Slizova et al., 2015). Moreover, adherence of bacteria to mucin proteins could lead to growth of microcolonies that could further develop into biofilms (Kleessen and Blaut, 2007). Biofilms could also be formed on the surface of intestinal or gastric epithelia and interact with the secreted or membrane-bound mucins.
Alteration of the mucus layer occurs in cases of inflammatory bowel diseases (Bodger et al., 2006; Baumgart et al., 2007; Sheng et al., 2012). The increased presence of B. fragilis group and Enterobacteriaceae and their ability to form biofilms could play a role in these diseases (Swidsinski et al., 2005b, 2009). Within the Enterobacteriaceae family, the adherent-invasive E. coli (AIEC) strains associated with Crohn's disease (Masseret et al., 2001; Darfeuille-Michaud et al., 2004; Eaves-Pyles et al., 2008; Martinez-Medina et al., 2009a), are shown to be higher biofilm producers than non-AIEC strains (Martinez-Medina et al., 2009b). As with inflammatory bowel diseases, impaired mucin production is related to colorectal cancer (Weiss et al., 1996; Kim and Ho, 2010) that is also linked to the presence of bacterial biofilms (Dejea et al., 2014). Altogether, these studies show that biofilms could play a key role in bacterial colonization of the healthy gut and in intestinal diseases.
Role of mucin components in modulation of bacterial virulence
In addition to acting as a carbon source or as receptors, mucin glycoprotein can influence the expression of different genes implicated in colonization and pathogenicity (Vogt et al., 2015). As example, MUC2 in the mucus layer can play a modulatory role in the pathogenesis of pathogens. Indeed, the ability of S. enterica serotype Typhimurium to cause cecal pathology in muc2−/− mice is more dependent on its invA gene, coding a Salmonella inner membrane protein component of the SPI-1 type 3 secretion system, than it is in wild-type mice (Zarepour et al., 2013). C. jejuni can also utilize mucin proteins as a signal to modulate the expression of its virulence factors. Many virulence genes of this pathogen are upregulated in the presence of MUC2 glycoprotein (Tu et al., 2008). Another example is the ability of V. cholerae to downregulate the expression of vps, coding for its polysaccharide, in response to mucosal signaling and inversely promoting motility in the mucus (Liu et al., 2015). Mucin also activates the two-component sensor histidine kinase ChiS in V. cholera. ChiS is the regulator of the chitinases and the chitin utilization pathway, but also plays a role in the virulence of the bacteria since the mutant strain is hypovirulent (Chourashi et al., 2016). Released monosaccharides from mucin O-glycans degradation can also act as a chemical cue to help pathogens to sense their environment and adapt accordingly. As such, sialic acid and GlcNAc are signals that regulate type 1 fimbriae gene expression and curli activity in E. coli (Barnhart et al., 2006; Konopka, 2012). GlcNAc and sialic acid also play roles in the virulence of EHEC. In aerobic condition, these mucin-derived sugars inhibit EHEC adhesion to epithelial cells. These amino sugars also repress the expression of genes of the locus of enterocyte effacement (LEE) via the transcriptional regulator NagC involved in the regulation of NAG catabolism (Le Bihan et al., 2017). In contrast, as the sole carbon sources under microaerobic conditions, sialic acid and NAG were shown to stimulate the production of EspB, an effector of the LEE (Carlson-Banning and Sperandio, 2016). EHEC and C. rodentium also sense fucose by a two-component system FusKR. It represses the expression of virulence genes while promoting growth (Pacheco et al., 2012; Keeney and Finlay, 2013). Moreover, it was also shown that fucose influences chemotaxis and biofilm formation of C. jejuni that are important during infection (Dwivedi et al., 2016). Thus, mucus and its derived sugars can play a role in the expression of virulence genes by pathogens.
Modulation of mucin composition by bacteria
Microbial molecular exchange with the host influences mucin composition. Several bacterial effectors can modulate the expression of mucin by mucus-producing cells (Table 2). Studies using germ-free rats revealed that the presence of microflora through the gastro intestinal tract has a strong and positive influence on the thickness and composition of the mucin (Szentkuti et al., 1990; Enss et al., 1992; Sharma et al., 1995). Different probiotic agents, such as Lactobacillus species, can stimulate the production of MUC2 and thereby the secretion of mucin in the intestine, improving pathogen resistance (Mack et al., 1999; Mattar et al., 2002; Caballero-Franco et al., 2007). Other commensal bacteria, such as B. thetaiotaomicron can increase the differentiation of goblet cells and their mucus-related gene expression (Wrzosek et al., 2013). Moreover, bacterial fermentation products, such as short-chain fatty acids (SCFAs) like butyrate and propionate enhance the production of MUC2 by the goblet cell in the gut (Barcelo et al., 2000; Burger-van Paassen et al., 2009). This could explain the therapeutic effect of butyrate in colitis where the mucin layer is altered (Finnie et al., 1995). Therefore, commensal residents are important in the maintenance of the mucus layer integrity.
Table 2.
Effects of bacterial effectors on mucin.
| Bacteria | Effector | Target | Effect on mucin | References | |
|---|---|---|---|---|---|
| Campylobacter jejuni | Distal colonic biopsies | Increased expression of MUC1 | Linden et al., 2008 | ||
| Clostridium difficile | Distal colonic biopsies | Increased expression of MUC1 | Linden et al., 2008 | ||
| ToxA | HT-29 cells | Decrease of mucin exocytosis | Kelly et al., 1994; Branka et al., 1997 | ||
| E. coli | EAEC | Secreted protein Pic | Hog gastric, bovine sub-maxillary and crude mouse large intestine mucin | Mucinase activity / Degradation | Henderson et al., 1999; Harrington et al., 2009 |
| Goblet cells | Secretagogue activity/Hypersecretion | Navarro-Garcia et al., 2010 | |||
| ETEC | Secreted EatA | Purified MUC2 | Degradation of MUC2 | Kumar et al., 2014 | |
| AIEC (LF82) | T84 cells | Diminished expression of MUC2 and MUC5A | Elatrech et al., 2015 | ||
| EHEC (O157:H7) | Adhesion | HT-29 cells | Increased expression of MUC2 | Xue et al., 2014 | |
| Lacto-bacillus | plantarum 299v | HT-29 cells | Increased MUC2 secretion | Mack et al., 1999 | |
| rhamnosus GG | HT-29 cells | Increased MUC2 secretion | Mack et al., 1999 | ||
| casei GG | Caco-2 cells | Increased MUC2 secretion | Mattar et al., 2002 | ||
| Listeria monocytogenes | Listeriolysin O (LLO) | HT29-MTX cells | Increased transcription of MUC3, MUC4 and MUC12 Increased secretion of MUC5A | Coconnier et al., 1998; Lievin-Le Moal et al., 2002, 2005 | |
| Salmonella St Paul | Distal colonic biopsies | Increased expression of MUC1 | Linden et al., 2008 | ||
| Shigella flexneri | SST3 | Mucin-producing polarized human intestinal epithelial cells | Alteration of glycosylation/ Increased permeability | Sperandio et al., 2013 | |
| Secreted protein Pic | Hog gastric, bovine sub-maxillary, crude mouse large-intestine mucin | Mucinase activity / Degradation | Henderson et al., 1999; Harrington et al., 2009 | ||
| Goblet cells | Secretagogue activity / Hypersecretion | Navarro-Garcia et al., 2010 | |||
| Vibrio cholerae | Toxin CT | Goblet cells | Increased mucin secretion | Lencer et al., 1990; Epple et al., 1997 | |
| Secreted TagA | LS174T goblet cell surface mucin | Cleaves mucin glycoproteins | Szabady et al., 2011 | ||
| Yersinia enterocolitica | Virulence Plasmid | Rabbit small intestinal mucin | Degradation/Solubilisation | Mantle and Rombough, 1993 | |
Modulation of mucin by pathogens
Pathogens have also adapted mechanisms to modulate mucin secretion to enhance pathogenesis by acting on the mucin-secreting cells, altering or inhibiting mucin production (Table 2). One of them is S. flexneri that alters the mucus layer through a type III secretion system-dependent manner. This pathogen will act on different elements, such as gene expression, mucin glycosylation and secretion, leading to a less effective mucus barrier (Sperandio et al., 2013). C. difficile produces a toxin, ToxA that is responsible for barrier dysfunction and causes severe inflammatory enteritis. ToxA also decreases the mucin exocytosis of colonic mucus-producing cells (Kelly et al., 1994; Branka et al., 1997). The recognition of bacterial components by these cells can also lead to an increased production and secretion of mucin in order to harm the present pathogen. As example, the adhesion of the EHEC O157:H7 to human colon cells HT-29 leads to an increased expression of MUC2 (Xue et al., 2014). Moreover, the cholera toxin of V. cholerae and lysteriolysin O of L. monocytogenes enhance the secretion of mucin by goblet cells and HT29-MTX cells, respectively (Lencer et al., 1990; Epple et al., 1997; Coconnier et al., 1998; Lievin-Le Moal et al., 2002, 2005). Surprisingly, the Pic protein secreted by S. flexneri and enteroaggregative E. coli (Henderson et al., 1999; Harrington et al., 2009) is known for its mucolytic activity, but is also a potent mucus secretagogue that induced hypersecretion of mucus by goblet cells (Navarro-Garcia et al., 2010). These studies show how pathogens can affect the behavior of mucus-producing cells in their advantage.
Mucin degradation by pathogens
Pathogens also developed specific mechanisms to subvert and penetrate the mucus barrier. Some bacteria can directly act on the mucin through a mucinase activity. During enterotoxigenic E. coli infections, the autotransporter A (EatA) is involve in mucin degradation and this participate to the delivery of E. coli toxins to the cell surface (Kumar et al., 2014). Another example is the adherent and invasive E. coli strain LF82, associated with Crohn's disease. LF82 possesses a protease called Vat-AIEC that is implicated in the degradation of mucins and therefore decreases mucus viscosity (Gibold et al., 2016). The Pic autotransporter found in enteroaggregative E. coli and Shigella flexneri can also degrade various glycoproteins including mucins (Henderson et al., 1999; Harrington et al., 2009). Moreover, the plasmid-bearing Yersinia enterocolitica, which contain mucin-degrading enzyme(s), will increase the permeability of the mucus gel layer, allowing the bacteria to move more easily through the mucin (Mantle and Rombough, 1993). V. cholerae also produces a secreted protease called TagA that is encoded by the Vibrio pathogenicity island (VPI). TagA specifically cleaves mucin glycoproteins and may directly modify host cell surface molecules during V. cholerae infection (Szabady et al., 2011). Therefore, to facilitate their infection process, pathogens can directly modify the mucus.
Inflammation and mucins
Pathogens associated molecular patterns, such as lipopolysaccharide (LPS) and peptidoglycan are also known to stimulate mucin production (Petersson et al., 2011). This stimulation can occur directly on secreting cells, but also be through proinflammatory cytokine production. Recognition of LPS by LPS-binding protein (LBP), CD14, and TLR4 (Toll-Like Receptor) leads to a strong pro-inflammatory response in mammalian cells. LPS has been shown to induce mucin gene expression by binding to TLR4 and LBP (Dohrman et al., 1998; Smirnova et al., 2003). LPS and flagellin from Gram-negative bacteria as well as lipoteichoic acid, a component of the cell wall of Gram-positive bacteria, induce mucin upregulation through the Ras pathway (McNamara and Basbaum, 2001; Theodoropoulos and Carraway, 2007). LPS also increases the production of IL-8 by goblet cells, which leads to secretion of mucin (Smirnova et al., 2003). In addition, pro-inflammatory cytokine IL-6 and TNF-α increase secretion of MUC2, MUC5A, MUC5B, and MUC6 by the intestinal cell line LS180 despite a reduced glycosylation (Enss et al., 2000). Inflammation could be one of the aspects affecting the integrity of the mucus layer in inflammatory bowel diseases. Furthermore, the AIEC strain LF82 is able to alter the expression of the mucin gene and IL-8 of colonic cells T84 that could also lead to a defective mucus layer (Elatrech et al., 2015). Thus, pathogens can also alter the mucus production indirectly, through inflammation.
Conclusion
Intestinal bacteria have adapted to colonize the mucus layer by adhering to intestinal mucus components, using mucus-derived nutrients and sensing chemical cues for adaptation. In many ways, pathogenic bacteria have used these strategies for successful infection. There has been growing recognition of the important role played by the mucus barrier and microbiota and their interaction with the pathogens in regulating the severity of infectious diseases. But, the precise mechanisms by which enteric bacterial pathogens interact with mucus components in combination with the microbiota activity are being investigated. As the mucus layer acts as a first line of defense against enteric bacteria, further investigations are needed to understand the interactions between pathogens, microbiota and the mucus layer, in order to develop efficient therapeutic strategies. Identifying and characterizing specific mucin signal(s) and corresponding regulatory adaptation and virulence responses could contribute to the development of new anti-infective strategies. In doing so, other weapons could be added to the arsenal against intestinal pathogens.
Author contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication. The manuscript was written by J-FS and JH and was duly revised by GLB, PV and MJ.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Judith Kashul for editing the manuscript. This research was supported by a Team grant from the Fonds de Recherche du Québec, Nature et Technologies (FRQNT PT165375), to JH and MJ and by the Discovery grant program of the Natural Sciences and Engineering Research Council of Canada (RGPIN-2015-05373 to JH and RGPIN-2016-04203 to MJ). J-FS is a recipient of a scholarship from the NSERC Collaborative Research and Training Experience Program in Milk Quality; and PV is a recipient of a scholarship from the FRQNT Québec Wallonie program.
Glossary
Abbreviations
- MLG
Mucus gel layer
- A/E
Attaching and effacing
- NAG
N-acetyl-D-glucosamine
- NANA
N-acetylneuraminic acid
- EHEC
Enterohemorrhagic E. coli
- MUB
Mucus-binding proteins
- LAB
Lactic acid bacteria
- HBGA
Histoblood group antigen
- SIgA
Secretory IgA
- AIEC
Adherent invasive E. coli
- LPB
LPS-binding protein
- TLR
Toll-like receptor
- VPI
Vibrio pathogenicity island.
References
- Alemka A., Corcionivoschi N., Bourke B. (2012). Defense and adaptation: the complex inter-relationship between Campylobacter jejuni and mucus. Front. Cell Infect. Microbiol. 2:15. 10.3389/fcimb.2012.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro-Moreno S., Pruss K., Taylor R. K. (2015). Intestinal colonization dynamics of Vibrio cholerae. PLoS Pathog. 11:e1004787. 10.1371/journal.ppat.1004787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni L., Nuding S., Wehkamp J., Stange E. F. (2014). Intestinal barrier in inflammatory bowel disease. World J. Gastroenterol. 20, 1165–1179. 10.3748/wjg.v20.i5.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arike L., Hansson G. C. (2016). The densely o-glycosylated MUC2 mucin protects the intestine and provides food for the commensal bacteria. J. Mol. Biol. 428, 3221–3229. 10.1016/j.jmb.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcelo A., Claustre J., Moro F., Chayvialle J. A., Cuber J. C., Plaisancie P. (2000). Mucin secretion is modulated by luminal factors in the isolated vascularly perfused rat colon. Gut 46, 218–224. 10.1136/gut.46.2.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart M. M., Lynem J., Chapman M. R. (2006). GlcNAc-6P levels modulate the expression of Curli fibers by Escherichia coli. J. Bacteriol. 188, 5212–5219. 10.1128/JB.00234-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart M., Dogan B., Rishniw M., Weitzman G., Bosworth B., Yantiss R., et al. (2007). Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 1, 403–418. 10.1038/ismej.2007.52 [DOI] [PubMed] [Google Scholar]
- Baumler A. J., Sperandio V. (2016). Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535, 85–93. 10.1038/nature18849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom K. S., Kissoon-Singh V., Gibson D. L., Ma C., Montero M., Sham H. P., et al. (2010). MUC2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 6:e1000902. 10.1371/journal.ppat.1000902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin Y., Chaucheyras-Durand F., Robbe-Masselot C., Durand A., de la Foye A., Harel J., et al. (2013). Carbohydrate utilization by enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. Environ. Microbiol. 15, 610–622. 10.1111/1462-2920.12019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick R., Ghosal A., Das B., Koley H., Saha D. R., Ganguly S., et al. (2008). Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect. Immun. 76, 4968–4977. 10.1128/IAI.01615-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H., Sabet C., Personnic N., Cossart P. (2007). Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 9, 1156–1166. 10.1016/j.micinf.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Bjursell M. K., Martens E. C., Gordon J. I. (2006). Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J. Biol. Chem. 281, 36269–36279. 10.1074/jbc.M606509200 [DOI] [PubMed] [Google Scholar]
- Blomberg L., Krivan H. C., Cohen P. S., Conway P. L. (1993). Piglet ileal mucus contains protein and glycolipid (galactosylceramide) receptors specific for Escherichia coli K88 fimbriae. Infect. Immun. 61, 2526–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodger K., Halfvarson J., Dodson A. R., Campbell F., Wilson S., Lee R., et al. (2006). Altered colonic glycoprotein expression in unaffected monozygotic twins of inflammatory bowel disease patients. Gut 55, 973–977. 10.1136/gut.2005.086413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhorst J., Helmer Q., Kleerebezem M., Siezen R. J. (2006). Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 152(Pt 1), 273–280. 10.1099/mic.0.28415-0 [DOI] [PubMed] [Google Scholar]
- Bollinger R. R., Barbas A. S., Bush E. L., Lin S. S., Parker W. (2007). Biofilms in the normal human large bowel: fact rather than fiction. Gut 56, 1481–1482. [PMC free article] [PubMed] [Google Scholar]
- Bollinger R. R., Everett M. L., Palestrant D., Love S. D., Lin S. S., Parker W. (2003). Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 109, 580–587. 10.1046/j.1365-2567.2003.01700.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger R. R., Everett M. L., Wahl S. D., Lee Y. H., Orndorff P. E., Parker W. (2006). Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: role of type 1 pili. Mol. Immunol. 43, 378–387. 10.1016/j.molimm.2005.02.013 [DOI] [PubMed] [Google Scholar]
- Branka J. E., Vallette G., Jarry A., Bou-Hanna C., Lemarre P., Van P. N., et al. (1997). Early functional effects of Clostridium difficile toxin A on human colonocytes. Gastroenterology 112, 1887–1894. 10.1053/gast.1997.v112.pm9178681 [DOI] [PubMed] [Google Scholar]
- Buisine M. P., Desreumaux P., Leteurtre E., Copin M. C., Colombel J. F., Porchet N., et al. (2001). Mucin gene expression in intestinal epithelial cells in Crohn's disease. Gut 49, 544–551. 10.1136/gut.49.4.544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger-van Paassen N., Vincent A., Puiman P. J., van der Sluis M., Bouma J., Boehm G., et al. (2009). The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem. J. 420, 211–219. 10.1042/BJ20082222 [DOI] [PubMed] [Google Scholar]
- Caballero-Franco C., Keller K., De Simone C., Chadee K. (2007). The VSL#3 probiotic formula induces mucin 554 gene expression and secretion in colonic epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G315–G322. 10.1152/ajpgi.00265.2006 [DOI] [PubMed] [Google Scholar]
- Carlson-Banning K. M., Sperandio V. (2016). Catabolite and oxygen regulation of enterohemorrhagic Escherichia coli virulence. MBio 7:e01852-16. 10.1128/mBio.01852-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. E., Smalley D. J., Tucker D. L., Leatham M. P., Norris W. E., Stevenson S. J., et al. (2004). Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U.S.A. 101, 7427–7432. 10.1073/pnas.0307888101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessa D., Winter M. G., Jakomin M., Baumler A. J. (2009). Salmonella enterica serotype Typhimurium Std fimbriae bind terminal α(1,2)fucose residues in the cecal mucosa. Mol. Microbiol. 71, 864–875. 10.1111/j.1365-2958.2008.06566.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourashi R., Mondal M., Sinha R., Debnath A., Das S., Koley H., et al. (2016). Role of a sensor histidine kinase ChiS of Vibrio cholerae in pathogenesis. Int. J. Med. Microbiol. 306, 657–665. 10.1016/j.ijmm.2016.09.003 [DOI] [PubMed] [Google Scholar]
- Coconnier M. H., Dlissi E., Robard M., Laboisse C. L., Gaillard J. L., Servin A. L. (1998). Listeria monocytogenes stimulates mucus exocytosis in cultured human polarized mucosecreting intestinal cells through action of listeriolysin O. Infect. Immun. 66, 3673–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Cohen P. S. (2015). Commensal and pathogenic Escherichia coli metabolism in the gut. Microbiol. Spectr. 3, 343–362. 10.1128/microbiolspec.MBP-0006-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crociani F., Alessandrini A., Mucci M. M., Biavati B. (1994). Degradation of complex carbohydrates by Bifidobacterium spp. Int. J. Food Microbiol. 24, 199–210. 10.1016/0168-1605(94)90119-8 [DOI] [PubMed] [Google Scholar]
- Crost E. H., Tailford L. E., Le Gall G., Fons M., Henrissat B., Juge N. (2013). Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS ONE 8:e76341. 10.1371/journal.pone.0076341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A., Boudeau J., Bulois P., Neut C., Glasser A. L., Barnich N., et al. (2004). High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127, 412–421. 10.1053/j.gastro.2004.04.061 [DOI] [PubMed] [Google Scholar]
- Dejea C. M., Wick E. C., Hechenbleikner E. M., White J. R., Mark Welch J. L., Rossetti B. J., et al. (2014). Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. U.S.A. 111, 18321–18326. 10.1073/pnas.1406199111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B., Gaskins H. R. (2001). Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 73, 1131s–1141s. [DOI] [PubMed] [Google Scholar]
- Derrien M., van Passel M. W., van de Bovenkamp J. H., Schipper R. G., de Vos W. M., Dekker J. (2010). Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 1, 254–268. 10.4161/gmic.1.4.12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Vaughan E. E., Plugge C. M., de Vos W. M. (2004). Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54(Pt 5), 1469–1476. 10.1099/ijs.0.02873-0 [DOI] [PubMed] [Google Scholar]
- de Vos W. M. (2015). Microbial biofilms and the human intestinal microbiome. NPJ Biofilms Microbiomes 1:15005. 10.1038/npjbiofilms.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrman A., Miyata S., Gallup M., Li J. D., Chapelin C., Coste A., et al. (1998). Mucin gene (MUC 2 and MUC 5AC) upregulation by Gram-positive and Gram-negative bacteria. Biochim. Biophys. Acta 1406, 251–259. 10.1016/S0925-4439(98)00010-6 [DOI] [PubMed] [Google Scholar]
- Dorofeyev A. E., Vasilenko I. V., Rassokhina O. A., Kondratiuk R. B. (2013). Mucosal barrier in ulcerative colitis and Crohn's disease. Gastroenterol. Res. Pract. 2013:431231. 10.1155/2013/431231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard F. P., Ribbera A., Jarvinen H. M., Kant R., Pietila T. E., Randazzo C., et al. (2013). Comparative genomic and functional analysis of Lactobacillus casei and Lactobacillus rhamnosus strains marketed as probiotics. Appl. Environ. Microbiol. 79, 1923–1933. 10.1128/AEM.03467-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi R., Nothaft H., Garber J., Xin Kin L., Stahl M., Flint A., et al. (2016). L-fucose influences chemotaxis and biofilm formation in Campylobacter jejuni. Mol. Microbiol. 101, 575–589. 10.1111/mmi.13409 [DOI] [PubMed] [Google Scholar]
- Eaves-Pyles T., Allen C. A., Taormina J., Swidsinski A., Tutt C. B., Jezek G. E., et al. (2008). Escherichia coli isolated from a Crohn's disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells. Int. J. Med. Microbiol. 298, 397–409. 10.1016/j.ijmm.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Elatrech I., Marzaioli V., Boukemara H., Bournier O., Neut C., Darfeuille-Michaud A., et al. (2015). Escherichia coli LF82 differentially regulates ROS production and mucin expression in intestinal epithelial T84 cells: implication of NOX1. Inflamm. Bowel Dis. 21, 1018–1026. 10.1097/MIB.0000000000000365 [DOI] [PubMed] [Google Scholar]
- Enss M. L., Cornberg M., Wagner S., Gebert A., Henrichs M., Eisenblatter R., et al. (2000). Proinflammatory cytokines trigger MUC gene expression and mucin release in the intestinal cancer cell line LS180. Inflamm. Res. 49, 162–169. 10.1007/s000110050576 [DOI] [PubMed] [Google Scholar]
- Enss M. L., Grosse-Siestrup H., Schmidt-Wittig U., Gartner K. (1992). Changes in colonic mucins of germfree rats in response to the introduction of a “normal” rat microbial flora. Rat colonic mucin. J. Exp. Anim. Sci. 35, 110–119. [PubMed] [Google Scholar]
- Epple H. J., Kreusel K. M., Hanski C., Schulzke J. D., Riecken E. O., Fromm M. (1997). Differential stimulation of intestinal mucin secretion by cholera toxin and carbachol. Pflugers Arch. 433, 638–647. [DOI] [PubMed] [Google Scholar]
- Erdem A. L., Avelino F., Xicohtencatl-Cortes J., Giron J. A. (2007). Host protein binding and adhesive properties of H6 and H7 flagella of attaching and effacing Escherichia coli. J. Bacteriol. 189, 7426–7435. 10.1128/JB.00464-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzold S., Kober O. I., Mackenzie D. A., Tailford L. E., Gunning A. P., Walshaw J., et al. (2014). Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ. Microbiol. 16, 888–903. 10.1111/1462-2920.12377 [DOI] [PubMed] [Google Scholar]
- Fabich A. J., Jones S. A., Chowdhury F. Z., Cernosek A., Anderson A., Smalley D., et al. (2008). Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76, 1143–1152. 10.1128/IAI.01386-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnie I. A., Dwarakanath A. D., Taylor B. A., Rhodes J. M. (1995). Colonic mucin synthesis is increased by sodium butyrate. Gut 36, 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstner G. (1995). Signal transduction, packaging and secretion of mucins. Annu. Rev. Physiol. 57, 585–605. [DOI] [PubMed] [Google Scholar]
- Gagnon M., Zihler Berner A., Chervet N., Chassard C., Lacroix C. (2013). Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J. Microbiol. Methods 94, 274–279. 10.1016/j.mimet.2013.06.027 [DOI] [PubMed] [Google Scholar]
- Garrido D., Kim J. H., German J. B., Raybould H. E., Mills D. A. (2011). Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS ONE 6:e17315. 10.1371/journal.pone.0017315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibold L., Garenaux E., Dalmasso G., Gallucci C., Cia D., Mottet-Auselo B., et al. (2016). The Vat-AIEC protease promotes crossing of the intestinal mucus layer by Crohn's disease-associated Escherichia coli. Cell. Microbiol. 18, 617–631. 10.1111/cmi.12539 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rodriguez I., Sanchez B., Ruiz L., Turroni F., Ventura M., Ruas-Madiedo P., et al. (2012). Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl. Environ. Microbiol. 78, 3992–3998. 10.1128/AEM.08024-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gum J. R., Jr., Hicks J. W., Toribara N. W., Siddiki B., Kim Y. S. (1994). Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor. J. Biol. Chem. 269, 2440–2446. [PubMed] [Google Scholar]
- Hanski C., Born M., Foss H. D., Marowski B., Mansmann U., Arasteh K., et al. (1999). Defective post-transcriptional processing of MUC2 mucin in ulcerative colitis and in Crohn's disease increases detectability of the MUC2 protein core. J. Pathol. 188, 304–311. [DOI] [PubMed] [Google Scholar]
- Harel J., Fairbrother J., Forget C., Desautels C., Moore J. (1993). Virulence factors associated with F165-positive Escherichia coli strains isolated from piglets and calves. Vet. Microbiol. 38, 139–155. [DOI] [PubMed] [Google Scholar]
- Harrington S. M., Sheikh J., Henderson I. R., Ruiz-Perez F., Cohen P. S., Nataro J. P. (2009). The pic protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. Infect. Immun. 77, 2465–2473. 10.1128/IAI.01494-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I. R., Czeczulin J., Eslava C., Noriega F., Nataro J. P. (1999). Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67, 5587–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L. V., Gordon J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292, 1115–1118. 10.1126/science.1058709 [DOI] [PubMed] [Google Scholar]
- Hoskins L. C., Agustines M., McKee W. B., Boulding E. T., Kriaris M., Niedermeyer G. (1985). Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J. Clin. Invest. 75, 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer L. L., Hamilton A. C., Steenbergen S. M., Vimr E. R. (1992). Cloning, sequencing and distribution of the Salmonella typhimurium LT2 sialidase gene, nanH, provides evidence for interspecies gene transfer. Mol. Microbiol. 6, 873–884. [DOI] [PubMed] [Google Scholar]
- Huang J. Y., Lee S. M., Mazmanian S. K. (2011). The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe 17, 137–141. 10.1016/j.anaerobe.2011.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. E., Gustafsson J. K., Holmen-Larsson J., Jabbar K. S., Xia L., Xu H., et al. (2014). Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 63, 281–291. 10.1136/gutjnl-2012-303207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. E., Jakobsson H. E., Holmen-Larsson J., Schutte A., Ermund A., Rodriguez-Pineiro A. M., et al. (2015). Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 18, 582–592. 10.1016/j.chom.2015.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. E., Larsson J. M., Hansson G. C. (2011). The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4659–4665. 10.1073/pnas.1006451107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. E., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G. C. (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 15064–15069. 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. E., Thomsson K. A., Hansson G. C. (2009). Proteomic analyses of the two mucus layers of the colon barrier reveal that their main component, the Muc2 mucin, is strongly bound to the Fcgbp protein. J. Proteome Res. 8, 3549–3557. 10.1021/pr9002504 [DOI] [PubMed] [Google Scholar]
- Jonckheere N., Skrypek N., Frenois F., Van Seuningen I. (2013). Membrane-bound mucin modular domains: from structure to function. Biochimie 95, 1077–1086. 10.1016/j.biochi.2012.11.005 [DOI] [PubMed] [Google Scholar]
- Juge N. (2012). Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 20, 30–39. 10.1016/j.tim.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Kankainen M., Paulin L., Tynkkynen S., von Ossowski I., Reunanen J., Partanen P., et al. (2009). Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. U.S.A. 106, 17193–17198. 10.1073/pnas.0908876106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney K. M., Finlay B. B. (2013). Microbiology: EHEC downregulates virulence in response to intestinal fucose. Curr. Biol. 23, R108–R110. 10.1016/j.cub.2012.12.027 [DOI] [PubMed] [Google Scholar]
- Kelly C. P., Becker S., Linevsky J. K., Joshi M. A., O'Keane J. C., Dickey B. F., et al. (1994). Neutrophil recruitment in Clostridium difficile toxin A enteritis in the rabbit. J. Clin. Invest. 93, 1257–1265. 10.1172/JCI117080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. S., Ho S. B. (2010). Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr. Gastroenterol. Rep. 12, 319–330. 10.1007/s11894-010-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleessen B., Blaut M. (2007). Modulation of gut mucosal biofilms. Br. J. Nutr. 93, S35–S40. 10.1079/BJN20041346 [DOI] [PubMed] [Google Scholar]
- Konopka J. B. (2012). N-acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica (Cairo) 2012:489208. 10.6064/2012/489208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin N. M., Cameron E. A., Martens E. C. (2012). How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10, 323–335. 10.1038/nrmicro2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Luo Q., Vickers T. J., Sheikh A., Lewis W. G., Fleckenstein J. M. (2014). EatA, an immunogenic protective antigen of enterotoxigenic Escherichia coli, degrades intestinal mucin. Infect. Immun. 82, 500–508. 10.1128/IAI.01078-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J. M., Karlsson H., Crespo J. G., Johansson M. E., Eklund L., Sjovall H., et al. (2011). Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm. Bowel Dis. 17, 2299–2307. 10.1002/ibd.21625 [DOI] [PubMed] [Google Scholar]
- Larsson J. M., Karlsson H., Sjovall H., Hansson G. C. (2009). A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology 19, 756–766. 10.1093/glycob/cwp048 [DOI] [PubMed] [Google Scholar]
- Le D. T., Tran T. L., Duviau M. P., Meyrand M., Guerardel Y., Castelain M., et al. (2013). Unraveling the role of surface mucus-binding protein and pili in muco-adhesion of Lactococcus lactis. PLoS ONE 8:e79850. 10.1371/journal.pone.0079850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan G., Sicard J. F., Garneau P., Bernalier-Donadille A., Gobert A. P., Garrivier A., et al. (2017). The NAG sensor NagC regulates LEE gene expression and contributes to gut colonization by Escherichia coli O157:H7. Front. Cell. Infect. Microbiol. 7:134. 10.3389/fcimb.2017.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencer W. I., Reinhart F. D., Neutra M. R. (1990). Interaction of cholera toxin with cloned human goblet cells in monolayer culture. Am. J. Physiol. 258(1 Pt 1), G96–G102. [DOI] [PubMed] [Google Scholar]
- Li H., Limenitakis J. P., Fuhrer T., Geuking M. B., Lawson M. A., Wyss M., et al. (2015). The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 6:8292. 10.1038/ncomms9292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht T. R., Christensen B. B., Krogfelt K. A., Molin S. (1999). Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology 145(Pt 9), 2615–2622. 10.1099/00221287-145-9-2615 [DOI] [PubMed] [Google Scholar]
- Lievin-Le Moal V., Huet G., Aubert J. P., Bara J., Forgue-Lafitte M. E., Servin A. L., et al. (2002). Activation of mucin exocytosis and upregulation of MUC genes in polarized human intestinal mucin-secreting cells by the thiol-activated exotoxin listeriolysin O. Cell. Microbiol. 4, 515–529. 10.1046/j.1462-5822.2002.00210.x [DOI] [PubMed] [Google Scholar]
- Lievin-Le Moal V., Servin A. L., Coconnier-Polter M. H. (2005). The increase in mucin exocytosis and the upregulation of MUC genes encoding for membrane-bound mucins induced by the thiol-activated exotoxin listeriolysin O is a host cell defence response that inhibits the cell-entry of Listeria monocytogenes. Cell. Microbiol. 7, 1035–1048. 10.1111/j.1462-5822.2005.00532.x [DOI] [PubMed] [Google Scholar]
- Linden S. K., Florin T. H., McGuckin M. A. (2008). Mucin dynamics in intestinal bacterial infection. PLoS ONE 3:e3952. 10.1371/journal.pone.0003952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wang Y., Liu S., Sheng Y., Rueggeberg K. G., Wang H., et al. (2015). Vibrio cholerae represses polysaccharide synthesis to promote motility in mucosa. Infect. Immun. 83, 1114–1121. 10.1128/IAI.02841-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane G. T., Gibson G. R. (1991). Formation of glycoprotein degrading enzymes by Bacteroides fragilis. FEMS Microbiol. Lett. 61, 289–293. 10.1111/j.1574-6968.1991.tb04363.x [DOI] [PubMed] [Google Scholar]
- Macfarlane S., McBain A. J., Macfarlane G. T. (1997). Consequences of biofilm and sessile growth in the large intestine. Adv. Dent. Res. 11, 59–68. 10.1177/08959374970110011801 [DOI] [PubMed] [Google Scholar]
- Macfarlane S., Woodmansey E. J., Macfarlane G. T. (2005). Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl. Environ. Microbiol. 71, 7483–7492. 10.1128/AEM.71.11.7483-7492.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D. R., Michail S., Wei S., McDougall L., Hollingsworth M. A. (1999). Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am. J. Physiol. 276(4 Pt 1), G941–G950. [DOI] [PubMed] [Google Scholar]
- MacKenzie D. A., Tailford L. E., Hemmings A. M., Juge N. (2009). Crystal structure of a mucus-binding protein repeat reveals an unexpected functional immunoglobulin binding activity. J. Biol. Chem. 284, 32444–32453. 10.1074/jbc.M109.040907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi J., Pirinccioglu N., Oldfield N. J., Carlsohn E., Stoof J., Aslam A., et al. (2014). A novel O-linked glycan modulates Campylobacter jejuni major outer membrane protein-mediated adhesion to human histo-blood group antigens and chicken colonization. Open Biol. 4:130202. 10.1098/rsob.130202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltby R., Leatham-Jensen M. P., Gibson T., Cohen P. S., Conway T. (2013). Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS ONE 8:e53957. 10.1371/journal.pone.0053957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantle M., Rombough C. (1993). Growth in and breakdown of purified rabbit small intestinal mucin by Yersinia enterocolitica. Infect. Immun. 61, 4131–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A., Southwick A. M., Earle K. A., Sonnenburg J. L. (2013). A refined palate: bacterial consumption of host glycans in the gut. Glycobiology 23, 1038–1046. 10.1093/glycob/cwt040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariscotti J. F., Quereda J. J., Garcia-Del Portillo F., Pucciarelli M. G. (2014). The Listeria monocytogenes LPXTG surface protein Lmo1413 is an invasin with capacity to bind mucin. Int. J. Med. Microbiol. 304, 393–404. 10.1016/j.ijmm.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Martens E. C., Chiang H. C., Gordon J. I. (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457. 10.1016/j.chom.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Medina M., Aldeguer X., Lopez-Siles M., Gonzalez-Huix F., Lopez-Oliu C., Dahbi G., et al. (2009a). Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm Bowel Dis. 15, 872–882. 10.1002/ibd.20860 [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M., Naves P., Blanco J., Aldeguer X., Blanco J. E., Blanco M., et al. (2009b). Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC). BMC Microbiol. 9:202. 10.1186/1471-2180-9-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Sosa S., Martin M. J., Hueso P. (2002). The sialylated fraction of milk oligosaccharides is partially responsible for binding to enterotoxigenic and uropathogenic Escherichia coli human strains. J. Nutr. 132, 3067–3072. [DOI] [PubMed] [Google Scholar]
- Masseret E., Boudeau J., Colombel J. F., Neut C., Desreumaux P., Joly B., et al. (2001). Genetically related Escherichia coli strains associated with Crohn's disease. Gut 48, 320–325. 10.1136/gut.48.3.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar A. F., Teitelbaum D. H., Drongowski R. A., Yongyi F., Harmon C. M., Coran A. G. (2002). Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediatr. Surg. Int. 18, 586–590. 10.1007/s00383-002-0855-7 [DOI] [PubMed] [Google Scholar]
- McGuckin M. A., Linden S. K., Sutton P., Florin T. H. (2011). Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9, 265–278. 10.1038/nrmicro2538 [DOI] [PubMed] [Google Scholar]
- McNamara N., Basbaum C. (2001). Signaling networks controlling mucin production in response to Gram-positive and Gram-negative bacteria. Glycoconj. J. 18, 715–722. 10.1023/A:1020875423678 [DOI] [PubMed] [Google Scholar]
- Moehle C., Ackermann N., Langmann T., Aslanidis C., Kel A., Kel-Margoulis O., et al. (2006). Aberrant intestinal expression and allelic variants of mucin genes associated with inflammatory bowel disease. J. Mol. Med. 84, 1055–1066. 10.1007/s00109-006-0100-2 [DOI] [PubMed] [Google Scholar]
- Moran A. P., Gupta A., Joshi L. (2011). Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut 60, 1412–1425. 10.1136/gut.2010.212704 [DOI] [PubMed] [Google Scholar]
- Naughton J. A., Marino K., Dolan B., Reid C., Gough R., Gallagher M. E., et al. (2013). Divergent mechanisms of interaction of Helicobacter pylori and Campylobacter jejuni with mucus and mucins. Infect. Immun. 81, 2838–2850. 10.1128/IAI.00415-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Garcia F., Gutierrez-Jimenez J., Garcia-Tovar C., Castro L. A., Salazar-Gonzalez H., Cordova V. (2010). Pic, an autotransporter protein secreted by different pathogens in the Enterobacteriaceae family, is a potent mucus secretagogue. Infect. Immun. 78, 4101–4109. 10.1128/IAI.00523-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K. M., Ferreyra J. A., Higginbottom S. K., Lynch J. B., Kashyap P. C., Gopinath S., et al. (2013). Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99. 10.1038/nature12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson H. E., Ambort D., Backstrom M., Thomsson E., Koeck P. J., Hansson G. C., et al. (2014). Intestinal MUC2 mucin supramolecular topology by packing and release resting on D3 domain assembly. J. Mol. Biol. 426, 2567–2579. 10.1016/j.jmb.2014.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwerkerk J. P., de Vos W. M., Belzer C. (2013). Glycobiome: bacteria and mucus at the epithelial interface. Best Pract. Res. Clin. Gastroenterol. 27, 25–38. 10.1016/j.bpg.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Pacheco A. R., Curtis M. M., Ritchie J. M., Munera D., Waldor M. K., Moreira C. G., et al. (2012). Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117. 10.1038/nature11623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palestrant D., Holzknecht Z. E., Collins B. H., Parker W., Miller S. E., Bollinger R. R. (2004). Microbial biofilms in the gut: visualization by electron microscopy and by acridine orange staining. Ultrastruct. Pathol. 28, 23–27. 10.1080/01913120490275196 [DOI] [PubMed] [Google Scholar]
- Petersson J., Schreiber O., Hansson G. C., Gendler S. J., Velcich A., Lundberg J. O., et al. (2011). Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 300, G327–G333. 10.1152/ajpgi.00422.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png C. W., Linden S. K., Gilshenan K. S., Zoetendal E. G., McSweeney C. S., Sly L. I., et al. (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 105, 2420–2428. 10.1038/ajg.2010.281 [DOI] [PubMed] [Google Scholar]
- Pullan R. D., Thomas G. A., Rhodes M., Newcombe R. G., Williams G. T., Allen A., et al. (1994). Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut 35, 353–359. 10.1136/gut.35.3.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pultz N. J., Hoskins L. C., Donskey C. J. (2006). Vancomycin-resistant Enterococci may obtain nutritional support by scavenging carbohydrate fragments generated during mucin degradation by the anaerobic microbiota of the colon. Microb. Drug Resist. 12, 63–67. 10.1089/mdr.2006.12.63 [DOI] [PubMed] [Google Scholar]
- Raouf A. H., Tsai H. H., Parker N., Hoffman J., Walker R. J., Rhodes J. M. (1992). Sulphation of colonic and rectal mucin in inflammatory bowel disease: reduced sulphation of rectal mucus in ulcerative colitis. Clin. Sci. 83, 623–626. 10.1042/cs0830623 [DOI] [PubMed] [Google Scholar]
- Rho J. H., Wright D. P., Christie D. L., Clinch K., Furneaux R. H., Roberton A. M. (2005). A novel mechanism for desulfation of mucin: identification and cloning of a mucin-desulfating glycosidase (sulfoglycosidase) from Prevotella strain RS2. J. Bacteriol. 187, 1543–1551. 10.1128/JB.187.5.1543-1551.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos S., Jonsson H. (2002). A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148(Pt 2), 433–442. 10.1099/00221287-148-2-433 [DOI] [PubMed] [Google Scholar]
- Ruiz-Palacios G. M., Cervantes L. E., Ramos P., Chavez-Munguia B., Newburg D. S. (2003). Campylobacter jejuni binds intestinal H(O) antigen (Fuc α1, 2Gal β1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 278, 14112–14120. 10.1074/jbc.M207744200 [DOI] [PubMed] [Google Scholar]
- Sharma R., Schumacher U., Ronaasen V., Coates M. (1995). Rat intestinal mucosal responses to a microbial flora and different diets. Gut 36, 209–214. 10.1136/gut.36.2.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y. H., Hasnain S. Z., Florin T. H., McGuckin M. A. (2012). Mucins in inflammatory bowel diseases and colorectal cancer. J. Gastroenterol. Hepatol. 27, 28–38. 10.1111/j.1440-1746.2011.06909.x [DOI] [PubMed] [Google Scholar]
- Slizova M., Nemcova R., Mad'ar M., Hadryova J., Gancarcikova S., Popper M., et al. (2015). Analysis of biofilm formation by intestinal lactobacilli. Can. J. Microbiol. 61, 437–446. 10.1139/cjm-2015-0007 [DOI] [PubMed] [Google Scholar]
- Smirnova M. G., Guo L., Birchall J. P., Pearson J. P. (2003). LPS up-regulates mucin and cytokine mRNA expression and stimulates mucin and cytokine secretion in goblet cells. Cell. Immunol. 221, 42–49. 10.1016/S0008-8749(03)00059-5 [DOI] [PubMed] [Google Scholar]
- Sonnenburg E. D., Zheng H., Joglekar P., Higginbottom S. K., Firbank S. J., Bolam D. N., et al. (2010). Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252. 10.1016/j.cell.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg J. L., Xu J., Leip D. D., Chen C. H., Westover B. P., Weatherford J., et al. (2005). Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959. 10.1126/science.1109051 [DOI] [PubMed] [Google Scholar]
- Sperandio B., Fischer N., Joncquel Chevalier-Curt M., Rossez Y., Roux P., Robbe Masselot C., et al. (2013). Virulent Shigella flexneri affects secretion, expression, and glycosylation of gel-forming mucins in mucus-producing cells. Infect. Immun. 81, 3632–3643. 10.1128/IAI.00551-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strugala V., Dettmar P. W., Pearson J. P. (2008). Thickness and continuity of the adherent colonic mucus barrier in active and quiescent ulcerative colitis and Crohn's disease. Int. J. Clin. Pract. 62, 762–769. 10.1111/j.1742-1241.2007.01665.x [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Loening-Baucke V., Herber A. (2009). Mucosal flora in Crohn's disease and ulcerative colitis-an overview. J. Physiol. Pharmacol. 60(Suppl. 6), 61–71. [PubMed] [Google Scholar]
- Swidsinski A., Loening-Baucke V., Lochs H., Hale L. P. (2005a). Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J. Gastroenterol. 11, 1131–1140. 10.3748/wjg.v11.i8.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A., Weber J., Loening-Baucke V., Hale L. P., Lochs H. (2005b). Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43, 3380–3389. 10.1128/JCM.43.7.3380-3389.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabady R. L., Yanta J. H., Halladin D. K., Schofield M. J., Welch R. A. (2011). TagA is a secreted protease of Vibrio cholerae that specifically cleaves mucin glycoproteins. Microbiology 157(Pt 2), 516–525. 10.1099/mic.0.044529-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentkuti L., Riedesel H., Enss M. L., Gaertner K., Von Engelhardt W. (1990). Pre-epithelial mucus layer in the colon of conventional and germ-free rats. Histochem. J. 22, 491–497. 10.1007/BF01007234 [DOI] [PubMed] [Google Scholar]
- Tasteyre A., Barc M. C., Collignon A., Boureau H., Karjalainen T. (2001). Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69, 7937–7940. 10.1128/IAI.69.12.7937-7940.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoropoulos G., Carraway K. L. (2007). Molecular signaling in the regulation of mucins. J. Cell. Biochem. 102, 1103–1116. 10.1002/jcb.21539 [DOI] [PubMed] [Google Scholar]
- Trabucchi E., Mukenge S., Baratti C., Colombo R., Fregoni F., Montorsi W. (1986). Differential diagnosis of Crohn's disease of the colon from ulcerative colitis: ultrastructure study with the scanning electron microscope. Int. J. Tissue React. 8, 79–84. [PubMed] [Google Scholar]
- Troge A., Scheppach W., Schroeder B. O., Rund S. A., Heuner K., Wehkamp J., et al. (2012). More than a marine propeller–the flagellum of the probiotic Escherichia coli strain Nissle 1917 is the major adhesin mediating binding to human mucus. Int. J. Med. Microbiol. 302, 304–314. 10.1016/j.ijmm.2012.09.004 [DOI] [PubMed] [Google Scholar]
- Tu Q. V., McGuckin M. A., Mendz G. L. (2008). Campylobacter jejuni response to human mucin MUC2: modulation of colonization and pathogenicity determinants. J. Med. Microbiol. 57(Pt 7), 795–802. 10.1099/jmm.0.47752-0 [DOI] [PubMed] [Google Scholar]
- van der Waaij L. A., Harmsen H. J., Madjipour M., Kroese F. G., Zwiers M., van Dullemen H. M., et al. (2005). Bacterial population analysis of human colon and terminal ileum biopsies with 16S rRNA-based fluorescent probes: commensal bacteria live in suspension and have no direct contact with epithelial cells. Inflamm. Bowel Dis. 11, 865–871. 10.1097/01.mib.0000179212.80778.d3 [DOI] [PubMed] [Google Scholar]
- Vimal D. B., Khullar M., Gupta S., Ganguly N. K. (2000). Intestinal mucins: the binding sites for Salmonella typhimurium. Mol. Cell. Biochem. 204, 107–117. 10.1023/A:1007015312036 [DOI] [PubMed] [Google Scholar]
- Vogt S. L., Pena-Diaz J., Finlay B. B. (2015). Chemical communication in the gut: effects of microbiota-generated metabolites on gastrointestinal bacterial pathogens. Anaerobe 34, 106–115. 10.1016/j.anaerobe.2015.05.002 [DOI] [PubMed] [Google Scholar]
- Weiss A. A., Babyatsky M. W., Ogata S., Chen A., Itzkowitz S. H. (1996). Expression of MUC2 and MUC3 mRNA in human normal, malignant, and inflammatory intestinal tissues. J. Histochem. Cytochem. 44, 1161–1166. 10.1177/44.10.8813081 [DOI] [PubMed] [Google Scholar]
- Wrzosek L., Miquel S., Noordine M. L., Bouet S., Joncquel Chevalier-Curt M., Robert V., et al. (2013). Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 11:61. 10.1186/1741-7007-11-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurpel D. J., Totsika M., Allsopp L. P., Hartley-Tassell L. E., Day C. J., Peters K. M., et al. (2014). F9 fimbriae of uropathogenic Escherichia coli are expressed at low temperature and recognise Galbeta1-3GlcNAc-containing glycans. PLoS ONE 9:e93177. 10.1371/journal.pone.0093177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Bjursell M. K., Himrod J., Deng S., Carmichael L. K., Chiang H. C., et al. (2003). A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076. 10.1126/science.1080029 [DOI] [PubMed] [Google Scholar]
- Xue Y., Zhang H., Wang H., Hu J., Du M., Zhu M. J. (2014). Host inflammatory response inhibits Escherichia coli O157:H7 adhesion to gut epithelium through augmentation of mucin expression. Infect. Immun. 82, 1921–1930. 10.1128/IAI.01589-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Song L., Liu Y., Pan Q., Zhong X., Li S., et al. (2015). Core 2 Mucin-Type O-Glycan is related to EPEC and EHEC O157:H7 adherence to human colon carcinoma HT-29 epithelial cells. Dig. Dis. Sci. 60, 1977–1990. 10.1007/s10620-015-3548-5 [DOI] [PubMed] [Google Scholar]
- Zarepour M., Bhullar K., Montero M., Ma C., Huang T., Velcich A., et al. (2013). The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect. Immun. 81, 3672–3683. 10.1128/IAI.00854-13 [DOI] [PMC free article] [PubMed] [Google Scholar]