Abstract
The ability of micro-organisms to degrade isosaccharinic acids (ISAs) while tolerating hyperalkaline conditions is pivotal to our understanding of the biogeochemistry associated within these environs, but also in scenarios pertaining to the cementitious disposal of radioactive wastes. An alkalitolerant, ISA degrading micro-organism was isolated from the hyperalkaline soils resulting from lime depositions. Here, we report the first whole-genome sequence, ISA degradation profile and carbohydrate preoteome of a Macellibacteroides fermentans strain HH-ZS, 4.08 Mb in size, coding 3,241 proteins, 64 tRNA, and 1 rRNA.
Keywords: Macellibacteroides fermentans, obligate anaerobe, alkalitolerant, whole-genome sequence, Bacteroidetes, isosaccharinic acid
Introduction
Alkaline environments have consistently been studied for the isolation of novel alkaliphilic and alkalitolerant microbial species (Duckworth et al. 1996), with particular focus on biotechnological applications of the enzymes produced by these organisms (Mamo and Mattiasson 2016). These alkaline environments are either natural, such as soda lakes or anthropogenic as a result of land contamination. One example of an anthropogenic alkaline environment can be found in Harpur Hill, Derbyshire, UK (Milodowski et al. 2015). Here, historical in-valley deposition of lime wastes generated through lime kiln workings has resulted in soil pH values of >11.0 following interactions between the wastes with rainwaters resulting in the generation of Ca(OH)2 (Burke et al. 2012).
The conditions are somewhat analogous to those expected within the deep geological disposal concept for the UK radioactive waste inventory, where anaerobic, alkaline conditions dominate (NDA 2010; Rout, Charles, Garratt, et al. 2015). Under these conditions isosaccharinic acids (ISAs) are generated and are a key consideration for safety assessments due to their ability to mobilise radioelements such as plutonium (Greenfield et al. 1997). Microbial degradation of ISAs is therefore a component of this analysis. To date, mixed cultures have demonstrated ISA degradation (Bassil et al. 2014; Kuippers et al. 2015; Rout, Charles, Doulgeris, et al. 2015), despite this, degradation by pure cultures has received limited attention and has been limited to historical descriptions of aerobic isolates (Pekarovičová and Mikulášová 1991; Strand et al. 1984).
The isolation of an ISA degrading strain is therefore of significant interest and the availability of a whole-genome sequence of such a strain would allow for the biochemistry of ISA degradation to be further investigated. A novel strain, designated HH-ZS, was isolated from the soil at Harpur Hill and identified via 16S rRNA profiling as Macellibacteroides fermentans, distinct from strain LIND7H (Jabari et al. 2012), of the Phylum Bacteroidetes. This study describes the ISA degradation profile and draft whole-genome sequence of this organism.
Materials and Methods
Culturing, Phenotypic, and Genomic DNA Isolation
Hyperalkaline soil was obtained from Harpur Hill, Derbyshire, and diluted in anoxic mineral media (B.S.I. 2005) with ISA containing cellulose degradation products (as per Rout et al. 2014) used as a carbon source. After anoxic incubation within this microcosm for 4 weeks at 25 °C, 10 µl of the reaction fluid was sub cultured onto fastidious anaerobic agar (LabM, UK) adjusted to pH 9 using 4 M NaOH. Growth of single colonies were observed following incubation at 25 °C in an anaerobic workstation (10% H2, 10% CO2, 80% N2; DW Scientific) for 3 days. Single colonies were further purified by sub-culture. Cell morphology was determined via Gram staining and SEM via a Quanta FEG 250 scanning electron microscope. Biochemical capabilities of the strain were determined using API 20 A kit (bioMérieux, USA), and through culture in mineral media at pH 9 containing 4 mM Ca(ISA)2, with ISA concentration and volatile fatty acid production monitored using HPAEC-PAD and GC as previously described (Rout et al. 2014). The pH profile of the isolate was determined via BioscreenC technology (Growth Curves USA, USA) with optical density of the isolate measured at 450–600 nm in fastidious anaerobic broth adjusted to a pH range of 4–12 in 1 pH unit increments using 4 M HCl or 4 M NaOH accordingly. Replicates were prepared in 100 well plates which were then sealed using gas proof tape and incubated for 62 h at 30 °C. Genomic DNA was extracted from harvested cells using an UltraClean Microbial Isolation Kit (Qiagen, USA). The concentration and purity of the extracted DNA was then assessed using a NanoDrop ND1000 spectrophotometer. This genomic DNA was then used to prepare a paired-end library.
Genome Sequencing, Assembly, and Annotation
Paired end sequence reads were prepared using an Illumina HiSeq 2500 system (Illumina, US) with sequencing carried out by BaseClear, NL. FASTQ sequence files were generated using the Illumina Casava pipeline version 1.8.3. Initial quality assessment was based on data passing the Illumina Chastity filtering. PhiX control signal associated reads were removed using an in house filtering protocol. Reads containing (partial) adapter sequence were trimmed up to a minimum read length of 50 bp. FASTQC quality control tool version 0.10.0 was used to provide a second quality assessment of the remaining reads. Low quality bases were trimmed to enhance the quality of FASTQ sequences using the “trim sequences” option of CLC Genomics Workbench version 8.5.1. Contiguous sequences were then assembled using the “De novo assembly” option of the CLC genomics workbench and mis-assemblies corrected using Pilon (Walker et al. 2014). Orientation of contigs within scaffolds was performed using SSPACE premium scaffolder v2.3 (Boetzer et al. 2011) and any gaps within these scaffolds were closed using GapFiller v1.10 (Boetzer and Pirovano 2012). These gaps were then filled using Sanger sequencing technology (MWG-Eurofins, Germany), primers were designed using NCBI Primer-Blast (Ye et al. 2012) and can be seen in supplementary table S1, Supplementary Material online.
Phylogenetic, Genome, and Proteome Analysis
In addition to whole-genome sequence, a partial fragment of the 16S rRNA gene was amplified by PCR using universal primers 8F and 1512R, with the fragment purified using a Qiagen QIAquick PCR purification kit (Qiagen, USA) prior to Sanger sequencing (Eurofins, Germany). The partial sequence was used to identify the isolate using the EzTaxon-e database (Kim et al. 2012) then used for phylogenetic comparison with other members of the Bacteroidales. These sequences were extracted from the National Center for Biotechnology Information (NCBI) and aligned using the MUSCLE module of MEGA 7.0.14 (Kumar et al. 2016) and a maximum likelihood tree generated in the same package (model: Tamura-Nei; bootstrap; 1,000).
For initial annotations sequences were submitted to the NCBI Prokaryotic Genome Annotation Pipeline tools (Tatusova et al. 2013) then further annotated on the RAST server (Overbeek et al. 2014). COG category assignment was performed using the WebMGA server (Wu et al. 2011) and Pfam domain predictions carried out using the NCBI conserved domain database (CDD) (Marchler-Bauer et al. 2014). For the prediction of signal peptides and transmembrane domains, SignalP 4.1 server (Petersen et al. 2011) and the TMHMM server v. 2.0 (Krogh et al. 2001) were used, respectively. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) were found online using the CRISPRfinder tool searching against a CRISPR database (Grissa et al. 2007).
Results and Discussion
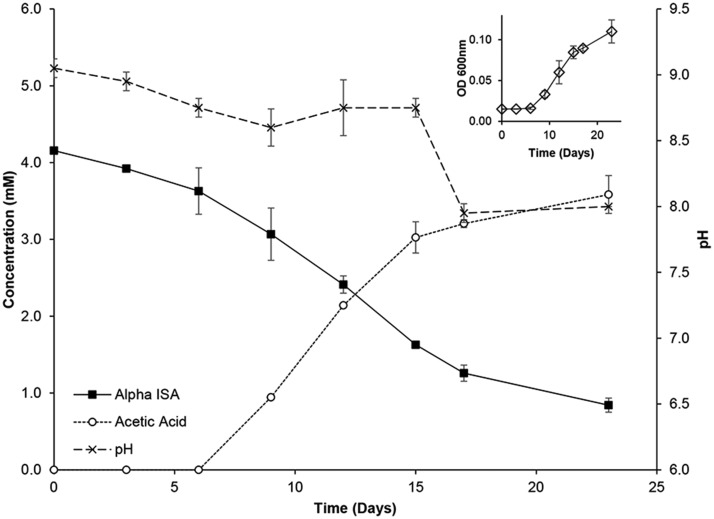
Macellibacteroides fermentans strain HH-ZS was a found to be a Gram negative rod, with rods being ∼1 µm in length (supplementary fig. S1, Supplementary Material online). The isolate was aerotolerant, but only capable of growth in anaerobic conditions at pH 5.0–10.0 with an optimum of 8.0 (supplementary fig. S2, Supplementary Material online). When grown in minimal medium with Ca(ISA)2 as the sole carbon source, there was a subsequent generation of acetic acid, coupled to a reduction in pH to 8.0 and increase in OD at 620 nm (fig. 1). This degradation presented a first order rate of 5.0 × 10−2 (± 1.5 × 10−3) day−1, and is the first rate presented for any microbial species for the degradation of ISA. The isolate was also capable of fermenting a range of other sugars (supplementary table S2, Supplementary Material online). Inference of phylogeny using partial 16S rRNA sequencing placed strain HH-ZS within the Bacteroidales showing homology to M. fermentans strain LIND7H (supplementary fig. S3, Supplementary Material online), with a 99.89% similarity observed using EzTaxon-e identification. This coupled to the alkaliphilitolerant nature of the isolate marked it as being a novel strain of M. fermentans.
Fig. 1.
—Isosaccharinic acid degradation profile of Macellibacteroides fermentans strain HH-ZS.
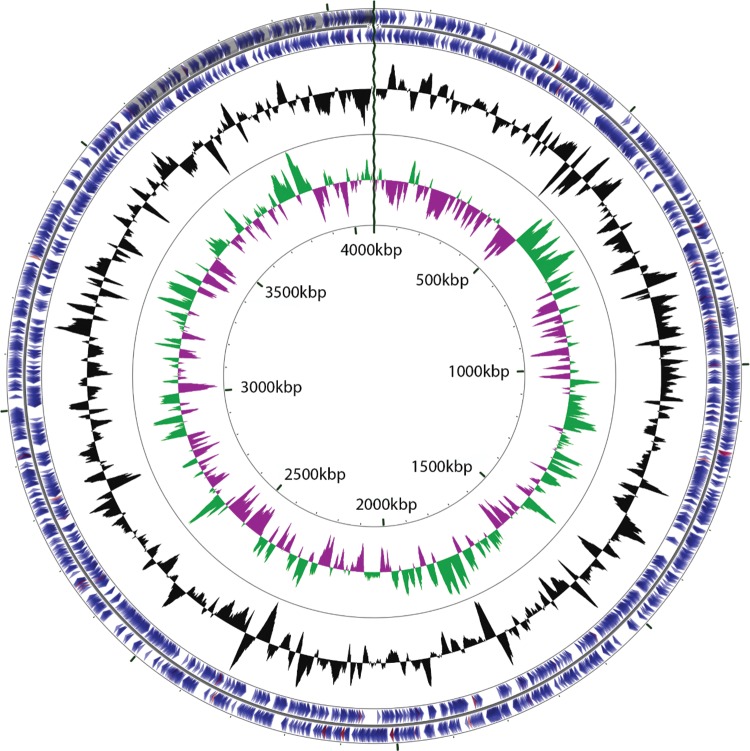
The assembly was based upon 2,264,150 reads yielding 524 MB of data with an average phred quality score of 37.67 following Cassava and FastQC pipeline analysis. The final assembly consisted of 4,081,835 bp within 67 scaffolds, representing a coverage of 127× and GC content of 41.71% (fig. 2). Following assembly, five gaps were remaining, resulting in a draft of the genome sequence. The genome contained a total of 3,345 genes, of which 3,241 were protein coding, 69 were RNA coding and 35 were pseudogenes and one CRISPR repeat. The majority of protein-coding genes (82.3%) were assigned a putative function, 2,318 were found to have Pfam domains, 757 to transmembrane helicases, and 422 signal peptides. The distribution of genes into COGs functions shown in supplementary table S3, Supplementary Material online. COG assignment associated 219 genes with carbohydrate metabolism and transport and further annotation using the CAZyme analysis toolkit (Park et al. 2010) identified a further 69 genes and placed all of these into CAZY domains (supplementary fig. S4, Supplementary Material online).
Fig. 2.
—Circular representation of the Macellibacteroides fermentans strain HH-ZS complete genome. Circles (from inside to outside) 1 and 2 (protein coding sequences on the forward and reverse strand, blue indicates CDS, peach shows tRNAs, pink rRNAs, and gray other). Circle 3 shows GC content % and Circles 4 and 5 show positive (green) and negative (magenta) GC skew.
When compared with other available whole-genome sequences available for Parabacteroides sp., M. fermentans HH-ZS was found to have a greater number of proteins associated with the glycoside hydrolase family. With respect to the organic material present in the soil, the genome also contained a number of carbohydrate binding module families, in particular CBM4, are calcium dependent and are implicated in the degradation of a number of cellulosic structures, with the exception of crystalline cellulose (Kataeva et al. 2001). This may be compensated by the presence of a predicted family-30 CBM, which has been shown to bind less available cellulose (Malburg et al. 1997; Raut et al. 2015).
Conclusions
Here, we present the first genome of Macellibacteroides. fermetans (strain HH-ZS), isolated from hyperalkaline soil, and demonstrating a wider pH tolerance range than previously described strain LIND7H and other closely related Parabacteroides sp. The isolate is also the first obligate anaerobe described as a degrader of ISA, and here we present the first rates for its degradation. CAZy analysis revealed that the strain harbored a number of carbohydrate degrading enzymes, which merit further investigation to determine the metabolic pathways associated with ISA degradation.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Acknowledgment
The efforts of Zohier B Salah were funded by a Libyan Government Ph.D. Scholarship.
Supplementary Material
Literature Cited
- B.S.I (2005) BS ISO 14853: 2005 Plastics: determination of the ultimate anaerobic biodegradation of plastic materials in an aqueous system—method by measurement of biogas production. BS ISO 14853: 2005, British Standards Institute, London, UK.
- Bassil NM, Bryan N, Lloyd JR.. 2015. Microbial degradation of isosaccharinic acid at high pH. ISME J. 9(2):310–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W.. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27(4):578–579. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W.. 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13(6):R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke IT, et al. 2012. Biogeochemical reduction processes in a hyper-alkaline leachate affected soil profile. Geomicrobiol J. 29(9):769–779. [Google Scholar]
- Duckworth A, Grant W, Jones B, Van Steenbergen R.. 1996. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol Ecol. 19:181–191. [Google Scholar]
- Greenfield B, Hurdus M, Spindler M, Thomason H. (1997). The effects of the products from the anaerobic degradation of cellulose on the solubility and sorption of radioelements in the near field. NSS/R375. AEA Technology Report for UK Nirex Ltd. [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C.. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35(Web Server):W52–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabari L, et al. 2012. Macellibacteroides fermentans gen. nov., sp. nov., a member of the family Porphyromonadaceae isolated from an upflow anaerobic filter treating abattoir wastewaters. Int J Syst Evol Microbiol. 62(Pt 10):2522–2527. [DOI] [PubMed] [Google Scholar]
- Kataeva IA, Seidel RD, Li X-L, Ljungdahl LG.. 2001. Properties and mutation analysis of the CelK cellulose-binding domain from the Clostridium thermocellum cellulosome. J Bacteriol. 183(5):1552–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O-S, et al. 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 62(Pt 3):716–721. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kuippers G, Bassil NM, Boothman C, Bryan N, Lloyd JR.. 2015. Microbial degradation of isosaccharinic acid under conditions representative for the far field of radioactive waste disposal facilities. Mineral Mag. 79(6):1443–1454. [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malburg SR, Malburg LM, Liu T, Iyo AH, Forsberg CW.. 1997. Catalytic properties of the cellulose-binding endoglucanase F from Fibrobacter succinogenes S85. Appl Environ Microbiol. 63:2449–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamo G, Mattiasson B. (2016) Alkaliphilic microorganisms in biotechnology. Biotechnology of extremophiles. Switzerland: Springer International Publishing AG.. [Google Scholar]
- Marchler-Bauer A, et al. 2014. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milodowski AE, Shaw RP, Stewart DI. (2015) The Harpur Hill Site: its geology, evolutionary history and a catalogue of materials present. Keyworth, Nottingham: British Geological Survey.
- NDA (2010) Near-field evolution status report. Harwell, Didcot, Oxfordshire: Nuclear Decommissioning Authority. [Google Scholar]
- Overbeek R, et al. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42(D1):D206–D214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BH, Karpinets TV, Syed MH, Leuze MR, Uberbacher EC.. 2010. CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 20(12):1574–1584. [DOI] [PubMed] [Google Scholar]
- Pekarovičová A, Mikulášová M, Černáková L.. 2007. Biodegradation of black liquor hydroxyacids by Micrococcus lylae. J Chem Technol Biotechnol. 52(4):539–543. [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10):785–786. [DOI] [PubMed] [Google Scholar]
- Raut MP, Karunakaran E, Mukherjee J, Biggs CA, Wright PC, Desvaux M.. 2015. Influence of substrates on the surface characteristics and membrane proteome of Fibrobacter succinogenes S85. PLoS One 10(10):e0141197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout SP, Charles CJ, Doulgeris C, McCarthy AJ, Rooks DJ.. 2015. Anoxic biodegradation of isosaccharinic acids at alkaline pH by natural microbial communities. PLoS One 10:e0137682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout SP, Charles CJ, Garratt EJ, et al. 2015. Evidence of the generation of isosaccharinic acids and their subsequent degradation by local microbial consortia within hyper-alkaline contaminated soils, with relevance to intermediate level radioactive waste disposal. PLoS One 10:e0119164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout SP, et al. 2014. Biodegradation of the alkaline cellulose degradation products generated during radioactive waste disposal. PLoS One 9(9):e107433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand SE, Dykes J, Chiang V.. 1984. Aerobic Microbial Degradation of Glucoisosaccharinic Acid. Appl Environ Microbiol. 47(2):268–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, et al. (2013) Prokaryotic genome annotation pipeline. Bethesda (MD): NCBI, National Center for Biotechnology Information. [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhu Z, Fu L, Niu B, Li W.. 2011. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics 12(1):444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, et al. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.