Abstract
RNA is central to the flow of biological information. From transcription to splicing, RNA localization, translation, and decay, RNA is intimately involved in regulating every step of the gene expression program, and is thus essential for health and understanding disease. RNA has the unique ability to base-pair with itself and other nucleic acids to form complex structures. Hence the information content in RNA is not simply its linear sequence of bases, but is also encoded in complex folding of RNA molecules. A general chemical functionality that all RNAs have is a 2’-hydroxyl group in the ribose ring, and the reactivity of the 2'-hydroxyl in RNA is gated by local nucleotide flexibility. In other words, the 2'-hydroxyl is reactive at single-stranded and conformationally flexible positions but is unreactive at nucleotides constrained by base pairing. Recent efforts have been focused on developing reagents that modify RNA as a function of RNA 2’ hydroxyl group flexibility. Such RNA structure probing techniques can be read out by primer extension in experiments termed RNA SHAPE (Selective 2’ Hydroxyl Acylation and Primer Extension). Herein we describe the efforts devoted to the design and utilization of SHAPE probes for characterizing RNA structure. We also describe current technological advances that are being used to utilize SHAPE chemistry with deep sequencing to probe many RNAs in parallel. The merger of chemistry with genomics is sure to open the door to genome-wide exploration of RNA structure and function.
RNA and its importance in biology
The survival of every cell depends on precise regulation of gene expression. For about three decades it was believed that gene expression was primarily controlled at the level of transcription. In this model the flow of genetic information would be primarily controlled by transcription factors, which would serve to either inhibit or promote the access of DNA to RNA polymerase. However, this model has changed dramatically with the increasing evidence that RNA molecules play key roles in nearly all biological processes1.
Studies on molecular evolution suggest that RNA molecules contributed significantly to the development of modern organisms by mediating the flow of genetic information between DNA and proteins. RNA has been shown to directly participate in transcription and alternative splicing of RNA contributes to human complexity. Metabolite-sensing RNAs and small non-coding RNAs can control the efficiency of gene translation2,3. Recently, RNAs have been implicated in chromatin remodeling and gene expression control.4–6 Additional modes of gene regulation at the level of RNA structure remain to be discovered. Despite the importance and biological necessity of these interactions, our understanding of how the physical and molecular properties of RNA control these events inside the cell is at best rudimentary.
Decades of in vitro analysis on the physical characteristics of RNA have shed light on the idiosyncrasies that make RNA such a diverse biopolymer. Despite being composed of only four chemically similar nucleotides, RNAs can form into complex structures and can adopt multiple alternative structures that can have similar energetic characteristics.7,8 RNA structures have been shown to be catalytic and provide exquisite scaffolds for the binding of metal ions, small molecules, trans-RNA sequences, and proteins.5,9,10 Extensive in vitro characterization of RNA structure and folding has revealed that RNA structure is modular and hierarchical, and folding proceeds by marching through a rugged free energy landscape.11–13 However, the structural characteristics of RNA in vivo are likely to be more complex. In vivo RNA structure is easily influenced by the rate of transcription, local solution conditions, temperature, the binding of small molecules, and through interactions with RNA-binding proteins.14–16 Further, it has been shown that functional RNA structures can require proteins for efficient catalysis and folding, underscoring the importance of protein binding.17,18 These observations hint that the physical state of RNA within the cell is very important for its function, but we know next to nothing of how intracellular RNA structure can contribute to the specificity of these binding events or the biological role of RNA molecules.
Methods used to probe RNA structure
RNA structure mapping has been used for decades to understand the physical conformations of a few RNAs. To obtain high confidence maps of RNA structure several reagents have been developed. RNases that require specific structural and sequence motifs are used to obtain blunt measurements of RNA secondary structure.19,20 Alternatively, chemical mapping, by direct alkylation of the RNA through reaction with dimethylsulfate, radical cleavage of solvent exposed backbone regions, or by metal catalyzed cleavage (both Lead (II) and Terbium (III)) are widely used for high-resolution determination of secondary and tertiary RNA structure.20–23 While these methods are quite useful for traditional probing techniques, many of these chemical methods are not generalizable and often have biases in their reactivities; this limits the ability to obtain structural information for every nucleotide. A relatively new methodology has emerged that can overcome many of these shortcomings. Within the review we highlight the genesis of SHAPE. We focus on RNA SHAPE (Selective 2’-Hydroxyl Acylation and Primer Extension), its use to understand RNA biology in vitro, and finally, our extension of the technique to probing RNA structure in living cells and the potential for its use for genome-wide measurement of RNA structure.
SHAPE Chemistry
Each RNA has different parts that have unique physical properties that contribute to the biological function. For example, single-stranded regions often serve as landing pads for proteins. Loops have been shown to be critical in the recognition of small-molecules as well as controlling the formation of long-distance interactions within complex RNAs24. Extended stem structures have been implicated in RNA-based diseases. Therefore, to really understand the total structure and function of RNAs, there needs to be a way to distinguish and assign these motifs to an RNA molecule of interest.
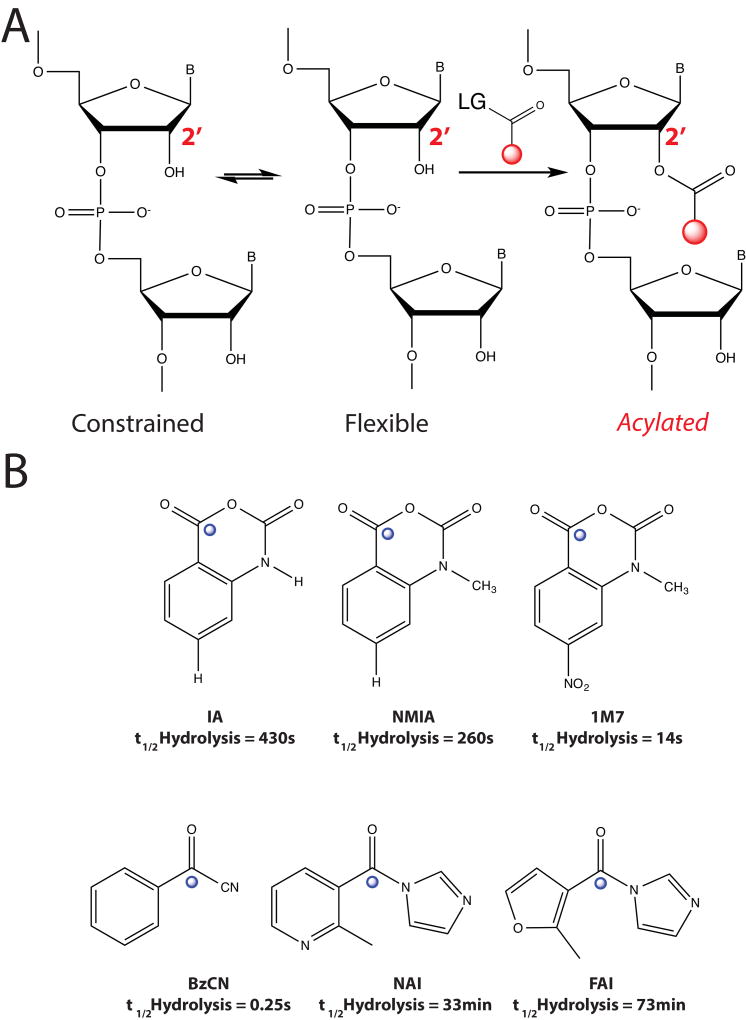
A general chemical functionality that all RNAs have is a 2’-hydroxyl group in the ribose ring, and the reactivity of the 2'-hydroxyl in RNA is gated by local nucleotide flexibility25. In other words, the 2'-hydroxyl is reactive at single-stranded and conformationally flexible positions but is unreactive at nucleotides constrained by base pairing. Thus, reagents that modify RNA as a function of RNA 2’-hydroxyl (2’-OH) groups can read out RNA structure. The generalizable nature of the 2’-OH makes it an ideal candidate for chemical interrogation of RNA structure; however, it is relatively inert in comparison to other functional groups on RNA. In solution, 2’-OH functional groups have pKa values that range from 12–14.26 Nevertheless, it has been shown that RNA functional groups can alter their pKa values to approach biological conditions, and these changes are dependent on RNA structure.27,28 In the case of SHAPE reactivity and RNA structure, the 2’-OH pKa and reactivity is modulated by the propensity for a flexible 2’-OH to rarely sample a conformation that renders it active for reaction with a solution electrophile (Figure 1, A).
Figure 1. Mechanisms of SHAPE Chemistry and Electrophiles Used for Acylation.
(A). Reaction pathway of RNA 2’-O-acylation. LG is short for leaving group. (B) Examples of SHAPE electrophiles developed for RNA structure probing. Blue spheres mark the site of 2’-OH attack.
Extensive studies with SHAPE reagents have shown that adenosine and guanine react approximately 1.6-fold more rapidly than do cytosine and uridine to SHAPE electrophiles. Also, experiments on a variety of RNA structures has shown the lack of dependence on solvent accessibility for SHAPE reactivity. Therefore, SHAPE electrophiles can sample even the interior of complex RNA structures29. Nevertheless, the unique environment surrounding the 2’-hydroxyl can contribute to its intrinsic reactivity. For example, localized general base catalysts, from multiple RNA functional groups, specific orientations of the bridging 3'-phosphate group can accelerate nucleophilic attack toward SHAPE electrophiles30. Overall, these studies have revealed the extensive work that has been put into establishing SHAPE reagents as an integral part of chemical probing experiments.
The First SHAPE Reagent
Biomolecules in the cell are comprised on redundant chemical functional groups that control diverse biological functions. Similar to the 2’-OH, hydroxyl groups can be found in proteins (serine, tyrosine) and in small molecules. Following the observation that hydroxyls can be activated to weakly attack esters31, several acylation electrophiles were tested against a series of nucleotide analogs with predicted differences in their ability to become reactive. The most promising of these was NMIA, or N-methylisatoic anhydride32. NMIA reacts with hydroxyl groups to release CO2 to form a 2-methylaminobenzoic acid ester (Figure 1, B). NMIA was shown previously to from an adduct with the catalytic serine residue in chymotrypsin. This adduct rendered the enzyme inactive, and thus removed the ability to break peptide bonds.33 A unique functionality in NMIA that makes it an ideal RNA structure probe is that the product of the acylation reaction creates a handle that is inert to the reversible deacylation mechanism. This is controlled by the nearby lone-pair of electrons on the secondary amine that is δ to the carbonyl carbon attached to reactive hydroxyl34. The stability of the acylation adduct makes it ideal for manipulations to the RNA after modification.
The initial strength of SHAPE was demonstrated by characterizing flexible regions in tRNA. Reactive nucleotides lie almost exclusively in the conserved T-, D-, and anti-codon loops and at the 3′-amino acid attachment terminus. Impressively, NMIA also was able to score relative reactivities. This was exemplified by the lower overall acylation of nucleotides in the T- and D- loops, but higher reactivities in the anti-codon and 3’-terminus. This is not surprising, given the T- and D- loops are in structural contacts with the rest of the tRNA molecule, whereas the anti-codon and 3’-terminus are naked. Further, hydroxyl acylation by NMIA was shown to robustly differentiate between flexible and constrained linkages in RNA. Importantly, these were not correlated with solvent-accessibility, which further underscores the sensitivity of the hydroxyl acylation chemistry. The ability to score reactivities, and relate them to nucleotide flexibility that is dependent on structural context is a major strength of SHAPE in comparison to other chemicals used to read out RNA structure. Further exploration into hydroxyl reactivity showed that acylation is relatively insensitive to nucleobase identity29. These initial experiments demonstrated the power of SHAPE chemistry to read folded RNA structures, yet RNA is quite dynamic folding and changing its structure on several time-scales. Tailoring the chemistry of SHAPE electrophiles pushed the boundaries of what can be measured by such reagents.
Timed SHAPE reactions
RNA folding has been shown to proceed heirachically, and through many transitions with different time scales13. This is especially true for RNAs that proceed through folding intermediates co-transcriptionally or through their interactions with other biological macromolecules35. These observations necessitate the need to develop RNA structure probes that can be used to interrogate structural intermediates or that are highly reactive with RNAs that have not reached their ground structure state due to long incubation times. Measuring RNA hydroxyl acylation is uniquely primed to fill these gaps because the hydroxyl reactivity corresponds to the local flexibility and structural state of RNA. In addition, the dynamic nature of the 2’-OH closely correlates with its acylation reactivity36.
A few modified SHAPE reagents have been designed to measure RNA structure at different time scales. These reagents are more reactive to 2’-OH nucleophilicity by alterations to the electronic nature of the carbonyl carbon electrophile (Figure 1, B). These modified SHAPE reagents are outfitted with functional groups that serve to pull away electrons, thus making these reagents more susceptible to hydroxyl attack. Such reagents can be used to distinguish between residues that are highly flexible during RNA folding and those that are reactive on shorter time scales.
The first fast-reacting SHAPE reagent was 1M7, a para-nitro derivative of NMIA (Figure 1, B). Importantly, 1M7 is significantly more labile to hydrolysis than NMIA. The half-life of 1M7 was reported to be 14 seconds, in comparison to 20 minutes for NMIA. This drastic change in reactivity allows the experimenter to complete a SHAPE experiment in as little as a few minutes. Structure probing on the RNaseP RNA domain showed that 1M7 and NMIA both give similar results of RNA modification37. 1M7 has been used to understand the structure of many RNAs, in particular those that undergo conformational changes due to protein binding or the interaction with small molecules. This is discussed in greater detail below.
RNA structural transitions often occur on the time scale of minutes, and these events occur on a continuum. It would be beneficial to study RNA structure over extended periods of time, while observing time-resolved changes in RNA structure. The implementation of reagents with well-defined reactivity times would fulfill this need. Mortimer et al. showed that benzoyl cyanide (BzCN) reacts with hydroxyl functional groups to yield a stable ester38. Formation of the stable cyanide ion leaving group renders the reaction irreversible. Structural reactivities were very similar to those observed for 1M7, suggesting the two reagents read fast structural transitions on the same time-scale. Importantly, in aqueous solutions BzCN undergoes hydrolysis in as little as 1 second at 37° C, making it an ideal reagent for time-resolved RNA structure probing experiments.
Studying the tertiary folding of the RNaseP RNA was used to demonstrate the power of BzCN as a fast reactive SHAPE reagent. Folding reactions of RNaseP RNA were initiated and aliquots were removed and reacted with BzCN. Analysis of adduct formation by reverse-transcription revealed that BzCN is able to read out kinetically distinct hydroxyl fluctuations, further supporting the hypothesis that RNA structure folding follows a hierarchical pathway, with tertiary contacts falling into the slowest phase before the structural ground state is reached. These results further support the notion that SHAPE is an ideal way to read out structural transitions in RNA folding, and because the reagents can be tailored for the experiment SHAPE represents a flexible method to dive into several forms of structural interrogation of RNA with itself and other biological macromolecules.
SHAPE and protein footprinting
Within the cell RNA is rarely without a protein partner. RNA-protein interactions are important for splicing, co-transcriptional folding of RNA, RNA localization, and RNA metabolism (degradation)35,39. Studying such interactions can shed light on how complex formation and RNA-protein interactions control these processes. Characterizing RNA-protein interactions using SHAPE chemistry has been exclusively conducted on in vitro, isolated complexes. A few examples we highlight here show the strength of SHAPE as a way to measure changes in RNA dynamics or alterations in RNA structure as a function of protein binding.
Characterizing RNA-protein complexes can often be difficult if there is no a priori knowledge of the interaction. As SHAPE chemicals measure local nucleotide flexibility, and given that protein binding to RNA likely effects the local dynamics of the bound nucleotides, SHAPE measurements offer a potentially straightforward approach to interrogate specific secondary structure conformational changes that occur with protein binding. Work utilizing this concept has explored RNA-protein interactions in the context of viral and bacterial RNAs interacting with viral, bacterial, and eukaryotic proteins. In bacteria, specifically Pseudomonas, understanding specifically how mRNA leader sequences regulate gene has been an outstanding question. The Crc protein is known to bind leader sequences and act as a translational repressor that helps to regulate Pseudomonas’ ability to assimilate sugars, nitrogenized compounds, and other molecules important for metabolism40 Moreno and colleagues hypothesized that detailed analysis of the secondary structure of the leader sequences might reveal the mechanism of Crc action. Indeed, examining the conformational changes in the mRNA leader sequence of two Pseudomonas’ mRNAs revealed that the addition of recombinant Crc protein could induce a specific protection of the RNA from modification with 1M7 (and DMS). The binding of Crc was localized to a region near the AUG codon for many mRNAs, leading to a model where Crc acted to inhibit translation of its target mRNAs by lowering the efficiency of functional ribosomal formation on the target RNAs40. In this example, SHAPE chemistry was combined with in vitro protein binding to directly reveal the location of a RNA-protein interaction.
In another study focused on understanding how proteins may help or guide RNA folding the Weeks laboratory examined the bI3 RNP, which is comprised of a group I intron, a monomer of the bI3 maturase, and two dimers of the Mrs1 protein. Previous work had shown that the bI3 RNA in solution, without its protein partners, is grossly misfolded relative to the known catalytically active structure, suggesting bI3 must undergo significant remodeling to become active. Applying SHAPE analysis to the bI3 RNA in the context of maturase and Mrs1 binding reveled protein specific conformational changes41. Importantly the authors observed that both proteins were critical for the bI3 RNA to fully rearrange into the catalytically active conformation. These results suggest the folding mechanism of group I introns is both highly cooperative but nonhierarchical, implicating protein cofactors as critical partners in establishing tertiary contacts in the RNA important for functional activity.
The Weeks laboratory has also employed SHAPE to explore the impact of protein binding on the secondary structure of viral RNAs. The Moloney MuLV Gag protein is known to assemble viral particles inside host cells, but can also target host mRNA molecules. Mechanistically it was not understood how specificity is achieved by Gag given its known simple tetranucleoide binding motif. SHAPE measurements of the viral genome, both in vitro and in virio (within an intact virion), revealed two regions, each 3’ to the two PAL domains in the viral genome, which were most impacted by the binding of the Gag protein42. The structural motif elucidated from this analysis showed that while the Gag protein prefers a short, ssRNA motif, there is a specific structural context within which the tetranucleotide motif must be presented to properly interact with Gag..
In addition to viral packaging, the capacity for viruses to hijack the host’s cellular machinery in order to replicate its self is critical in the viral life cycle. Translation of the viral genome is an important step during infection and unlike most host mRNAs, many viral genomes use an IRES sequence to initiate translation. The host protein Gemin5 has been shown to directly interact with IRES sequences, however what controls the binding of Gemin5 to host or viral RNA was not known. Pineiro et al. used SHAPE to explore the interaction between the foot-and-mouth disease virus IRES domain and Gemin5. The authors observed that Gemin5 binds a specific region of the IRES domain and is able to induce conformational changes after binding to the IRES RNA. Exploring the structural changes induces with the addition of another protein factor PTB, which is known to enhance translation initiation of IRES containing RNAs, revealed that the binding sites of Gemin5 and PTB are spatially near each other43. Overall these and other studies detail the usefulness making SHAPE measurement in the presence or absence of proteins that bind to the RNA being analyzed. SHAPE allows for easy prototyping of new experiments as the standard reaction conditions require minimal modification. Additionally, because SHAPE is quantitative, using SHAPE to analyze the binding of proteins to RNA can yield single-nucleotide resolution and quantitative information about the biding characteristics of that interaction.
SHAPE on RNA-Small Molecule Interactions
RNA structure is often viewed as being malleable and dynamic, where many alternative structures can be adopted without dramatically changing the energetic landscape. The ability to alternatively switch from one conformation, with ease suggests that interactions could tip the equilibrium of RNA structure to control biological functions, such as transcription or translation44. Recently, it has been discovered that RNA has the ability to bind endogenous metabolites to modulate gene expression. These metabolite-binding RNA macromolecules are termed ‘riboswitches’. To date, metabolite-sensing riboswitches such as thiamine pyrophosphate, vitamin B12, lysine, glycine, S-adenosyl methionine, flavin mononucleotide, guanine, adenine, glucosamine-6-phosphate, magnesium2+ and preQ1 have been identified that bind metabolites, coenzymes or nucleobases. Remarkably, the number of genes controlled by binding to these cellular metabolites is ~3–4% within their respective genomes, and is comparable to the quantity regulated by metabolite-sensing proteins2.
Riboswitches can also be RNA-based platforms located in the non-coding regions of mRNA. Canonically, they consist of two domains, an aptamer domain and an expression platform. The evolutionarily conserved aptamer domain functions to bind the metabolite when its concentration has reached a threshold. Most of the identified riboswitch sequences appear to function by transitioning between two structural states. In the metabolite-free conformation, the expression platform interacts with the cellular machinery to turn on either translation or transcription. Metabolite-binding prompts a structural transition which sequesters the coding portion of the mRNA into the folded tertiary complex. In this state, the folded complex serves to turn off the expression platform, sequestering the flow of genetic information.
Because SHAPE can robustly read out RNA dynamics it represents an ideal method to understand how small-molecule binding can influence RNA structure. A clear example of the strength of SHAPE probing was the evaluation of both bound and free states of riboswitch aptamer domains. While many structures of ligand-bound riboswitches have revealed the details of bimolecular recognition, their unliganded structures remain poorly characterized. Characterizing the molecular details of the unliganded state is crucial for understanding the riboswitch's mechanism of action because it is this state that actively interrogates the cellular environment and helps direct the regulatory outcome. A nice example of using SHAPE to understand the contributions of metal ions and ligands to the mature structure of a riboswitch was with the S-adenosylmethionine (SAM) riboswitch45. SHAPE probing revealed that in the presence of SAM, almost the entire RNA becomes protected from modification over a narrow magnesium concentration range (0.5–1 mM MgCl2) (Figure 2, A). Furthermore, in a manner distinct from modification patterns in the absence of ligand, a complete population shift from the free to bound state modification signature occurs in a conserved pseudoknot and kink-turn. These observations suggested these tertiary structure parts rarely sample structures lacking formation of tertiary architecture in the presence of ligand (SAM). This is supported by the observation that the Tm for many of the RNA residues increased in the presence of SAM, consistent with the hypothesis that SAM binding is responsible for thermodynamic stability of the aptamer structure (Figure 2, B). These results demonstrate that at physiological magnesium concentrations (0.5–1 mM), magnesium and SAM binding is coupled to fully stabilize the tertiary structure of the aptamer. Additional detailed SHAPE reactivity patterns suggest that in the absence of ligand, the RNA exists as an ensemble of conformations ranging from minimal tertiary structure to the bound-like state. Increasing the concentration of magnesium favors the formation of structures that are increasingly reminiscent of the SAM-bound structure, as indicated by Nucleotide Analog Interference Mapping (Figure 2, C). Second, the addition of ligand globally stabilizes the architecture of the RNA. This study was equally important because SHAPE was coupled with other biophysical methods (small angle X-ray scattering and X-ray crystallography) to establish that conformational heterogeneity in riboswitches can be limited by adopting a bound-state, such interactions have a profound effect on riboswitch function. It also clearly establishes the use of SHAPE in different conditions to identify interactions that control RNA structure and function.
Figure 2. SHAPE Probing on the SAM-I Riboswitch.
(A) Denaturing gel electrophoresis diagram for SHAPE probing, as a function of increasing magnesium (−/+) S-adenosyl methionine (SAM) (B) Ligand dependent changes in apparent melting temperatures mapped on the secondary structure of the SAM-I riboswitch. (C). Additional nucleotide analog interference mapping of guanine (inosine, yellow) and adenine (2-amino purine, red; di-amino purine, cyan) identifies specific nucleotide positions as important for folding and/or binding. Image is reprinted with permission from Cell Press, Structure Journal.
Studies of riboswitches focused on only isolated aptamer domains have provided a great deal of our understanding of these molecular toggles, however some riboswitches function through contacts that are peripheral to the aptamer, but are equally as important. SHAPE RNA structure probing of riboswitches has also been used to study the structure of peripheral elements found outside the aptamer-binding domain. An example of how SHAPE probing can elucidate additional RNA interactions that are important for function is with studies on the Bacillus subtilis lysC lysine riboswitch46,47.
The lysC riboswtich modulates its own gene expression upon lysine binding through a transcription attenuation mechanism. The riboswitch aptamer is organized around a single five-way junction that provides the scaffold for two long-range tertiary interactions (loop L2–loop L3 and helix P2–loop L4) to form the lysine binding site. SHAPE analysis of the riboswitch, as a function of Magnesium2+ concentration, revealed that the lysine riboswitch aptamer exhibited clear changes in NMIA reactivity. This was most apparent in regions involved in L2–L3 and P2–L4 long-range interactions, also demonstrating that the P2–L4 is particularly important for the organization of the ligand-binding site and for the riboswitch transcription attenuation control. Additional analysis with complementary techniques revealed that there is a folding synergy between domains that are in close juxtaposition involving two stems for riboswitch docking (P1 and P5). The P1-P5 interaction is directly facilitated by the concentration of lysine. Thus, SHAPE structure probing, and complimentary techniques revealed that riboswitches have extensive structural interactions that are removed from the aptamer domain. This is a clear demonstration of how SHAPE probing can be used to identify and study structural elements within functional RNAs that may be presently overlooked. Overall, these two instances and many others display the strength of chemical probing in analyzing small-molecule interactions with RNA.
Extending SHAPE Chemical Probing In Vivo
Nearly all structural work utilizing SHAPE chemistry has been conducted in vitro, with the above in virio work on the HIV genome as a notable exception, however, the structural characteristics of RNA in vivo are predicted to be different than those in vitro. In vivo RNA structure can be influenced by the rate of transcription, local solution conditions, the binding of small molecules, and through interactions with RNA-binding proteins35,39,48,49. These observations hint that the physical state of RNA within the cell is very important for its function, but little is known of how intracellular RNA structure can contribute to the specificity of these binding events or the biological functions of newly described non-coding RNAs.
Despite these exciting efforts, SHAPE had the major limitation of not being able to be applied to RNA structure in living cells. Potential limitations of SHAPE reagents are their short half-lives and limited solubility. Indeed, SHAPE electrophiles are approximately 20-fold less soluble than dimethyl sulfate, which is used to probe RNA structure in cells at greater than 200mM final concentration (the solubility limit of SHAPE reagents is ~10mM final)34,50,51. The environment inside a cell complicates these experiments because of the competing environment of electrophiles. Proteins, nucleic acid polymers and even nucleotide triphosphates, which can have concentrations of 2 µM to 8 mM52, depending on the cell type can compete with SHAPE reactions. To overcome these obstacles we recently reported the design and utilization of novel SHAPE reagents that can be used to probe RNA structure in living cells.
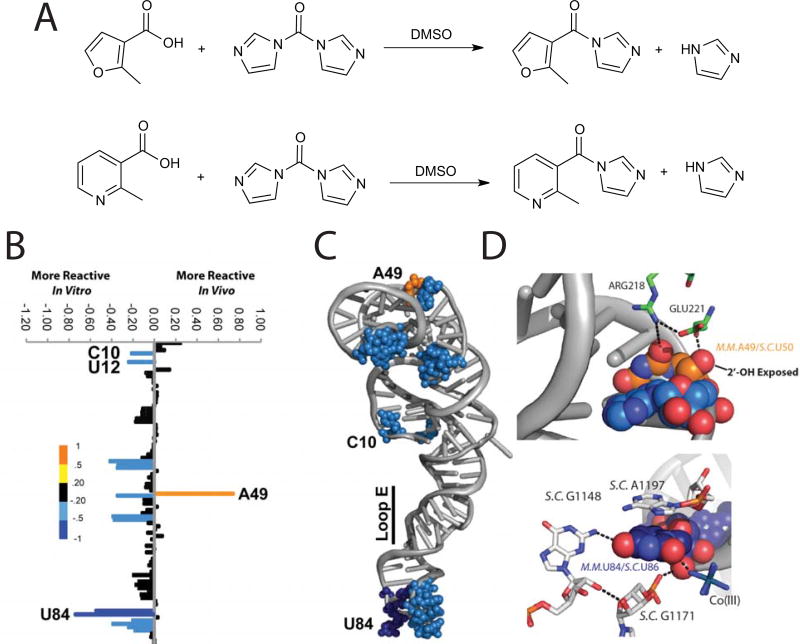
Spitale et al. designed two SHAPE electrophiles, one of which is named NAI, that read RNA structure with similar efficacy to the canonical SHAPE electrophiles (Figure 3, A). However, the newly designed reagents had much longer half-lives (~30 minutes, Figure 1, A&B) and have solubility of up to 200mM in aqueous solutions. For our initial analysis we focused on the 5S ribosomal RNA (5S rRNA). We chose 5S rRNA because of its abundant nature, highly characterized structure and ability to fold into a stable structure without the need for protein cofactors53. Residues modified by SHAPE probing corresponded to regions of 5S rRNA that are in loops or bulged (Figure 3, B & C). Our comparisons between in vitro and in vivo SHAPE measurements reveal unique characteristics of 2’-OH flexibility that are due to protein binding and tertiary contacts within the mature ribosome (this is highlighted in Figure 3 C & D), where residues 49 and 84 are held in unique conformations due to their interactions with other RNAs and proteins in the context of the mature ribosome). Residues with lower B-factors (a measurement of flexibility) in the crystal structure were shown to be less reactive in vivo. Further, residues with the highest B-factor (highly dynamic in the crystal), displayed the largest differences in the cell, with a marked increase in reactivity. This result further suggests that NAI is able to distinguish structure-specific RNA dynamics that are the result of a mature cellular complex and can be used to compare X-ray structures to interrogate cellular RNA complexes. Our analysis was the first comparison of 5S rRNA structure in vitro versus in vivo. Further, we were able to read out the secondary structure of the 5S rRNA across 5 different species and of less-abundant nuclear RNAs mammalian cells.
Figure 3. Synthesis and Utilization of the First In Vivo SHAPE Reagents.
(A). Synthesis of FAI and NAI as 1.0 M stock solution of 1:1 compound:imidazole in DMSO. Note that CO2 is also produced during the reaction but it evolves from solution. (B) SHAPE modification patterns with differences between in vivo and in vitro calculated. (C) Differential SHAPE modifications mapped onto the crystal structure of 5S rRNA. (D) Zoomed in view of residue A49. (E) Zoomed in view of reside U84.
Additionally, there is a recent paper by the Weeks lab54 describing the use of the fast-acting 1M7 SHAPE electrophile to probe the structure of a non-native RNA expressed in Escherichia coli cells. This paper studied the structure of the add adenine riboswitch. SHAPE probing in vivo and in vitro revealed that the structure of the adenine-bound aptamer-tRNA riboswitch construct is similar inside and outside of cells. However, the ligand-free form is much more compact and stable in vivo. Overall, this manuscript suggests that the in-cell environment may have a strong influence on stabilizing RNA structure elements. These two examples show that SHAPE reagents are generalizable structure probes capable of measuring RNA structure in the complex environment of the cell.
Decades of structural and biophysical work have shed light on the intricate details of RNA structure and function in vitro and most of these investigations have been done on single RNAs at a time. While the accuracy of such measurements, when coupled to detailed mutagenesis work has proven to be quite powerful, these experiments can be quite cumbersome and time-consuming. Recent efforts have tried to overcome this barrier by merging RNA structure probing with deep sequencing.
SHAPE on genomes
The majority of research done to understand RNA structure has come from one-RNA-at-a-time analysis. When investigating the structure of long RNAs, specific regions of an RNA are often analyzed in isolation, outside the context of the full-length RNA molecule. While this has been useful to probe many types of RNA structures there is always a concern that this will reveal a potential false-positive RNA structure signature that may not be present in the actual full-length transcript. Recent efforts have been focused on studying RNA structure on mature, complete RNAs. The first such endeavor was to study the entirety of the HIV genome.
HIV is a single stranded virus that is the causative agent for AIDS and other serious health problems55. HIV, as well as other viruses, has its life cycle intimately controlled by the RNA structure of the single-stranded genome. To study the structure of the HIV genome, RNA was gently extracted from purified virions in the presence of buffers containing monovalent and divalent ions, consistent with maintaining RNA secondary and tertiary structure. The HIV genomic RNA was not denatured by heat, chemical denaturants, magnesium chelation, or removal of monovalent cations during this process. To characterize the entire genome SHAPE signature was read out by tiling primers, a total of 31, along the HIV genome.
A keen observation from this data was that the abundance of RNA secondary structure stability modulated protein activity through the progression of ribosome translation. This model was further corroborated by ribosome toeprinting to show that highly structured RNA at protein-domain junctions facilitates native folding of HIV proteins by allowing time for domains to fold independently during translation. Several unstructured RNA regions, including splice site acceptors and hypervariable regions were found to have potentially important RNA structure elements. Overall, this work laid the foundation for understanding how RNA structure can be measured in its intact biological state. And, by doing so illuminates potentially new aspects of RNA structure and function that can play important roles in genome regulation.
High-Throughput Sequencing Approaches to RNA Structure
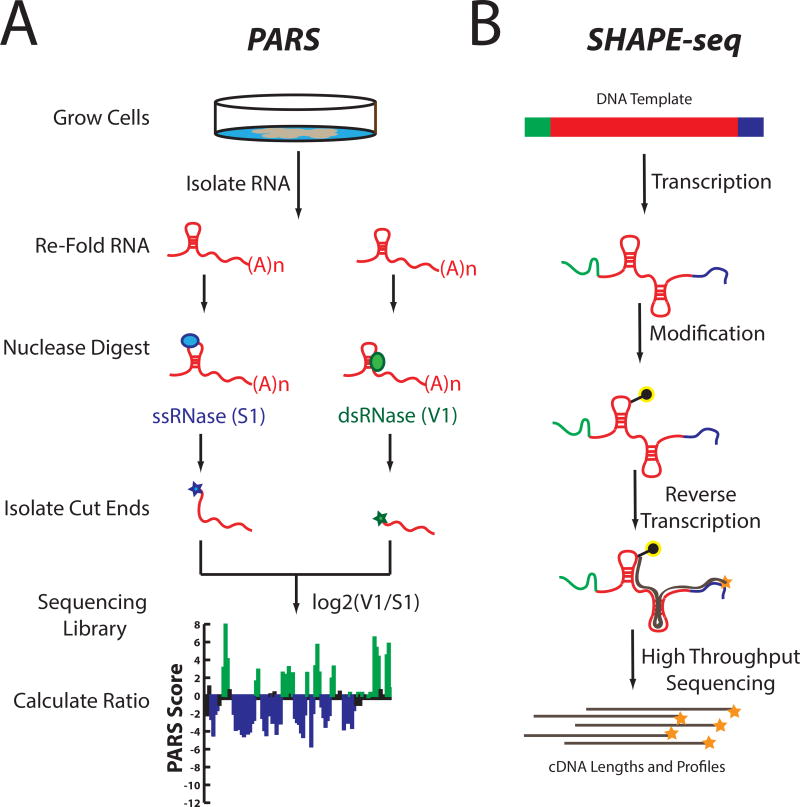
Understanding how the structure of RNA can be used to control its biological function remains a key challenge in molecular biology. Measuring RNA’s structure with chemical or enzymatic probing methods are often limited to the measurement of one RNA per experiment. Additionally, depending on whether the assay is using standard gel or capillary electrophoresis, only ~100–600 nucleotides can be analyzed per reverse transcription primer. Recently, conventional RNA structure probing using structure-specific nuclease digestion of RNA has been paired with deep sequencing to read out secondary structure. We recently developed one such technology, Parallel Analysis of RNA Structure, or PARS. Similar to low-throughput enzymatic probing, PARS starts by treating an in vitro folded RNA pool with structure-specific enzymes. However, PARS then uses deep sequencing to map the resulting cleavage sites at single nucleotide resolution, allowing it to simultaneously profile thousands of RNAs of various lengths. PARS has been used to characterize the entire yeast transcriptome in vitro and to understand the thermodynamic stabilities of RNAs, transcriptome-wide56–58 (Figure 4). Thus, transcriptome-wide measurements of RNA structure can identify functional elements that contribute to RNA biology, thereby revealing unique roles of RNA structure in biology. These investigations, as well as others involving the marriage of deep sequencing and enzyme probing of RNA structure59,60 have opened a new window with which to view the transcriptome.
Figure 4. Methods to Merge RNA Structure Probing with Deep Sequencing.
(A) Experimental outline of Parallel Analysis of RNA Structure (PARS). (B) Experimental outline of SHAPE-seq
The development of a complimentary technique, termed FragSeq, was used to generate the structures of noncoding transcripts from mouse cells. FragSeq was implemented by quantifying reads generated after digestion by RNase P1, a single-strand specific nuclease59. FragSeq was able to recapitulate the known secondary structure of well known ncRNAs U1a, U3b and U5, as well as several other ncRNAs whose secondary structures have been previously examined. To extend their structure studies, the authors also reported the structures of previously unprobed RNAs using conventional techniques, particularly long (>120 nt) C/D box snoRNAs. Together, PARS and FragSeq have demonstrated the utility and feasibility of marrying RNase footprinting with sequencing to study the structures of large pools of RNAs, in vitro. Together, these new methods have permitted RNA structure analysis of large cohorts of RNA molecules. Nevertheless, methods using RNase molecules for structure probing suffer from two key limitations: low overall resolution and the inability to probe RNA structure in vivo.
Merging Chemical-Based Probing of RNA Structure With Deep Sequencing
There has been a substantial amount of knowledge learned about RNA structure, folding, and mechanisms involving RNA through interrogation of isolated functional motifs. However, the separation of RNA domains, or the removal of RNAs from their cellular millieu may introduce biases in the final structure state or present an artifact of removing the domain from the fully formed RNA. Therefore, there is a need to study RNAs in their full, mature form, within their biological context. Some effort has been focused on studying RNA structure inside cells. However, these investigations have been limited to well studied RNAs, using function as a proxy for folding. These results have suggested that unique characteristics about the environment of the cell result in different RNA structure states in comparison to in vitro, although additional studies are needed to make broad claims about how different RNA behaves, in vivo15,16,61,62. For studying RNA structure inside cells, it is clear that chemical methods (such as in vivo SHAPE) are to be merged with recent efforts to clone cDNA molecules that arise from chemical-induced reverse transcription stops. Although not using in vivo SHAPE chemistry, recent papers have been published toward such goals.
Merging deep sequencing with chemical probing is based on the ability to identify cDNA stops and map them back to a reference RNA sequence. SHAPE-Seq (Figure 4, B) combines SHAPE chemistry with multiplexed paired-end deep sequencing of primer extension products. Identified RT stops, generated by SHAPE modifications, are analyzed using a fully automated data analysis pipeline, based on a rigorous maximum likelihood model of the SHAPE-Seq experiment63–65. These reports are in good agreement with manual footprinting experiments, demonstrating the ability of these methods to accurately model secondary structures even in a high-throughput manner. These efforts, ultimately culminate in a nice merger of biochemical and computational techniques that can be used to understand the structure of RNAs both in vitro and inside the cell, on a gene-specific basis. Further, such a powerful chemical-genomic pipeline can be used to characterize rationally designed RNAs and should permit screening by structure for desired functional outputs.
Additional efforts have pushed the field even further ahead, with a particular focus on taking chemical modification probing experiments to in vivo, whole transcriptome analyses. Two recent examples have both focused on using dimethylsulfate (DMS) sequencing. DMS modifies single-stranded A and C bases and leads to a block in reverse transcription, thus yielding structural information on two out of four bases. In the first example, application of this method to Arabidopsis thaliana seedlings yielded the first in vivo genome-wide RNA structure map at nucleotide resolution for any organism, with quantitative structural information across more than 10,000 transcripts66. A key observation made is that consistent with in vitro analyses there is a strong correlation of secondary structure near the start codon and translation efficiency. Remarkably, a global comparison of RNA structure in the context of gene ontology analyses revealed that mRNAs associated with stress response proteins have more single-strandedness to their transcripts, which opens up the suggestion that these RNAs can be easily manipulated to alter protein expression.
A second analysis of RNA structure was conducted in both yeast and human cell types67. The key finding from this study was that RNA structural elements seem to be denatured in vivo, in comparison to in vtro refolded RNAs. These data suggest that RNAs may constantly be remodeled by proteins within the cell. To test this hypothesis, RNA structure was probed under ATP-depleting conditions (where RNA chaperons would be inactive), revealing an energy-dependent process assosicated with RNA unfolding, in vivo. Overall, these studies highlight further the importance of RNA structure in the context of the cell and suggest that RNAs are rather dynamic and their thermodynamic state has a biological role on controlling protein expression.
Overall Perspective and Future Opportunities
RNA structure probing has continued to evolve from using blunt measurements through RNA cleavage to high-resolution analysis with chemical probes. The exquisite precision of chemistry provides RNA researchers with a toolbox to interrogate even the most complex of RNA structural traits under a variety of conditions. 2’-hydroxyl acylation has emerged as a leading chemical probing technique due to its generality and quantitative output. The extension of this technology to in vivo measurements presents a mandate that researchers focused on understanding RNA structure and function should begin to study RNA structure within the native environment of the cell. Enabling in vivo SHAPE measurements transcriptome-wide to yield an RNA structure-ome that measures all four nucleobases will be a major milestone in the near future.
Sequencing technologies are extremely powerful at extrapolating quantitative information about biology. The recent use of sequencing to understand the entire yeast transcriptome, the secondary structure of metazoan transcripts and mammalian nuclear RNAs have set a benchmark for the ability to understand RNA structure on a global level. The next step is to join emerging chemical technologies with sequencing to understand how RNA structure forms and changes within a living cell. Much remains to be done and learned from studying RNA structure inside living cells. In vivo and dynamic RNA structure maps will yield a crucial understanding of how RNA structures may change to regulate different biological states. Structure probing can also be performed under different conditions, such as alterations in temperature or the presence of specific proteins or small-molecule ligands, to probe the impact of these perturbations on RNA structure. Further, investigating RNA viruses in their native context, through different parts of their life cycle may provide powerful inroads to the design of novel therapeutics. Global interrogation of RNA structure is sure to assist computational strategies to better model RNA structure and predict RNA function. Cross-comparison of genomic RNA structure maps with high-resolution maps of RNA–protein interactions will be one immediate avenue whereby such integrative analyses can yield useful biological insights.
Box1. Practical considerations for chemical probe selection.
Key to choosing the right structural probe is knowingl the practical considerations of the reagents. First, each of these chemicals should be handled with care. They are highly reactive and modification (damage)68 of nucleic acids is a known cause of DNA mutations and disease. NMIA, DMS, and metals used for RNA cleavage are currently commercially available at many chemical supply companies. Both 1M7 and NAI can be synthesized in a single synthetic step. 1M7 is purified by crystallization37, whereas NAI does not need to be purified before experiments.53 Special caution should be used when handling 1M7, as it is highly reactive and will hydrolyze. NAI can be stored form months at −80°C in anhydrous dimethyl sulfoxide.
Acknowledgments
This work is supported by NIH R01-HG004361 (H.Y.C.). H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute. E.T.K. acknowledges support from the U.S. National Institutes of Health (GM068122). R.C.S. is supported by the Giannini Foundation.
References
- 1.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- 4.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. doi:S0092-8674(07)00659-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. doi:S1097-2765(11)00645-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. doi:S0092-8674(09)00083-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gartland WJ, Sueoka N. Two interconvertible forms of tryptophanyl sRNA in E. coli. Proc Natl Acad Sci U S A. 1966;55:948–956. doi: 10.1073/pnas.55.4.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams A, Lindahl T, Fresco JR. Conformational differences between the biologically active and inactive forms of a transfer ribonucleic acid. Proc Natl Acad Sci U S A. 1967;57:1684–1691. doi: 10.1073/pnas.57.6.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dann CE, 3rd, et al. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. doi:S0092-8674(07)00901-4 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Keene JD. Biological clocks and the coordination theory of RNA operons and regulons. Cold Spring Harb Symp Quant Biol. 2007;72:157–165. doi: 10.1101/sqb.2007.72.013. [DOI] [PubMed] [Google Scholar]
- 11.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–684. doi: 10.1038/nature08717. doi:nature08717 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG. A rugged free energy landscape separates multiple functional RNA folds throughout denaturation. Nucleic Acids Res. 2008;36:7088–7099. doi: 10.1093/nar/gkn871. doi:gkn871 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thirumalai D, Lee N, Woodson SA, Klimov D. Early events in RNA folding. Annual review of physical chemistry. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 14.Yadava RS, Mahen EM, Fedor MJ. Kinetic analysis of ribozyme-substrate complex formation in yeast. Rna. 2004;10:863–879. doi: 10.1261/rna.5234204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahen EM, Harger JW, Calderon EM, Fedor MJ. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Mol Cell. 2005;19:27–37. doi: 10.1016/j.molcel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 16.Watson PY, Fedor MJ. Determination of intracellular RNA folding rates using self-cleaving RNAs. Methods Enzymol. 2009;468:259–286. doi: 10.1016/S0076-6879(09)68013-7. [DOI] [PubMed] [Google Scholar]
- 17.Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajkowitsch L, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. doi:5445 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Trang P, Hsu AW, Liu F. Nuclease footprint analyses of the interactions between RNase P ribozyme and a model mRNA substrate. Nucleic Acids Res. 1999;27:4590–4597. doi: 10.1093/nar/27.23.4590. doi:gkc674 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. doi:nprot.2007.380 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hargittai MR, Musier-Forsyth K. Use of terbium as a probe of tRNA tertiary structure and folding. Rna. 2000;6:1672–1680. doi: 10.1017/s135583820000128x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 23.Pan T. Probing RNA structure by lead cleavage. Curr Protoc Nucleic Acid Chem. 2001;Chapter 6(Unit 6):3. doi: 10.1002/0471142700.nc0603s00. [DOI] [PubMed] [Google Scholar]
- 24.Mauger DM, Siegfried NA, Weeks KM. The genetic code as expressed through relationships between mRNA structure and protein function. FEBS Lett. 2013;587:1180–1188. doi: 10.1016/j.febslet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 26.Velikyan I, Acharya S, Trifonova A, Foldesi A, Chattopadhyaya J. The pK(a)'s of 2'-hydroxyl group in nucleosides and nucleotides. J Am Chem Soc. 2001;123:2893–2894. doi: 10.1021/ja0036312. [DOI] [PubMed] [Google Scholar]
- 27.Guo M, et al. Direct Raman measurement of an elevated base pKa in the active site of a small ribozyme in a precatalytic conformation. J Am Chem Soc. 2009;131:12908–12909. doi: 10.1021/ja9060883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryder SP, et al. Investigation of adenosine base ionization in the hairpin ribozyme by nucleotide analog interference mapping. RNA. 2001;7:1454–1463. [PMC free article] [PubMed] [Google Scholar]
- 29.Weeks KM, Mauger DM. Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res. 2011;44:1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGinnis JL, Dunkle JA, Cate JH, Weeks KM. The mechanisms of RNA SHAPE chemistry. J Am Chem Soc. 2012;134:6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chamberlin SI, Merino EJ, Weeks KM. Catalysis of amide synthesis by RNA phosphodiester and hydroxyl groups. Proc Natl Acad Sci U S A. 2002;99:14688–14693. doi: 10.1073/pnas.212527799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2 '-hydroxyl acylation and primer extension (SHAPE) Journal of the American Chemical Society. 2005;127:4223–4231. doi: 10.1021/Ja043822v. doi: [DOI] [PubMed] [Google Scholar]
- 33.Moorman AR, Abeles RH. A New Class of Serine Protease Inactivators Based on Isatoic Anhydride. Journal of the American Chemical Society. 1982;104:6785–6786. doi: 10.1021/Ja00388a053. doi: [DOI] [Google Scholar]
- 34.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 35.Schroeder R, Grossberger R, Pichler A, Waldsich C. RNA folding in vivo. Curr Opin Struct Biol. 2002;12:296–300. doi: 10.1016/s0959-440x(02)00325-1. [DOI] [PubMed] [Google Scholar]
- 36.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S-2) in RNA. Journal of the American Chemical Society. 2008;130:12244. doi: 10.1021/Ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 38.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry. J Am Chem Soc. 2008;130:16178–16180. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- 39.Liebeg A, Mayer O, Waldsich C. DEAD-box protein facilitated RNA folding in vivo. RNA Biol. 2010;7:803–811. doi: 10.4161/rna.7.6.13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno R, Marzi S, Romby P, Rojo F. The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation initiation. Nucleic Acids Res. 2009;37:7678–7690. doi: 10.1093/nar/gkp825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan CD, Weeks KM. Nonhierarchical ribonucleoprotein assembly suggests a strain-propagation model for protein-facilitated RNA folding. Biochemistry. 2010;49:5418–5425. doi: 10.1021/bi100267g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gherghe C, et al. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc Natl Acad Sci U S A. 2010;107:19248–19253. doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pineiro D, Fernandez N, Ramajo J, Martinez-Salas E. Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res. 2013;41:1017–1028. doi: 10.1093/nar/gks1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoddard CD, et al. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blouin S, Chinnappan R, Lafontaine DA. Folding of the lysine riboswitch: importance of peripheral elements for transcriptional regulation. Nucleic Acids Res. 2011;39:3373–3387. doi: 10.1093/nar/gkq1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antal M, Boros E, Solymosy F, Kiss T. Analysis of the structure of human telomerase RNA in vivo. Nucleic Acids Res. 2002;30:912–920. doi: 10.1093/nar/30.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurg A, Sommer G, Metspalu A. An RNA stem-loop structure involved in the packaging of bovine leukemia virus genomic RNA in vivo. Virology. 1995;211:434–442. doi: 10.1006/viro.1995.1425. [DOI] [PubMed] [Google Scholar]
- 50.Lindell M, Romby P, Wagner EG. Lead(II) as a probe for investigating RNA structure in vivo. Rna. 2002;8:534–541. doi: 10.1017/s1355838201020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wells SE, Hughes JM, Igel AH, Ares M., Jr Use of dimethyl sulfate to probe RNA structure in vivo. Methods Enzymol. 2000;318:479–493. doi: 10.1016/s0076-6879(00)18071-1. [DOI] [PubMed] [Google Scholar]
- 52.Forrester T. An estimate of adenosine triphosphate release into the venous effluent from exercising human forearm muscle. J Physiol. 1972;224:611–628. doi: 10.1113/jphysiol.1972.sp009915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spitale RC, et al. RNA SHAPE analysis in living cells. Nat Chem Biol. 2013;9:18–20. doi: 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry. 2013;52:8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wertheim JO, et al. The Global Transmission Network of HIV-1. The Journal of infectious diseases. 2013 doi: 10.1093/infdis/jit524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan Y, Qu K, Ouyang Z, Chang HY. Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nat Protoc. 2013;8:849–869. doi: 10.1038/nprot.2013.045. [DOI] [PubMed] [Google Scholar]
- 57.Wan Y, et al. Genome-wide measurement of RNA folding energies. Mol Cell. 2012;48:169–181. doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Underwood JG, et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng Q, et al. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 2010;6:e1001141. doi: 10.1371/journal.pgen.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watson PY, Fedor MJ. The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo. Nat Struct Mol Biol. 2011;18:359–363. doi: 10.1038/nsmb.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mahen EM, Watson PY, Cottrell JW, Fedor MJ. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010;8:e1000307. doi: 10.1371/journal.pbio.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mortimer SA, Trapnell C, Aviran S, Pachter L, Lucks JB. SHAPE-Seq: High-Throughput RNA Structure Analysis. Curr Protoc Chem Biol. 2012;4:275–297. doi: 10.1002/9780470559277.ch120019. [DOI] [PubMed] [Google Scholar]
- 64.Aviran S, et al. Modeling and automation of sequencing-based characterization of RNA structure. Proc Natl Acad Sci U S A. 2011;108:11069–11074. doi: 10.1073/pnas.1106541108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lucks JB, et al. Multiplexed RNA structure characterization with selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci U S A. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2013 doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 67.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2013 doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maluf SW, Marroni NP, Heuser VD, Pra D. DNA damage and oxidative stress in human disease. Biomed Res Int. 2013;2013:696104. doi: 10.1155/2013/696104. [DOI] [PMC free article] [PubMed] [Google Scholar]