Abstract
The mtDNA mutator mouse lacks the proofreading capacity of the sole mtDNA polymerase, leading to accumulation of somatic mtDNA mutations, and a profound premature aging phenotype including elevated oxidative stress and apoptosis, and reduced mitochondrial function. We have previously reported that endurance exercise alleviates the aging phenotype in the mutator mice, reduces oxidative stress, and enhances mitochondrial biogenesis. Here we summarize our findings, with the emphasis on the central role of p53 in these adaptations. We demonstrate that mtDNA in sedentary and exercised PolG mice carry similar amounts of mutations in muscle, but in addition to that sedentary mice have more non-mutational damage, which is mitigated by exercise. It follows therefore that the profound alleviation of the mtDNA mutator phenotype in muscle by exercise may not require a reduction in mtDNA mutational load, but rather a decrease of mtDNA damage and/or oxidative stress. We further hypothesize that the observed ‘alleviation without a reduction of mutational load’ implies that the oxidative stress in PolG muscle is maintained, at least in part, by the ‘malicious cycle’, a hypothetical positive feedback potentially driven by the ‘transcriptional mutagenesis’, that is the conversion of chemically modified nucleotides into mutant RNA bases by the mitochondrial RNA polymerase.
Introduction: the role of mtDNA mutations in the progeroid phenotype of the mtDNA ‘mutator’ mouse
The mtDNA mutator (PolG) mouse harbors a proofreading exonuclease-deficient mtDNA polymerase (polymerase gamma, or POLG1) [1•,2,3], and therefore accumulates very high levels of somatic mutations in the mtDNA. This mouse is remarkable in that it accurately recapitulates many features of human aging [2,4••,3]. Crucial components of the PolG phenotype include mitochondrial insufficiency [5•,2], and high rates of apoptosis [6,3]. While mtDNA point mutations are ostensibly the culprit of the mutator phenotype (see however [7,8]), the mechanism(s) whereby they cause the phenotype are a matter of intense research.
One view is that point mutations are causing problems by reducing the activity of mtDNA-encoded enzymes [9]. Interestingly, despite massive amounts of mutations in mtDNA of mutator mice (a couple dozen per mtDNA molecule), the accompanying decrease in the activity of mitochondrial enzymes in the mutator mice is relatively modest, with surprisingly high residual activity (occasionally about 35% but typically much more) [9,4••]. This perhaps reflects the fact that mtDNA mutations are typically ‘recessive’, that is need to reach a high percentage in the cell (the ‘phenotypic threshold’) before any phenotype shows up. Importantly, mutations that affect different mtDNA functions do not cooperate in reaching the threshold [10]. Nascent mutations in the mtDNA mutator mouse constitute a complex mixture of diverse sequence changes, which may not have enough time for ‘clonal expansion’, Thus they may not reach, despite their huge total presence, individual intracellular thresholds and thus will fail to cause respiratory defects.
Another view is that the PolG phenotype could be due in part to mutations with ‘dominant’ effects. For example, for decades, mtDNA mutations were considered a likely source of increased levels of reactive oxygen species (ROS) [11]. Indeed at least some mtDNA mutations are known to increase ROS levels, for example [12]. Increased ROS production is a gain-of-function change, every enzyme molecule with such a mutation adds to ROS level independently of other enzyme molecules, which is the meaning of ‘dominant’ in the mtDNA sense. Conceivably, dominant mutations should have a good chance to contribute to the PolG mouse phenotype because their effects are additive, proportional to the total mutational load, which is high in PolG mice, and are not subject to phenotypic thresholds.
In this opinion paper we review current evidence in support of the role of ROS in the PolG phenotype. The detailed mechanism whereby ROS contribute to the phenotype is revealed by the molecular dissection of the effect of endurance exercise, which completely reverses the mutator phenotype [13]. We discuss a hypothetical mechanism that appears to account for the current observations, including some contradictory ones, and leads to testable predictions.
PolG phenotype and oxidative stress
Initial characterization of PolG mice illustrated an absence of increased oxidative damage despite significant accumulation of mtDNA point mutations [2,3]. However, recent studies using more sensitive ROS assays have provided support for the presence of oxidative stress-induced damage in both the somatic and stem cell pool of PolG mice [14••,5•,4••]. Furthermore, oral administration of antioxidants has been shown to ameliorate stem cell defects, and transgenic over-expression of mitochondrial-targeted catalase has been shown to partially ameliorate cardiomyopathy in PolG mice [14••,15], implicating oxidative stress in the development of the PolG phenotype.
Increased oxidative stress in the mitochondria of the PolG mouse also induces nuclear DNA damage, as represented by telomere erosion [16••], which in turn facilitates translocation of the tumor suppressor protein, p53, into the nucleus from its cytoplasmic depots [17]. Indeed, we clearly observed elevated nuclear p53 levels in the sedentary PolG mouse, in tandem with nuclear DNA damage [16••]. Nuclear p53 has been shown to activate senescence and apoptosis programs in both somatic and stem cells [17]. This naturally explains the pro-apoptotic state characteristic of many PolG tissues [18,3,16••]. Furthermore, p53 represses the transcription of PGC-1α, the master regulator of mitochondrial biogenesis and cellular antioxidant response [19••]. We believe that this may add to the general downregulation of mitochondrial bioenergetic capacity, reduced mtDNA copy number, and overall decrease in mitochondrial electron transport chain complex assembly in the sedentary PolG mouse [3,15,9,4••,16••], which probably also depends o the high levels of mtDNA mutations. We believe that the nuclear relocalization of p53, reduces the availability of p53 for the mitochondria, which subsequently attenuates p53/TFAM, and p53/POLG1 complex formation in the mitochondria of PolG mice [20•,16••].
Exercise profoundly alleviates the progeroid phenotype and non-mutational mtDNA damage, but does not reduce mutational load of mtDNA
Recent work by our group showed that the systemic mitochondrial dysfunction and multisystem pathology in PolG mice is efficiently mitigated by endurance exercise [4••,16••]. Not surprisingly, the hunt for molecular mechanism(s) underlying the systemic rescue of pathology in PolG mice as executed by endurance exercise has garnered much attention.
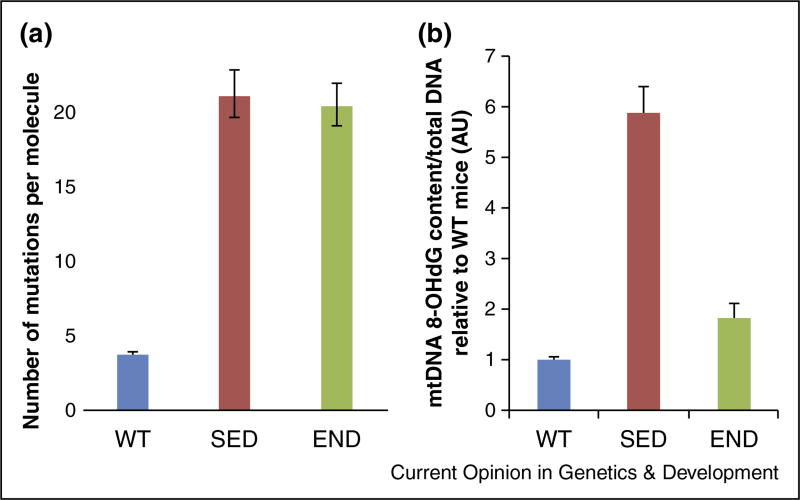
The profound amelioration of the PolG phenotype by exercise allows us to explore the role of mtDNA mutations in maintaining this phenotype. If we agree that the progeroid phenotype of the PolG mouse is caused by mtDNA mutations, then the simplest hypothesis would be that exercise ameliorates PolG phenotype by reducing the mutational load. However, our extensive measurements of mutational load (~700 kb of high quality sequence analyzed, ~800 mutations scored) using the single molecule full length mtDNA mutational analysis [21,22] revealed no difference in the muscle of sedentary and exercised PolG mice (Figure 1a). If not decrease in mutations, what could have caused amelioration by exercise? Interestingly, unlike mtDNA mutations, the non-mutational oxidative DNA damage (i.e. 8-OHdG) is clearly decreased by exercise (Figure 1b). Of note, the sequencing of shorter mtDNA fragments does show a lower mutation load in exercised mice [4••]. We interpret this as additional evidence of the decrease of non-mutational mtDNA damage and unrepaired mismatches in the exercised PolG mice (see Supplement 1 for full discussion).
Figure 1.
Oxidative damage (a) is alleviated by exercise (b) but not mutational load (WT: wild type littermate control; SED: sedentary PolG; END: endurance exercised PolG mouse muscle). WT are daughters of heteroplasmic PolG mothers, hence relatively high mutant background. See Supplement 2 for experimental details.
Hypothesis: mtDNA damage contributes to the mutator phenotype via ‘transcriptional mutagenesis’
Given the connection of non-mutational mtDNA damage with the PolG phenotype, it is tempting to speculate that non-mutational mtDNA damage might in fact be contributing to the phenotype. Adding weight to this hypothesis, the lack of p53 in the muscle-specific p53 knockout abolishes the amelioration of the PolG phenotype by exercise. Importantly, p53 is a mtDNA repair protein [23••,24••,20•,25•], particularly relevant to the repair of oxidative damage [26]. Therefore lack of p53 is expected to prevent the alleviation of non-mutational mtDNA damage. Indeed ‘short-fragment mutations’, which we believe are a measure of the non-mutational damage, are not decreased by exercise in muscle-specific p53 knockout PolG mice [16••], which is entirely consistent with the hypothesis. Of course, lack of p53 may have other endpoints and this hypothesis by no means implies to negate other potential pathways as responsible for the PolG phenotype. However, these observations are intriguingly compatible with mtDNA damage being one of them.
What could be a possible mechanism relating non-mutational mtDNA damage to the mtDNA mutator phenotype? We would like to propose the ‘transcriptional mutagenesis’, specifically, the conversion of damaged DNA into mutated transcripts by the mitochondrial RNA polymerase as it incorrectly transcribes chemically modified nucleotides. These mutated transcripts will be translated into mutated polypeptides which, in turn, are expected to promote oxidative (and also perhaps proteotoxic) stress just like (or even more efficiently than) mutated polypeptides originating from true mtDNA mutations (see discussion in the Introduction). Note that this hypothesis has a solid foundation. The ‘transcriptional mutagenesis’, that is conversion of DNA damage into mutated transcripts by RNA polymerase [27], is a well-established phenomenon. This includes conversion of oxidative damage, for example, 8-OHdG [28] and has been established for various RNA polymerases, including the mitochondrial RNA polymerase [29••]. ‘Transcriptional mutagenesis’ has been recently implicated in cellular stress and premature cellular aging [30••].
Attractiveness of a hypothesis depends on the competitiveness of alternative explanations. What mechanism other than mitigation of mtDNA damage could mediate the effect of exercise? A potentially attractive explanation is based on the PGC-1α, which is robustly up-regulated by exercise [31,32], PGC-1α promotes mitochondrial biogenesis and increases expression of cytosolic and mitochondrial antioxidants, thus decreasing oxidative stress [33,34]. This explanation, however, appears insufficient, because such mechanism would have worked even better in the absence of p53, a known PGC-1α suppressor [19••].
However, in contradiction with this prediction, in muscle-specific p53 knockout PolG mice, exercise entirely fails to alleviate oxidative stress and ameliorate the mutator phenotype in muscle-specific p53 knockout PolG mice [16••]. A solution to this striking controversy would be that p53 works to suppress ROS via a PGC-1α-independent pathway, such as the mtDNA repair pathway, as proposed above.
Another competing hypothesis would be that mtDNA damage contributes to the mutator phenotype via a pathway other than ‘transcriptional mutagenesis’. For example, the mere presence of mtDNA damage might be expected to elicit toxic stress response. While this is a plausible possibility, we note that p53 is considered a master DNA damage sensor of the cell. It therefore would be surprising, if this hypothesis were correct, that this effect does not disappear and in fact is even enhanced in the muscle-specific p53 knockout PolG mice.
A hypothetical model: the ‘malicious cycle’ and its demise by exercise
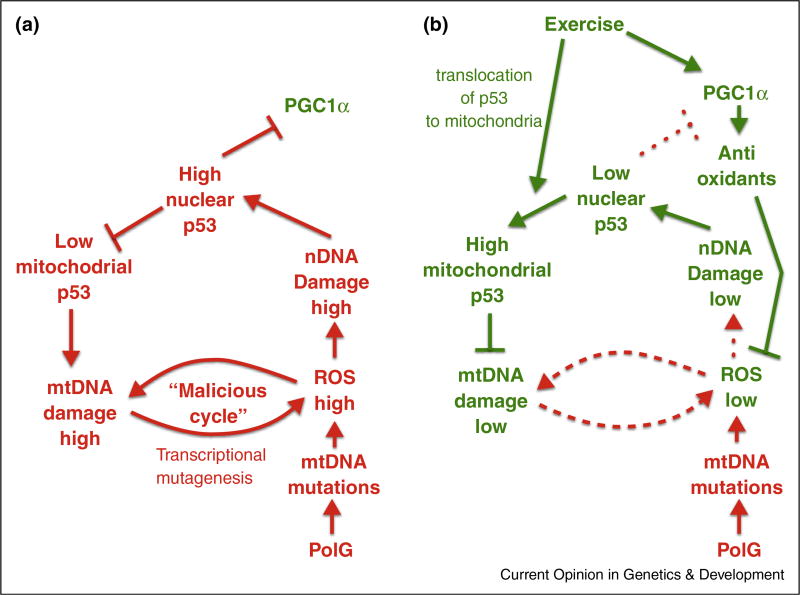
The findings and hypotheses discussed above are summarized in Figure 2, which presents a hypothetical model of the maintenance of the progeroid phenotype in the sedentary PolG mouse (Figure 2a) and its amelioration by endurance exercise (Figure 2b).
Figure 2.
Hypothetical mechanism underpinning the amelioration of premature aging in the mtDNA mutator mouse by endurance exercise. (a) Sedentary PolG mouse: ‘a double cycle’. Defective polymerase gamma (bottom) creates an influx of de novo mutations in mtDNA. The accumulating mtDNA mutations cause increase of ROS production, which leads to increased oxidative mtDNA and nuclear DNA damage. mtDNA damage adds to the ROS production caused by mtDNA mutations, presumably via transcription mutagenesis, which completes the hypothetical ‘malicious cycle’. Moreover, nuclear DNA damage triggers the translocation of the p53 into the nucleus, which reduces the available pool of p53 for mitochondria. The paucity of p53, a likely DNA repair enzyme, results in an even higher level of DNA damage, resulting in another self-sustaining cycle. (b) Exercised PolG mouse: Exercise intercepts the ‘double cycle’ in two ways: First, exercise promotes the expression of PGC-1α, which in turn activates an antioxidant gene expression network. This serves to attenuate overall oxidative stress and nuclear DNA damage, and results in the subsequent release of p53 from the nucleus, making the p53 pool available for the mitochondria. PGC-1α upregulation also results in an increased mitochondrial biogenesis. Second, the translocation of p53 into mitochondria, where p53 serves as a repair enzyme, reduces the mtDNA damage and thus disrupts the ‘malicious cycle’. This ultimately sets the system in a different equilibrium with lower mtDNA damage.
Of note, if our hypothesis that mtDNA damage increases ROS levels via ‘transcriptional mutagenesis’, is correct, then one can predict the existence of a self-sustained mtDNA damage cycle. It is dubbed here the ‘malicious cycle’: more mtDNA damage — more oxidative stress — more mtDNA damage (Figure 2a). We use the term ‘malicious cycle’ to distinguish this model from the classical ‘vicious cycle’ model, which assumes real DNA mutations rather than non-mutational damage. In addition to the malicious cycle, the data discussed in the previous points to another self-sustaining circle: mtDNA mutations cause ROS increase, which causes nDNA damage. This prompts translocation of p53 to the nucleus, which represses PGC-1α, and depletes mitochondria of p53, preventing the repair of mtDNA damage, which increases ROS thus completing the ‘double circle’ (Figure 2a).
While this model appears to accurately capture the molecular phenotype of the sedentary PolG, its major advantage is in presenting an attractive mechanism(s) for the disruption of the ‘double cycle’ and amelioration of the sedentary PolG phenotype by exercise. There are two points at which exercise interrupts the cycle.
First, endurance exercise has been shown to robustly upregulate PGC1α [31,32], which in turn promotes mitochondrial biogenesis and increases expression of cytosolic and mitochondrial antioxidants [33,34]. This increase in global antioxidant capacity normalizes the cellular redox status, which subsequently attenuates nuclear DNA damage, allowing p53 to leave the nucleus [35••], resulting in reduced apoptosis and senescence [16••]. Similar results were obtained by others [36]. This change in the cellular location of p53 alleviates the abrogation of PGC-1α repression caused by nuclear p53, which may further increases PGC-1α expression (Figure 2b).
Second, we believe that its translocation to mitochondria results in repair of the mtDNA damage and disruption of the ‘malicious cycle’, because, as argued in ‘Hypothesis: mtDNA damage contributes to the mutator phenotype via “transcriptional mutagenesis’”, p53 is a mtDNA repair enzyme. Note that both inhibitory arms in Figure 2b are needed to interrupt the cycle. Indeed, in the p53 KO mouse, amelioration of the phenotype fails, probably because the ‘malicious cycle’ remains active despite increased antioxidants due to mtDNA damage remaining unrepaired.
Generalization of the model to other tissues
While detailed studies of the role of p53 in the amelioration of the PolG phenotype were done in muscle, we believe that similar principles should underlie the pathology and its exercise-driven amelioration in other tissues. It has been proposed that haemopoetic deficiencies of the PolG mouse are caused by oxidative stress early in development of hematopoietic stem and progenitor cells (HSC) [14••]. Furthermore, exercise induces an increase in PGC-1α expression in a variety of tissues other than muscle, for example, heart, muscle satellite cells and HSC [16••]. Additionally, we have previously reported systemic increase in mitochondrial activity in exercised PolG mice, a mitochondrial characteristic regulated by PGC1 family members [4••]. In addition to muscle, translocation of p53 to mitochondria upon exercise is observed in the heart and is yet to be explored in other tissues. It is tempting to speculate that the same model is applicable to other tissues and may be important to maintaining the stem cell pool in exercised PolG mice.
Thus this hypothesis may explain various aspects of amelioration by exercise in the mutator mouse. Importantly, p53 also translocates to mitochondria in normal exercised mice [36••], and it is tempting to speculate that p53 may regulate mtDNA protective functions beyond merely reducing the rate of point mutations in the mtDNA mutator mouse.
Concluding remarks and perspectives
We have previously argued that because mutation levels in PolG mice are much higher than in normal aging mice or humans, this mouse does not adequately emulate the aging process [37]. While we believe that this argument is still correct, paradoxically, the PolG mouse does capture many fine details of natural aging, including the amelioration by exercise. Our results imply that this controversy has a solution. If indeed mtDNA mutations per se are not the direct drivers of premature aging in PolG mice, which instead is driven by some downstream endpoints like mtDNA damage and/or oxidative stress, then PolG phenotype may closely emulate human aging/disease simply because it is, for example, driven by a similar level of oxidative stress (see, however [38]). In relation to this, interestingly, pigmented neurons of substantia nigra in early stages of Parkinson’s disease, have been reported to carry up to 10 point mutations per mtDNA molecule [39] This is approaching mutational levels in the PolG mouse. Of note, these measurements were done using short PCR fragments and Taq-based thermostable polymerase. Thus these apparent high mutation loads may in fact reflect high non-mutational mtDNA damage in these neurons (converted into mutations by the PCR). This would imply that levels of mtDNA damage in the polG mice may be comparable to the levels in some human tissues and thus be relevant to the human case.
The proposed model makes a number of testable predictions. We would expect that blocking the movement of p53 to the mitochondrion using appropriate pharmacological inhibitors [40] should abrogate the alleviation of mtDNA damage if our proposed model holds true. Alternatively, a transgenic mutator mouse model with mitochondrial-targeted p53 will repair mtDNA damage, and thus prevent aspects of progeroid aging. In fact, mitochondrial-targeted p53 may present as a novel therapeutic modality for diseases of mitochondrial etiology in the future.
Supplementary Material
Acknowledgments
The authors thank Ms. Jenny Nguyen for artwork. This work is supported by the Canadian Institutes of Health Research Banting Fellowship and American Federation for Aging Research Fellowship (A Safdar), Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (A Saleem), NIH grant R37-AG012279 to JT, and the Ellison Medical Foundation Senior Scholar Award to KK.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.gde.2016.06.011.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
• • of outstanding interest
- 1•.Zhang D, Mott JL, Chang SW, Denniger G, Feng Z, Zassenhaus HP. Construction of transgenic mice with tissue-specific acceleration of mitochondrial DNA mutagenesis. Genomics. 2000;69:151–161. doi: 10.1006/geno.2000.6333. This is the first study that detailed the generation of a heart-specific mitochondrial polymerase gamma proofreading mutant transgenic mouse model and linked increase in mtDNA mutations to age-associated cardiomyopthy. [DOI] [PubMed] [Google Scholar]
- 2.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 3.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 4••.Safdar A, Bourgeois JM, Ogborn DI, Little JP, Hettinga BP, Akhtar M, Thompson JE, Melov S, Mocellin NJ, Kujoth GC, et al. Endurance exercise rescues progeroid aging and induces systemic mitochondrial rejuvenation in mtDNA mutator mice. Proc Natl Acad Sci. 2011;108:4135–4140. doi: 10.1073/pnas.1019581108. A seminal study that first described the effects of endurance exercise in mediating systemic mitochondrial rejuvenation, which in turn mitigated multisystem pathology, and rescued progeroid aging phenotypes in mtDNA mutator mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Kolesar JE, Safdar A, Abadi A, MacNeil LG, Crane JD, Tarnopolsky MA, Kaufman BA. Defects in mitochondrial DNA replication and oxidative damage in muscle of mtDNA mutator mice. Free Radic Biol Med. 2014;75:241–251. doi: 10.1016/j.freeradbiomed.2014.07.038. This is the first study that comprehensively demonstrated the presence of mtDNA replication defect, and oxidative damage in mtDNA mutator mice. [DOI] [PubMed] [Google Scholar]
- 6.Hiona A, Sanz A, Kujoth GC, Pamplona R, Seo AY, Hofer T, Someya S, Miyakawa T, Nakayama C, Samhan-Arias AK, et al. Mitochondrial DNA mutations induce mitochondrial dysfunction, apoptosis and sarcopenia in skeletal muscle of mitochondrial DNA mutator mice. PLoS One. 2010;5:e11468. doi: 10.1371/journal.pone.0011468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- 8.Kraytsberg Y, Simon DK, Turnbull DM, Khrapko K. Do mtDNA deletions drive premature aging in mtDNA mutator mice? Aging Cell. 2009;8:502–506. doi: 10.1111/j.1474-9726.2009.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgar D, Shabalina I, Camara Y, Wredenberg A, Calvaruso MA, Nijtmans L, Nedergaard J, Cannon B, Larsson NG, Trifunovic A. Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metab. 2009;10:131–138. doi: 10.1016/j.cmet.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 10.Khrapko K, Turnbull D. Mitochondrial DNA mutations in aging. Prog Mol Biol Transl Sci. 2014;127:29–62. doi: 10.1016/B978-0-12-394625-6.00002-7. [DOI] [PubMed] [Google Scholar]
- 11.Bandy B, Davison AJ. Mitochondrial mutations may increase oxidative stress: implications for carcinogenesis and aging? Free Radic Biol Med. 1990;5(8):3–539. doi: 10.1016/0891-5849(90)90152-9. [DOI] [PubMed] [Google Scholar]
- 12.Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, MacGregor GR, Wallace DC. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Safdar A, Bourgeois JM, Ogborn DI, Little JP, Hettinga BP, Akhtar M, Thompson JE, Melov S, Mocellin NJ, Kujoth GC, et al. Endurance exercise rescues progeroid aging and induces systemic mitochondrial rejuvenation in mtDNA mutator mice. Proc Natl Acad Sci U S A. 2011;108:4135–4140. doi: 10.1073/pnas.1019581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Ahlqvist KJ, Hämäläinen RH, Yatsuga S, Uutela M, Terzioglu M, Götz A, Forsström S, Salven P, Angers-Loustau A, Kopra OH, et al. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. An informative piece of work that details the role of mitochondrial dysfunction in somatic stem cells as one of the underlying mechanisms of progeroid phenotype in mtDNA mutator mice. [DOI] [PubMed] [Google Scholar]
- 15.Dai D-F, Chen T, Wanagat J, Laflamme M, Marcinek DJ, Emond MJ, Ngo CP, Prolla TA, Rabinovitch PS. Age-dependent cardiomyopathy in mitochondrial mutator mice is attenuated by overexpression of catalase targeted to mitochondria. Aging Cell. 2010;9:536–544. doi: 10.1111/j.1474-9726.2010.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Safdar A, Khrapko K, Flynn JM, Saleem A, De Lisio M, Johnston APW, Kratysberg Y, Samjoo IA, Kitaoka Y, Ogborn DI, et al. Exercise-induced mitochondrial p53 repairs mtDNA mutations in mutator mice. Skeletal Muscle. 2016;6:520–618. doi: 10.1186/s13395-016-0075-9. A comprehensive study that deciphered interplay of ROS-p53-PGC1α axis in endurance exercise-mediated maintenance of the mtDNA genome in the context of aging. Exercise failed to prevent mitochondrial dysfunction, reduce mtDNA mutations, and ultimately preventing progeroid sarcopenea in mtDNA mutator mice with muscle-specific deletion of p53. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 18.Zhang D, Mott JL, Farrar P, Ryerse JS, Chang SW, Stevens M, Denniger G, Zassenhaus HP. Mitochondrial DNA mutations activate the mitochondrial apoptotic pathway and cause dilated cardiomyopathy. Cardiovasc Res. 2003;57:147–157. doi: 10.1016/s0008-6363(02)00695-8. [DOI] [PubMed] [Google Scholar]
- 19••.Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. An informative piece of work that detailed how the telomere-p53-PGC axis functions to regulate telomere erosion and mitochondrial dysfunction, subsequently causing organ and metabolic failure followed by senescence and cell death. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Bakhanashvili M, Grinberg S, Bonda E, Simon AJ, Moshitch-Moshkovitz S, Rahav G. p53 in mitochondria enhances the accuracy of DNA synthesis. Cell Death Differ. 2008;15:1865–1874. doi: 10.1038/cdd.2008.122. A comprehensive report on the role of mitochondrial-resident p53 as a component of the mtDNA error-repair pathway, thereby functioning as the guardian of the mitochondrial genome. [DOI] [PubMed] [Google Scholar]
- 21.Greaves LC, Beadle NE, Taylor GA, Commane D, Mathers JC, Khrapko K, Turnbull DM. Quantification of mitochondrial DNA mutation load. Aging Cell. 2009;8:566–572. doi: 10.1111/j.1474-9726.2009.00505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraytsberg Y, Nicholas A, Caro P, Khrapko K. Single molecule PCR in mtDNA mutational analysis: genuine mutations vs. damage bypass-derived artifacts. Methods. 2008;46:269–273. doi: 10.1016/j.ymeth.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.de Souza-Pinto NC, Harris CC, Bohr VA. p53 functions in the incorporation step in DNA base excision repair in mouse liver mitochondria. Oncogene. 2004;23:6559–6568. doi: 10.1038/sj.onc.1207874. This is the first study that demonstrated the effect of p53 on mtDNA stability, through its base excision repair capability, in vivo using murine liver mitochondria. [DOI] [PubMed] [Google Scholar]
- 24••.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. A formative paper that reported a novel role of p53 in enhancing mtDNA genomic stability through its ability to translocate to mitochondria, and interact with mitochondrial polymerase gamma in response to ROS-mediated mtDNA damage in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Bakhanashvili M, Grinberg S, Bonda E, Rahav G. Excision of nucleoside analogs in mitochondria by p53 protein. Aids. 2009;23:779–788. doi: 10.1097/QAD.0b013e328329c74e. This study extensively characterized the intrinsic 3′ → 5′ exonuclease proofreading activity of purified wild-type recombinant p53. Using in vitro biochemical assays, the authors illustrated that p53 preferentially repairs transversion mutations, and enhances DNA replication fidelity. [DOI] [PubMed] [Google Scholar]
- 26.Park J-H, Zhuang J, Li J, Hwang PM. p53 as guardian of the mitochondrial genome. FEBS Lett. 2016 doi: 10.1002/1873-3468.12061. http://dx.doi.org/10.1002/1873-3468.12061. [DOI] [PMC free article] [PubMed]
- 26.Viswanathan A, You HJ, Doetsch PW. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284:159–162. doi: 10.1126/science.284.5411.159. [DOI] [PubMed] [Google Scholar]
- 28.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc Acad Sci U S A. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Nakanishi N, Fukuoh A, Kang D, Iwai S, Kuraoka I. Effects of DNA lesions on the transcription reaction of mitochondrial RNA polymerase: implications for bypass RNA synthesis on oxidative DNA lesions. Mutagenesis. 2013;28:117–123. doi: 10.1093/mutage/ges060. An important paper that suggest that mitochondrial RNA polymerase can bypass non-bulky oxidative DNA lesions induced by mitochondrial reactive oxygen species generation. [DOI] [PubMed] [Google Scholar]
- 30••.Vermulst M, Denney AS, Lang MJ, Hung C-W, Moore S, Moseley MA, Mosely AM, Thompson JW, Thompson WJ, Madden V, et al. Transcription errors induce proteotoxic stress and shorten cellular lifespan. Nat Commun. 2015;6:8065. doi: 10.1038/ncomms9065. A seminal study that details the novel role of transcriptional errors in aging-associated loss in proteostasis and shortened lifespan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Handschin C, Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature. 2008;454:463–469. doi: 10.1038/nature07206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Safdar A, Little JP, Stokl AJ, Hettinga BP, Akhtar M, Tarnopolsky MA. Exercise increases mitochondrial PGC-1alpha content and promotes nuclear-mitochondrial cross-talk to coordinate mitochondrial biogenesis. J Biol Chem. 2011;286:10605–10617. doi: 10.1074/jbc.M110.211466. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jäger S, Handschin C, Zheng K, Lin J, Yang W, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 34.Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Acad Sci U S A. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35••.Saleem A, Hood DA. Acute exercise induces tumour suppressor protein p53 translocation to the mitochondria and promotes a p53-Tfam-mitochondrial DNA complex in skeletal muscle. J Physiol. 2013;591:3625–3636. doi: 10.1113/jphysiol.2013.252791. This is the first study that reported the mitochondrial translocation of p53 and its interaction with Tfam and mtDNA in response to an acute bout of endurance exercise in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Granata C, Oliveira RSF, Little JP, Renner K, Bishop DJ. Training intensity modulates changes in PGC-1α and p53 protein content and mitochondrial respiration, but not markers of mitochondrial content in human skeletal muscle. FASEB J. 2016;30:959–970. doi: 10.1096/fj.15-276907. [DOI] [PubMed] [Google Scholar]
- 37.Khrapko K, Kraytsberg Y, de Grey ADNJ, Vijg J, Schon EA. Does premature aging of the mtDNA mutator mouse prove that mtDNA mutations are involved in natural aging? Aging Cell. 2006;5:279–282. doi: 10.1111/j.1474-9726.2006.00209.x. [DOI] [PubMed] [Google Scholar]
- 38.Maslov AY, Ganapathi S, Westerhof M, Quispe-Tintaya W, White RR, Van Houten B, Reiling E, Dolle MET, van Steeg H, Hasty P, et al. DNA damage in normally and prematurely aged mice. Aging Cell. 2013 doi: 10.1111/acel.12071. http://dx.doi.org/10.1111/acel.12071. [DOI] [PMC free article] [PubMed]
- 39.Lin MT, Cantuti-Castelvetri I, Zheng K, Jackson KE, Tan YB, Arzberger T, Lees AJ, Betensky RA, Beal MF, Simon DK. Somatic mitochondrial DNA mutations in early Parkinson and incidental Lewy body disease. Ann Neurol. 2012;71:850–854. doi: 10.1002/ana.23568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787:414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.