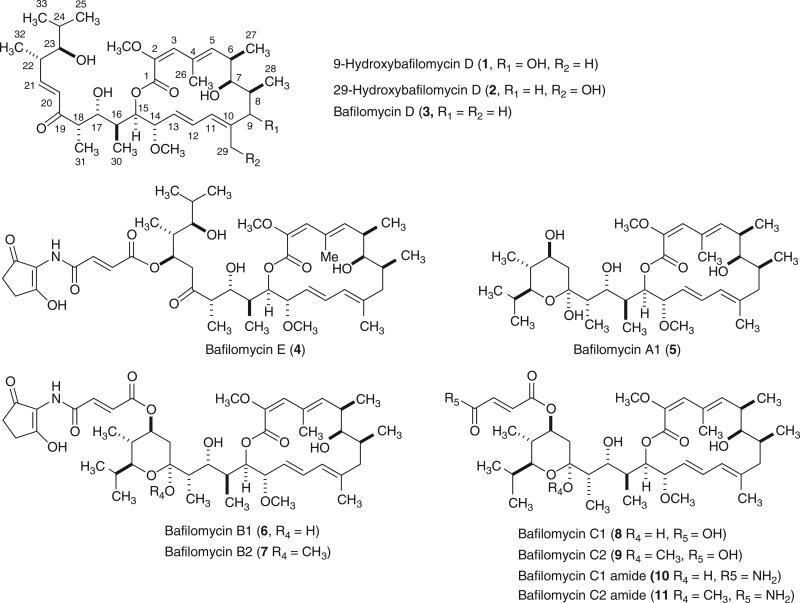
Bafilomycins (Figure 1), macrolide antibiotics with a 16-membered lactone ring as their defining structural scaffold, are produced by a variety of streptomycetes.1–5 These macrolides exhibit a variety of biological activities, including antitumor,6 antifungal,7 antiparasitic8 and immunosuppressant activities.9 In particular, bafilomycin A1 is an extremely potent and specific inhibitor of the vacuolar ATPases10 and has also been found to inhibit the release of β-amyloid11 and mitogen-induced DNA synthesis.12 These features provide only a few examples of biological activities that have drawn considerable interest to the bafilomycins.
Figure 1.
Structures of new (1 and 2) and known (3–11) bafilomycins isolated from Streptomyces sp. YIM56209.
In our on-going effort to search for new and biologically active secondary metabolites produced by actinomycetes from unexplored and underexplored ecological niches,13–16 an endophytic actinomycete Streptomyces sp. YIM56209 was isolated from a healthy stem of Drymaria cordata. Initial screening of the crude extract showed potent cytotoxicity against selected cancer cell lines as well as moderate inhibitory effect on prolactin (PRL)-initiated phosphorylation of ERK1/2 in MCF-7 breast cancer cells. Large-scale fermentation and subsequent fractionation of the crude extract led to the isolation of two new bafilomycins, named 9-hydroxybafilomycin D (1) and 29-hydroxybafilomycin D (2), together with nine known bafilomycins identified as bafilomycin D (3),3 bafilomycin E (4),3 bafilomycin A1 (5),1 bafilomycin B1 (6),1,7 bafilomycin B2 (7),1 bafilomycin C1 (8),1 bafilomycin C2 (9),1 bafilomycin C1 amide (10)17 and bafilomycin C2 amide (11)17 (Figure 1). The structures of 1 and 2 were established by comprehensive spectroscopic analyses, whereas the structures of 3–11 were confirmed by comparing their 1H and 13C NMR data with those in the literature. We present herein the isolation, structural elucidation and bioactivity assessment of these new and known bafilomycins.
The ethyl acetate extract of the Streptomyces sp. YIM56209 culture was subjected to sequential silica gel and Sephadex LH-20 chromatography, followed by further purification with reversed-phase HPLC to give 11 bafilomycins (1–11) (Figure 1).
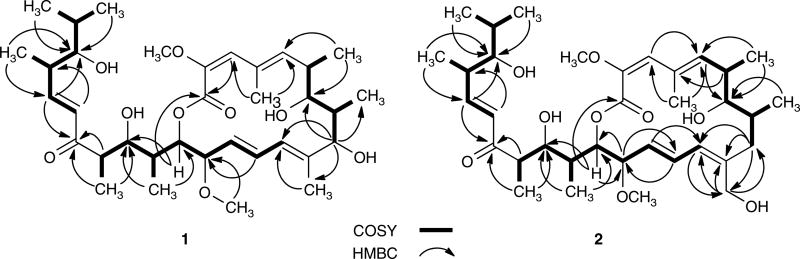
Compound 1 was isolated as a white amorphous powder. High-resolution MALDI-FTMS analysis of 1 afforded an [M+Na]+ ion at m/z 643.38197, consistent with a molecular formula of C35H56O9 (calculated [M+Na]+ ion at m/z 643.38165), which contained an extra oxygen atom in comparison with the molecular formula of 3 (C35H56O8). The 13C NMR spectrum of 1 (Table 1) shows 34 signals, representative of 35 carbons (two, C-7 and C-23, have the same chemical shift at δC 80.0). In conjunction with the gHMQC spectrum, the presence of two methoxy groups (2-OCH3 and 14-OCH3), nine Me groups (C-25, C-26, C-27, C-28, C-29, C-30, C-31, C-32 and C-33), six methine carbons (C-6, C-8, C-16, C-18, C-22 and C-24), seven olefinic carbons (C-3, C-5, C-11, C-12, C-13, C-20 and C-21), six oxymethine carbons (C-7, C-9, C-14, C-15, C-17 and C-23), three olefinic quaternary carbons (C-2, C-4 and C-10) and two carbonyl groups (C-1 and C-19) was confirmed (Table 1 and Figure 1). Compared with a similar data set obtained from 3 (Supplementary Tables S1 and S2), the shift of the methylene carbon signal (δC 41.6 at C-9 of 3) to the oxygenated methine signal (δC 77.2 at C-9 of 1), together with its MS data, suggested that 1 was likely a C-9 hydroxylated congener of 3. This conclusion was further supported by gHMBC correlation of H-9 (δH 3.85) with C-7 (δC 80.0), C-11 (δC 125.5), C-28 (δC 18.2) and C-29 (δC 18.8) (Figure 2). Accordingly, 1 was finally established as 9-hydroxybafilomycin D with the stereochemistry of C-9 remaining undetermined because of the limited quantities of compound presently available (Figure 1).
Table 1.
Summary of 1H and 13C NMR spectroscopic data for 1 and 2 (in CDCl3)
| 1 (J in Hz) | 2 (J in Hz) | |||
|---|---|---|---|---|
|
|
|
|||
| Position | δH mult | δc | δH mult | δc |
| 1 | — | 166.7 | — | 166.4 |
| 2 | — | 141.8 | — | 141.6 |
| 3 | 6.63 (s) | 133.2 | 6.60, s | 132.6 |
| 4 | — | 133.4 | — | 133.2 |
| 5 | 5.73, d (9.0) | 141.0 | 5.81, d (9.0) | 142.1 |
| 6 | 2.54, m | 37.2 | 2.54, m | 36.6 |
| 7 | 3.41, d (7.0) | 80.0 | 3.43, d (7.0) | 81.4 |
| 8 | 2.31, m | 38.8 | 1.91, m | 40.6 |
| 9 | 3.85, d (10.5) | 77.2 | 2.30, m; 2.10, m | 36.5 |
| 10 | — | 147.2 | — | 143.7 |
| 11 | 6.02, d (10.5) | 125.5 | 5.90, d (11.0) | 128.1 |
| 12 | 6.51, dd (14.5, 11.0) | 133.3 | 6.64, dd (15.0, 11.0) | 132.0 |
| 13 | 5.29, dd (15.0, 9.5) | 130.5 | 5.23, dd (15.0, 9.5) | 129.5 |
| 14 | 3.81, dd (8.5, 8.0) | 83.4 | 3.81, dd (8.5, 8.0) | 83.0 |
| 15 | 5.05, d (8.5) | 76.7 | 5.05, d (9.0) | 76.2 |
| 16 | 2.06, m | 38.8 | 2.07, m | 38.3 |
| 17 | 3.77, m | 72.9 | 3.76, m | 72.2 |
| 18 | 2.97, m | 46.6 | 2.97, m | 46.4 |
| 19 | — | 203.4 | — | 203.0 |
| 20 | 6.28, d (16.0) | 129.6 | 6.29, d (16.0) | 129.3 |
| 21 | 6.90, dd (15.5, 8.0) | 148.9 | 6.90, dd (16.0, 8.0) | 148.5 |
| 22 | 2.54, m | 40.3 | 2.52, m | 40.0 |
| 23 | 3.18, t (6.0) | 80.0 | 3.18, t (6.0) | 79.8 |
| 24 | 1.72, m | 31.2 | 1.73, m | 30.9 |
| 25 | 0.92, d (7.0) | 17.0 | 0.93, d (7.0) | 16.8 |
| 26 | 1.98, s | 14.2 | 1.98, s | 14.1 |
| 27 | 1.07, d (7.0) | 17.4 | 1.07, d (7.0) | 17.0 |
| 28 | 0.95, d (7.0) | 18.2 | 0.97, d (7.0) | 22.0 |
| 29 | 2.03, s | 18.8 | 4.58, d (12.5); 4.07, d (12.5) | 63.1 |
| 30 | 0.95, d (7.0) | 10.9 | 0.95, d (7.0) | 10.6 |
| 31 | 1.21, d (7.0) | 10.6 | 1.21, d (7.0) | 10.2 |
| 32 | 1.08, d (6.5) | 16.9 | 1.08, d (6.5) | 16.6 |
| 33 | 0.95, d (7.0) | 19.9 | 0.95, d (7.0) | 19.7 |
| 2-OCH3 | 3.68, s | 60.5 | 3.67, s | 60.1 |
| 14-OCH3 | 3.22, s | 56.0 | 3.21, s | 55.8 |
Figure 2.
Key HMBC and H, H-COSY correlations for the two new bafilomycin congeners 1 and 2.
Compound 2 was isolated as a white amorphous powder. The molecular formula of 2 was also determined by high-resolution MALDI-FTMS analysis, which yielded an [M+Na]+ ion at m/z 643.38151, consistent with a molecular formula of C35H56O9 (calculated [M+Na]+ ion at m/z 643.38165). The 1H and 13C NMR data (Table 1) of 2 were similarly compared with those of 3 as well as 2 (Supplementary Tables S1 and S2), and the shift of the Me group signal [δC 20.2 and δH 1.91(s) at C-29 of 3] to the oxymethylene signal [δC 63.1 and δH 4.58(d), 4.07(d) at C-29 of 2], together with its MS data, readily established that 2 is most likely a C-29 hydroxylated analog of 3. This conclusion was further supported by gHMBC correlations of H2-29 [δH 4.58 (d), 4.07 (d)] with C-9 (δC 36.5), C-10 (δC 143.7) and C-11 (δC 128.1), H-11 [δH 5.90 (d)] with C-29 (δC 63.1), and H2-9 [δH 2.30 (m), 2.10 (m)] with C-29 (δC 63.1) (Figure 2). Thus, the structure of 2 was deduced to be 29-hydroxybafilomycin D (Figure 1).
The extreme cytotoxicity of the bafilomycins has limited their practical utility as both molecular probes and potential therapeutics. We therefore assessed the cytotoxicity of the two new bafilomycin analogs (1 and 2) together with the nine known ones (3–11) using A-549 human lung adenocarcinoma and HT-29 human colorectal adenocarcinoma cancer cell lines. Although 3–11 exhibited potent cytoxicity in general, under the conditions tested, the two new analogs 1 and 2 are, on average, two to three orders of magnitude less toxic, a property that could potentially be explored for the utilities of bafilomycin family (Table 2).
Table 2.
Summary of biological activities for 1 and 2 in comparison with 3–11
| Cytotoxicity (IC50 in nm unless otherwise noted) | Inhibition of ERK1/2 phosphorylation (IC50 in µm)a | |||
|---|---|---|---|---|
|
|
|
|||
| Compound | A549 | HT29 | EGF mediated | PRL mediated |
| 1 | 7600 ± 400 | 9000 ± 400 | NAb | NAb |
| 2 | 3900 ± 200 | 3400 ± 100 | NAb | 31.6 |
| 3 | 1.3 ± 0.5 | 2.2 ± 0.4 | NAb | 30.0 |
| 4 | 10.9 ± 0.9 | NDc | Slight stimulationd | NAb |
| 5 | 70.2 ± 2.3 | 90.4 ± 3.6 | NAb | 41.5 |
| 6 | 20.9 ± 1.2 | 110 ± 7.3 | NAb | 49.2 |
| 7 | 17.0 ± 0.7 | 148 ± 35 | NAb | 51.0 |
| 8 | 1.4 ± 0.1 | 44.3 ± 11 | Toxice | Toxice |
| 9 | 1.5 ± 0.1 | 2000 ± 500 | Toxice | Toxice |
| 10 | 70.2 ± 2.3 | NDc | NAb | 12.3 |
| 11 | 70.2 ± 2.3 | NDc | NAb | NAb |
Abbreviations: EGF, epidermal growth factor; PRL, prolactin.
EGF- or PRL-mediated phosphorylation of ERK1/2 after 15 min determination as described in Supplementary Information.
No activity was observed up to 100 µm.
Not detectable. Activity was either not detected up to 100 µm or reliable standard derivations were not attainable.
Stimulated activity was found to be ~150% at 100 µm.
Toxicity to cells abrogated accurate data acquisition.
The ability of 5 to inhibit vacuolar-type H+-ATPases, resulting in reduced acidification of sorting endosomes and subsequent interference with post-internalization receptor trafficking, is well-established.10,18 In light of the links between trafficking and signal transduction for many receptors,19 we attempted to probe the effect of 1–11 on phosphorylation of the mitogen-activated protein kinases, ERK1/2, initiated by two well-characterized breast cancer mitogens, PRL20 and epidermal growth factor (EGF),21 in the breast cancer cell line MCF-7. As summarized in Table 2, the relative activities of bafilomycin family members in this assay differed substantially. None of the bafilomycins tested was able to reduce EGF-initiated signals, although 4 slightly amplified these signals. The relative lack of activity in this assay resembles the failure of bafilomycin to inhibit EGF signals to c-fos in fibroblasts, despite inhibition of EGF-induced mitosis.12 In contrast, six of the analogs, 2, 3, 5, 6, 7 and 10, were able to reduce PRL-mediated signals to ERK1/2; 10 displayed optimum activity with an IC50 of ~12 µm. The relative specificity of bafilomycins for PRL signaling pathway inhibition, compared with EGF, may reflect differences in receptor trafficking or other biological actions of these compounds; the susceptibility of receptor trafficking to reduced acidification has been reported to vary among cell types.22
In summary, we have isolated two new bafilomycin analogs, 9-hydroxybafilomycin D (1) and 29-hydroxy-bafilomycin D (2), together with nine known bafilomycin congeners (3–11) from fermentation culture of the endophyte actinomycete Streptomyces sp. YIM56209 (Figure 1). The structures of 1 and 2 were elucidated by MS and NMR techniques. Hydroxylation at either the C-9 or C-29 positions of 3, both integral components of the 16-membered lactone scaffold, profoundly impacts the biological activity of 1 and 2 relative to 3, as well as other previously known bafilomycins. For example, 1 was found to be ~5000 times less cytotoxic to A549 cells and ~4000 times less toxic to HT-29 cells than 3, whereas 2 was found to be ~3000 times less cytotoxic to A549 cells and ~1500 times less toxic to HT-29 cells relative to 3. As the bafilomycins are unable to be used clinically because of their fatal toxicity,10 this discovery opens a new path for the practical application of bafilomycins. Moreover, the ability to inhibit PRL-mediated signaling pathways may be useful as few reagents are available to probe the activities of this hormone/cytokine in normal physiology and pathology, including breast cancer.
EXPERIMENTAL SECTION
Optical rotations were measured in CHCl3 on a Perkin-Elmer 241 instrument (Perkin-Elmer, Waltham, MA, USA) at the sodium D line (589 nm). 1H and 13C NMR spectra were recorded at 25 °C on a Varian Unity Inova 500 instrument (Agilent Technologies, Inc., Santa Clara, CA, USA) operating at 500 MHz for 1H and 125 MHz for 13C nuclei. High-resolution mass spectral analyses were acquired on an IonSpec HiResMALDI FT-MS with a 7 T superconducting magnet (IonSpec, Inc., Lake Forest, CA, USA). Semi-preparative HPLC was performed on a Varian HPLC system with an Alltima C18 column (5 µ, 10.0×250 mm, Alltech Associates, Inc., Deerfield, IL, USA). Column chromatography was performed either on silica gel (230–400 mesh, Natland International, Research Triangle Park, NC, USA), or Sephadex LH-20 (Pharmacia, Kalamazoo, MI, USA). Chemical regents were purchased from Sigma-Aldrich Corporation (St Louis, MO, USA).
The producing stain YIM56209 was isolated from a healthy stem of traditional Chinese medicinal plant D. cordata from Xishuangbanna, Yunnan, China, which was used to treat hepatitis and nephritis, and was identified as Streptomyces sp. using a polyphasic taxonomy approach. Pure strain was permanently stored in 50% glycerol at −80 °C. The fermentation medium consisted of 2.0 g yeast extract, 5.0 g malt extract and 2.0 g dextrose in 1.0 l Milli-Q water (Millipore Corporation, Billerica, MA, USA), pH 7.2. A three-stage fermentation procedure was adopted: (1) seed culture in 10 ml medium at 28 °C for 4 days, (2) 1.0 ml of the resultant culture as inoculum to 50 ml medium in 250-ml Erlenmeyer flasks at 28 °C for 3 days and (3) 20 ml the resultant culture as inoculum to 400 ml medium in 2.0-l Erlenmeyer flasks at 28 °C for 7 days, all on a rotary shaker at 250 r.p.m.
A total of 9.6 l of fermentation culture was collected and extracted with ethyl acetate. The combined extracts were concentrated under reduced pressure to give the crude residue (3.1 g), which was subjected to silica gel chromatography eluted with a hexane-ethyl acetate gradient (0–100%). Six fractions, A–F, were monitored by TLC analysis and collected. Fraction E (103 mg) was further chromatographed over Sephadex LH-20 column and eluted with MeOH to give three subfractions E1–E3. Subfraction E2 was finally purified by semi-preparative HPLC to afford 1 (1.0 mg), 2 (1.9 mg), 4 (2.1 mg), 10 (1.5 mg), 8 (1.8 mg), 11 (2.1 mg) and 9 (2.4 mg). Similarly, 3 (11.3 mg) and 5 (9.8 mg) were obtained from fraction C (260 mg) and 6 (12.8 mg) and 7 (21.2 mg) were obtained from fraction D (271 mg).
9-hydroxybafilomycin (1): white amorphous powder; (c 0.17, CHCl3); UV (MeOH) λmax nm, 285 and 246; 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3), see Table 1; MALDI-FTMS (positive ion), [M+Na]+ ion at m/z 643.38197 for C35H56O9 (calcd for [M+Na]+, 643.38165).
29-hydroxybafilomycin (2): white amorphous powder; (c 0.13, CHCl3); UV (MeOH) λmax nm, 285 and 246; 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3), see Table 1; MALDI-FTMS (positive ion), [M+Na]+ ion at m/z 643.38151 for C35H56O9 (calcd for [M+Na]+, 643.38165).
Experimental procedures for cytotoxicity and ERK1/2 phosphorylation assays as well as 1H and 13C NMR data and spectra for 1 and 2 are provided as Supplementary Information.
Supplementary Material
Acknowledgments
We thank the Analytical Instrumentation Center of the School of Pharmacy, University of Wisconsin-Madison, USA for providing support in obtaining MS and NMR data and the University of Wisconsin Paul P Carbone Comprehensive Cancer Center SMSF for assistance with the activity analyses. This work was supported in part by the NIH grant CA113297 (BS), T32 GM08349 (KCC), the Wisconsin Alumni Research Foundation and the Chinese Ministry of Education 111 Project B08034 (YD).
Footnotes
Supplementary Information accompanies the paper on The Journal of Antibiotics website (http://www.nature.com/ja)
References
- 1.Werner G, Hagenmaier H, Albert K, Kohlshorn H, Drautz H. The structure of the bafilomycins, a new group of macrolide antibiotics. Tetrahedron Lett. 1983;24:5193–5196. [Google Scholar]
- 2.Werner G, et al. Metabolic products of microorganisms. 224, bafilomycins, a new group of macrolide antibiotics production, isolation, chemical structure and biological activity. J. Antibiot. 1984;37:110–117. doi: 10.7164/antibiotics.37.110. [DOI] [PubMed] [Google Scholar]
- 3.Kretschmer A, Dorgerloh M, Deeg M, Hagenmaier H. The structures of novel insecticidal macrolides: bafilomycins D and E, and oxohygrolidin. Agric. Biol. Chem. 1985;49:2509–2511. [Google Scholar]
- 4.Carr G, et al. Bafilomycins produced in culture by Streptomyces spp. Isolated from marine habitats are potent inhibitors of autophagy. J. Nat. Prod. 2010;73:422–427. doi: 10.1021/np900632r. [DOI] [PubMed] [Google Scholar]
- 5.Ndejouong B, et al. Hygrobafilomycin, a cytotoxic and antifungal macrolide bearing a unique monoalkylmaleic anhydride moiety, from Streptomyces varsoviensis. J. Antibiot. 2010;63:359–363. doi: 10.1038/ja.2010.52. [DOI] [PubMed] [Google Scholar]
- 6.Wilton JH, Hokanson GC, French JC. PD 118 576: a new antitumor macrolide antibiotic. J. Antibiot. 1985;38:1449–1452. doi: 10.7164/antibiotics.38.1449. [DOI] [PubMed] [Google Scholar]
- 7.Frändberg E, et al. Streptomyces halstedii K122 produces the antifungal compounds bafilomycin B1 and C1. Can. J. Microbiol. 2000;46:753–757. [PubMed] [Google Scholar]
- 8.Goetz MA, Mccormick PA, Monaghan RL, Ostlind DA. L-155, 175: a new antiparasitic macrolide fermentation, isolation and structure. J. Antibiot. 1985;38:161–168. doi: 10.7164/antibiotics.38.161. [DOI] [PubMed] [Google Scholar]
- 9.Vanek Z, Mateju J, Curdova E. Immunomodulators isolated from microorganisms. Folia Microbiol. 1991;36:99–111. doi: 10.1007/BF02814487. [DOI] [PubMed] [Google Scholar]
- 10.Bowman EJ, Siebers A, Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant. Proc. Natl Acad. Sci. USA. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knops J, et al. Cell-type and amyloid precursor protein-type specific inhibition of Aβ release by bafilomycin A1, a selective inhibitor of vacuolar ATPases. J. Biol. Chem. 1995;270:2419–2422. doi: 10.1074/jbc.270.6.2419. [DOI] [PubMed] [Google Scholar]
- 12.Saurin AJ, Hamllet J, Clague MJ, Penington R. Inhibition of mitogen-induced DNA synthesis by bafilomycin A1 in Swiss 3T3 fibrobasts. Biotechnol. J. 1996;313:65–69. doi: 10.1042/bj3130065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, et al. Erythronolides H and I, new erythromycin congeners from a new halophilic actinomycete Actinopolyspora sp. YIM90600. Org. Lett. 2009;11:1353–1356. doi: 10.1021/ol900143j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S, et al. Discovery and total synthesis of a new estrogen receptor heterodimerizing actinopolymorphol A from Actinopolymorpha rutilus. Org. Lett. 2010;12:3525–3527. doi: 10.1021/ol1013526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powell E, et al. Identification and characterization of a novel estrogenic ligand actinopolymorphol A. Biochem. Pharmacol. 2010;80:1221–1229. doi: 10.1016/j.bcp.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang S, et al. Cycloheximide and congeners as inhibitors of eukaryotic protein synthesis from endophytic actinomycetes Streptomyces sps. YIM56132 and YIM56141. J. Antibiot. 2011;64:163–166. doi: 10.1038/ja.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon S, Hwang W, Chung YR, Shin J. New cytotoxic bafilomycin C1-amide produced by Kitasatospora cheerisanensis. J. Antibiot. 2003;56:856–861. doi: 10.7164/antibiotics.56.856. [DOI] [PubMed] [Google Scholar]
- 18.Beyenbach KW, Wieczorek H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J. Exp. Biol. 2006;209:577–589. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- 19.Von Zastrow M, Sorkin A. Signaling on the endocytic pathway. Curr. Opin. Cell Biol. 2007;19:436–445. doi: 10.1016/j.ceb.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carver KC, Arendt LM, Schuler LA. Complex prolactin crosstalk in breast cancer: new therapeutic implications. Mol. Cell. Endocrinol. 2009;307:1–7. doi: 10.1016/j.mce.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009;21:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 22.Presley JF, Mayor S, McGraw TE, Dunn KW, Maxfield FR. Bafilomycin A1 treatment retards transferrin receptor recycling more than bulk membrane recycling. J. Biol. Chem. 1997;272:13929–13936. doi: 10.1074/jbc.272.21.13929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.