Abstract
Dysfunctional mitochondria and generation of reactive oxygen species (ROS) promote chronic diseases, which have spurred interest in the molecular mechanisms underlying these conditions. Previously, we have demonstrated that disruption of post-translational modification of proteins with β-linked N-acetylglucosamine (O-GlcNAcylation) via overexpression of the O-GlcNAc-regulating enzymes O-GlcNAc transferase (OGT) or O-GlcNAcase (OGA) impairs mitochondrial function. Here, we report that sustained alterations in O-GlcNAcylation either by pharmacological or genetic manipulation also alter metabolic function. Sustained O-GlcNAc elevation in SH-SY5Y neuroblastoma cells increased OGA expression and reduced cellular respiration and ROS generation. Cells with elevated O-GlcNAc levels had elongated mitochondria and increased mitochondrial membrane potential, and RNA-sequencing analysis indicated transcriptome reprogramming and down-regulation of the NRF2-mediated antioxidant response. Sustained O-GlcNAcylation in mouse brain and liver validated the metabolic phenotypes observed in the cells, and OGT knockdown in the liver elevated ROS levels, impaired respiration, and increased the NRF2 antioxidant response. Moreover, elevated O-GlcNAc levels promoted weight loss and lowered respiration in mice and skewed the mice toward carbohydrate-dependent metabolism as determined by indirect calorimetry. In summary, sustained elevation in O-GlcNAcylation coupled with increased OGA expression reprograms energy metabolism, a finding that has potential implications for the etiology, development, and management of metabolic diseases.
Keywords: bioenergetics, mitochondria, nuclear factor 2 (erythroid-derived 2-like factor) (NFE2L2) (Nrf2), O-GlcNAcylation, reactive oxygen species (ROS), O-GlcNAcase, OGT
Introduction
Chronically impaired mitochondrial respiration and uncontrolled reactive oxygen species (ROS)2 generation contribute to complex chronic diseases (1). Interventions that improve mitochondrial function and reduce uncontrolled ROS production will have widespread clinical applications in the treatment of chronic disease. Prolonged treatment with glucosamine (GlcN), a common amino-sugar, improved mitochondrial function and extended the life span in both Caenorhabditis. elegans and mice. GlcN treatment increased mitochondrial respiration and transiently induced mitochondrial ROS generation followed by a significant reduction in ROS (2). The initial transient ROS induction promotes mitochondrial biogenesis and improves anti-oxidant response resulting in life-span extension (2). Of note, GlcN feeds into the hexosamine biosynthetic pathway (HBP) after the pathway rate-limiting enzyme glutamine fructose-6-phosphate aminotransferase 1 (GFAT) leading to increased O-GlcNAcylation; thus, we hypothesize that O-GlcNAc regulates mitochondrial function, the oxidative stress response, and can influence chronic disease progression.
O-GlcNAcylation is the addition of a single O-linked β-d-N-acetylglucosamine sugar moiety to serine or threonine residues in nuclear, cytoplasmic, and mitochondrial proteins (3). O-GlcNAcylation is facilitated by two functionally opposing enzymes, O-GlcNAc transferase (OGT) catalyzes the addition of O-GlcNAc and O-GlcNAcase (OGA) hydrolyzes the modification (3). During catalysis, OGT uses the nucleotide sugar UDP-GlcNAc generated from the HBP. GlcN feeds into the HBP downstream of GFAT1 and increases cellular O-GlcNAcylation by elevating the synthesis of UDP-GlcNAc (4). Therefore, GlcN influence on mitochondrial function and life-span extension could be mediated by O-GlcNAc. Because UDP-GlcNAc synthesis relies on the integration of multiple metabolic pathways, O-GlcNAcylation can sense changes in nutrients (5). Importantly, chronic disruptions in O-GlcNAcylation due to nutrient imbalance influence metabolic diseases (6, 7).
O-GlcNAc is a key regulator of mitochondrial function (8, 9). Both OGT (8) and OGA localize to the mitochondria (10), and the pyrimidine nucleotide carrier 1 (PNC1) transports UDP-GlcNAc into the mitochondria providing substrate for OGT (10). Several electron transport chain (ETC) subunits are modified by O-GlcNAc (11) suggesting that O-GlcNAc modulates ETC activity. For example, short-term high-glucose treatment elevates O-GlcNAcylation but suppresses ETC activity and ATP production in rat cardiomyocytes (12); however, short-term OGA inhibition in rats increased oxygen consumption in isolated mitochondria (11). OGT or OGA overexpression in SH-SY5Y neuroblastoma cells alters the mitochondrial proteome, including several TCA cycles and ETC proteins (9). The loss of ETC protein expression adversely affected mitochondrial shape and impaired respiration. These data indicate O-GlcNAc is essential for the proper regulation of mitochondrial function and energy metabolism.
Here, we report that increasing O-GlcNAcylation in cell lines and mice by inhibiting OGA or with GlcN supplementation reprograms mitochondrial function and changes energy metabolism. Sustained elevation in O-GlcNAc lowered respiration and reduced ROS generation. These changes were followed by transcriptional reprogramming of the proteome and a decrease in the transcription factor nuclear factor (erythroid-derived)-like 2 (NRF2)-mediated oxidative stress response. In contrast, OGT liver knockdown severely impaired mitochondrial function, increased ROS levels, and activated the NRF2 antioxidant response. Together, these data demonstrate distinct roles for O-GlcNAc in modulating mitochondrial function and provide new insights into how pharmacological targeting of O-GlcNAc protects against aging and metabolic diseases.
Results
Thiamet-G (TMG) or glucosamine (GlcN) treatment alters mitochondrial bioenergetic capacity
Prolonged treatment with GlcN improves mitochondrial function and extends the life span in both C. elegans and mice (2). We hypothesized that sustained elevation of O-GlcNAcylation would alter energy metabolism. To determine how O-GlcNAcylation affects energy metabolism, we treated SH-SY5Y neuroblastoma cells with either TMG (10 μm) or GlcN (0.35 mm) for at least 3 weeks prior to harvesting these cells. TMG is a highly selective OGA inhibitor (13), whereas GlcN feeds into the hexosamine biosynthetic pathway after GFAT1 increasing cellular pools of UDP-GlcNAc (4). In contrast to GlcN, which can influence a variety of metabolic pathways, TMG precisely manipulates O-GlcNAcylation by targeting only OGA.
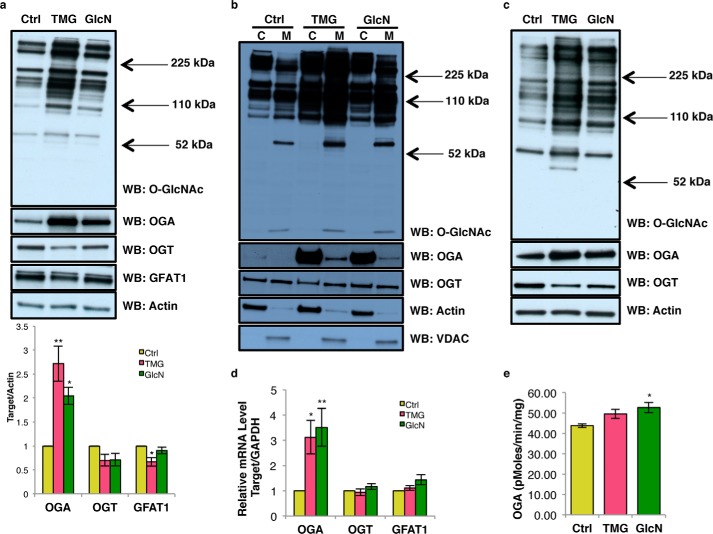
Both TMG- and GlcN-treated SH-SY5Y cells increased total and mitochondrial O-GlcNAc levels (Fig. 1, a and b). Although not significant, OGT protein and transcript levels were trending lower; while OGA protein and transcript levels were significantly increased in response to elevated O-GlcNAcylation (Fig. 1, a and d) (14). GFAT1 protein level was decreased in TMG-treated cells (Fig. 1a). OGA-specific activity was increased in GlcN-treated SH-SY5Y cells (Fig. 1e). Similar results were found in NT2 human teratocarcinoma cells treated with TMG or GlcN (Fig. 1c).
Figure 1.
Prolonged TMG or GlcN treatment increases O-GlcNAcylation and alters O-GlcNAc cycling. O-GlcNAc, OGT, OGA, and GFAT1 levels of whole-cell (quantitation of bands is below the blot) (a), cytoplasm and mitochondrial content in SH-SY5Y cells (b), and NT2 whole cell (c) are shown. For whole-cell or cytoplasmic extract, actin was used as loading control, whereas VDAC and citrate synthase (CS) was used as a loading control for mitochondrial extracts (n = 3). d, OGA, OGT (MGEA5), and GFAT1 transcript levels were determined using qRT-PCR (n = 3). e, OGA activity (n = 3). * indicates significance p < 0.05. ** indicates significance p < 0.01. WB, Western blotting; C, cytoplasmic; M, mitochondrial..
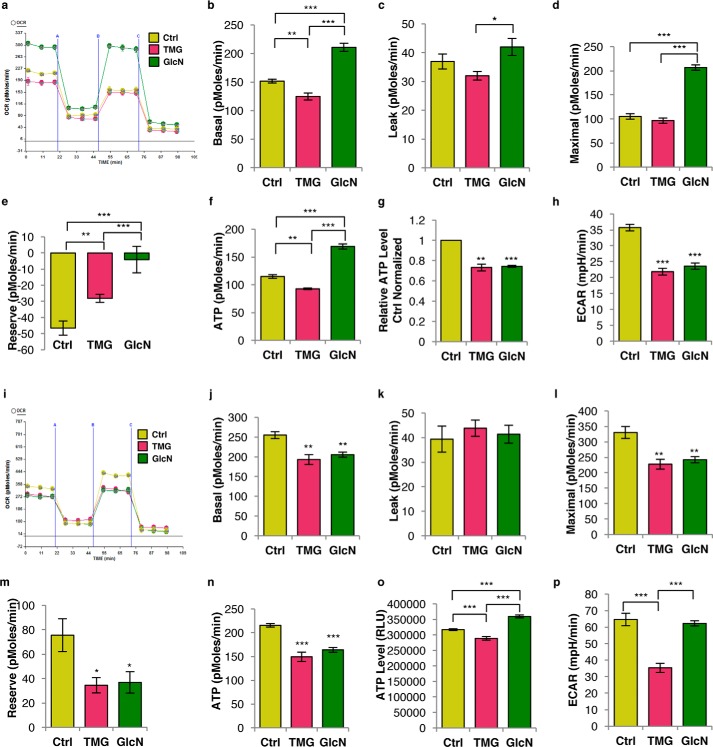
Prolonged elevation of O-GlcNAc altered cellular respiration in SH-SY5Y cells (Fig. 2a). TMG lowered baseline oxygen consumption rates (OCR), whereas GlcN increased baseline OCR (Fig. 2b). Under basal conditions, in vivo ATP production was lower in the presence of TMG- but higher in GlcN-treated cells (Fig. 2f). Neither treatment affected the proton leak rate (Fig. 2c). The maximum respiratory rate was increased in GlcN cells, whereas TMG was similar to control cells (Fig. 2d). SH-SY5Y cells grown in oxidative media regardless of TMG or GlcN treatment have no reserve capacity (Fig. 2e), showing that these cells were respiring at their maximum capacity. To eliminate any cell type-specific effects, we performed the respiration assays in NT2 cells. TMG-treated NT2 cells replicated the data from SH-SY5Y cells (Fig. 2, i–n). In NT2 cells, GlcN-treated mirrored the TMG-treated results from SH-SY5Y/NT2 cells showing lower respiration rates (Fig. 2, i–n). NT2 cell mitochondria had much more reserve capacity than the SH-SY5Y cells suggesting that these cells were not respiring at a maximal rate. The total cellular ATP levels in both TMG- and GlcN-treated SH-SY5Y cells and in TMG-treated NT2 cells were lower (Fig. 2, g and o). TMG treatment caused both SH-SY5Y and NT2 cells to produce significantly less ATP. Likely, GlcN-treated SH-SY5Y cells produced and consumed more energy due to increased phosphorylation of GlcN to GlcN-6-P by hexose kinase (4).
Figure 2.
Prolonged TMG or GlcN treatment alters mitochondrial bioenergetics capacity. a, representative graph of the bioenergetics profile in SY5Y cells. b, basal respiration; c, leak rate; d, maximal respiration rate; e, reserve capacity rate; and f, ATP production rate in O-GlcNAc elevated SH-SY5Y cells were determined using XF24 analyzer (average ± S.E., n = 6). g, relative ATP level representative plot (average ± S.E., n = 4) showed both TMG- (10 μm) and GlcN (0.35 mm)-treated SH-SY5Y cells maintained at low cellular ATP levels. Displayed relative ATP levels were average value of each replicated result normalized with their control. h, ECAR was measured using XF24 analyzer (average ± S.E., n = 6). i, bioenergetics profile of NT2 cells; j, basal respiration rate; k, leak rate; l, maximal respiration rate; m, reserve capacity rate; and n, ATP production rate (average ± S.E., n = 6). NT2 cell representative plots showing cellular ATP levels (average ± S.E., n = 9) (o) and glycolysis rate (n = 6) (p). * indicates significance p < 0.05. ** indicates significance p < 0.01. *** indicates significance p < 0.001.
Because oxidative phosphorylation and glycolysis are interdependent energy-producing pathways (15), we examined glycolytic energy production by measuring cellular glycolytic flux. Both TMG- and GlcN-treated SH-SY5Y cells had lower basal glycolytic rates (Fig. 2h), and TMG-treated NT2 cells had a lower basal glycolytic rate. GlcN-treated NT2 cells showed no changes (Fig. 2p).
TMG or GlcN treatment increased complex I and complex IV activity and elevated cellular NAD+/NADH ratio
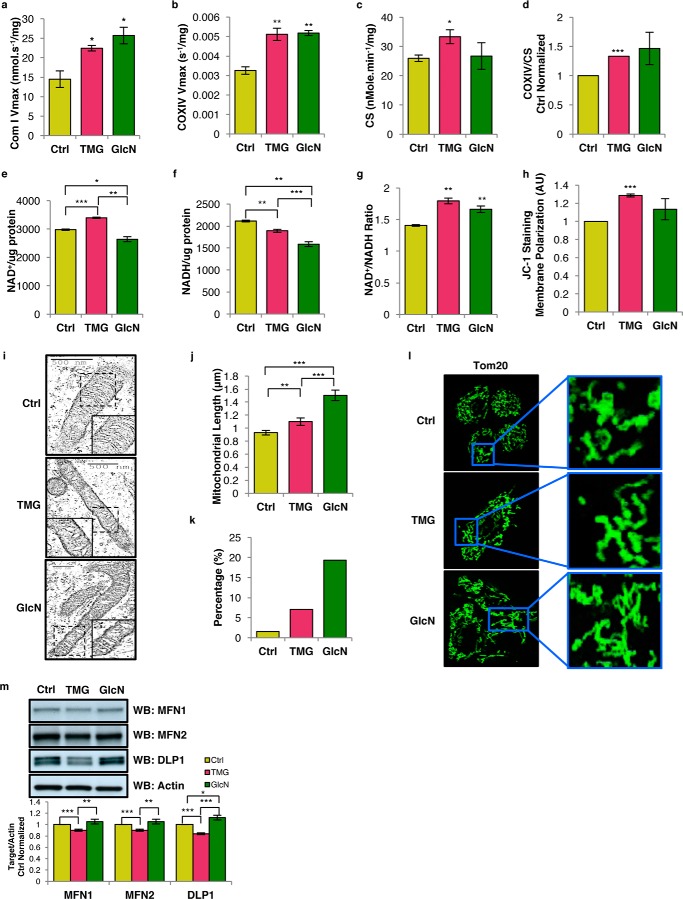
Because prolonged TMG treatment lowered cellular respiration, we postulated that the ETC complex activity was lower; however, mitochondria isolated from both TMG- and GlcN-treated SH-SY5Y cells had increased complex I activity (Fig. 3a). TMG also elevated complex IV activity (Fig. 3, b–d). Because isolated ETC complex activity did not correspond to the in vitro respiration measurements, we considered whether the observed decrease in OCR was due to reduced concentration of NADH available for respiration. The NAD+ concentration was elevated in TMG but reduced in GlcN-treated SH-SY5Y cells (Fig. 3e), whereas the NADH concentration was decreased in both TMG- and GlcN-treated SH-SY5Y cells (Fig. 3f). The NAD+ and NADH ratio skewed toward NAD+ for both TMG- and GlcN-treated cells (Fig. 3g). To ascertain whether altered cellular respiration was due to changes in mitochondrial membrane potential, we probed for mitochondrial membrane polarization differences in TMG- or GlcN-treated SH-SY5Y cells. TMG significantly enhanced mitochondrial hyperpolarization (Fig. 3h).
Figure 3.
TMG or GlcN treatment elevates cellular NAD+/NADH ratio, enhances mitochondrial hyperpolarization, and promotes longer mitochondria. a–d, representative plots showing SH-SY5Y complex I Vmax (average ± S.E., n = 3) normalized with total protein concentration (a), complex IV (COXIV) Vmax (b), citrate synthase (CS) activity after being normalized with total protein concentration (c), and COXIV Vmax normalized with CS activity (average ± S.E., n = 3) (d). e–g, NAD+ (e) and NADH levels (f) were altered NAD+/NADH ratio (average ± S.E., n = 4) (g) normalized with protein concentration. h, membrane potential was measured using JC-1 (average ± S.E., n = 3). i, transmission electron microscopy. j, mitochondria lengths (average ± S.E., n = 120) were measured; k, percentage of mitochondria longer than 2 μm was calculated. l, confocal staining with TOM20. m, mitofusin 1 (MFN1), mitofusin 2 (MFN2), and dynamin-like protein (DLP1) protein levels. Protein levels were first normalized to actin level and then normalized to control cells (n = 3). * indicates significance p < 0.05. ** indicates significance p < 0.01. *** indicates significance p < 0.001. WB, Western blotting.
Prolonged TMG or GlcN treatment promotes longer mitochondria
To assess whether mitochondrial morphology was changed in TMG- or GlcN-treated SH-SY5Y cells, we used transmission electron microscopy to visualize the mitochondria. The approximate average length of mitochondria from both treated cells was longer compared with control cells (Fig. 3, i and j). A typical length of a mitochondrion ranges from ∼0.5 to 2 μm (16). We found that both treatments increased the percentage of mitochondria that were longer than 2 μm (Fig. 3k). Confocal imaging data agreed with the electron microscopy data showing longer mitochondria in TMG- or GlcN-treated cells (Fig. 3l). Next, we measured the proteins involved in regulating mitochondrial fission and fusion. Mitofusin 1 (MFN1), mitofusin 2 (MFN2), and dynamin-like proteins (DLP1), which facilitate mitochondrial fusion and fission, were lower in TMG-treated cells; however, GlcN treatment increased the DLP1 protein levels, whereas MFN1 and MFN2 levels were unaffected (Fig. 3m).
Prolonged TMG or GlcN altered the transcriptome
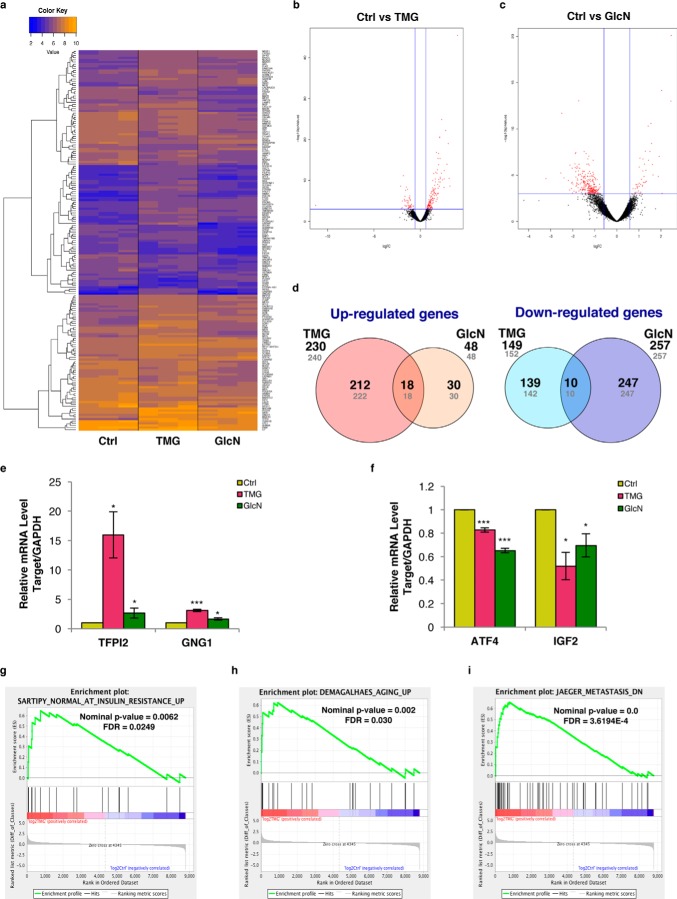
We considered whether the observed mitochondrial changes were mediated by alterations in the transcriptome. Our Next Generation RNA-sequencing (RNA-seq) analysis found 8793 genes in TMG- or GlcN-treated SH-SY5Y cells with high sequencing counts (cpm of >10 in at least two samples) (Fig. 4, a–c, and supplemental Table S1; RNA-seq data are available on the Gene Expression Omnibus). A total of 240 genes for TMG treatment and 48 genes for GlcN treatment were elevated, whereas 152 genes for TMG treatment and 257 genes for GlcN treatment were reduced (p value < 0.01) (Fig. 4d). Among these genes, 230 genes from TMG and 48 genes from GlcN treatment were increased 1.5-fold or more, whereas 149 genes from TMG and 257 genes from GlcN decreased 1.5-fold or more (Fig. 4d). The tissue factor pathway inhibitor 2 (TFPI2), guanine nucleotide-binding protein, γ subunit 1 (GNG1), activating transcription factor 4 (ATF4), and insulin-like growth factor 2 (IGF2) were selected for orthogonal validation and agreed with the RNA sequencing results (Fig. 4, e and f). As a positive control (Fig. 1d), the RNA-seq data showed that OGA (MGEA5) gene expression was increased in both TMG and GlcN treatment (supplemental Tables S2 and S3) (14). Using gene set enrichment analysis (GSEA) (17) to compare TMG-treated with control gene expression data sets, we identified genes from the insulin-signaling pathway, aging, and cancer metastasis enriched in prolonged TMG-treated cells (Fig. 4, g–i, and supplemental Table S4). These data suggested that prolonged O-GlcNAc elevation modulates the progression of chronic metabolic disease.
Figure 4.
Transcriptome alterations from sustained TMG or GlcN treatment. a, heat map of top 100 genes with counts per million of >10 in at least two of the three replicate samples generated from Next Generation RNA-seq analysis. Volcano plots for TMG (b) and GlcN (c) compared with control cells. d, representative pie charts showing number of differentially expressed genes up-regulated or down-regulated after TMG or GlcN treatment. Shown in gray are number of genes in which their expression was changed (p < 0.01) compared with control cells. Numbers in bold represent p < 0.01 genes changed more than 1.5-fold (n = 3). e and f, orthogonal validation of TFPI2 and GNG1 transcript levels (e) and ATF4 and IGF2 transcript levels (average ± S.E., n = 3) (f). g–i, plots obtained from c2 gene sets from GSEA analysis comparing long-term TMG treatment to insulin resistance (Sartipy_Normal_At_Insulin_Resistance_Up gene set) (g), aging (Demagalhaes_Aging_Up gene set) (h), and metastasis (Jaeger_Metastasis_DN gene set) gene sets (i). * indicates significance p < 0.05. ** indicates significance p < 0.01. *** indicates significance p < 0.001.
NRF2-mediated oxidative stress response was down-regulated
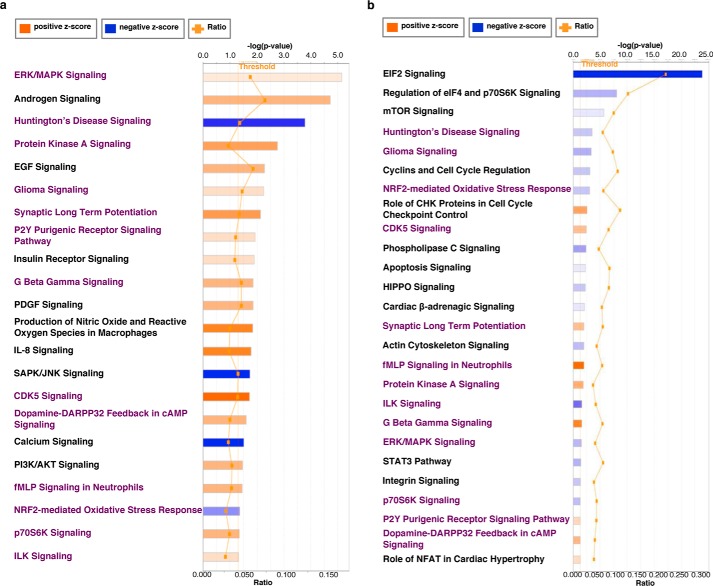
To define biological functions of the altered genes, we performed Ingenuity Pathway Analysis (IPA). A stringent threshold (cpm of >100) was applied to the gene set to prevent false positives. 22 canonical pathways for TMG-treated (Fig. 5a) and 26 canonical pathways for GlcN-treated cells (Fig. 5b) were identified (supplemental Table S5). 13 pathways overlapped in both TMG and GlcN-treated cells and changed in the same predicted manner (see Fig. 5, a and b). Interestingly, the NRF2-mediated oxidative stress response was identified as down-regulated in both TMG- and GlcN-treated SH-SY5Y cells. These data suggest that both TMG and GlcN treatments could modulate ROS levels.
Figure 5.
TMG or GlcN treatment alters the transcriptome and down-regulated NRF2-mediated oxidative stress response. a and b, canonical pathways significantly associated with changes in genes expression after long-term TMG (a) and GlcN treatment (b).
Prolonged TMG- or GlcN-treated cells generated less ROS
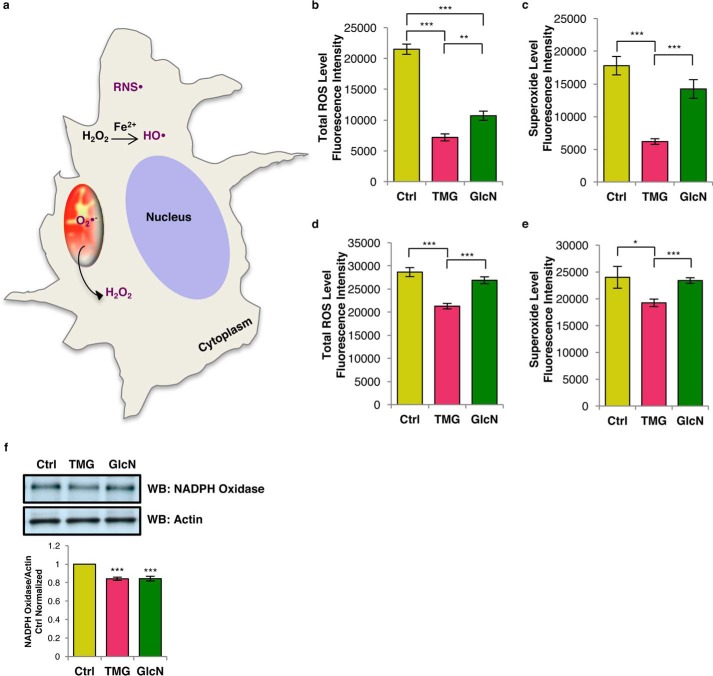
TMG or GlcN could affect cellular ROS levels, including reactive nitrogen species (RNS), hydroxyl radical (OH•), hydrogen peroxide (H2O2), and superoxide (O2˙̄) (Fig. 6a). Total ROS (RNS•, OH•, and H2O2) was reduced in both TMG- and GlcN-treated SH-SY5Y cells (Fig. 6b). Superoxide levels were reduced in TMG-treated cells (Fig. 6c). Total ROS and superoxide levels were lower in TMG-treated NT2 cells, whereas GlcN-treated NT2 cells trended lower (Fig. 6, d and e). Because NADPH oxidase is a major source of cytoplasmic ROS (18), we measured and found that both TMG and GlcN treatments decreased NADPH oxidase protein levels in SH-SY5Y cells (Fig. 6f). Together, these data indicated that prolonged elevations of O-GlcNAcylation suppress ROS production.
Figure 6.
Prolonged TMG or GlcN treatment reduces ROS. a, representative schematic displaying cytoplasmic ROS (total ROS: HO•, H2O2, and RNS•) and mitochondrial ROS (superoxide: O2˙̄. b and c, total ROS (b) and superoxide level (average ± S.E., n = 9) (c) in SY5Y cells were decreased. d and e, total ROS (d) and superoxide level (e) in NT2 cells (average ± S.E., n = 9). ROS assay results have been replicated in at least three independent experiments. f, NADPH oxidase levels after prolonged TMG or GlcN treatment in SH-SY5Y cells (average ± S.E., n = 3). * indicates significance p < 0.05. ** indicates significance p < 0.01. *** indicates significance p < 0.001. WB, Western blotting.
O-GlcNAc regulates NRF2 function
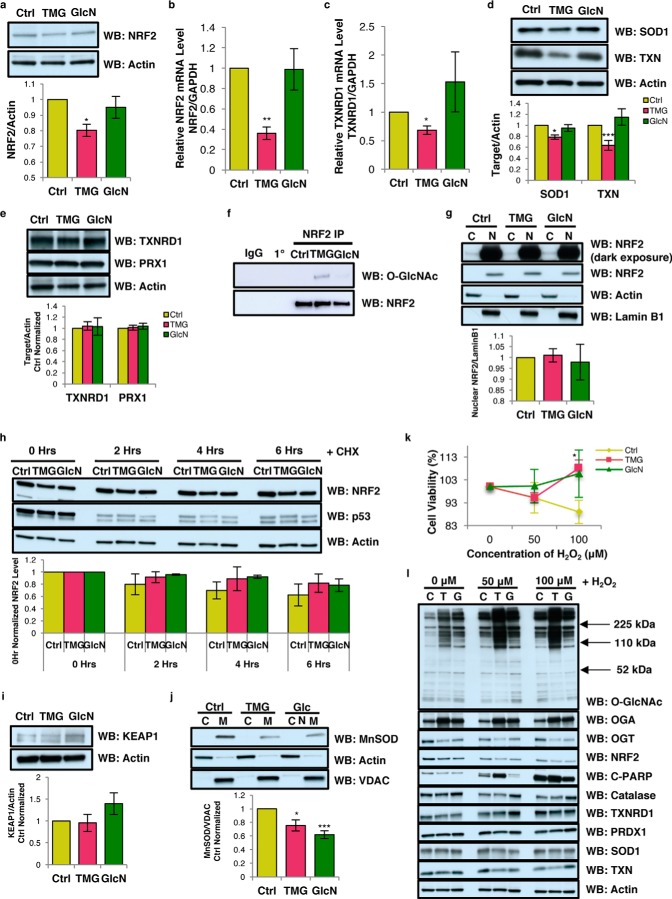
We hypothesized that O-GlcNAc could be directly modulating NRF2 function. TMG treatment lowered NRF2 protein and NRF2 transcript levels (Fig. 7, a and b). Additionally, TMG treatment lowered the NRF2 target gene thioredoxin reductase 1 (TXNRD1) transcript level (Fig. 7c) and reduced superoxide dismutase 1 (SOD1) and thioredoxin (TXN) protein levels (Fig. 7d). However, TXNRD1 and peroxidase 1 (PRX1) protein expression was not altered (Fig. 7e). Interestingly, both TMG and GlcN treatments increased NRF2 O-GlcNAcylation (Fig. 7f).
Figure 7.
O-GlcNAc regulates NRF2 function but NRF2 localization, protein stability, and acute oxidative stress response were not altered by TMG or GlcN treatment. a–d, representative results shown for NRF2 protein level (n = 3) (a), NRF2 (n = 5) (b), TXNRD1 (n = 4) transcript levels (c), and SOD1 (n = 7) and TXN (n = 4) protein levels (d). Protein levels were first normalized with loading control actin and then normalized to control samples. Transcript levels were measured by qRT-PCR and normalized to GAPDH levels. e, TXNRD1 and PRX1 protein levels in TMG- or GlcN-treated SY5Y cells. f, immunoprecipitation results showed increased NRF2 O-GlcNAcylation (n = 3). g, NRF2 localization was not altered in TMG- or GlcN-treated SH-SY5Y cells. h, NRF2 protein stability after cycloheximide treatment with p53 as positive control for the treatment. Actin was used as loading control, and the protein levels were further normalized with the control. i, KEAP1 protein levels in TMG- or GlcN-treated SY5Y cells. j, mitochondrial MnSOD1 protein level is significantly decreased after TMG or GlcN treatment in SH-SY5Y cells. Cells were separated into cytoplasm and mitochondrial fractions. VDAC was used as the loading control for mitochondrial fraction, and actin was used as mitochondrial fraction negative control (average ± S.E., n = 3). k, cell viability plot after acute oxidative stress test in TMG- or GlcN-treated SH-SY5Y cells (average, ± S.E., n = 3). l, O-GlcNAc, OGA, OGT, NRF2, C-PARP, catalase, TXNRD1, PRDX1, SOD1, and TXN protein levels after 7 h of various concentrations H2O2 treatment (n = 3) from control (C), TMG (T), and GlcN (G)-treated cells. Actin was used as loading control, and the protein levels were further normalized with the control. * indicates significance p < 0.05. ** indicates significance p < 0.01. *** indicates significance p < 0.001. WB, Western blotting.
Because O-GlcNAcylation of transcription factors influences their cytolocalization (19), we fractionated the nuclear extract from the cytoplasm, and we found increased O-GlcNAcylation did not alter the nuclear–NRF2 protein levels (Fig. 7g). Because O-GlcNAcylation can promote protein stability (20, 21), we pretreated SH-SY5Y cells with cycloheximide (CHX) to inhibit protein translation (22) and harvested the cells at different time points after CHX treatment. We observed no change in NRF2 protein levels in O-GlcNAc-elevated cells between 0 h and up to 6 h after CHX treatment in relation to control cells (Fig. 7h). The NRF2-binding partner KEAP1–E3–ligase complex (Kelch-like ECH associated protein 1) promotes degradation of the transcription factor. However, KEAP1 protein levels did not change after TMG and GlcN treatment (Fig. 7i), suggesting that the TMG-mediated decrease in NRF2 protein levels was not due to altered stability of the protein but due to the decrease in the transcript.
MnSOD, a non-NRF2 controlled antioxidant enzyme, is a major antioxidant enzyme present in mitochondria and is essential for the removal of superoxide generated by the ETC complexes (23). MnSOD protein level was decreased in both TMG- and GlcN-treated SH-SY5Y cells (Fig. 7j). Because several antioxidant response pathways were down-regulated with TMG and GlcN treatment, we treated SH-SY5Y cells with 50 or 100 μm H2O2 for upwards of 7 h to induce oxidative stress (24), and we measured cell viability. TMG increased cell viability in 100 μm H2O2 (Fig. 7k). The total O-GlcNAcylation level was robustly increased after oxidative stress induction in control cells (Fig. 7l), and the apoptosis marker, cleaved poly(ADP-ribose) polymerase (PARP) protein, was increased after oxidative stress (Fig. 7l). Catalase, TXNRD1, PRDX1, and TXN protein levels were not significantly altered (Fig. 7l). Likely, the increased cell viability in TMG-treated cells was due to increased O-GlcNAcylation mediating other stress-response pathways (25).
Prolonged elevations of O-GlcNAcylation decreased mitochondrial respiration and ROS generation in mouse tissue
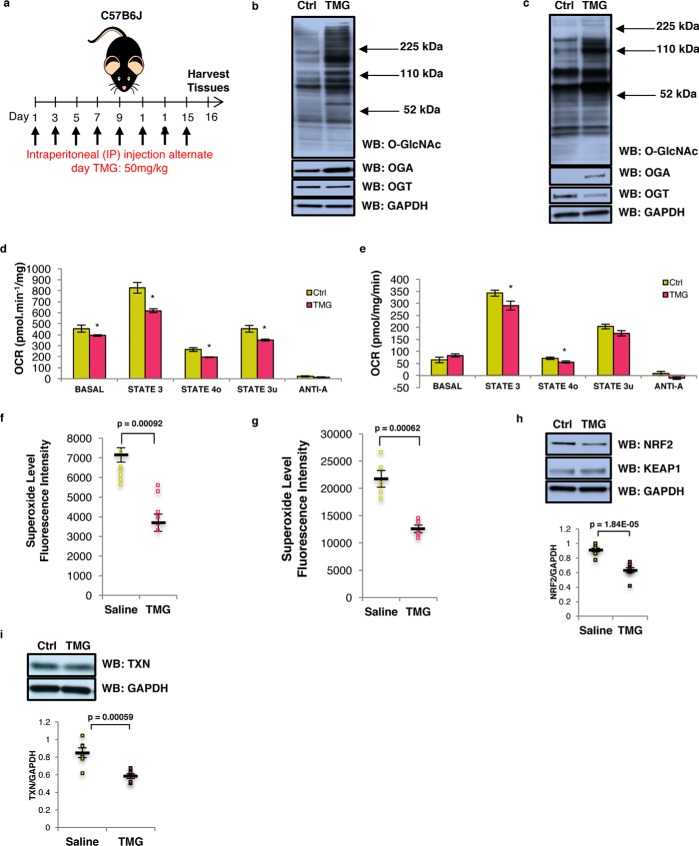
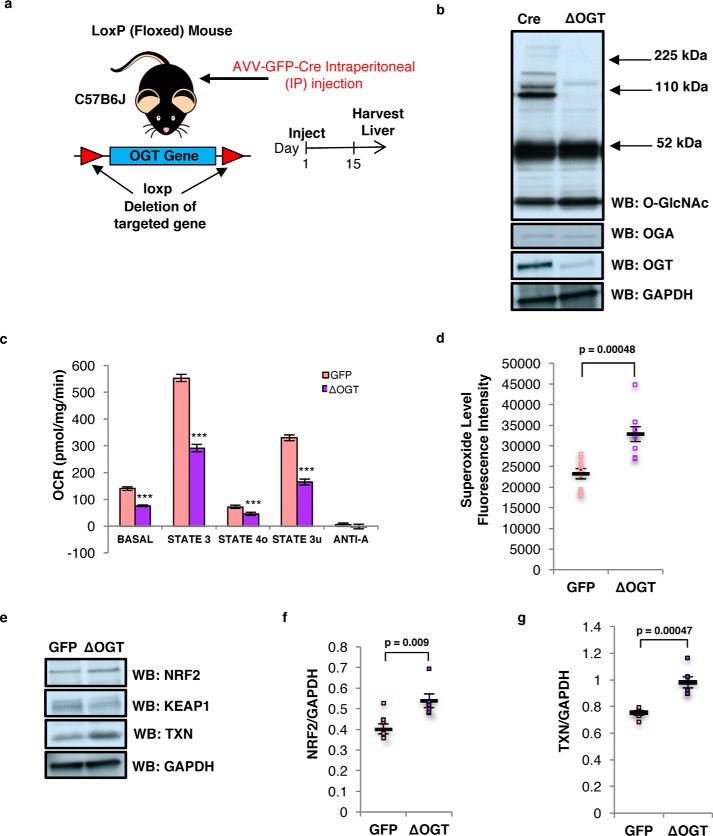
Because long-term TMG treatment reduced mitochondrial respiration in cell lines, we asked the questions whether TMG-treated mice replicated the cell line data. We intraperitoneally injected TMG into C57B6J mice at a concentration of 50 mg/kg (13) every other day for up to 15 days prior to sacrificing the animals. Liver and brain tissues were then used for subsequent experiments (Fig. 8a). TMG robustly elevated brain O-GlcNAcylation and OGA expression and reduced OGT protein levels (Fig. 8b). Similar results were obtained in the livers of TMG-treated mice (Fig. 8c).
Figure 8.
Elevated O-GlcNAcylation decreases mitochondrial respiration and ROS generation. a, experimental design of TMG dosed mice. b and c, total O-GlcNAcylation, OGA, OGT, and GAPDH (loading control) levels in TMG-treated mouse brain (b) and liver (n = 6) (c). d and e, OCR profile on isolated mitochondria from long-term TMG-treated mouse brain (d) and liver (e). The assay was done using pooled mitochondria from five mice for each condition, and the results have been replicated in three independent experiments. f and g, mitochondrial superoxide level (n = 6) in both TMG-treated mouse brain (f) and liver (g) showed decreased superoxide levels. h and i, NRF2 and KEAP1 (h), TXN (i), and GAPDH (loading control) levels for TMG-treated mice (n = 6) are shown. Average protein level displayed was normalized to GAPDH. * indicates significance p < 0.05. ** indicates significance p < 0.01. *** indicates significance p < 0.001. WB, Western blotting.
Next, we purified and pooled mitochondria from four TMG and four control mouse brains and performed ex vivo respiration assays (26). TMG lowered basal respiration (basal, in the presence of substrate but no ADP) as well as phospho-relating respiration (State 3, + ADP), resting respiration (State 40, proton leak rate, + oligomycin), and uncoupled respiration (State 3u, maximal respiratory rate, + FCCP). Anoxic respiration (+ antimycin A) was the same between control and TMG-treated mouse brain (Fig. 8d). TMG reduced liver mitochondrial respiration of State 3 and State 40 (Fig. 8e). The superoxide level was lower in TMG-treated brain (Fig. 8f) and liver (Fig. 8g). Brain NRF2 and TXN protein level was decreased in TMG-injected mice (Fig. 8, h and i). Sustained inhibition of OGA in mouse brain and liver tissue replicated the data from cell lines suggesting that the metabolic changes seen with TMG were independent of cell type and were an adaptive mechanism to OGA inhibition.
OGT knockdown impaired mitochondrial respiration and increased ROS generation in mouse liver
To understand how the loss of OGT affects cellular energetics, we generated conditional OGT liver knock-out C57B6J mice by intraperitoneal injection of the AVV-GFP-Cre virus. Fifteen days after viral injection, liver tissue was harvested (Fig. 9a). Liver O-GlcNAcylation and OGT protein levels were reduced (Fig. 9b). OGT knockdown decreased ex vivo basal liver respiration, as well as State 3, State 40, and State 3u. No difference was measured in anoxic respiration (Fig. 9c). In contrast to TMG treatment, the mitochondrial superoxide levels in the OGT knockdown mouse livers were increased (Fig. 9d). NRF2 protein level and its targeted gene TXN were elevated in OGT knockdown livers (Fig. 9, e–g).
Figure 9.
OGT liver knockdown impaired mitochondrial respiration and increased ROS generation. a, experimental design of liver-specific OGT knockdown mice. b, total O-GlcNAcylation, OGA, OGT, and GAPDH (loading control) levels in OGT-knockdown mouse liver (n = 10) are shown. c, OCR profile on isolated mitochondria from OGT-knockdown mouse liver. The assay was done using pooled mitochondria from five mice for each condition, and the results have been replicated in three independent experiments. d, mitochondrial superoxide level in OGT-knockdown mouse liver was increased (n = 8). e–g, NRF2, KEAP1, TXN, and GAPDH (loading control) levels OGT-knockdown mice (n = 6). Average protein level displayed was normalized to GAPDH. * indicates significance p < 0.05. ** indicates significance p < 0.01. *** indicates significance p < 0.001. WB, Western blotting.
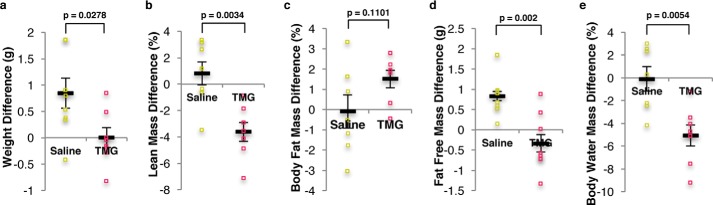
Prolonged O-GlcNAc elevation promoted weight loss, including reduction of lean mass, free fat mass, and water mass
Next, we determined whether long-term elevation of O-GlcNAc altered whole-body composition. Total body weight of TMG-treated mice was lower after the 2-week treatment period (supplemental Table S6); however, control mice gained weight (Fig. 10a). TMG treatment resulted in a reduction in lean mass percentage compared with control mice (Fig. 10b). Although we did not find a significant difference in the body fat mass percentage (Fig. 10c), the free fat mass percentage (all other mass other than fat mass) in TMG-treated mice was lower (Fig. 10d). Furthermore, TMG treatment reduced body water mass percentage (Fig. 10e).
Figure 10.
O-GlcNAc elevation altered body mass in mice. a–e, representative plots shown weight (a), lean mass (b), body fat mass difference percentage (c), free fat mass difference (d), and body water mass difference percentage (n = 8) (e), for mice after 2 weeks of TMG treatment (50 mg/kg). Body fat mass differences percentage were calculated from the percentage difference before and after injection (saline or TMG). Body water mass difference percentage calculated by taking the percentage of the differences of water mass before and after injection (saline or TMG).
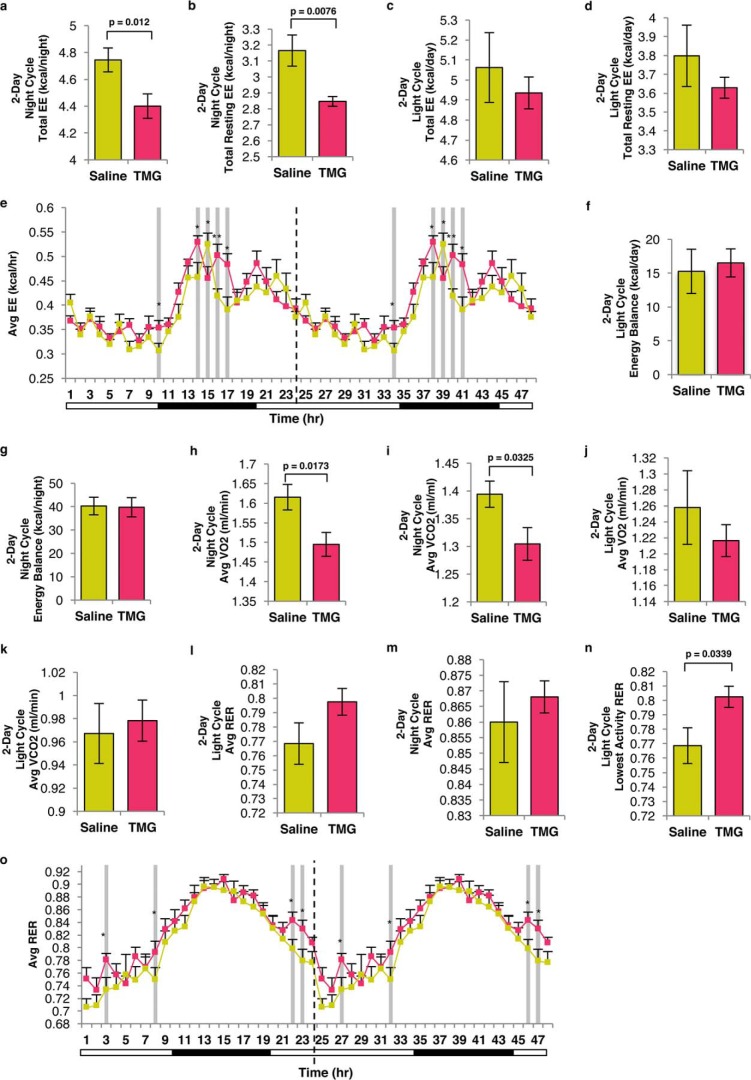
Long-term TMG treatment lowered energy expenditure and oxygen consumption, skewing mice toward carbohydrate metabolism
Because TMG down-regulated mitochondrial respiration and reduced free fat mass body composition, we postulated that the whole-body energy metabolism of the TMG-treated mice would be altered. A 2-week TMG treatment reduced total energy expenditure (EE) and resting EE at nighttime (most active) (Fig. 11, a and b), but no changes were measured during the daytime (less active) (Fig. 11, c and d). When these mice entered the night cycle, the average EE pattern of TMG-treated mice was higher and then declined back to control levels when they re-entered the light cycle (Fig. 11e). The energy balance in TMG-treated mice (calories left over from food consumption) was not changed during the light or night cycles (Fig. 11, f and g).
Figure 11.
Sustained elevation in O-GlcNAc alters energy metabolism in mice. a and b, representative plots illustrating nocturnal total EE (a) and total resting EE (b). c and d, representative plots illustrating diurnal total EE (c) and total resting EE (d). e, double-plotted average EE for the last 24 h after TMG injection was plotted (n = 8, * indicates p < 0.05; ** indicates significance p < 0.01). Dotted line represents blot replication. f and g, plot showing diurnal energy balance (f) (total energy intake − total energy expenditure) and nocturnal (g) energy balance. h and i, plot showing nocturnal average oxygen consumption rate (VO2) (h) and carbon dioxide exhaling rate (VCO2) (i). j and k, plot showing diurnal average VO2 (j) and VCO2 (k). l and m, average RER (VCO2/VO2) plot showing diurnal (l) and nocturnal (m). n, diurnal lowest activity RER. o, double-plotted hourly RER (VCO2/VO2) calculated on the last day of TMG injection. Dotted line represents blot replication.
TMG treatment resulted in a significantly lower OCR especially during the nighttime (O2 consumption and CO2 elimination, Fig. 11, h and i). The OCR was not changed significantly during daytime (Fig. 11, j and k). The respiratory exchange ratio (RER, ratio of whole-body CO2 production to O2 consumption) determines the relative contribution of carbohydrates and lipids to overall energy expenditure (27). The average RER in TMG-treated mice was increased during either the daytime (Fig. 11l) or nighttime (Fig. 11m) cycle but not significantly. However, basal RER during the daytime cycle was increased in the TMG-treated mice (Fig. 11n). The hourly RER at day 15 following TMG injection was higher during the daytime cycle in TMG-treated mice, whereas nighttime RER in both control and TMG-treated animals were equivalent. Again, when TMG-treated mice re-entered the light cycle, their average hourly RER remained significantly higher compared with control mice (Fig. 11o). These data showed that TMG-treated mice utilized carbohydrates as their predominant source for energy throughout a 24-h cycle, even during the inactive daytime cycle when lipid oxidation is preferred.
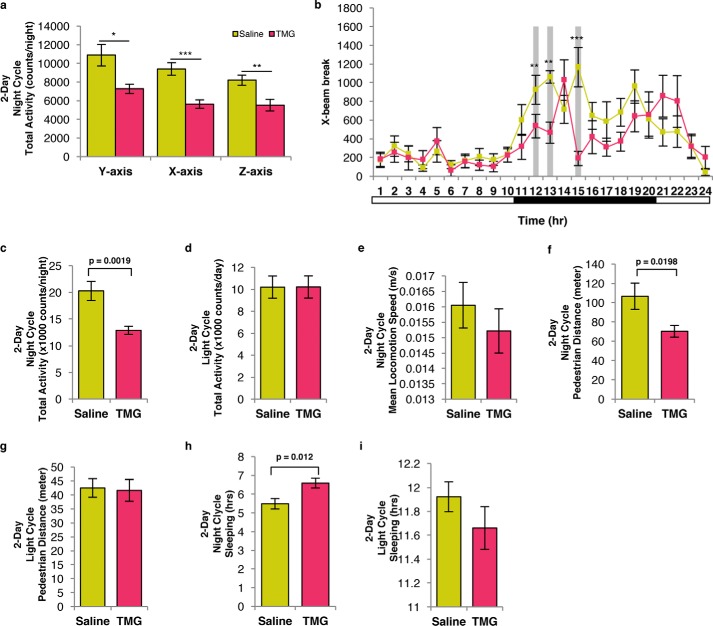
TMG-treated mice were less active than control mice
Next, we assessed the physical activity of the TMG-treated mice. During the nighttime, TMG-treated mice had less x, y, and z axis beam break activity (Fig. 12, a and b) and total activity (sum of x and y axis beam breaks) compared with control mice (Fig. 12c). No changes between TMG- and saline-treated mice during the daytime cycle were found (Fig. 12, d and b). Although locomotion speed was unchanged (Fig. 12e), TMG-treated mice walked a significantly shorter distance than control mice during the nighttime cycle (Fig. 12f). The distance traveled during the daytime cycle was not different (Fig. 12g). Finally, TMG treatment increased the sleep duration during the nighttime cycle compared with saline-treated mice (Fig. 12h), but no change in sleep duration was measured during the daytime cycle (Fig. 12i).
Figure 12.
TMG-treated mice were less active than control mice. a and b, total activity based on y axis, x axis, and z axis beam breaks (a), and hourly X-beam break plot (b). Permutation test (10,000 cycles) of hourly RER during light cycle (p = 0.034) and X-beam break during night cycle (p = 0.0002). c and d, representative plots of total activity (x + y) nocturnal (c) and diurnal (d) total activity. g, representative plot of nocturnal (e) mean locomotion speed. Representative plots of nocturnal (f) and diurnal pedestrian distance. h and i, representative plots illustrating nocturnal (h) and diurnal (i) sleep duration.
Discussion
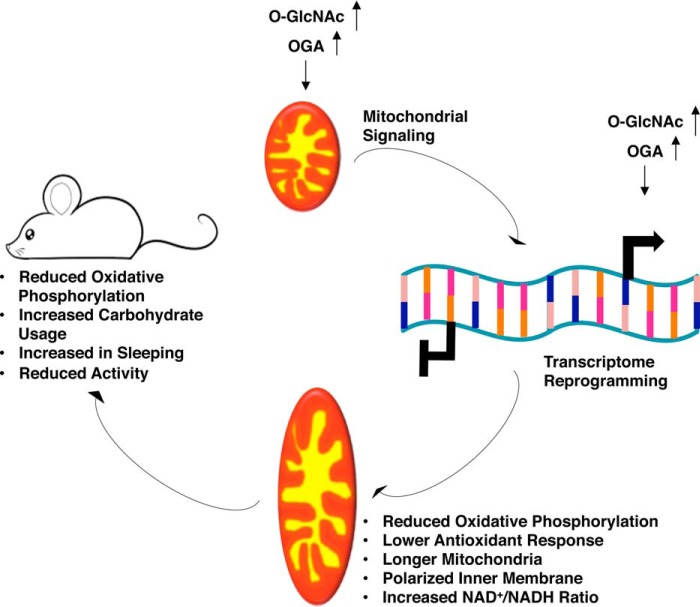
Our data demonstrate a fundamental role for O-GlcNAcylation in regulating mitochondrial function and whole-body energy metabolism. GlcN supplementation increased O-GlcNAcylation and induced a phenotype of increased mitochondrial respiration in SH-SY5Y cells and reduced ROS production. In contrast to GlcN, TMG-treated cells (SH-SY5Y or NT2) or mice became less ETC-dependent maintaining lower oxygen consumption rates due to reduced respiration. Prolonged TMG-treated animals shifted their metabolism toward carbohydrate utilization and reduced their physical activity. The decreased respiration caused by prolonged TMG treatment did not impair mitochondrial function because less ROS is generated. Loss of O-GlcNAcylation by liver-specific OGT knockdown lowered and impaired mitochondrial respiration, robustly elevating superoxide levels. Importantly, these results support O-GlcNAcylation in maintaining proper mitochondrial function, whereas complete loss of O-GlcNAc negatively impacts cellular energetics.
Prolonged inhibition of OGA or elevation in OGT substrate availability led to dramatic changes in mitochondrial respiration duplicating results from prolonged GlcN treatment in C. elegans and mice (2). However, genetic ablation of OGT dramatically lowers the basal level of O-GlcNAc producing severe metabolic phenotypes (28–30), and OGT or OGA overexpression impairs respiration (9). Recently, RNAi-mediated knock-out of the mitochondrial form of OGT (mOGT), which is only expressed in a few mammals, including primates (31), in HeLa cells caused mitochondrial fragmentation and reduced membrane potential, whereas knock-out of both mOGT and the nuclear and cytoplasmic form of OGT led to compensatory increase in mitochondrial respiration partially due to the decrease in mitochondrial content (32). Even though TMG treatment lowered mitochondrial respiration in this study, ROS was not increased suggesting an intervention that raises both O-GlcNAc and OGA expression promotes a healthier metabolic phenotype than genetic manipulation of OGT or OGA alone.
Curiously, the metabolic effect TMG had on cells and tissue was opposite to the effects seen in several studies with the overexpression or deletion of OGT or OGA. These differences could be due to alterations in the rate of O-GlcNAc cycling. O-GlcNAc cycling is the addition followed by the removal of the modification. The rate of cycling on a given protein might be more important to the function of the protein than the addition or removal in isolation. Previously, we demonstrated that overexpression of both OGT or OGA causes the same mitotic progression phenotype and increased aneuploidy in cells (33, 34). However, OGT overexpression increased O-GlcNAc levels, whereas OGA overexpression decreased O-GlcNAc levels; hence, how could the different changes in O-GlcNAc cause the exact same phenotype. We argued that the rate of O-GlcNAc cycling was the key determinant to the phenotypical changes, and we demonstrated that TMG treatment could reverse the mitotic phenotypes seen with OGT and OGA overexpression (34). Both OGT and OGA overexpressions would induce an increase in O-GlcNAc cycling considering that both OGT and OGA overexpressions increase the endogenous expression of the other enzyme (33). TMG treatment produced two unique effects. The sustained elevation in O-GlcNAc signals through a transcriptional network, possibly through the E2F transcription factors (35), to induce the expression of OGA while inhibiting activity of OGA, which in turn slows O-GlcNAc removal. The cells have both higher O-GlcNAcylation and higher OGA expression. Cells and tissue appear to adapt their O-GlcNAc cycling rate to these changes potentially producing the metabolic and transcriptional changes seen with the sustained treatment with TMG.
We contend that TMG and to some extent the GlcN treatment produced a two-stage adaptive process. First, total cellular and mitochondrial O-GlcNAc levels increased, promoting increased respiration (10) and signaling from the mitochondria to the nucleus via increases ROS production (36). The initial metabolic changes from GlcN treatment in both worms and animals caused a spike in ROS production; however, ROS and ATP levels declined with sustained GlcN treatment (2). Furthermore, many ETC complex subunits are heavily modified by O-GlcNAc (11), and short-term elevation of O-GlcNAc increased mitochondrial respiration and ATP production in rat cardiomyocytes (10, 11). The initial induction of respiration caused by increased O-GlcNAcylation transmits a signal to the nucleus altering gene transcription. We argue under short-term TMG treatment the increase in O-GlcNAc levels induced OGA transcription and a rapid rise in OGA protein levels to restore the proper rate of O-GlcNAc cycling (14). Second, under conditions of sustained OGA inhibition, elevated O-GlcNAc and OGA levels mediate the changes in the transcriptome resulting in longer mitochondria, reduced energy demand, and ROS generation. Thus, adaptation to sustained elevations in O-GlcNAc led to mitochondrial respiration decreases. The changes in respiration were due to a combination of altered gene expression and ETC function. We observed that both in vivo complex I and IV activity was increased after TMG treatment demonstrating that the decreased respiration was not solely due to ETC activity suppression. The mitochondrial membrane was hyperpolarized suggesting that the ETC was functioning at capacity; however, proton recycling from the intermembrane space into the inner mitochondrial space was decreased. Potentially, the slower proton-recycling rate was due to suppression of ATP synthase caused by a decrease in ATP synthase subunit expression (ATP6) or O-GlcNAcylation of ATP synthase subunits (11). Certainly, identification of the mitochondrial O-GlcNAcome after sustained TMG treatment would identify ETC proteins regulated by O-GlcNAc and warrants further investigation.
Although we cannot rule out other metabolic or mitochondrial changes affecting ROS production, the combination of lower mitochondrial respiration, no change in proton leak rate, and the hyperpolarization of the mitochondrial membrane would reduce mitochondrial ROS production. Consequently, the decrease in ROS signaling led to lower expression of antioxidant response genes. Under normal conditions, the generation of mitochondrial ROS is needed to induce antioxidant and growth programs in response to changes in the environment (36). TMG or GlcN treatment does not preclude the generation of ROS, but the basal ROS level was lowered. Because the cell did not need a robust antioxidant response, NRF2 gene programs were reduced. Under low ROS conditions, KEAP1 constantly degrades NRF2; however, elevated ROS oxidizes cysteine residues on KEAP1 leading to the release of NRF2, translocation into the nucleus, and initiation of antioxidant gene expression, including NRF2 itself. The decrease in NRF2 mRNA expression was due to long-term TMG treatment reprogramming NRF2 transcription. Interestingly, the NRF2 analogue in C. elegans is modified by O-GlcNAc, and increased NRF2 O-GlcNAcylation increased life span (37). Although we see O-GlcNAc on NRF2, TMG treatment reduced NRF2 expression and function suggesting that the TMG-induced changes in the antioxidant response were not due to the increased O-GlcNAcylation of NRF2 but through the reduced expression of the NRF2 transcript.
Likely, the transcriptome reprogramming by TMG or GlcN treatment was due to changes in O-GlcNAc-mediated transcriptional and epigenetic regulation. O-GlcNAc not only influences chromatin but also chromatin readers, writers, and erasers, thus complicating the specific roles of long-term O-GlcNAc elevation in transcriptional reprogramming (38). Of note, our IPA suggests that only specific gene networks were affected by long-term increased O-GlcNAcylation and argues that the combination of elevated O-GlcNAc and OGA expression must target these networks in some manner. Future investigation into these mechanisms could lead to novel mechanistic insight into the roles of O-GlcNAc in regulating gene networks.
The cellular reprogramming caused by elevations in both O-GlcNAc and OGA has a consequence on the energetic needs of animals. TMG-treated mice shifted their energy metabolism away from fatty acid oxidation and toward carbohydrate usage. The TMG-treated mice did not show a need for greater energy demand since sleep time was increased, movement was reduced, and energy expenditure was lower. Potentially, O-GlcNAc is impacting circadian rhythms. O-GlcNAcylation of both BMAL1 and CLOCK (key proteins of the circadian response) blocks ubiquitination and degradation of the proteins leading to increased expression of circadian genes (39). The expression of circadian genes is tied to several metabolic processes, including insulin sensitivity (40), which could promote the increase in carbohydrate usage and sleep time in TMG-treated mice.
TMG treatment-induced metabolic changes in the mouse would impact chronic diseases. Previously, mitochondrial function was impaired under diabetic conditions (12, 41) with O-GlcNAc levels higher in rat diabetic cardiomyocytes without the concomitant increase in OGA expression. Importantly, adenoviral overexpression of OGA led to improved mitochondrial function and calcium handling in diabetic cardiomyocytes and improved coronary endothelial cell function in diabetic mice (42, 43). Furthermore, OGA overexpression lowered O-GlcNAc levels and improved outcomes in mouse models of stretch-induced cardiac hypertrophy (44). Together, these data demonstrate that sustained elevations in O-GlcNAc without the concurrent increase in OGA expression was detrimental to cells; however, cellular function could be restored with OGA overexpression.
The combination of increased O-GlcNAcylation and OGA expression could restore some metabolic function lost in chronic metabolic diseases such as Alzheimer's disease (AD). TMG and TMG derivatives are actively being investigated as potential therapeutics for AD (45). The argument for inhibiting OGA in AD patients is that the treatment will increase O-GlcNAcylation of Tau, preventing Tau phosphorylation and subsequent Tau tangles (46–48). Treatment with TMG does increase Tau O-GlcNAcylation and improve outcomes in animal models of tauopathies (46). Our data argue for additional metabolic benefits for brain health after prolonged TMG treatment. Mitochondrial dysfunction is a characteristic of AD, and the mitochondrial cascade hypothesis argues that loss of mitochondrial function over time alters metabolite production and increases the generation of ROS causing neuronal damage and cell death (49). Screening of adult children of parents with AD demonstrates a decrease in mitochondrial function prior to the onset of any cognitive impairment or increased production of β-amyloid (50). The TMG-treated animals had lower mitochondrial respiration and ROS production but with a shift toward carbohydrate usage, which could slow AD development.
Long-term TMG treatment showed no adverse effects on animal health (51) nor does TMG treatment promote insulin resistance like less selective OGA inhibitors (52, 53). O-GlcNAcylation dampens insulin signaling by modifying several proteins in the insulin signaling pathway (54, 55), whereas OGT can relocate to the plasma membrane after insulin induction contributing to the ablation of insulin signaling (56, 57). O-GlcNAcylation is increased in diabetic tissue suggesting that the increase in glucose availability induces more flux through the HBP, increases O-GlcNAcylation of insulin signaling proteins, and leads to a decrease in responsiveness to insulin (58). Yet TMG treatment of animals increases O-GlcNAc levels without producing diabetes (51). We suggest that the combination of increased carbohydrate usage, O-GlcNAcylation, and OGA expression seen with long-term TMG treatment describes why TMG treatment does not induce diabetes and could potentially make these animals more sensitive to insulin.
In conclusion, long-term elevation in O-GlcNAcylation coupled with an increase in OGA expression modulates mitochondrial function and reprograms the transcriptome to favor a reduction in antioxidant response while lowering ATP production. These profound changes in metabolism caused mice to sleep more and move less, reduced body mass, and increased carbohydrate metabolism (Fig. 13). These physiological changes suggest that long-term increases in O-GlcNAcylation followed by a concurrent increase in OGA expression changes the metabolic program of animals without causing adverse effects. Potentially, progression of chronic metabolic diseases such as diabetes and Alzheimer's would be positively impacted by a combination of increased O-GlcNAcylation and OGA expression.
Figure 13.
O-GlcNAc regulates energy metabolism by reprogramming mitochondrial function. Representative model for prolonged O-GlcNAcylation and OGA expression leading to transcriptome reprogramming of mitochondrial function and energy metabolism.
Experimental procedures
Cell culture
Both SH-SY5Y and NT2 cells were cultured in DMEM (Sigma), 44 mm sodium bicarbonate (Sigma), 5 mm galactose (Fisher), 15 mg/liter phenol red (Sigma) and supplemented with 10% fetal bovine serum (FBS; Gemini), 1% penicillin/streptomycin (Sigma), 1% GlutaMAX (Gibco). Cells were adapted to the 5 mm galactose media for 2 weeks prior to treatment with 10 μm TMG (SD Specialty Chemicals) or 0.35 mm glucosamine (GlcN, Sigma) for at least 3 weeks prior to experiments. Media were replenished daily.
Animal protocols and models
The University of Kansas Medical Center Animal Care and Use Committee approved all experiments in this study. Two-month-old male C57Bl/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All mice were housed using a standard 12-h light/dark cycle with access to chow and water ad libitum. Mice were treated with a 50 mg/kg thiamet-G intraparietal injection every other morning for 15 days for a total of 8 doses per mouse. After completion of dosing, mice were fasted for 16 h before isoflurane (Fisher) anesthesia assisted cervical dislocation on day 16.
Ogttm1Gwh mice were obtained from Natasha Zachara, John Hopkins University, and liver-specific KO of OGT was performed by treating male mice with 5.0 × 1011 virus particles of AAV8.TBG.PI.Cre.rBG vector (University of Pennsylvania vector core) dissolved in 200 μl of sterile PBS via i.p. injection.
Cell lysis and immunoblotting
Cells were lysed, and samples were prepared for SDS-PAGE and immunoblotted as described previously (9). At least three independent experiments were repeated for all immunoblotting. Relative protein levels were analyzed using ImageJ 3.2 (National Institutes of Health) and were normalized to actin or GAPDH (34).
Cellular respiration, isolated mitochondrial respiration, and glycolysis measurement
An XF24 analyzer (Agilent Seahorse XF Technology, Santa Clara, CA) was used to measure cellular respiration and glycolysis. 75,000 control, TMG-, or GlcN-treated cells were seeded per well in a Seahorse 24-well cell culture plate 24 h prior to experimental assay. For respiration assays, cells were incubated in unbuffered DMEM (Sigma), 25 mm glucose (Sigma), phenol red (Sigma), 200 mm GlutaMax-1 (Gibco), NaCl (Fisher) at 37 °C in a CO2-free incubator for 1 h prior to loading. OCR was measured over a period of 100 min. Drugs oligomycin (0.5 μm, Sigma), FCCP (0.5 μm, Sigma), antimycin A (0.2 μm, Sigma), and rotenone (0.1 μm, Sigma) were added to each well sequentially at various time points during the assay (9).
For animal tissues, 15 μg per well of mouse liver or brain mitochondria was prepared in mitochondrial assay solution (MAS) (26). Respiration was initiated by adding mitochondria, followed by sequential addition of 6 mm ADP (Sigma), 1 μm FCCP (Sigma), 1 μm oligomycin (Sigma), and 4 μm antimycin A (Sigma) (26).
For glycolysis assays, ECAR was measured over a period of 100 min. Glucose (25 mm), oligomycin (1 μm), and 2-deoxy-d-glucose (100 mm, Sigma) were added at specific time points during the assay (9).
Statistical analysis
Statistical significance of all results, with the exception of the 24-h time point Avg RER, locomotion, and Avg EE, was assessed using a two-sample t test with p < 0.05 considered to be statistically significant. All data were generated using at least three independent experiments and were represented as means ± S.E. The 24-h time point Avg RER, locomotion, and Avg EE were assessed using permutation test with p < 0.05 considered to be statistically significant.
Antibodies
All primary and secondary antibodies used for immunoblotting were used at a concentration of 1:1000 and 1:10,000 dilution accordingly. Anti-O-linked N-acetylglucosamine antibody (RL2, ab2739) was purchased from Abcam. Both anti-OGT (AL-34) and OGA (345) were gracious gifts from the Laboratory of Gerald Hart in the Department of Biological Chemistry, The Johns Hopkins University School of Medicine. Anti-actin (A2066), NRF2 (ab62352), mTFA (ab47517), GAPDH (ab9484), oxidative stress defense (catalase, SOD1, TRX, and smooth muscle actin) mixture (ab179843), PRX pathway (TRX, TXNRD1, and PRX1) mixture (ab184868), citrate synthase (ab96600), and NADPH oxidase 4 (ab79971) were purchased from Abcam. Anti-DLP1 (611113) was purchased from BD Biosciences; anti-Mfn1 (sc-50330) and anti-p53 (sc-126) were purchased from Santa Cruz Biotechnology. Anti-cleaved PARP (9542) and anti-lamin B1 (D4Q4Z) were purchased from Cell Signaling Technology, and anti-Mfn2 (M6444) was purchased from Sigma.
Mitochondrial purification from cells and tissues
Mitochondria were isolated from SH-SY5Y cells using the modified cavitation method described previously (9). Briefly, 2 × 108 cells were trypsin digested off the plate and washed twice with pre-chilled PBS and then resuspended into 3 ml of the mitochondrial isolation medium. The cell suspensions were transferred into a pre-chilled cavitation chamber (nitrogen bomb; Parr Instrument Co., Moline, IL) and subjected to 900 p.s.i. for 15 min. Subsequently, the pressurized cell suspension was collected from the cavitation chamber, followed by centrifugation at 1000 × g for 10 min to pellet the cell debris. The clear supernatant was collected and centrifuged at 12,000 × g for 15 min. Crude mitochondrial pellet was washed three times with 500 μl of isolation medium. Washed pellet was lysed with Nonidet P-40 lysis buffer.
Both mouse brain and liver mitochondria were isolated using the modified method of two differential centrifugations (26). Promptly after euthanasia (brain <30 s, liver <1 min), tissues were rinsed twice with ice-cold PBS, homogenized (Teflon glass homogenizer with 15 strokes), and extracted in ∼10 volumes of mitochondrial isolation buffer (MSHE + BSA). All subsequent steps were performed on ice. Homogenate was centrifuged at 800 × g for 10 min at 4 °C. Supernatant was decanted through two layers of cheesecloth to a new tube followed by further centrifugation at 8,000 × g for 10 min at 4 °C. Mitochondrial pellet was washed once with MSHE + BSA, and the final pellet was resuspended with s minimal volume of MSHE + BSA. Total protein concentration was determined using the BCA protein assay kit (Thermo Fisher Scientific) per the manufacturer's instruction.
Total RNA isolation and reverse transcription (RT)-PCR
Total RNA was isolated by using TRI Reagent® solution (Ambion) per the manufacturer's instruction. Approximately 2 × 106 cells were resuspended in 1 ml of TRI Reagent solution, and then 200 μl of chloroform (Fisher) was added to extract RNA. Upon centrifugation, an equal amount of isopropyl alcohol (Fisher) was added to the upper layer solution containing RNA. RNA pellets were precipitated by centrifugation, washed once with 70% ethanol, and air-dried, and the pellet was dissolved in nuclease-free water (Life Technologies, Inc.) (14).
RNA concentration was measured using a Nanodrop 2000c (Thermo Fisher Scientific), and 1 μg of total RNA was reverse-transcribed using iScript Reverse Transcription Supermix (Bio-Rad) per the manufacturer's instruction. Total volume of 20 μl of each completed reaction mix was incubated in a thermal cycler (Model 2720, Applied Biosystems). Protocol was as follows: priming for 5 min at 25 °C, room temperature for 30 min at 42 °C, and room temperature inactivation for 5 min at 85 °C. cDNA products were diluted with nuclease-free water, 1:10 dilution, and analyzed by qPCR (14).
qPCR assay
qPCR assay on cDNA samples were done using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) per the manufacturer's instruction. 2 μl of cDNA were used with a total reaction volume of 20 μl. Primers used for OGT were F, CATCGAGAATATCAGGCAGGAG, and R, CCTTCGACACTGGAAGTGTATAG (14); for OGA were F, TTCACTGAAGGCTAATGGCTCCCG, and R, ATGTCACAGGCTCCGACCAAGT (14); for GAPDH were F, CCACATCGCTCAGACACCAT, and R, CCAGGCGCCCAATACG; for GFAT1 were F, AGCCCTCTGTTGATTGGTGT, and R, TCCATCTGGAGTGTTTGCAC; for Nrf2 were F, CCAGCACATCCAGTCAGAAA, and R, GCATGCAGTCATCAAAGTACAAA; for TXNRD1 were F, GCAATCCAGGCAGGAAGATT, and R, AGGCCACAAGCACCATATTC; for GNG1 were F, GAGCAGCTTCGCAAAGAA, and R, AGGATCCTCTCCAGAACG; for TFPI2 were F, CAGGTTTCCAGATGAAGCTACT, and R, GAGTCACATTGGCAGAGCA; for ATF4 were F, GAGAGAAGATGGTAGCAGCAAA, and R, TTCTTCTGGCGGTACCTAGT; and for IGF2 were F, CCTGGAGACGTACTGTGCTA, and R, GACTGCTTCCAGGTGTCATATT. A 96-well PCR plate (Midsci) with the reaction mixture was loaded onto CFX96 Touch real-time PCR detection system (Bio-Rad). The protocol was as follows: polymerase activation and DNA denaturation for 30 s at 95 °C, amplification denaturation for 5 s at 95 °C, and annealing for 30 s at 60 °C with 40 cycles, and melt curve at 65–95 °C with 0.5 °C increment 5 s/step.
OGA activity assay
Whole-cell extract samples (60 μg/reaction) were lysed using Nonidet P-40 lysis buffer. O-GlcNAcase activity was assayed in a total reaction volume of 200 μl (includes 10 μl of 500 mm sodium cacodylate (Sigma), pH 6.4, 3% BSA (Sigma), 2 μl of 100 mm p-nitrophenyl-N-acetyl-β-d-glucosaminide (Sigma), water, and cell extract) at 37 °C for up to 2 h. Reactions were stopped with 100 μl of 500 mm sodium carbonate (Sigma) every 30 min subsequently, and the absorbance at 400 nm was measured using a plate reader (BioTek Instrument). One unit is the amount of enzyme required to catalyze the release of p-nitrophenyl from 1 pmol of p-nitrophenyl-O-GlcNAc/min at 37 °C (59). These samples were assayed in a 96-well plate (pure Grade S-781671, BRANDplates®).
JC-1 staining for mitochondrial membrane potential
Mitochondrial membrane potential was analyzed with the potential-dependent fluorescent dye JC-1 (Life Technologies, Inc.). 2 μm JC-1 in Hanks' balanced salt solution (Sigma) was added to 2 × 106 cells per sample at 37 °C, 5% CO2 for 30 min. Cells were washed once and resuspended in 500 μl of warm Hanks' balanced salt solution. Cells were analyzed by a flow cytometer with 488 nm excitation and gated for singlet cells excluding cell debris. JC-1 dye exhibits a potential-dependent accumulation in mitochondria via a fluorescence emission shift from green (∼529 nm) to red (∼590 nm). Mitochondrial polarization was determined by red/green intensity ratio.
NAD+/NADH assay
NAD+/NADH ratios were measured using the NAD+/NADH detection kit (Cell Technology Inc., Mountain View, CA) per the manufacturer's instruction. 2 × 106 cells were in the assay, and fluorescence was measured with excitation/emission spectra of 570 and 600 nm, respectively. Assay results were normalized to protein amount by determining protein concentration of supernatant via DC Assay (Bio-Rad).
Transmission electron microscopy
Long-term TMG- and GlcN-treated SY5Y cells were prepared for electron microscopic experiments as described previously (9). Cells were seeded and cultured on Thermanox (Fisher) coverslips 24 h prior to fixation. Cells were fixed with 2% glutaraldehyde in 0.1 m cacodylate buffer, pH 7.4, and post-fixed with 1% osmium tetraoxide, 0.3% potassium ferricyanide. Cells were dehydrated in a graded series of 50, 70, 80, 95, and 100% ethanol. Coverslips were embedded in 100% epoxy resin, and the resin was cured overnight at 60 °C. Thin sections of 70–90 nm were cut on a Reichert Ultracut-S (Reichert Technologies, Buffalo, NY) and stained with uranyl acetate and lead citrate. Images were captured at 80 kV using a JEOL JEM-1400 transmission electron microscope (JEOL USA, Peabody, MA) equipped with a Lab6 gun at the University of Kansas Medical Center EM Core Laboratory.
Mitochondrion length
Measurement of mitochondria length was performed using ImageJ 3.2 software (National Institutes of Health) from multiple experiments.
Complex I activity assay
Crude isolated mitochondria were used for the complex I activity. A modified method following the decrease in NADH absorbance at 340 nm occurs when ubiquinone (CoQ1, Sigma), reduced to ubiquinol, was used for the complex I activity assay. CoQ1 (50 μm) was used to initiate the reaction at 30 °C. After 5 min, rotenone (10 μm) was added to obtain non-complex I activity. Total protein concentration was measured using the DC assay kit (Bio-Rad) as instructed by the manufacturer. Finally, complex I activity was expressed as nanomoles/min/mg protein (60).
Cytochrome c oxidase and citrate synthase activity assays
Complex IV (COXIV) and citrate synthase (CS) Vmax activities in TMG- or GlcN-treated SH-SY5Y cells were evaluated in both whole-cell and isolated mitochondria. Whole cells were trypsinized and washed twice with ice-cold 1× PBS, and then pelleted by centrifuging at 1000 × g for 3 min. The whole-cell pellets were resuspended in ice-cold PBS at a concentration of 30 × 106 cells/ml. Mitochondria were isolated as described before. A total of 40 × 106 cells were used for the mitochondrial isolation and the mitochondria pellets were resuspended in 400 μl of the mitochondrial isolation buffer. Both whole-cell and isolated mitochondrial COXIV and CS Vmax activities in non-treated and TMG- or GlcN-treated SH-SY5Y cells were determined as described previously (61). COXIV Vmax activity was normalized both to protein concentration and to CS activity.
Immunofluorescence microscopy
Cells were cultured on microscope slides (Fisher), washed with 1× PBS, and fixed with 4% paraformaldehyde (Sigma) for 20 min. Cells were permeabilized with 0.1% Triton X-100 (Sigma) in ice-cold PBS and then blocked with a TBST blocking solution (0.2% azide, 0.2% powdered dry milk, 12% chicken serum, 1% BSA, 100 mm glycine, 0.1% Triton X-100 in TBST, pH 7.5) for 1 h. Tom20 (1:500) in TBST blocking buffer was applied overnight. Slides were washed with TBST and incubated with fluorescent secondary antibodies (1:500 mouse Alexa-Fluor 488 nm Life Technologies, Inc.) for 1 h. DAPI solution (PBS, 0.01% Triton X-100, 0.001% DAPI, Sigma) was applied for 20 min. Finally, slides were washed and mounted on coverslips using ProLong Gold Anti-fade (Life Technologies, Inc.) (34).
cDNA library preparation
cDNA library was prepared using Illumina TruSeq Stranded mRNA sample preparation kit (Illumina) as per the manufacturer's instruction. Total RNA was isolated using the same method as described previously, and 800 ng of the total RNA per reaction was used to initiate the protocol.
RNA sequencing analysis
The quality of RNA sequencing results was first assessed using FastQC (0.11.2). RSEM (1.2.22) was utilized to align the reads to the human reference genome HG38 and to calculate gene expression values. EdgeR (3.14.0) was then used to normalize the expression values using the TMM-method (weighted trimmed mean of M-values) and for differential expression analyses. First, the negative binomial conditional common likelihood was maximized to estimate a common dispersion value across all genes (estimate CommonDisp). Next, tag-wise dispersion values were estimated by an empirical Bayes method based on weighted conditional maximum likelihood (estimateTagwiseDisp). Finally, the differential gene expression was calculated by computing gene-wise exact tests for differences in the means between two groups of negative-binomially distributed counts. Hierarchical clustering analysis was determined using Euclidean distance. The following R-packages were utilized for calculations and visualizations: gplots and edgeR.
To reduce the burden of multiple testing in differential gene expression analyses, a filter was initially applied to reduce the number of genes. Genes were removed if they did not present a meaningful gene expression across all samples; only genes with counts/million of >10 in at least two samples were considered in differential expression analyses involving the heat map, volcano plots, and pie chart. A counts/million of >100 threshold was used for the IPA analysis. The Benjamini and Hochberg procedure was used to control the false discovery rate.
Pathway and GSEA
IPA software was used to compare biological functions or canonical pathways in non-treated, long-term TMG- or GlcN-treated SH-SY5Y cells. Flexible format and Illumina Human Ref-8 version 3.0 array platform were the only parameters altered from default IPA settings for the Core analyses used to evaluate up-regulated and down-regulated genes (fold change >1.3, p < 0.05).
GSEA analysis was done using the javaGSEA desktop software. The default settings was used, except the permutation type was set to Gene_set with 1000 permutation types, and the metric for ranking genes was set to Diff_of_Classes (normalized expression data used were log2 transformed) when executing GSEA. All five gene set collections (c1–c6, h1) were queried from the Molecular Signatures Database (MSigDB) version 3.1.
ROS detection assay
Cells were analyzed for reactive oxygen species levels using a ROS-ID total ROS detection kit (Enzo Life Sciences) per the manufacturer's protocol. Briefly, 2 × 105 cells/well of control, prolonged TMG-, or GlcN-treated cells were seeded in a 96-well plate (pure Grade BRANDplates Midsci) a day prior to the assay. Cells were treated with ROS detection reagent at 37 °C for 1 h, and ROS level was measured via a fluorescence plate reader (TECAN Infinite M200, Switzerland) and quantified as level of fluorescence per cell. For both liver and brain isolated mitochondria, superoxide levels were determined using MitoSOX (Life Technologies, Inc.) as instructed by the manufacturer. Briefly, 150 μg/well isolated mitochondria was resuspended in ice-cold MAS (without rotenone) added with substrates (5 mm succinate, 5 mm malate, 5 mm glutamate) and then stained with MitoSOX at a concentration of 3.5 μm at 37 °C for 10 min. The mitochondria were washed once with MAS and transferred into a 96-well plate (pure Grade BRANDplates Midsci). Superoxide level was detected via fluorescence plate reader with excitation wavelength at 510 nm and emission wavelength at 590 nm.
ATP level determination
Cellular ATP level was determined by using a CellTiter-Glo luminescent cell viability assay kit (Promega) per the manufacturer's instruction. Briefly, 2 × 105 cells/well concentration of cells were seeded in a 96-well plate (pure Grade BRANDplates Midsci) a day prior to the assay. Cells were treated with ATP detection reagent at room temperature for 10 min. Cellular ATP levels were determined by measuring the luminescent signal on a plate reader (TECAN Infinity M200).
Nuclear extraction
Cells were harvested, pelleted, and then resuspended in hypotonic buffer (20 mm HEPES, 50 mm NaF, 5 mm Na2P2O7, 50 mm N-acetylglucosamine, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 1 mm PMSF, protease inhibitor mixture, pH 7.5) followed by centrifugation. Supernatant (cytoplasmic content) was saved, and nuclear pellet was washed with hypotonic buffer and then lysed with Nonidet P-40 lysis buffer and centrifuged for 20 min at 20,000 × g. Resulting supernatants were used for protein determination via Bradford assay (Bio-Rad). The protein levels were analyzed via immunoblotting, and anti-lamin B1 was used for nuclear content loading control, and actin was used for cytoplasmic content.
Oxidative stress and cell viability assay
Prolonged TMG- or GlcN-treated cells were incubated with different concentrations of H2O2 (50 μm, 100 μm, Sigma) for 7 h prior to cell viability determination via annexin V (Life Technologies, Inc.) and propidium iodide (PI, Life Technologies) staining. Annexin V-FITC/PI double labeling method was used per the manufacturer's instructions to determine cell viability after acute oxidative stress. Cells were digested with 2% BSA trypsin, washed once with 1× PBS, and incubated with annexin V/PI mixture and then subjected to an LSR II flow cytometer (BD Biosciences) and analyzed with FACSDiva software (BD Biosciences) for cell viability determination.
Cycloheximide treatment
Cells were treated with 50 μg/ml CHX (Sigma) and harvested at different time points for up to 6 h. Immunoblotting was performed to determine NRF2 protein stability after prolonged TMG or GlcN treatment. p53 was used as positive control for the CHX treatment.
Body composition and indirect calorimetry measurement
Fat mass, lean mass, and water mass were measured with an EchoMRI 4 in 1–1100 analyzer (EchoMRI; Houston, TX). Prior to the indirect calorimetry measurements, mice were housed individually for 5 days. Subsequently, mice were transferred and housed in the calorimetric chamber (Promethion High Definition Continuous Respiratory system for mice, Sable Systems Inc.) for 3–5 days accordingly. The chamber measured food intake, energy expenditure, substrate utilization, and various outcomes of the mice for baseline measurement calibration. Then, mice were transferred back to the normal individual cage. 50 mg/kg TMG or saline dosage was delivered to each mouse via peritoneal injection every other day for up to 2 weeks. During the last 5 days of injection, mice were transferred back into the calorimetric chamber and parameters measured as mentioned previously (saline versus TMG injection). These experiments were repeated twice, and each repeat included eight mice.
Author contributions
E. P. T. performed most of the experiments and wrote the paper. S. R. M. assisted E. P. T. in performing the animal experiments. R. T. optimized the protocol for the isolated mitochondrial respiration study. S. J. K. performed the NAD+/NADH assay. S. G. and P. D. performed the RNA-seq analysis, and D C. K. provided guidance for the analysis. Z. Z. and M. M. assisted E. P. T. experimentally. J. P. T. provided guidance for the indirect calorimetric study. L. D. assisted in the Monte Carlo simulation. R. H. S. and P. K. provided conceptual guidance for the mitochondrial functional study. All mouse studies were performed under the supervision of U. A., K. R. P., and N. E. Z. C. S. provided overall guidance and contributed to manuscript preparation. All authors approved the manuscript.
Supplementary Material
Acknowledgments
We thank Christy Hagan, Kay Minn, Heather Wilkins, and Ian Weilding for their technical support. We thank both Barbara Fegley and Pat St. John for their help and the support of the Electron Microscope (EM) Core Facility, sponsored in part by National Institutes of Health NIGMS COBRE Grant P20GM104936 and National Institutes of Health Grant 1S10RR027564. We also thank Richard Hasting and the Flow Cytometry Laboratory, sponsored by the National Institutes of Health NIGMS COBRE Grant P30GM103326.
This work was supported by National Institutes of Health Grant R01DK100595 from NIDDK (to C. S. and K. R. P.), University of Kansas Alzheimer's Disease Center pilot grant (to C. S.), National Institutes of Health Grants R01DK098414 (to U. A.) and P30AG035982 (to R. S.), a Mabel A. Woodyard fellowship, and University of Kansas Medical Center Biomedical Research Training Program (to E. P. T.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1–S6.
- ROS
- reactive oxygen species
- O-GlcNAc
- N-acetylglucosamine
- OGT
- O-GlcNAc transferase
- OGA
- O-GlcNAcase
- HBP
- hexosamine biosynthetic pathway
- TMG
- thiamet-G
- PI
- propidium iodide
- PARP
- poly(ADP-ribose) polymerase
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- MAS
- mitochondrial assay solution
- CHX
- cycloheximide
- RER
- respiratory exchange ratio
- qPCR
- quantitative PCR
- F
- forward
- R
- reverse
- SOD
- superoxide dismutase
- ETC
- electron transport chain
- OCR
- oxygen consumption rate
- RNA-seq
- RNA-sequencing
- EE
- energy expenditure
- mOGT
- mitochondrial form of OGT
- AD
- Alzheimer's disease
- Avg
- average
- IPA
- ingenuity pathway analysis
- GSEA
- gene set enrichment analysis
- RNS
- reactive nitrogen species
- CS
- citrate synthase
- TXN
- thioredoxin
- ECAR
- extracellular acidification rate
- TRX
- thioredoxin.
References
- 1. Lezi E., and Swerdlow R. H. (2012) Mitochondria in neurodegeneration. Adv. Exp. Med. Biol. 942, 269–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weimer S., Priebs J., Kuhlow D., Groth M., Priebe S., Mansfeld J., Merry T. L., Dubuis S., Laube B., Pfeiffer A. F., Schulz T. J., Guthke R., Platzer M., Zamboni N., Zarse K., and Ristow M. (2014) d-Glucosamine supplementation extends life span of nematodes and of ageing mice. Nat. Commun. 5, 3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hart G. W., Slawson C., Ramirez-Correa G., and Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marshall S., Nadeau O., and Yamasaki K. (2004) Dynamic actions of glucose and glucosamine on hexosamine biosynthesis in isolated adipocytes: differential effects on glucosamine 6-phosphate, UDP-N-acetylglucosamine, and ATP levels. J. Biol. Chem. 279, 35313–35319 [DOI] [PubMed] [Google Scholar]
- 5. Hart G. W. (2014) Three decades of research on O-GlcNAcylation–a major nutrient sensor that regulates signaling, transcription and cellular metabolism. Front. Endocrinol. (Lausanne) 5, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bond M. R., and Hanover J. A. (2013) O-GlcNAc cycling: a link between metabolism and chronic disease. Annu. Rev. Nutr. 33, 205–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dias W. B., and Hart G. W. (2007) O-GlcNAc modification in diabetes and Alzheimer's disease. Mol. Biosyst. 3, 766–772 [DOI] [PubMed] [Google Scholar]
- 8. Love D. C., Kochan J., Cathey R. L., Shin S. H., Hanover J. A., and Kochran J. (2003) Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J. Cell Sci. 116, 647–654 [DOI] [PubMed] [Google Scholar]
- 9. Tan E. P., Villar M. T., E L., Lu J., Selfridge J. E., Artigues A., Swerdlow R. H., and Slawson C. (2014) Altering O-linked β-N-acetylglucosamine cycling disrupts mitochondrial function. J. Biol. Chem. 289, 14719–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Banerjee P. S., Ma J., and Hart G. W. (2015) Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc. Natl. Acad. Sci. U.S.A. 112, 6050–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ma J., Liu T., Wei A. C., Banerjee P., O'Rourke B., and Hart G. W. (2015) O-GlcNAcomic profiling identifies widespread O-linked β-N-acetylglucosamine modification (O-GlcNAcylation) in oxidative phosphorylation system regulating cardiac mitochondrial function. J. Biol. Chem. 290, 29141–29153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu Y., Suarez J., Fricovsky E., Wang H., Scott B. T., Trauger S. A., Han W., Hu Y., Oyeleye M. O., and Dillmann W. H. (2009) Increased enzymatic O-GlcNAcylation of mitochondrial proteins impairs mitochondrial function in cardiac myocytes exposed to high glucose. J. Biol. Chem. 284, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuzwa S. A., Macauley M. S., Heinonen J. E., Shan X., Dennis R. J., He Y., Whitworth G. E., Stubbs K. A., McEachern E. J., Davies G. J., and Vocadlo D. J. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4, 483–490 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Z., Tan E. P., VandenHull N. J., Peterson K. R., and Slawson C. (2014) O-GlcNAcase expression is sensitive to changes in O-GlcNAc Homeostasis. Front. Endocrinol. (Lausanne) 5, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fernie A. R., Carrari F., and Sweetlove L. J. (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7, 254–261 [DOI] [PubMed] [Google Scholar]
- 16. Baffou G., Rigneault H., Marguet D., and Jullien L. (2014) A critique of methods for temperature imaging in single cells. Nat. Methods 11, 899–901 [DOI] [PubMed] [Google Scholar]
- 17. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mueller C. F., Laude K., McNally J. S., and Harrison D. G. (2005) ATVB in focus: redox mechanisms in blood vessels. Arterioscler. Thromb. Vasc. Biol. 25, 274–278 [DOI] [PubMed] [Google Scholar]
- 19. Housley M. P., Rodgers J. T., Udeshi N. D., Kelly T. J., Shabanowitz J., Hunt D. F., Puigserver P., and Hart G. W. (2008) O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 283, 16283–16292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruan H. B., Han X., Li M. D., Singh J. P., Qian K., Azarhoush S., Zhao L., Bennett A. M., Samuel V. T., Wu J., Yates J. R. 3rd, and Yang X. (2012) O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 16, 226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., and Cho J. W. (2006) Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8, 1074–1083 [DOI] [PubMed] [Google Scholar]
- 22. Obrig T. G., Culp W. J., McKeehan W. L., and Hardesty B. (1971) The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J. Biol. Chem. 246, 174–181 [PubMed] [Google Scholar]
- 23. Sullivan L. B., and Chandel N. S. (2014) Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tangmansakulchai K., Abubakar Z., Kitiyanant N., Suwanjang W., Leepiyasakulchai C., Govitrapong P., and Chetsawang B. (2016) Calpastatin overexpression reduces oxidative stress-induced mitochondrial impairment and cell death in human neuroblastoma SH-SY5Y cells by decreasing calpain and calcineurin activation, induction of mitochondrial fission and destruction of mitochondrial fusion. Mitochondrion 30, 151–161 [DOI] [PubMed] [Google Scholar]
- 25. Groves J. A., Lee A., Yildirir G., and Zachara N. E. (2013) Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis. Cell Stress Chaperones 18, 535–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogers G. W., Brand M. D., Petrosyan S., Ashok D., Elorza A. A., Ferrick D. A., and Murphy A. N. (2011) High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS ONE 6, e21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pendergast D. R., Leddy J. J., and Venkatraman J. T. (2000) A perspective on fat intake in athletes. J. Am. Coll. Nutr. 19, 345–350 [DOI] [PubMed] [Google Scholar]
- 28. Lagerlöf O., Slocomb J. E., Hong I., Aponte Y., Blackshaw S., Hart G. W., and Huganir R. L. (2016) The nutrient sensor OGT in PVN neurons regulates feeding. Science 351, 1293–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Donnell N., Zachara N. E., Hart G. W., and Marth J. D. (2004) Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang A. C., Jensen E. H., Rexach J. E., Vinters H. V., and Hsieh-Wilson L. C. (2016) Loss of O-GlcNAc glycosylation in forebrain excitatory neurons induces neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 113, 15120–15125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trapannone R., Mariappa D., Ferenbach A. T., and van Aalten D. M. (2016) Nucleocytoplasmic human O-GlcNAc transferase is sufficient for O-GlcNAcylation of mitochondrial proteins. Biochem. J. 473, 1693–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sacoman J. L., Dagda R. Y., Burnham-Marusich A. R., Dagda R. K., and Berninsone P. M. (2017) Mitochondrial O-GlcNAc transferase (mOGT) regulates mitochondrial structure, function and survival in HeLa cells. J. Biol. Chem. 292, 4499–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Slawson C., Zachara N. E., Vosseller K., Cheung W. D., Lane M. D., and Hart G. W. (2005) Perturbations in O-linked β-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J. Biol. Chem. 280, 32944–32956 [DOI] [PubMed] [Google Scholar]
- 34. Tan E. P., Caro S., Potnis A., Lanza C., and Slawson C. (2013) O-Linked N-acetylglucosamine cycling regulates mitotic spindle organization. J. Biol. Chem. 288, 27085–27099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muthusamy S., Hong K. U., Dassanayaka S., Hamid T., and Jones S. P. (2015) E2F1 transcription factor regulates O-linked N-acetylglucosamine (O-GlcNAc) transferase and O-GlcNAcase expression. J. Biol. Chem. 290, 31013–31024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reczek C. R., and Chandel N. S. (2015) ROS-dependent signal transduction. Curr. Opin. Cell Biol. 33, 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li H., Liu X., Wang D., Su L., Zhao T., Li Z., Lin C., Zhang Y., Huang B., Lu J., and Li X. (2017) O-GlcNAcylation of SKN-1 modulates the life span and oxidative stress resistance in Caenorhabditis elegans. Sci. Rep. 7, 43601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Z., Costa F. C., Tan E. P., Bushue N., DiTacchio L., Costello C. E., McComb M. E., Whelan S. A., Peterson K. R., and Slawson C. (2016) O-Linked N-Acetylglucosamine (O-GlcNAc) transferase and O-GlcNAcase interact with Mi2β protein at the agamma-globin promoter. J. Biol. Chem. 291, 15628–15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li M. D., Ruan H. B., Hughes M. E., Lee J. S., Singh J. P., Jones S. P., Nitabach M. N., and Yang X. (2013) O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 17, 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chaves I., van der Horst G. T., Schellevis R., Nijman R. M., Koerkamp M. G., Holstege F. C., Smidt M. P., and Hoekman M. F. (2014) Insulin-FOXO3 signaling modulates circadian rhythms via regulation of clock transcription. Curr. Biol. 24, 1248–1255 [DOI] [PubMed] [Google Scholar]
- 41. Dassanayaka S., Readnower R. D., Salabei J. K., Long B. W., Aird A. L., Zheng Y. T., Muthusamy S., Facundo H. T., Hill B. G., and Jones S. P. (2015) High glucose induces mitochondrial dysfunction independently of protein O-GlcNAcylation. Biochem. J. 467, 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Makino A., Dai A., Han Y., Youssef K. D., Wang W., Donthamsetty R., Scott B. T., Wang H., and Dillmann W. H. (2015) O-GlcNAcase overexpression reverses coronary endothelial cell dysfunction in type 1 diabetic mice. Am. J. Physiol. Cell Physiol. 309, C593–C599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clark R. J., McDonough P. M., Swanson E., Trost S. U., Suzuki M., Fukuda M., and Dillmann W. H. (2003) Diabetes and the accompanying hyperglycemia impairs cardiomyocyte calcium cycling through increased nuclear O-GlcNAcylation. J. Biol. Chem. 278, 44230–44237 [DOI] [PubMed] [Google Scholar]
- 44. Facundo H. T., Brainard R. E., Watson L. J., Ngoh G. A., Hamid T., Prabhu S. D., and Jones S. P. (2012) O-GlcNAc signaling is essential for NFAT-mediated transcriptional reprogramming during cardiomyocyte hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 302, H2122–H2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stivers P. J., Harmonay L., Hicks A., Mehmet H., Morris M., Robinson G. M., Strack P. R., Savage M. J., Zaller D. M., Zwierzynski I., and Brandish P. E. (2015) Pharmacological inhibition of O-GlcNAcase does not increase sensitivity of glucocorticoid receptor-mediated transrepression. PLoS ONE 10, e0145151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yuzwa S. A., Shan X., Jones B. A., Zhao G., Woodward M. L., Li X., Zhu Y., McEachern E. J., Silverman M. A., Watson N. V., Gong C. X., and Vocadlo D. J. (2014) Pharmacological inhibition of O-GlcNAcase (OGA) prevents cognitive decline and amyloid plaque formation in bigenic tau/APP mutant mice. Mol. Neurodegener. 9, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yuzwa S. A., Cheung A. H., Okon M., McIntosh L. P., and Vocadlo D. J. (2014) O-GlcNAc modification of tau directly inhibits its aggregation without perturbing the conformational properties of tau monomers. J. Mol. Biol. 426, 1736–1752 [DOI] [PubMed] [Google Scholar]
- 48. Yuzwa S. A., Shan X., Macauley M. S., Clark T., Skorobogatko Y., Vosseller K., and Vocadlo D. J. (2012) Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol. 8, 393–399 [DOI] [PubMed] [Google Scholar]
- 49. Swerdlow R. H., Burns J. M., and Khan S. M. (2014) The Alzheimer's disease mitochondrial cascade hypothesis: progress and perspectives. Biochim. Biophys. Acta 1842, 1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mosconi L., de Leon M., Murray J., E L., Lu J., Javier E., McHugh P., and Swerdlow R. H. (2011) Reduced mitochondria cytochrome oxidase activity in adult children of mothers with Alzheimer's disease. J. Alzheimers Dis. 27, 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Macauley M. S., Shan X., Yuzwa S. A., Gloster T. M., and Vocadlo D. J. (2010) Elevation of global O-GlcNAc in rodents using a selective O-GlcNAcase inhibitor does not cause insulin resistance or perturb glucohomeostasis. Chem. Biol. 17, 949–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vosseller K., Wells L., Lane M. D., and Hart G. W. (2002) Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. U.S.A. 99, 5313–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Macauley M. S., Bubb A. K., Martinez-Fleites C., Davies G. J., and Vocadlo D. J. (2008) Elevation of global O-GlcNAc levels in 3T3-L1 adipocytes by selective inhibition of O-GlcNAcase does not induce insulin resistance. J. Biol. Chem. 283, 34687–34695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Klein A. L., Berkaw M. N., Buse M. G., and Ball L. E. (2009) O-Linked N-acetylglucosamine modification of insulin receptor substrate-1 occurs in close proximity to multiple SH2 domain binding motifs. Mol. Cell. Proteomics 8, 2733–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Whelan S. A., Dias W. B., Thiruneelakantapillai L., Lane M. D., and Hart G. W. (2010) Regulation of insulin receptor substrate 1 (IRS-1)/AKT kinase-mediated insulin signaling by O-Linked β-N-acetylglucosamine in 3T3-L1 adipocytes. J. Biol. Chem. 285, 5204–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang X., Ongusaha P. P., Miles P. D., Havstad J. C., Zhang F., So W. V., Kudlow J. E., Michell R. H., Olefsky J. M., Field S. J., and Evans R. M. (2008) Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451, 964–969 [DOI] [PubMed] [Google Scholar]
- 57. Whelan S. A., Lane M. D., and Hart G. W. (2008) Regulation of the O-linked β-N-acetylglucosamine transferase by insulin signaling. J. Biol. Chem. 283, 21411–21417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Copeland R. J., Bullen J. W., and Hart G. W. (2008) Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am. J. Physiol. Endocrinol. Metab. 295, E17–E28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Butkinaree C., Cheung W. D., Park S., Park K., Barber M., and Hart G. W. (2008) Characterization of β-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J. Biol. Chem. 283, 23557–23566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Esteves A. R., Lu J., Rodova M., Onyango I., Lezi E., Dubinsky R., Lyons K. E., Pahwa R., Burns J. M., Cardoso S. M., and Swerdlow R. H. (2010) Mitochondrial respiration and respiration-associated proteins in cell lines created through Parkinson's subject mitochondrial transfer. J. Neurochem. 113, 674–682 [DOI] [PubMed] [Google Scholar]
- 61. E L., and Swerdlow R. H. (2016) Lactate's effect on human neuroblastoma cell bioenergetic fluxes. Biochem. Pharmacol. 99, 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.