Abstract
The current understanding of the specificity of the bacterial class I release factors (RFs) in decoding stop codons has evolved beyond a simple tripeptide anticodon model. A recent molecular dynamics study for deciphering the principles for specific stop codon recognition by RFs identified Arg-213 as a crucial residue on Escherichia coli RF2 for discriminating guanine in the third position (G3). Interestingly, Arg-213 is highly conserved in RF2 and substituted by Ile-196 in the corresponding position in RF1. Another similar pair is Leu-126 in RF1 and Asp-143 in RF2, which are also conserved within their respective groups. With the hypothesis that replacement of Arg-213 and Asp-143 with the corresponding RF1 residues will reduce G3 discrimination by RF2, we swapped these residues between E. coli RF1 and RF2 by site-directed mutagenesis and characterized their preference for different codons using a competitive peptide release assay. Among these, the R213I mutant of RF2 showed 5-fold improved reading of the RF1-specific UAG codon relative to UAA, the universal stop codon, compared with the wild type (WT). In-depth fast kinetic studies revealed that the gain in UAG reading by RF2 R213I is associated with a reduced efficiency of termination on the cognate UAA codon. Our work highlights the notion that stop codon recognition involves complex interactions with multiple residues beyond the PXT/SPF motifs. We propose that the R213I mutation in RF2 brings us one step forward toward engineering an omnipotent RF in bacteria, capable of reading all three stop codons.
Keywords: Michaelis-Menten, mutagenesis, ribosome, translation, translation release factor, stop codon
Introduction
Peptide release during translation termination is the end of protein synthesis on the ribosome (for review see Ref. 1). Translation termination begins when one of the three stop codons (UAA, UAG, or UGA) is translocated into the ribosomal A site, and the peptidyl tRNA is translocated into the P site. The ester bond between the P site tRNA and polypeptide chain must be hydrolyzed for the release of the nascent peptide. In bacteria, class I release factors RF13 and RF2 recognize overlapping sets of three different termination signals (stop codons). Both factors recognize UAA, whereas UAG is read specifically by RF1 and UGA by RF2 (2). In eukaryotic protein synthesis, eRF1 is the single class I RF, facilitating peptide release at all three stop codons. It is an open question why bacteria have two RFs acting in a semispecific manner, whereas eukaryotes have only one RF. The accuracy with which RFs discriminate against the other 61 (sense) codons in bacteria is remarkably high. The reported error rate is 1 in 104 in vivo (3); the frequency of error for all single mismatches relative to stop codons ranges from 1 in 103 to 106 in vitro (4). According to the “tripeptide anticodon” hypothesis, conserved amino acid motifs, PXT in RF1 and SPF in RF2, mimic the tRNA triplet anticodon and determine codon specificity of the RFs. This simple mimicry model was supported by low, and later high, resolution crystal structures of various ribosomal release complexes (RCs) with different stop codons and RFs bound to the A site of the ribosome (5–9). In these structures the conserved amino acids of the tripeptide anticodon motif form a recognition loop that surrounds the cognate codons. The crystal structures were later used in MD-free energy perturbation simulations to demonstrate the mechanism of stop codon reading, first in bacteria (10) and later in mitochondria (11). This approach, although primarily aimed to clarify the role of the key residues of the tripeptide anticodon motif, identified additional side chains on RFs, possibly involved in specific stop codon recognition. Sund et al. (10) proposed that the following residues are involved: Gly-120, Glu-123, Leu-126, Gln-185, and Ile-196 on RF1 and Gly-137, Glu-140, Asp-143, Val-192, and Arg-213 on RF2 (Escherichia coli numbering, used throughout in this work) in specific reading of the second and third positions of the stop codon.
Differential reading of the third position of a stop codon by RFs attracts considerable interest because it underpins the specificity of RF2 for the UGA codon. RF1 accepts both an adenine and a guanine at the third position (A3 and G3, respectively), whereas RF2 has to discriminate against G3 reading, which would otherwise lead to misreading of the tryptophan codon (UGG) as a stop codon. Recent MD-free energy perturbation simulation (using the available structures of termination complexes) revealed that a water molecule is possibly introduced into the decoding site during RF1 binding to the UAG and UAA stop codons, which facilitates further hydrogen-bonding between G3 of the stop codon and U531 of rRNA, essential for G3 reading (10). In the case of RF2, however, a conserved residue, Arg-213, prevents the water molecule from entering the codon recognition site due to its long side chain, thereby preventing G3 reading by RF2 (10).
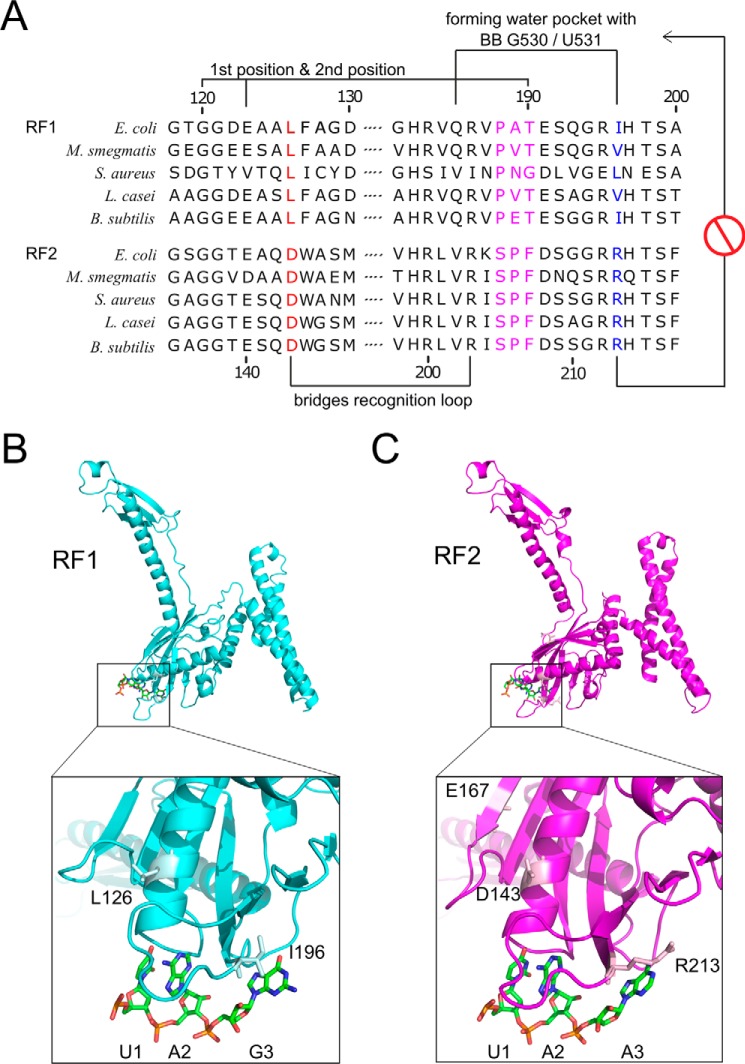
When RF1 and RF2 amino acid sequences from various bacteria are aligned together focusing mainly on the stop codon reading elements, a high degree of homology was observed between the two groups with some exceptions, which are moderately well conserved within the group. These include the PXT motif on RF1 and the SPF motif on RF2. Similarly, Ile-196 on RF1 and Arg-213 on RF2 were found in the corresponding positions (Fig. 1). Because Arg-213 was proposed to play a crucial role for RF2's discrimination for G3 from MD simulation (10), we hypothesized that exchanging Arg-213 with isoleucine (Ile) as in RF1 or with alanine (Ala), an even smaller side chain might allow RF2 to read G3 better than the WT because the water-blocking effect of Arg-213 would be removed. Alternatively, changing Ile-196 to Arg in RF1 might increase its discrimination against G3, whereas replacing Ile-196 with Ala would be less discriminative of G3 compared with the WT RF1.
Figure 1.
Sequence alignment of bacterial class I release factors RF1 and RF2 and their structural comparison. A Leu-126 in RF1 and Asp-143 in RF2 are shown in red; they bridge the recognition loop together with Arg-186 in RF1 and Arg-203 in RF2, respectively, as indicated. The tripeptide anticodon is shown in magenta. Ile-196 in RF1 and Arg-213 in RF2 are highlighted in blue. B, RF1 is shown in cyan, and the residues Leu-126 and Ile-196 are highlighted in light cyan. mRNA UAA is highlighted with green carbon atoms. See PDB 4V7P (6). C, RF2 is shown in magenta, and Asp-143, Glu-167, and Arg-213 are highlighted in light magenta. mRNA UAA is highlighted as in B. See PDB code 4V67 (5).
Other than the residues discussed above, Leu-126 in RF1 and Asp-143 in RF2 were also found in the corresponding positions, which are highly conserved in their respective groups (Fig. 1A). Interestingly, these residues are located in the α-helix, supporting the stop codon recognition loop (Fig. 1, B and C). Even though these residues may not be directly involved in stop codon recognition, they play supporting roles for the major residues. Thus, we reasoned that exchanging Asp-143 with Leu will also reduce RF2's discrimination for G3 and changing Leu-126 in RF1 to Asp would increase discrimination for G3.
Mutational analyses of the key residues on RF1/RF2, although crucial for complementing the available structural data, are limited. Mutations of the PXT and SPF motifs of RF1 and RF2 first illustrated the crucial role of these tripeptide motifs in specific reading of the stop codons (12). Besides those, biochemical data are available for the alanine mutants of few conserved residues within the decoding region of RF1 (Gln-185, Arg-186, Thr-190, His-197, and Thr-198) (13, 14). These mutations showed a modest decrease in the maximal rate of peptide release (kcat) but resulted in a large 100-fold increase in the dissociation constant (kD) when bound to the ribosome with a cognate stop codon in the A site. However, similar mutational analysis of RF2 is not available to date. Based on a fitness compensatory mutation (E167K) in the RF1 knock-out background, Glu-167 was suggested to play a crucial role in UAG reading by RF2 (12, 15). However, the structural basis of improved UAG reading by this RF2 mutant is not understood. Two earlier studies (12, 16) claimed that the R213I mutation is toxic for overexpression, and it gives a dominant lethal phenotype in the bacteria, but the actual reason for such behavior may not be this mutation alone (17, 18). Also, no mutational studies are available for Asp-143 in RF2.
Here, using site-directed mutagenesis, we generated several E. coli RF1/RF2 variants (RF1 I196A, RF1 I196R, RF1 L126D, RF2 R213A, RF2 R213I, and RF2 D143L) and characterized those in a competition assay for single-round peptide release (19) from ribosomal RCs containing major stop codon UAA (RCUAA) versus RCXXX carrying various codons (symbolized as XXX) in the A site. These include UAG and UGA stop codons and UGG (Trp), UCA (Ser), and AAA (Lys) codons. Among all the mutants tested, the R213I mutant RF2 showed the highest degree of improvement (5-fold) in UAG reading relative to UAA during translation termination while maintaining discrimination against UCA and UGG codons. To understand the basis of the altered sensitivity of the R213I mutant, we characterized it further with fast kinetic analysis for reading UAA and UAG codons and also in full-length protein synthesis assay using a reconstituted transcription-translation system.
Results
Competitive peptide release by the RF variants from RCUAA versus RCXXX
Ribosomal RCs were prepared with fMet-tRNAfMet in the P site and various codons in the A site (see the list of codons in Table 1). The fMet in the RCUAA was radioactively labeled with 3H. Other RCs (RCXXX) contained one of the five different codons (UAG, UGA, UGG, UCA, or AAA) in the A site, and [35S]fMet-tRNAfMet in the P site. All RF proteins RF1 and RF2 WT and mutants RF1 I196A, RF1 I196R, RF1 L126D, RF2 R213A, RF2 R213I, and RF2 D143L were prepared using standard laboratory protocols.
Table 1.
A values (as defined in Equation 1) determined from the competition assays using [35S]fMet-RCUAAversus [3H]fMet-RCXXX, where XXX represents the codon specified in the first row of the table
| UAG | UGA | UCA | UGG | AAA | |
|---|---|---|---|---|---|
| RF1 WT | 0.9 | 29.5 | 29.5 | 32.5 | –a |
| RF1 I196A | 1 | 27.7 | 68.2 | 135 | –a |
| RF1 I196R | 1.4 | 68.9 | 36.8 | 31.3 | –a |
| RF1 L126D | 1.3 | 9.2 | 16.1 | 12.1 | –a |
| RF2 WT | 33 | 1 | 36.7 | 33.9 | –a |
| RF2 R213A | 22.4 | 1.1 | 59 | 66.3 | –a |
| RF2 R213I | 6.5 | 1.4 | 61.5 | 20.2 | –a |
| RF2 D143L | 12.2 | 1.1 | 10.3 | 52.2 | –a |
a Beyond the detection limit.
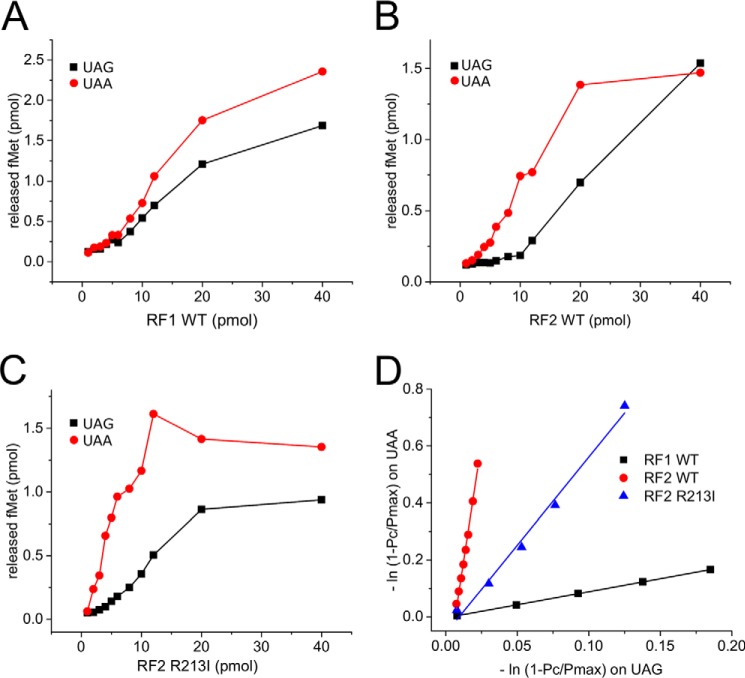
In the competition experiments, equal amounts of RCUAA carrying [3H]fMet-tRNAfMet and RCXXX carrying [35S]fMet-tRNAfMet were mixed together. To this mix, RF variants were added in increasing concentrations, and the amounts of [3H]fMet and [35S]fMet released in a single round reaction were measured (Fig. 2). From these data, a discrimination parameter A, reflecting the relative preference of the RFs for UAA over the other codon was determined as described under “Experimental procedures.” The higher the value of A, the higher is the discrimination against the competing codon with respect to UAA. Thus, decrease in the A value for a particular codon due to any mutation in a RF will indicate relatively improved reading of that codon by the mutant RF. The A values are listed in Table 1. All RF mutants recognized UAA and the respective cognate codons of their WT counterparts (UAG for RF1 or UGA for RF2 mutants) with similar efficiency (A values ranging from 0.9 to 1.4), thereby demonstrating no discrimination between UAA and the specific cognate codon (Fig. 2, A and B, Table 1). In contrast, no peptide release was detected with RCs with a sense codon AAA coding for lysine; consequently, the slope of the discrimination curve was infinite, and the A value could not be determined (Table 1).
Figure 2.
Representative examples of the competitive peptide release experiment. RFs in increasing concentration were subjected to competition for fMet release from [35S]fMet-RCUAA versus [3H]fMet-RCUAG added in equal proportion in the reaction. The plots indicate the amount of [35S]fMet (circles) and [3H]fMet (squares) released for a given concentration of RF1 WT (A), RF2 WT (B), and RF2 R213I (C). D, the negative natural logarithm of the fraction of fMet retained on one complex was plotted as a function of the same on the competing pair (using the initial points from the plots in A, B, and C) to generate plots, which could be fitted as straight lines and the slopes of which represent the discrimination factor for that pair of codons as described in Equation 1. The higher the slope, the higher is the discrimination for that RF for UAG relative to UAA. A values resulting from for all competition assays are listed in Table 1.
None of the RF1 variants displayed an improved reading of UGA respective to UAA while maintaining substantial levels of discrimination on other non-cognate codons tested here. The RF1 I196A produced a WT-like A value for the non-cognate UGA codon but showed 4- and 2-fold higher discriminations against UGG and UCA codons, respectively. The I196R mutant, on the other hand, showed a 2-fold increased discrimination against UGA but WT-like A values for UCA and UGG codons. Interestingly, the L126D mutation showed an ∼2-fold improved reading of UGA, but at the same time it also lost discrimination against UGG and UCA codons. Based on these results, we excluded all RF1 mutants from further investigation, as none of those showed improvement in omnipotent reading of the stop codons.
For RF2 mutants the same screening process was applied; in contrast to RF1, all RF2 variants showed a moderate to significant decrease in discrimination against RF1-specific UAG codon. The R213A mutation showed a 1.5× smaller A value for the UAG codon but ∼2-fold higher A values for both UCA and UGG codons. The D143L mutant showed a 3-fold smaller A value for the UAG codon, but it also lost discrimination against the UCA codon to the same extent, whereas its discrimination against UGG codon increased 1.5×. The most interesting mutant was RF2 R213I, as it displayed 5× less discrimination against the UAG codon than the RF2 WT (Fig. 2C) while having an almost 2-fold higher discrimination against UCA and a moderate (1.5×) loss of discrimination against UGG codon. Thus, our competition assay identified RF2 R213I as a promising candidate toward making an omnipotent release factor. Therefore, RF2 R213I was selected for further in-depth kinetic analysis.
Sense codon reading by RF2 R213I; tested in full-length protein synthesis assay in vitro
The competition assays were limited only to three sense codons. Thus, to test the ability of RF2 R213I to discriminate against a variety of sense codons in a cellular scenario, we performed an in vitro full-length protein synthesis experiment using a fully reconstituted transcription (T7 based)-translation system with individually purified components from E. coli (20). Recognition of any sense codon by RF2 R213I would lead to peptide release and, thus, would generate truncated products due to premature translation termination. Alternatively, a full-length protein band of expected size would signify its capacity in recognizing the stop codon accurately. E. coli dihydrofolate reductase (DHFR) was used as the reporter protein as it contains a broad range of sense codons (all except ACG, TCC, AGG) and UAA as the stop codon. As shown in Fig. 3, a single clear band of the full-length DHFR protein was seen with RF1 WT and RF2 WT as well as RF2 R213I (Fig. 3, lanes A, B, and C, respectively). The negative control reaction without RFs showed only a faint spot in the same place (Fig. 3, lane D). In addition, we evaluated the amount of synthesized protein by measuring radioactive count. The same counts per mm2 were noted for RF1 WT and RF2 WT samples (8600 and 8700 counts/mm2, respectively), whereas it was somewhat lower for RF2 R213I samples (7700 counts/mm2). The value for the negative control was ∼600 counts/mm2. The experiment was done in duplicates with similar outcomes. Because we did not detect any truncated products of DHFR with either RF2 WT or R213I mutant, we conclude that similar to RF2 WT, the RF2 R213I mutant does not induce a discrete premature peptide release or translation termination on any sense codon within the limits of this assay. It also means that RF2 R213I retains the specificity against the sense codons at the same level as the RF2 WT.
Figure 3.
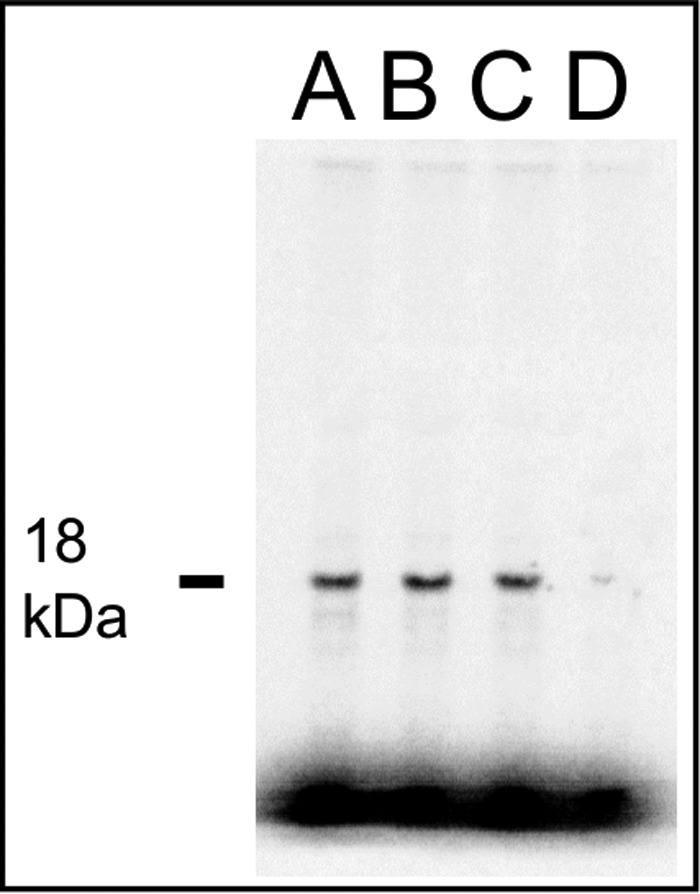
In vitro synthesis of full-length DHFR with RF1 WT (A), RF2 WT (B), RF2 R213I (C), and no RF (D) using a fully reconstituted transcription-translation system containing [35S]Met and individually purified translation components from E. coli. The production of only a single band of 18 kDa corresponding to the full-length DHFR suggests no occurrence of premature termination for any of the RF variants tested here (within the limits of the assay).
Determination of the kinetic parameters of RF2 R213I in peptide release
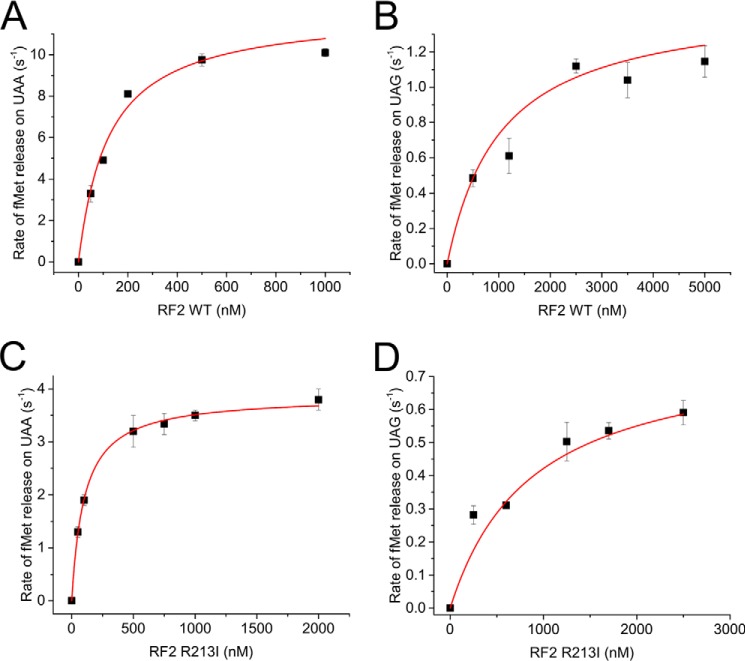
To investigate the kinetic properties of the RF2 R213I, we performed a single turnover fMet (peptide) release assay using fast kinetics (Fig. 4). RCUAA and RCUAG presenting UAA and UAG codons in the A site, respectively, were produced by binding [3H]fMet to AUG-UAA/UAG mRNA-programmed 70S ribosomes. RF variants at different concentrations were rapidly mixed with the RCs in the quench-flow, and the rate of [3H]fMet release was measured by plotting the amount of released [3H]fMet against time. The rates, when plotted against RF concentration, generated hyperbolic curves, from which Michaelis-Menten parameters, namely kcat (the maximal rate of peptide release) and Km (the RF concentration at half-maximal rate), were estimated (Fig. 4).
Figure 4.
Michaelis-Menten kinetics for peptide release by RF2 WT (A and B) and RF2 R213I (C and D) on UAA (A and C)- and UAG-programmed (B and D) RCs. The data were fitted with hyperbolic function, and kcat and Km for each combination were estimated using Michaelis-Menten equation.
At UAA codons, the RF2 WT released [3H]fMet with a kcat of 12.1 ± 1.1 s−1 and a Km of 123 ± 29 nm (Fig. 4A), leading to a translational efficiency (kcat/Km) of 98.2 × 10−6 μm−1 s−1, in good agreement with published results (4, 21). In comparison, RF2 R213I showed a 3-fold smaller kcat (3.8 ± 0.03 s−1) than WT, whereas the Km was similar to the WT (102 ± 3.5 nm); this resulted in a translational efficiency of 37.2 × 10−6 μm−1 s−1. At the non-cognate UAG stop codon, the RF2 WT showed a kcat equal to 1.49 ± 0.16 s−1 and a Km of 1045 ± 342 nm, resulting in kcat/Km values of 1.4 × 10−6 μm−1 s−1, almost 70× less than the UAA codon. For UAG reading by RF2 R213I, kcat was 0.79 ± 0.13 s−1, and Km was 875 ± 282 nm; thus, kcat/Km was 0.9 × 10−6 μm−1 s−1, ∼40 times less than the cognate UAA codon.
When compared with the WT, the translation termination efficiency (kcat/Km) of the RF2 R213I mutant was found to be 2.6× less at the cognate UAA codon but quite similar for the non-cognate UAG codon. Thus, the relatively improved UAG reading by RF2 R213I with respect to UAA in the competition assay (Fig. 2C) is essentially not due to its increased efficiency of UAG reading but, rather, due to its reduced efficiency in UAA reading. We also note that the reduced efficiency of RF2 R213I in UAA reading originated from the 3-fold decrease in the kcat value compared with the WT. However, on non-cognate UAG codon, a 2-fold decrease in the kcat value for RF2 R213I relative to the WT was partially compensated by a decrease in Km, thereby leading to termination efficiency at UAG codon comparable with the WT.
Discussion
During the last decade, it was shown that stop codon recognition by the class I release factors is far more complex than the simple tripeptide anticodon theory (22). The tRNA mimicry hypothesis is certainly valid from the structural viewpoint, as the RFs span from the decoding center to the peptidyl transferase center, like the tRNAs, and the tripeptide motifs PXT in RF1 and SPF in RF2 appear in the similar positions as the tRNA anticodons, but it oversimplifies the mechanism of stop codon recognition. Computational, biochemical, and structural studies unraveled a complex network (including, but not limited to the tripeptide anticodon) of hydrogen bonds between the RFs, the stop codon, and the ribosome, which are now believed to be essential for accurate recognition of the stop codons (5–10, 13, 14, 23). In this context, an early observation that a single point mutation E167K in RF2, acquired at the prfA-deleted background, enables it to read all three stop codons is worth mentioning (15). However, the basis of RF2 E167K to be an omnipotent RF in vivo is not well understood. This is not only because Glu-167 is located far from the decoding center but also because it showed significant loss of its activity for reading the cognate UAA codon. Thus, there could be additional fitness compensatory mutations (within the ribosome or elsewhere) in the RF2 E167K strain, which were not checked or characterized. Thus, in the current study we decided to try the reverse approach by incorporating mutations in the RFs based on structural and computational studies, which would direct us toward rational design of an omnipotent release factor.
The first step of designing a RF that would read both UAG and UGA codons other than UAA codon is either to reduce the discrimination for G3 by RF2 or for G2 by RF1. In the current study we focused mainly on modifying RF2 for improved G3 reading; for that we closely inspected the analysis of the third position reading by Sund et al. (10) using MD simulation based on high-resolution crystal structures of the RFs on the ribosome (7, 9). As mentioned in the introduction, the study by Sund et al. (10) suggested exclusion of a water molecule at the decoding center by the long side chain of Arg-213 as the reason for RF2's inability to read G3. They also suggested that in contrast, the corresponding residue Ile-196 on RF1 would allow inclusion of the water molecule in the same pocket, which would, in turn, facilitate formation of the hydrogen-bond network required for G3 reading by a RF. We hypothesized that replacing Arg-213 with Ile will reduce RF2's discrimination for G3, thereby resulting in improved reading of non-cognate UAG codon by RF2. However, improved G3 reading also brings risk that the modified RF2 would recognize Trp coding UGG codon, which is not desired. Thus, we checked all our RF1 and RF2 mutants for reading UAG, UGA, UGG, UCA, and AAA codons in competition for reading UAA codon.
The first position (U1) reading is shown to be strictly regulated by Gly-120 in RF1 and Gly-137 in RF2 (5, 7). Thus, as expected, none of our RF1 and RF2 variants showed any peptide release activity on AAA codon. The RF1 Ile-196 and Leu-126 mutants showed a varied degree of discrimination for different codons (Table 1). Although RF1 I196A showed increased discrimination against the sense codons, the RF1 I196R mutant showed higher discrimination for UGA. In comparison, RF1 L126D showed loss of discrimination against all non-cognate codons. The exact molecular mechanism behind these behaviors remains unclear. One could speculate that the size of the water pocket is important for the overall accuracy of the RFs. Changing hydrophobic Ile-196 to electropositive Arg with a longer side chain might repel the water molecule and alter the geometry of the pocket. Similarly, replacing Ile-196 with Ala, having a much smaller side chain, might disrupt its interactions with the decoding center, which would affect its accuracy. We also notice that replacing Leu-126 with Asp has a deleterious effect on the overall accuracy of RF1.
The Arg-213 in RF2 is proposed to obstruct a water molecule from entering the decoding center, which is essential for G3 reading (10). In agreement with this theory, when we changed Arg-213 to Ile we saw that in competition with UAA, the discrimination against UAG was been decreased five times. However, as expected from reduced G3 discrimination, RF2 R213I showed 1.5× higher reading of UGG compared with the WT. However, it showed higher discrimination against UCA codon; also, it did not show premature termination in any sense codon when tested in full-length DHFR synthesis (Fig. 3); these results altogether highlight its potential for being an omnipotent RF. When Arg-213 was replaced by Ala, only moderate loss of discrimination against UAG was seen, whereas the degree of discrimination increased for UCA and UGG codons.
Lastly, changing Asp-143 to Leu (as in RF1) resulted in a 3-fold decrease in discrimination for the UAG codon. It is likely that the missing aspartate residue leads to destabilization of Arg-213, which in turn leads to a higher acceptance of G3 reading. However, surprisingly RF2 D143L also showed activity on UCA codons and therefore was not pursued for further studies.
To understand the basis of improved UAG reading by RF2 R213I relative to UAA, we estimated the efficiency of termination by this factor on these two codons. As shown in Fig. 4, compared with the WT, the mutant RF2 R213I showed 3× less efficiency (kcat/Km) in reading cognate UAA codon while maintaining WT-like efficiency in reading the non-cognate UAG codon. This allowed the RF2 R213I mutant to be an improved reader of UAG relative to UAA as compared with RF2 WT. The loss of efficiency in reading UAA codon is primarily due to a 3× smaller maximal rate of peptide release. This observation is similar to the in vivo identified UAG-reading RF2 E167K mutant (15), which showed a 2× lower efficiency for reading UAA. Combining our results with RF2 R213I and the earlier study by Ito et al. (15) with RF2 E167K, we conclude that an improved UAG reading by RF2 seems to be linked with the loss of its efficiency in UAA reading. UAA is the major stop codon in E. coli and many other bacteria, whereas UAG is the universal minor stop codon (24). A reduced efficiency of UAA recognition would allow more RF2 to be available for termination on UAG codons. Additionally, the decrease in the Km value on UAG compared with UAA indicates a higher affinity of RF2 R213I for UAG-programmed release complexes, which would also aid in its comparatively improved UAG reading.
Can RF2 R213I read UAG in vivo? An earlier study with RF2 R213I claimed that it is toxic for overexpression in E. coli and an associated dominant lethal phenotype with it (12). In our hands, however, there was no problem in RF2 R213I overexpression. One difference between these two constructs is that the earlier construct did not have a T246A mutation, without which RF2 is known to be toxic for overexpression in E. coli (18). Thus, the toxicity might have originated from the Thr-246 residue instead of the actual Arg-213 mutation. However, all our attempts to replace a chromosomal copy of prfB gene encoding RF2 WT with plasmid borne RF2 R213I were unsuccessful. Also, deletion of prfA (encoding RF1) gene in E. coli BL21(DE3) led to a spectrum of mutations in the prfB gene and other parts of the bacterial genome.4 Thus, it is unlikely that an RF1 lacking bacterial strain will be able to sustain only with the RF2 R213I mutant.
In conclusion, swapping Arg-213 with Ile is one step forward for designing an omnipotent RF. Based on the current study, the earlier (15), and the unpublished results it can be predicted that several additional mutations will be required for creating a bacterial strain sustaining only with RF2. On a technical point, our study indicates that the competitive peptide release assay can be easily adapted for high-throughput screening of a large number of RF mutants for cognate versus non-cognate codon reading in a time- and cost-effective manner.
Experimental procedures
Reagents
70S ribosomes and tRNAfMet were purified from exponentially growing E. coli MRE600 cells as described earlier (25). mRNAs were transcribed with T7 RNA polymerase after PCR amplification from the hybridized DNA oligonucleotide templates, resulting in the following sequence: 5′-GGG AAU UCG GGC CCU UGU UAA CAA UUA AGG AGG UAU ACU AUG XXX CUG CAG A21-3′ (XXX indicates the codon in the A site, and the Shine-Dalgarno sequence and the start codon AUG in the P site are underlined), which was then purified using poly-dT affinity chromatography (25). RCs containing [3H]fMet-tRNAfMet in the P site and UAA (also [35S]fMet-tRNAfMet and UAG, UGA, UCA, UGG, or AAA in the A site) were prepared using the following mix: 12.5 μm 70S ribosomes, initiation factors (IFs) (5 μm IF1, 2 μm IF2, 3.75 μm IF3), 18.8 μm mRNA, 25 μm [3H]fMet-tRNAfMet (or [35S]fMet-tRNAfMet), 1× HEPES-Polymix buffer (pH 7.5) (20), and 1 mm GTP. The protocol was a minor modification of one published earlier (4, 26). The reaction was incubated for 10 min at 37 °C. The resulting RCs were separated from free [3H]fMet-tRNAfMet (or [35S]fMet-tRNAfMet) and other components by centrifugation over a 30% sucrose cushion (21). RC concentration was measured at A260 and stored in aliquots (10 μl) at −80 °C after snap-freezing in liquid nitrogen.
All RF mutants (listed in Table 1) were created by site-directed mutagenesis following a standard QuikChange protocol (Stratagene). Plasmids pTrc-prfA and pET11a-prfB were used to express RF1 and RF2, respectively, with hexahistidine tag fused at the C terminus. Both the constructs are under control of the lac promotor. In addition, the pET11a-prfB plasmid carried a point mutation T246A that enables an otherwise toxic overexpression of RF2 in E. coli (18). These plasmid constructs were transformed in heat-shock competent E. coli BL21-Gold(DE3) cells (Agilent Technologies) that already contained the plasmid pacDuet 1-prmC expressing HemK methylase, which methylates the essential Gln of the GGQ motif of the class-I RFs 21. RF overexpression was induced by adding isopropyl 1-thio-β-d-galactopyranoside (1 mm) to the log-phase cells, which were grown for a further 4 h. All RF proteins were purified on HisTrap columns (GE Healthcare) using standard protocol, which was followed by extensive dialysis against HEPES-Polymix buffer. Protein concentration was determined using the Bradford assay (27). Pure protein preparations were aliquoted (50 μl), snap-frozen in liquid nitrogen, and stored at −80 °C. Methylation of Gln-235 in RF1 and Gln-252 in RF2 was validated by mass spectrometry, as described previously (28). All other translation factors were purified as described elsewhere (25).
Competitive peptide release assay
All reactions were carried out at 37 °C in HEPES-Polymix buffer (pH 7.5). RC mixes (10 μl) contained 4 pmol of each of [35S]fMet-RCUAA and [3H]fMet-RCXXX (XXX indicates the codon in the A site). RF mixes contained various amounts of RFs (0–40 pmol in HEPES-Polymix buffer). RC and RF mixes were separately preincubated for 1 min at 37 °C; equal volumes of RF mix were then added to an RC mix and incubated at 37 °C for 10 s. Only one round of peptide release proceeds within this timeframe, and the RFs are not recycled (29, 30). The reactions were quenched with 20 μl (same as the reaction volume) of 50% formic acid. The precipitates were pelleted by centrifugation at 20,000 × g for 30 min. [35S]fMet and [3H]fMet released into the supernatant were quantified by scintillation counting (Beckman LS 6500 Scintillation Counter, Beckman Counter). The initial linear phase of peptide release was used to fit the following equation to derive a discrimination factor A described by Bouakaz et al. (19),
| (Eq. 1) |
where Pc is the amount of fMet released after one round of peptide release at a given RF concentration, and Pmax is the total releasable amount of fMet after incubation with saturating amounts of RF1/RF2 (100 pmol) and RF3 (500 pmol). For RCs with sense codons in the A site, Pmax was defined as the maximum amount of dipeptide formed (19). Practically, Pmax was usually in the range of 80% of the total radioactive counts. The residual amount of (∼20%) radioactivity was attributed to inactive RCs that are incapable of peptide release in one round (without RF3 and GTP). The spontaneous release of fMet (background reaction) was measured after the addition of the HEPES-Polymix buffer to the RC mix, and the value was subtracted before data processing. All competition assays were performed at least in duplicates.
Kinetic characterization of RF2 (WT and R213I) in single-turnover peptide release assay
Single turnover peptide release assay was carried out by rapid mixing of equal volumes (16 μl each) of [3H]fMet-RC (10 nm) and a RF stock (50 nm to 5 μm concentrations) and in RQF-3 quench-flow instrument (KinTek). The reaction was quenched with 16% formic acid (final concentration) at various time points. The released fMet was separated from unreacted RCs by centrifugation of the reaction mix at 20,000 × g. The ratio of the counts of the supernatant to the total count was used as a measure of peptide release. Alternatively, it was also estimated by subtracting the fraction of the peptides retained in the RC (count of the pellet/total count) from 1. All experiments were carried out using HEPES-Polymix (pH 7.5) and repeated three times.
In vitro full-length protein synthesis
Full-length protein synthesis experiments were performed as described in detail by Mandava et al. (20) using a fully reconstituted transcription-translation composed of individually purified components from bacteria E. coli and T7 RNA polymerase. The E. coli dhfr gene, encoding DHFR, was used as the DNA template. Briefly, all the required translation factors and T7-RNA polymerase were mixed together in HEPES-Polymix buffer containing energy pump components. The addition of DNA template initiated transcription of DHFR mRNA followed by translation. The synthesized protein incorporated radioactively labeled [35S]methionine included in the reaction mix and was imaged using a personal molecular imager ChemiDoc MP (Bio-Rad).
Author contributions
S. S. conceived the study. G. K. performed the experiments. Both authors analyzed and interpreted the results and wrote the paper.
Acknowledgment
We acknowledge Raymond Fowler for proofreading the manuscript and Santanu Dasgupta for critical comments.
This work was supported by research grants from Swedish Research Council 2016-06264, 2013-8778, and 2014-4423 (to S. S.) and 2008-6593 (Linnaeus grant to Uppsala RNA Research Center) and by the Knut and Alice Wallenberg Foundation to RiboCORE (KAW2011.0081). The authors declare that they have no conflicts of interest with the contents of this article.
X. Ge and S. Sanyal, unpublished results.
- RF
- release factor
- RC
- release complex
- DHFR
- dihydrofolate reductase
- IF
- initiation factor.
References
- 1. Youngman E. M., McDonald M. E., and Green R. (2008) Peptide release on the ribosome: mechanism and implications for translational control. Annu. Rev. Microbiol. 62, 353–373 [DOI] [PubMed] [Google Scholar]
- 2. Scolnick E., Tompkins R., Caskey T., and Nirenberg M. (1968) Release factors differing in specificity for terminator codons. Proc. Natl. Acad. Sci. U.S.A. 61, 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jørgensen F., Adamski F. M., Tate W. P., and Kurland C. G. (1993) Release factor-dependent false stops are infrequent in Escherichia coli. J. Mol. Biol. 230, 41–50 [DOI] [PubMed] [Google Scholar]
- 4. Freistroffer D. V., Kwiatkowski M., Buckingham R. H., and Ehrenberg M. (2000) The accuracy of codon recognition by polypeptide release factors. Proc. Natl. Acad. Sci. U.S.A. 97, 2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Korostelev A., Asahara H., Lancaster L., Laurberg M., Hirschi A., Zhu J., Trakhanov S., Scott W. G., and Noller H. F. (2008) Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl. Acad. Sci. 105, 19684–19689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Korostelev A., Zhu J., Asahara H., and Noller H. F. (2010) Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 29, 2577–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laurberg M., Asahara H., Korostelev A., Zhu J., Trakhanov S., and Noller H. F. (2008) Structural basis for translation termination on the 70S ribosome. Nature 454, 852–857 [DOI] [PubMed] [Google Scholar]
- 8. Petry S., Brodersen D. E., Murphy F. V. 4th, Dunham C. M., Selmer M., Tarry M. J., Kelley A. C., and Ramakrishnan V. (2005) Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123, 1255–1266 [DOI] [PubMed] [Google Scholar]
- 9. Weixlbaumer A., Jin H., Neubauer C., Voorhees R. M., Petry S., Kelley A. C., and Ramakrishnan V. (2008) Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322, 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sund J., Andér M., and Aqvist J. (2010) Principles of stop-codon reading on the ribosome. Nature 465, 947–950 [DOI] [PubMed] [Google Scholar]
- 11. Lind C., Sund J., and Aqvist J. (2013) Codon-reading specificities of mitochondrial release factors and translation termination at non-standard stop codons. Nat. Commun. 4, 2940. [DOI] [PubMed] [Google Scholar]
- 12. Uno M., Ito K., and Nakamura Y. (2002) Polypeptide release at sense and non-cognate stop codons by localized charge-exchange alterations in translational release factors. Proc. Natl. Acad. Sci. U.S.A. 99, 1819–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Field A., Hetrick B., Mathew M., and Joseph S. (2010) Histidine 197 in release factor 1 is essential for A Site binding and peptide release. Biochemistry 49, 9385–9390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trappl K., Mathew M. A., and Joseph S. (2014) Thermodynamic and kinetic insights into stop codon recognition by Release Factor 1. PLoS ONE 9, e94058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ito K., Uno M., and Nakamura Y. (1998) Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. Proc. Natl. Acad. Sci. U.S.A. 95, 8165–8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ito K., Ebihara K., Uno M., and Nakamura Y. (1996) Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA-protein mimicry hypothesis. Proc. Natl. Acad. Sci. U.S.A. 93, 5443–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dinçbas-Renqvist V., Engström A., Mora L., Heurgué-Hamard V., Buckingham R., and Ehrenberg M. (2000) A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 19, 6900–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Uno M., Ito K., and Nakamura Y. (1996) Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie 78, 935–943 [DOI] [PubMed] [Google Scholar]
- 19. Bouakaz L., Bouakaz E., Murgola E. J., Ehrenberg M., and Sanyal S. (2006) The role of ribosomal protein L11 in class I release factor-mediated translation termination and translational accuracy. J. Biol. Chem. 281, 4548–4556 [DOI] [PubMed] [Google Scholar]
- 20. Mandava C. S., Peisker K., Ederth J., Kumar R., Ge X., Szaflarski W., and Sanyal S. (2012) Bacterial ribosome requires multiple L12 dimers for efficient initiation and elongation of protein synthesis involving IF2 and EF-G. Nucleic Acids Res. 40, 2054–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Indrisiunaite G., Pavlov M. Y., Heurgué-Hamard V., and Ehrenberg M. (2015) On the pH dependence of class-1 RF-dependent termination of mRNA translation. J. Mol. Biol. 427, 1848–1860 [DOI] [PubMed] [Google Scholar]
- 22. Ito K., Uno M., and Nakamura Y. (2000) A tripeptide “anticodon” deciphers stop codons in messenger RNA. Nature 403, 680–684 [DOI] [PubMed] [Google Scholar]
- 23. He S. L., and Green R. (2010) Visualization of codon-dependent conformational rearrangements during translation termination. Nat. Struct. Mol. Biol. 17, 465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Korkmaz G., Holm M., Wiens T., and Sanyal S. (2014) Comprehensive analysis of stop codon usage in bacteria and its correlation with release factor abundance. J. Biol. Chem. 289, 30334–30342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johansson M., Bouakaz E., Lovmar M., and Ehrenberg M. (2008) The kinetics of ribosomal peptidyl transfer revisited. Mol. Cell 30, 589–598 [DOI] [PubMed] [Google Scholar]
- 26. Freistroffer D. V., Pavlov M. Y., MacDougall J., Buckingham R. H., and Ehrenberg M. (1997) Release factor RF3 in E. coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 16, 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 28. Heurgué-Hamard V., Champ S., Engström A., Ehrenberg M., and Buckingham R. H. (2002) The hemK gene in Escherichia coli encodes the N5-glutamine methyltransferase that modifies peptide release factors. EMBO J. 21, 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao H., Zhou Z., Rawat U., Huang C., Bouakaz L., Wang C., Cheng Z., Liu Y., Zavialov A., Gursky R., Sanyal S., Ehrenberg M., Frank J., and Song H. (2007) RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell 129, 929–941 [DOI] [PubMed] [Google Scholar]
- 30. Pallesen J., Hashem Y., Korkmaz G., Koripella R. K., Huang C., Ehrenberg M., Sanyal S., and Frank J. (2013) Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1. eLife 2, e00411. [DOI] [PMC free article] [PubMed] [Google Scholar]