Abstract
Systemic lupus erythematosus is an autoimmune disease that can affect numerous tissues and is characterized by the production of nuclear antigen-directed autoantibodies (e.g., anti-dsDNA). Using a combination of virtual and ELISA-based screens, we made the intriguing discovery that several HIV-protease inhibitors can function as decoy antigens to specifically inhibit the binding of anti-dsDNA antibodies to target antigens such as dsDNA and pentapeptide DWEYS. Computational modeling revealed that HIV-protease inhibitors comprised structural features present in DWEYS and predicted that analogues containing more flexible backbones would possess preferred binding characteristics. To address this, we reduced the internal amide backbone to improve flexibility, producing new small-molecule decoy antigens, which neutralize anti-dsDNA antibodies in vitro, in situ, and in vivo. Pharmacokinetic and SLE model studies demonstrated that peptidomimetic FISLE-412,1 a reduced HIV protease inhibitor analogue, was well-tolerated, altered serum reactivity to DWEYS, reduced glomeruli IgG deposition, preserved kidney histology, and delayed SLE onset in NZB/W F1 mice.
Graphical abstract
INTRODUCTION
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease that disproportionately affects young women. As the disease progresses, SLE adversely impacts essentially every organ system (e.g., skin, joints, kidneys, central nervous system (CNS), hematopoietic cells). The underlying features of the disease include the development of autoantibodies, including reactivity to double stranded (ds) DNA. Anti-dsDNA antibodies are of particular importance in lupus because (i) they are diagnostic of the disease, (ii) they contribute to tissue damage in the kidney and other organs, and (iii) their titers correlate with disease activity.2 Ultimately, complexes of DNA and anti-DNA antibody are internalized through Fc receptors (FcR) or carry DNA to Toll-like receptor (TLR) 9 to activate myeloid cells and other FcR and TLR9 expressing cells. A subset of these anti-dsDNA antibodies also binds to a peptide consensus sequence (D/E,W,D/E,Y,S/G)3,4 (herein shortened to “DWEYS”), found in the N-methyl d-aspartate receptor (NMDAR) expressed on neurons,5,6 and the first component of complement (C1q).7 Both a mouse and human monoclonal antibody with this specificity, termed R4A8 and G11, respectively bind to kidney and neuronal epitopes including DNA and NMDAR.1,7,9,10 Since up to 80% of SLE patients manifest cognitive dysfunction as part of neuropsychiatric lupus and renal disease occurs in approximately one-half of all SLE patients, accounting for one-third of all lupus deaths, the deposition of pathogenic antibodies in brain and kidney is of great interest.
Current lupus therapy is aimed primarily at reducing general inflammation (synthetic glucocorticoids), targeting inflammatory mediators by elimination of autoreactive lymphocytes (cytotoxic agents) or nonspecific B-cell inhibitors (belimumbab).11,12 While effective at reducing inflammation, these chronic treatments have undesirable side effects and reduce overall immunity (i.e., immunosuppression), leading to increased risk for infections and cancer.13–15 Thus, novel therapeutic approaches to lupus are greatly needed. Toward this end, we began a program to target specific autoantibody populations with small-molecule “decoy antigen” compounds, thereby rendering them unavailable to bind tissue antigen. We reasoned that this therapeutic approach should have fewer adverse side effects and would lead to a reduction in associated lupus pathology, particularly in the kidney and brain.
RESULTS
HIV Protease Inhibitors and Derivatives as Anti-dsDNA Antibody Decoy Antigens
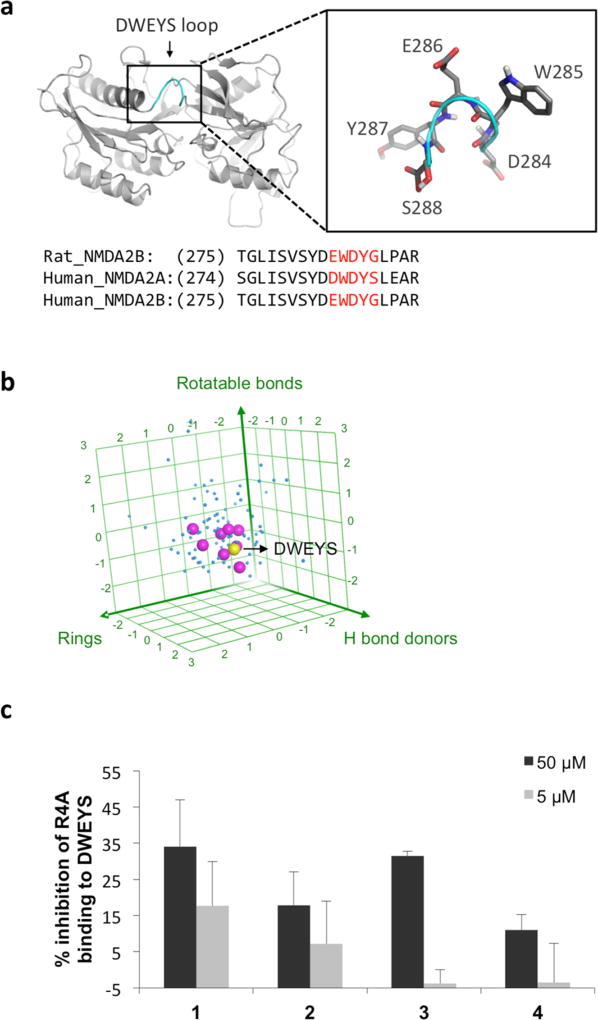
The DWEYS peptide consensus sequence, which binds the R4A autoantibody, is present in a loop on the NMDAR. We derived a consensus peptide structure for DWEYS (Figure 1a) by overlaying the corresponding residues in an experimental structure of NMDAR2B (rat, PDB code 3JPY16) and two homology models of human NMDAR2A and NMDAR2B (available in Modbase;17 see methods in Experimental Section). To identify compounds that might function as experimental decoy antigens, we carried out a DWEYS fingerprint similarity search against the DrugBank database (version 4.2),18 which comprises 5055 drug entries, including FDA approved drugs, nutraceuticals, and experimental compounds. Three fingerprint types were applied in the search, including MACCS structural keys (BIT_-MACCS), 3D shape (ESshape), and pharmacophore atom triangle (piDAPH3) fingerprints to examine different aspects of the consensus structure.19 The three-step screening gave 100 hits out of 5055 entries from the entire database (2%). Interestingly, 8 out of 100 hits belong to the HIV protease inhibitors catalog, including saquinavir20 (1), nelfinavir21 (2), lopinavir22(3), indinavir23(4), and related compounds. The molecular descriptors of the resulting 100 hits of our fingerprint search are displayed in a 3D plot for hydrogen donors, rotatable bonds, and ring numbers (Figure 1b). Our analysis suggested this subset of HIV protease inhibitors had features similar to the DWEYS peptide consensus structure. To assess whether these HIV protease inhibitors might function as anti-dsDNA antibody decoy antigens, we used a competitive ELISA to ask whether they would inhibit the binding of the R4A monoclonal antibody to DWEYS-coated plates1 and found several HIV protease inhibitors exhibited mild inhibition in the assay (Figure 1c).
Figure 1.
HIV protease inhibitors possess structural similarity to DWEYS consensus peptide and inhibit lupus anti-dsDNA antibodies. (a) DWEYS loop region (highlighted in enlargement box) in the NMDA receptor (NR2A) structural model derived from consensus sequences of known rat and human NMDA receptor structures. (b) 3D plot of hydrogen bond donors, rotatable bonds, and ring numbers of the top 100 hits from fingerprint similarity search against DWEYS. Note the clustering of HIV protease inhibitors (magenta dots) around the DWEYS peptide (yellow dot) relative to other top hits (blue dots). Axes were plotted with 3 standard deviations in both directions from the data mean. (c) Select HIV protease inhibitors reduce R4A monoclonal antibody binding to DWEYS in competitive ELISAs.
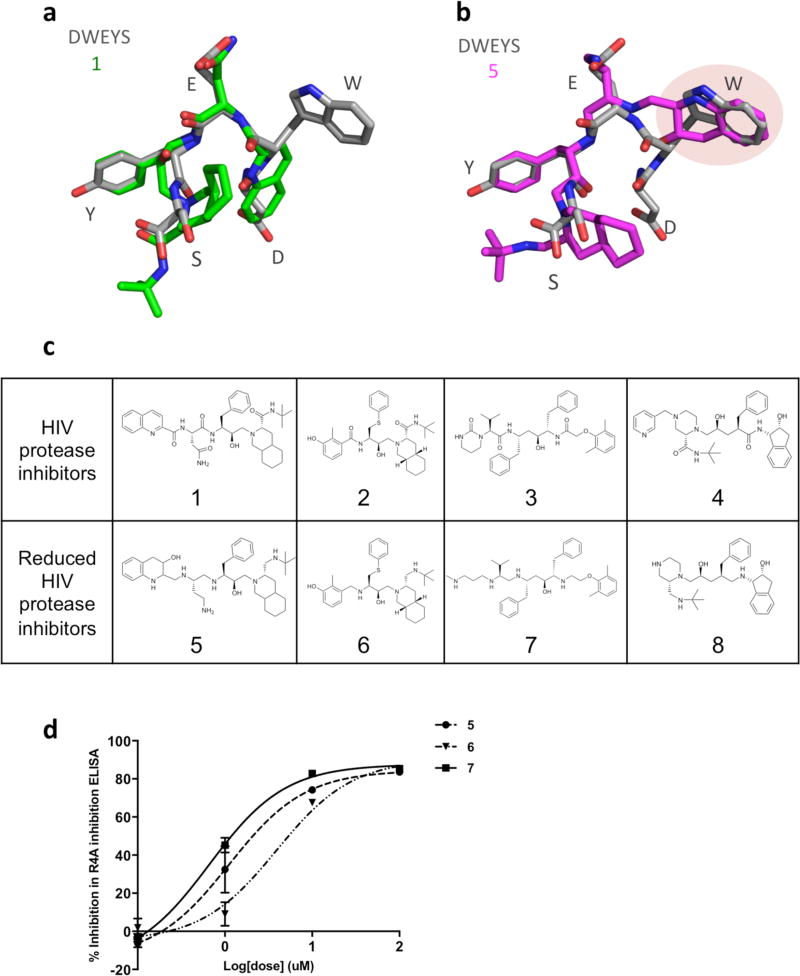
With an eye toward elucidating even higher affinity decoy antigens, we performed computational and structural activity analyses of the top three HIV protease inhibitors (1, 2, and 3) that yielded promising results in the ELISA assay (Figure 1c). Significantly, in these modeling analyses we could draw on a rich database of HIV protease inhibitor structures, since these drugs have been cocrystallized with multiple substrate proteins. We derived consensus structures for the HIV protease inhibitors, starting first with compound 1. Five distinct conformations of 1 were identified from cocrystal structures; these were modeled using MOE flexible alignment module24 (representative ligand conformations in PDB entries 1C6Z, 1FB7, 3PWW, 3S56, and 3UFN) and found to have good 3D structural overlap with the DWEYS consensus structural model (Figure 2a). Note that the hairpin-like conformation of 1 mimics the turn structure of the DWEYS peptide, with significant structural conservation between the backbones of the WEY sequence, and overlap of 1 hydrogen bonding and aromatic moiety of the glutamic acid (E) and tyrosine (Y) side chains (Figure 2a). However, due to the constrained amide bonds on 1, the decahydroisoquinoline (DIQ) group and the quinoline ring were not able to fit well with the peptide side chain and mimic the peptide conformation. Like compound 1, the other two HIV protease inhibitors, 2 and 3, were also found to mimic the DWEYS consensus loop structural model and have a structurally constrained internal amide backbone (Supporting Information, Figure S1). On the basis of these analyses, we reasoned that HIV protease inhibitors with reduced internal carbonyl groups would manifest increased structural flexibility and likely achieve better binding to the R4A autoantibody.
Figure 2.
HIV protease inhibitors and their derivatives possess 3D structural similarities with DWEYS and inhibit SLE anti-dsDNA antibodies. Structural overlay models of DWEYS (gray) with (a) 1 (green) and (b) 5 (magenta). Hydrogens are not displayed. (c) 2D Structures of HIV protease inhibitors and reduced derivatives. (d) IC50 curves of HIV protease inhibitor derivatives as assessed in ELISA-based screens with R4A monoclonal antibody and DWEYS-coated plates.
To test this prediction, we chemically reduced the backbone of the HIV protease inhibitors to increase the flexibility. Of the chemically modified HIV protease inhibitors, some were triaged due to stability issues. The newly formed compounds 5, 6, and 7, described as reduced HIV protease inhibitors (Figure 2c), were purified and characterized by mass spectrometry and nuclear magnetic resonance (NMR) spectra. Their purities were analyzed by HPLC and are above 95% (compound 7, >92%) (Experimental Section, 2. Chemistry, Table 1). Subsequently, we examined the structural overlay of newly formed compounds 5, 6, and 7 with the DWEYS consensus. The conformations of compounds 5, 6, 7 were constructed by modifying their parent HIV protease inhibitors’ protein database (PDB) structures and optimizing their geometry by energy minimization in the MMFF94× force field using the MOE software default parameters.25 Flexible alignments of 5, 6, and 7 to the DWEYS consensus peptide yield better fits compared to their parent HIV protease inhibitors. The reduction of the amide bonds introduced more rotatable bonds to compounds 5, 6, 7 and thus more flexibility. The resulting flexible alignment confirmed a higher degree of similarity of 5 to DWEYS as compared to the rigid compound 1, as evidenced by a better fit of the tetrahydroquinaldic moiety to the W side chain within the peptide (Figure 2b). In addition, the flexible backbone of 6 (reduced form of 2) had better hairpin mimicking conformation and facilitated side chain similarity alignment with the peptide D side chain compared to 2 (Supporting Information, Figure S1a,b). The borane reduction opened the 2-tetrahydropyrimidinone ring on 7; hence, the conformation similarity of 7 and DWEYS is lower compared to 3 (Supporting Information, Figure S1c,d). Overall, the average strain energy (U) of compounds 5, 6, and 7 decreased and the grand alignment score (S) improved (except for compound 7) in their alignment with DWEYS (Supporting Information, Table S1). Interestingly, the R4A autoantibody is known to bind dsDNA, the NMDAR receptor (containing the consensus sequence D/EWD/EYS/G),5 and the complement protein C1q (EADSV).7 It has been suggested that more cross-reactivity could be the result of increased flexibility in the binding regions of antibodies and/or their ligands.26–29 Taken together, these results strongly argue that reducing the structurally constrained HIV protease inhibitors’ internal backbone could produce decoy antigen therapeutics with preferred characteristics.
Table 1.
Structures and Analytical Data for Reduced HIV Protease Inhibitor Compounds
| Compounds | Structures |
1H/13C NMR |
MS [M+H]+ |
HPLC (purity) |
|---|---|---|---|---|
| 5 |
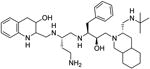
|
+ | 635.53 | 98% |
| 6 |
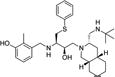
|
+ | 540.4 | 97.6% |
| 7 |
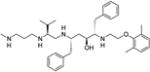
|
+ | 589.33 | 91.6% |
| 8 |
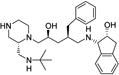
|
+ | 495.53 | 96.5% |
Previously, we have shown that compound 5, the reduced form of 1, inhibited autoantibody binding in vitro (ELISA), ex vivo (kidney glomeruli), and in vivo (hippocampal neurotoxicity).1 When assayed in parallel, compounds 5, 6 (reduced compound 3), and 7 (reduced compound 2) inhibited the ability of the anti-dsDNA monoclonal antibody R4A to bind DWEYS peptide in a dose-dependent manner (IC50 values of 4.5 ± 1.6 µM, 7.27 ± 0.5 µM, and 2.08 ± 0.8 µM, respectively) (Figure 2d). Reduced compound 4 (8) also showed moderate activity with an IC50 of 42.7 ± 19.7 µM. These results largely validate the computer modeling and hypothesis that introduction of a more flexible backbone into HIV protease inhibitors represents a rich genus of new scaffolds for use as potential lupus therapeutics.
Kidney Glomeruli R4A Binding Assay
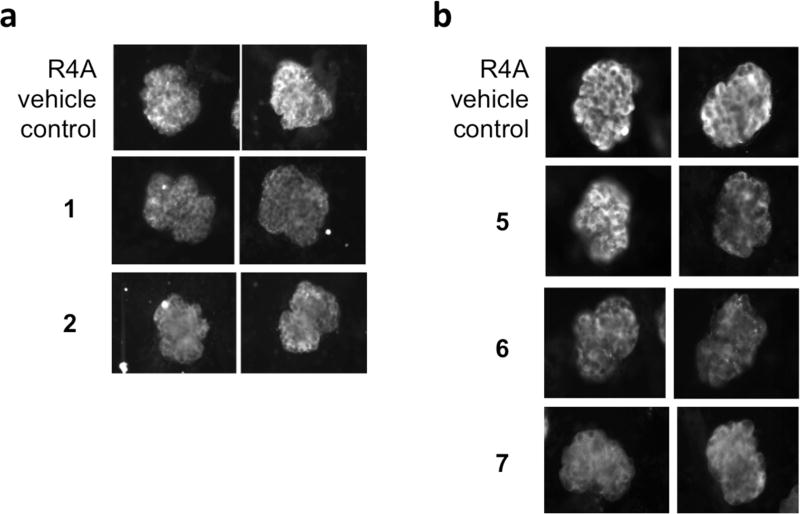
We next wished to test both unmodified (parent HIV protease inhibitors) and chemically reduced derivatives 5, 6, 7 in a more physiologically relevant setting, namely, the isolated kidney glomerulus. In this assay we incubated the R4A antibody with mouse glomeruli in the presence or absence of compound and assessed binding with fluorescently conjugated secondary antibodies. In agreement with previous work,1 R4A bound to glomeruli (Figure 3). In marked contrast, R4A pretreated with either 1 or 2 showed diminished binding to glomerular antigens (Figure 3a). Qualitatively similar results were observed when R4A was pretreated with 5, 6, or 7 (Figure 3b): each of these chemically reduced HIV protease inhibitor derivatives decreased the binding of R4A to kidney tissue. Since renal disease occurs in approximately one-half of all SLE patients and accounts for one-third of all lupus deaths, a compound that decreases the deposition of pathogenic antibodies in the kidney is a potentially valuable therapeutic.
Figure 3.
HIV protease inhibitors and derivatives suppress pathogenic SLE antibody deposition in the kidney ex vivo. Two representative images of murine glomerular binding of R4A antibody (20 µg/mL) preincubated with (a) HIV protease inhibitors (1 or 2) or (b) reduced derivatives (5, 6, and 7) at 50 µM.
Neurotoxicity Model
It is known that R4A or NMDAR-binding antibodies can elicit excitotoxic death of neurons in the mouse hippocampus and that this, in turn, contributes to cognitive impairment and memory loss.30–32 Accordingly, we next assayed whether the reduced HIV protease inhibitors would block the neurotoxicity of R4A in vivo. In this assay, R4A antibodies were stereotactically injected into the dorsal hippocampus of a living mouse in the presence or absence of compound or vehicle. One day later, the animal was sacrificed and apoptosis was revealed by TUNEL staining of the brain at the injection site. As expected,1,5,30,33 R4A alone caused neuronal apoptosis as indicated by positive TUNEL staining (Figure 4, left). In contrast, pretreatment with 5 and 6 abrogated this neurotoxic effect, while 7 did not, possibly due to lower solubility (Figure 4).
Figure 4.
HIV protease inhibitor derivatives (5, 6, 7) block neurotoxicity of R4A antibodies in vivo. CA1 region of the hippocampus is injected with R4A antibody (20 µg/mL) preincubated with vehicle or the indicated compound (50 µM). Methylene green stains living neuron cell bodies, and neurotoxicity is indicated by TUNEL positive staining (brown). Photomicrographs are at 20×.
Taken together, these glomerular ex vivo and brain in vivo experiments demonstrate that parent (unmodified) and chemically reduced HIV protease inhibitors can function as decoy antigens to protect the kidney and brain from the pathogenic effects of anti-dsDNA antibodies.
Preliminary Pharmacokinetic Studies
Having made significant investments in other small molecule therapeutic programs only to reach an impasse due to toxicology and/or pharmacokinetic (PK) issues, we next sought to use these criteria to assist in deciding which drug candidate to test in more rigorous in vivo disease models. All of the top three leads advanced through pilot toxicology studies, with no obvious behavioral changes or tissue abnormalities following a week of daily dosing. We next determined the pharmacokinetics of each compound after either intravenous (iv) or oral (gavage) administration at 1 mg/kg and 10 mg/kg, respectively (Supporting Information, Figure S2). In both cases, 5 exhibited the best PK profile and was well tolerated at the administered dose levels. On the basis of these criteria, we began testing 5 in the NZB/W F1 lupus mouse model.
NZB/W F1 Lupus Model: Efficacy Testing
NZB/W F1 mice spontaneously develop lupus-like phenotypes, including elevated serum anti-dsDNA autoantibodies and immunemediated glomerulonephritis that leads to kidney failure as the animals age. Like the human situation, disease in this strain is strongly biased toward females. In our study, we began dosing female NZB/W F1 mice with either vehicle or compound 5 (5 mg kg−1 day−1 in two daily ip injections) at 22 weeks of age, prior to onset of severe proteinuria. Body weights, blood, and urine were taken weekly. The blood was used to assess anti-dsDNA reactivity and urine used to assess proteinuria. Humane end points included ≥3+ proteinuria on two consecutive weeks, loss of ≥20% body weight, or prostration. Terminal blood and kidney tissues were taken for subsequent analysis.
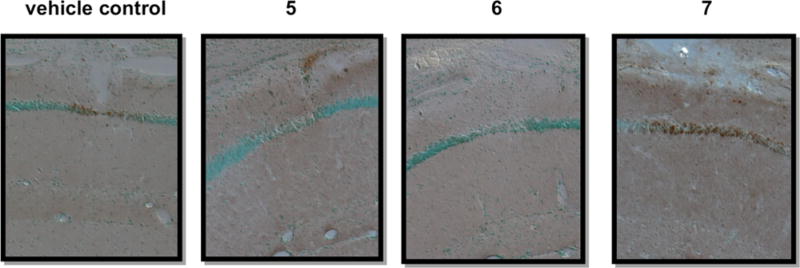
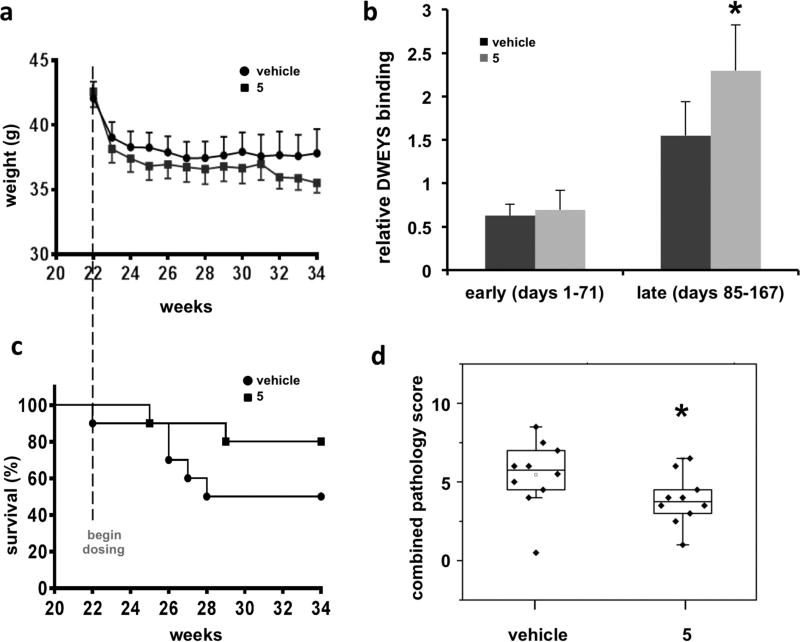
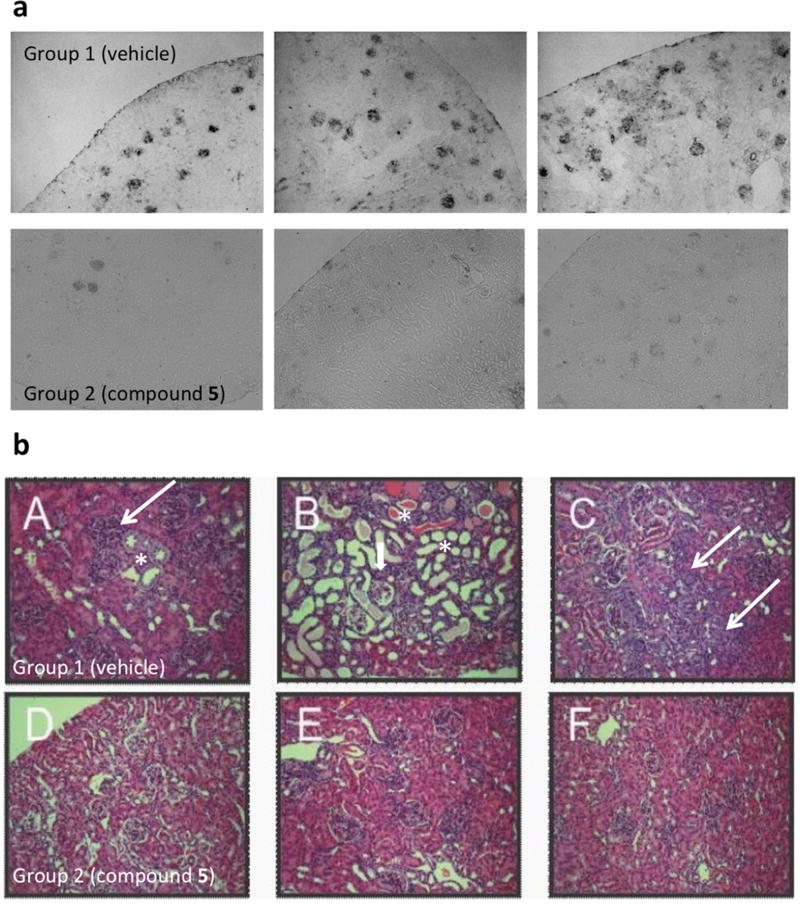
Body weights throughout the study were similar between the vehicle and 5-treated groups (Figure 5a), and no 5-treated animals had to be removed due to weight loss or prostration, suggesting that the decoy antigen was well tolerated. 5-treated mice showed delayed onset of disease (26–36 weeks) and a trend toward increased disease-free survival (Figure 5c), although it did not reach statistical significance (perhaps owing to small cohorts). Strikingly, the kidneys from 5-treated mice had significantly less staining for mouse IgG (autoantibody deposition) in the glomeruli and exhibited preserved histology compared to vehicle-treated mice (Figure 6). As displayed in representative sections from mice at 60 weeks, lymphocyte aggregates (thin arrows), tubule dilation and casts (*), and glomerulonephropathy (thick arrows) are prominent in vehicle-treated mice while minimal in 5-treated mice (Figure 6b). Conglomerate blinded analyses of kidney histopathology scores (60 weeks) resulted in significantly lower standard combined pathology scores in 5-treated mice (Figure 5d). This combined pathology score encompassed four criteria (glomerular sclerosis, glomerular cellularity, crescents, and interstitial inflammation). Of these criteria, the 5-treated animals showed improvement in all, suggesting that their deaths may not be due to impaired renal function. Finally, sera from 5-treated animals manifest higher anti-DWEYS reactivity relative to sera from vehicle-treated mice (Figure 5b), presumably due to reduced IgG deposition in target organs of 5-treated mice. Taken together, these results demonstrate that 5 represents a novel class of potential therapeutics that results in a trend toward increased disease-free survival and preserved kidney histology in the NZB/W F1 mouse model of SLE. The hypothesized decoy antigen mechanism of action was supported by the reduction of immune complex deposition and downstream pathology in kidneys from 5-treated mice compared to vehicle-treated mice.
Figure 5.
Compound 5 is well-tolerated, alters serum anti-dsDNA reactivity, delays SLE onset, and significantly reduces combined kidney pathology scores. (a) Daily body weight measurement in chronically dosed (compound 5 (5 mg/kg) or vehicle) NZB/W F1 mice. (b) Serum DWEYS reactivity in chronically dosed (5 mg/kg) NZB/W F1 mice. Note that late in the dosing (days 85–167) 5 treatment increased serum anti-dsDNA reactivity compared with vehicle treatment (p < 0.05). (c) Disease-free survival in compound 5 (5 mg kg−1 day−1 b.i.d., ip) or vehicle-treated NZB/W F1 mice (initially n = 10/group). (d) Combined kidney pathology scores in 5-treated NZB/W F1 mice. Horizontal line indicates median value, Z = 1.97 (p < 0.05).
Figure 6.
Compound 5 reduces glomeruli IgG deposition and preserves kidney histopathology in NZB/W F1 mice. (a) IgG deposition in kidneys from vehicle versus compound 5 chronically dosed NZB/W F1 mice. Formalin-fixed, paraffin-embedded sections from three representative mice per treatment group were stained for mouse IgG. (b) Kidney histopathology in vehicle or compound 5 chronically dosed mice. Formalin-fixed, paraffin-embedded sections from three representative mice per treatment group (group 1, vehicle, panels A–C; group 2, compound 5, 5 mg/kg, panels D–F) were stained with H&E.
DISCUSSION
In this manuscript we report a fingerprint similarity search of existing compounds/drugs (either FDA approved or experimental) with structural features common to the cross-reactive antigen peptide DWEYS3 that resulted in 100 hits, 8 of which belonged to the HIV protease inhibitor family. To further refine our “decoy antigens”, we employed computational modeling to investigate aspects of the HIV protease inhibitor compounds which could be modified to improve the three-dimensional fit to DWEYS. We discovered that reducing the structurally constrained HIV protease inhibitors’ internal backbone could improve their flexibility and enhance the fitting profile to DWEYS. Using an ELISA-based screen, we confirmed that many HIV protease inhibitors, including 1, 2, and 3, dose-dependently inhibited the binding of anti-dsDNA antibodies to target antigens. Next, we chemically reduced these HIV protease inhibitors and identified several more potent derivatives that could neutralize anti-dsDNA antibodies in vitro, in situ, and in vivo. Among these, one small molecule (5) previously described1 was well-tolerated in vivo and delayed SLE disease progression in NZB/W F1 mice. At necropsy, 5-treated animals displayed significantly less staining for mouse IgG (autoantibody deposition) in the glomeruli and preserved kidney histology compared to vehicle-treated mice.
Using a small molecule compound (MW under <700 Da) to target an autoimmune antibody is novel, and the strategy of targeting an anti-DNA antibody with a larger drug complex has only been tested once before to our knowledge. Abetimus (Abetimus sodium, LJP 394, Rentol, Riquent) is a tetrameric set of oligodeoxyribonucleotides attached to a proprietary nonimmunogenic PEG carrier (approximately 54 000 Da) that was tested in several clinical trials. It was found to lower anti-DNA Ab levels and reduce anti-DNA B-cell clones in animal studies, the latter causing it to be termed a “tolerogen”. Initial human trials were promising and advanced to the phase III level (LJP394-90-09 and LJP394-90-14). The last phase III trial was halted after it was determined that significance differences were not going to be achieved between placebo and drug-treated groups (results of this study have not been published to our knowledge). Poststudy analyses of the published trials led to the conclusions that there were significantly fewer renal flares, and time to renal flare in the drug-treated group was half that of placebo when a responding subset of patients (patients with high affinity DNA binding antibodies) were compared with the placebo group.34 There has been criticism of patient inclusion criteria, end points measured, and dosage of drug used in the study that was halted.35 Furthermore, the oligonucleotides present in the abetimus conjugate were not tested for TLR9 binding. It is possible that the immune complexes formed by anti-DNA antibody and abetimus were, in fact, proinflammatory, thereby exacerbating lupus symptoms. In contrast, compound 5 is monovalent and will not form immune complexes and is a mimetope of a peptide that does not bind TLR9 (unpublished data). Our data suggest that blocking anti-dsDNA antibody from binding tissue or oligonucleotides that constitute TLR9 ligands may still be a valid therapeutic approach in lupus but requires carefully planned testing. As our kidney evidence in a lupus model suggests, the use of small molecule agent 5, rather than a large drug-conjugate, to neutralize autoreactive antibodies may allow for less tissue binding and generate fewer side effects than the drug-complex used in the aforementioned study.
In summary, our investigation has led us to the discovery that clinically approved HIV drugs may be themselves repurposed, or modified to be made more flexible, to target autoantibodies prevalent in SLE. These compounds appear to function as decoy antigens, reducing IgG deposition and associated pathology in the kidney, although not neutralizing all contributors to early death in SLE. Our approach of increasing flexibility of structurally relevant drugs/compounds to increase binding affinity to pathogenic autoantibodies could serve as a valuable tool in structure–activity analyses going forward. The fact that our starting compounds have a proven safety profile in HIV patients who, like SLE patients, may be immune-compromised is a promising back-drop for the reductively modified offspring which we have termed “reduced HIV protease inhibitors”. We are continuing tests of our lead compounds for safety parameters and oral dosing using mouse models of lupus.
EXPERIMENTAL SECTION
1. General
Unless otherwise indicated chemicals and reagents were purchased from Sigma-Aldrich. D-DWEYS was acquired from Keck Biotechnology Resource Laboratory (New Haven, CT) and was greater than 99% pure as indicated by HPLC and MS (Supporting Information, page S9). Saquinavir, nelfinavir, and other HIV protease inhibitors were obtained from the pharmacy of North Shore University Hospital (Manhasset, NY), extracted from capsules, purified by flash chromatography, and confirmed by mass spectrometry (MS). Animals were housed under standard conditions and were assigned to groups by random selection upon arrival. All procedures were in accordance with the IACUC of The Feinstein Institute. Excel or Prism was used for graphing and statistical analyses. Results are reported as mean values with standard deviations unless otherwise indicated. Student’s t tests were used to distinguish significance with the criteria of p < 0.05 and are indicated in figure by an asterisk (*) symbol.
2. Chemistry
Chemicals were obtained from commercial suppliers and used without further purification unless otherwise indicated. All 1H NMR spectra were recorded on a Bruker Avance DRX 500 and 600 MHz. The 13C NMR spectra were recorded at 125 MHz. Chemical shifts are relative to the deuterated solvent peak and are in ppm. The coupling constants (J) are measured in Hz. The 1H signals are described as s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet), or br s (broad singlet). Low- and high-resolution mass spectrometry was carried out at the LCQ DECA XP MS. HPLC was performed using Waters system combining a 600 PUMP and a 996 PAD UV detector. The analytical column was a Phenomenex Luna C18 100 Å 250 mm × 4.6 mm.
General Procedure for the Reduction of Selected HIV Protease Inhibitors (HIVPIs)
To the solution of 100 mg of HIVPI in 20 mL of dry THF was added 20 mL of 1 M BH3 in THF. The solution was stirred at 65 °C under N2 overnight. The reaction was quenched by adding MeOH slowly, and then the solvent was removed and the crude residue was dried under vacuum pump to yield white solid, which was dissolved in 1 mL of MeOH and diluted with 10 mL of piperidine. The solution was stirred at 65 °C under N2 for another 1– 2 days. The solvent was concentrated, and the residue was dissolved in methanol, then using ether to precipitate the crude product which was purified by HPLC for the animal studies and characterizations. Information regarding the analytical data of the analogues is provided in Table 1 and Supporting Information, sections I and II.
Compound Characterization NMR Data. 2-{[3-Amino-1-({1-benzyl-3-[3-(tert-butylamino-methyl)-octahydro-isoquinolin-2-yl]-2-hydroxy-propylamino}-methyl)-propylamino]-methyl}-1,2,3,4-tetrahydro-quinolin-3-ol (5)
FISLE-412 (5) was published in 2011.1
3-({3-[3-(tert-Butylaminomethyl)octahydroisoquinolin-2-yl]-2-hydroxy-1-phenylsulfanylmethylpropylamino}methyl)-2-methylphenol (6)
White foam; MS (ESI) m/z 540.4 [M + H]+; 1H NMR (500 MHz, (CD3)2CO) δ 1.08 (s, 9H, C(CH3)3), 1.14–1.21 (m, 3H), 1.31–1.37 (m, 2H), 1.48–1.51 (m, 3H), 1.61–1.86 (m, 4H), 2.22 (s, 3H), 2.24 (m, 1H), 2.32 (dd, J = 3.4, 11.65 Hz, 1H), 2.46 (dd, J = 6.4, 13.7 Hz, 1H), 2.66 (dd, J = 3.9, 12 Hz, 1H), 2.73–2.78 (m, 2H), 2.87–2.90 (m, 1H), 3.04 (dd, J = 5.7, 13.7 Hz, 1H), 3.18 (dd, J = 7.2, 13.2 Hz, 1H), 3.56 (dd, J = 4.3, 13.2 Hz, 1H), 3.73 (dd, J = 12.3, 29.9 Hz, 2H), 3.87 (dd, J = 5.75, 11.5 Hz, 1H), 6.74 (dd, J = 7.95, 13.85 Hz, 2H), 6.91 (t, J = 7.75 Hz, 1H), 7.16–7.19 (m, 1H), 7.27–7.31 (m, 2H), 7.42–7.44 (m, 2H); 13C NMR (125 MHz, (CD3)2CO) δ 11.51, 21.48, 26.65, 27.35, 29.02, 32.12,32.23, 35.07, 35.56, 37.76, 46.96, 50.63, 50.90, 56.87, 60.58, 61.64, 64.04, 69.28, 114.46, 121.50, 123.94, 126.41, 126.60, 129.74, 129.89, 138.54, 141.15, 156.24
2-[2-(2,6-Dimethylphenoxy)ethylamino]-5-[3-methyl-2-(3-methylaminopropylamino)butylamino]-1,6-diphenylhexan-3-ol (7)
Colorless oil; MS (ESI) m/z 589.33 [M + H]+; 1H NMR (500 MHz, (pyridine-d5) δ 0.82 (d, 3H, J = 6.9 Hz), 0.87 (d, 3H, J = 6.9 Hz), 1.93 (m, 1H), 2.03–2.09 (m, 3H), 2.24 (s, 6H), 2.49 (s, 3H), 2.62–2.69 (m, 1H), 2.75–2.77 (m, 2H), 2.95–3.08 (m, 3H), 3.16 (t, 1H, J = 12 Hz), 3.28–3.32 (m, 3H), 3.35–3.45 (m, 3H), 3.62–3.64 (m, 1H), 3.73–3.77 (m, 1H), 3.84 (m, 1H), 4.04 (m, 1H), 4.13–4.17 (m, 1H), 4.26–4.31 (m, 1H), 4.36–4.39 (m, 1H), 6.93–7.01 (m, 2H), 7.16–7.29 (m, 9H), 7.4 (m, 2H). 13C NMR (125 MHz, (pyridine-d5) δ 16.83, 17.17, 19.78, 26.72, 28.8, 33.40, 35.17, 35.50, 38.93, 41.5, 44.18, 44.74, 46.96, 48.11, 59.65, 61.33, 65.22, 68.99, 70.04, 125.24, 127.77, 127.82, 129.68, 129.76, 129.80, 130.15, 130.29, 131.53, 137.85, 138.33, 156.22.
1-{2-Benzyl-5-[2-(tert-butylaminomethyl)piperazin-1-yl]-4-hydroxypentylamino}indan-2-ol (8)
White foam; MS (ESI) m/z 495.53 [M + H]+; 1H NMR (600 MHz, (MeOD-d4) δ 0.99 (s, 9H), 1.35–1.44 (m, 2H), 2.1–2.12 (m, 1H), 2.32–2.36 (m, 2H), 2.43–2.52 (m, 5H), 2.56–2.66 (m, 4H), 2.68–2.83 (m, 5H), 2.92 (dd, 1H, J = 5.22, 15.9 Hz), 3.76–3.78 (m, 1H), 3.86 (d, 1H, J = 4.9 Hz), 4.41 (td, 1H, J = 3.3, 5.1 Hz), 7.05–7.25 (m, 9H). 13C NMR (125 MHz, (MeOD-d4) δ 28.6, 38.31, 39.85, 40.60, 41.37, 42.63, 44.51, 48.33, 52.18, 52.18, 52.34, 60.03, 60.93, 67.4, 67.4, 72.94, 125.84, 126.39, 127.14, 127.69, 128.95, 129.51, 130.53, 142.17, 144.01.
3. DWEYS Peptide Structure Modeling
Four experimental three-dimensional (3D) structures of rat NR2B were identified in the Protein Data Bank (PDB codes 3JPW, 3JPY, 3QEL, 3QEM). All four sequences were aligned and 3D structures superimposed accordingly using default parameters of the MOE protein align and superpose tools (Chemical Computing Group25). The consensus sequence of the turn binding epitope was identified as EWDYG for this protein. Two homology models of the human NR2A and NR2B proteins were identified in ModBase, and their sequences were aligned onto the sequences of rat NR2B and their structures overlaid using the 3JPY structure as a template (most complete rat NR2B experimental structure). The rmsd of main chain atoms of overlaid 3JPY, human NR2A, and NR2B models is 0.352 Å. The DWEYS peptide model was constructed by mutating the central D residue of the human NR2A turn sequence DWDYS into an E. Rotamers of the E side chain were explored and the most stable rotamer was selected.
4. DWEYS Fingerprint-Based Similarity Search
We conducted a DWEYS fingerprint-based similarity search against the DrugBank database (version 4.3)18 which includes a collection of 5055 drug entries including FDA-approved drugs, nutraceuticals, and experimental compounds. The constructed DWEYS structure was used as the template. Database was prepared by energy minimization using MMFF94× force field. Three fingerprint types, MACC structure key (BIT_MACC), shape (ESsape3D), and pharmacophore atom triangle (piDAPH3), were calculated against each molecule in the database. The Tanimoto coefficient36 was applied to score compound similarity with BIT_MACCS. For ESshape3D and phDAPH3 fingerprints, the similarity scores are calculated as the inverse of the distances between two corresponding fingerprints.37 Nine-hundred-ninety-three compounds (20%) with more than 50% overlap of MACCS fingerprint were subjected to ESshape3D fingerprint search. Two-hundred-nineteen out of 993 (22%) compounds within the first round with 30% overlap of DWEYS in ESshape3D fingerprint were submitted to piDAPH3 fingerprint search and yielded 100 hits.
5. Compound Screening with Inhibition ELISA
D-DWEYS peptide (20 µg/mL) in phosphate buffered saline, pH 7.2 (PBS), was added to Costar 96-well plates (product no. 3690, Corning, Tewksbury, MA) and incubated at 37 °C overnight uncovered to allow coating to occur. Before blocking, plates were rinsed 1× with ddH2O and then blocked with 3% BSA in PBS for 1 h at 37 °C. Compounds at varying concentrations were preincubated with R4A antibody (20 µg/mL) in low-protein binding Costar plates (product no. 2797, Corning, Tewksbury, MA) for 1 h at 37 °C. Coated/blocked plates were washed 5 times with PBS/0.5% Tween 20, and then compound/controls plus R4A preincubated samples were added to wells for 1 h at 37 °C. Following a standard wash, a goat anti-mouse secondary IgG antibody conjugated to alkaline phosphatase (catalog no. 1030-04, Southern Biotech, Birmingham, AL) was added to plates at a 1:1000 dilution in PBS for 1 h at room temperature. After standard washes, phosphatase substrate pNPP (no. S0942, Sigma, St. Louis, MO) dissolved in 0.5 M Na2CO3, 0.01 M MgCl2 was incubated with ELISA plates for 30–60 min until color was developed and read at 405 nm.1
6. Structural Overlays of HIV Protease Inhibitors and Derivatives vs DWEYS Peptide
The Protein Data Bank was searched for protein-bound 1, 2, and 3 ligands, and 23, 7, and 8 structures bound to HIV protease were identified, respectively. Structures were downloaded and overlaid using MOE align and superposed default parameters. Proteins were removed and five (PDB codes 1C6Z, 1FB7, 3PWW, 3S56, and 3UFN), three (PDB codes 2QAK, 2R5Q, 2PYM), and four (PDB codes 4L1A, 2RKF, 2RKG, 1RV7) distinct conformations for 1, 2, and 3, respectively, were identified in the superimposed ligands by visual inspection. The five conformations of saquinavir were used as a starting point conformational database for flexible alignment of 1 onto the DWEYS structural model kept rigid using the MOE small molecule flexible alignment module (MMFF94× force field used for potentials). Resulting overlays of 1 to DWEYS were triaged and prioritized using the 3D similarity score generated by the similarity function of this module. The overlays of other HIV protease inhibitors and derivative compounds to DWEYS were generated using a similar methodology. A database of reduced derivative conformations was generated by structural modification of their parent HIV protease inhibitors initial conformations followed by geometry optimization of the resulting structure by energy minimization in the MMFF94× force field, leading to the nearest stable conformer.
7. Glomerular Binding with R4A Antibody
Murine glomeruli were isolated, adhered to glass slides, and the glomerular binding assay was carried out essentially as described.10 Monoclonal antibody R4A (mouse) was applied at 20 µg/mL for 1 h at room temperature and visualized with anti-mouse IgG antibody conjugated to FITC (Jackson ImmunoResearch Laboratories). Slides were mounted by using Vectashield. For inhibition of antibody binding, R4A was preincubated with test compound (50 µM) for 1 h at 37 °C before incubation with glomeruli, as was done for ELISA inhibition assays. Slides were washed with PBS and mounted with coverslips by using Vectashield mounting medium (catalog no. H-1000; Vector Laboratories). Images were acquired at room temperature on an upright microscope (Axioplan 2; Zeiss) with a Zeiss Plan-Neofluar 40× lens using the OpenLab, version 4.04, software (Improvision). For each experimental condition, images were acquired with a Hamamatsu ORCA-ER digital camera (model no. C4742-80) at the same exposure, imported into Adobe Photoshop-CS3, and then adjusted for contrast and brightness identically in all frames in each treatment group. Images were enlarged 2× for the figures.
8. Neurotoxicity Model
All animal procedures were in compliance with the Animal Welfare Act, the Public Health Service Policy on Humane Care and Use of Laboratory Animals, the U.S. Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research and Training, the National Institutes of Health Guide for the Use and Care of Laboratory Animals, and the Research Animal Resource Center Users Guide. Male C57BL/6 mice between 30 and 34 g were anesthetized with 2.5% avertin (ip), placed in a stereotaxic frame, and injected stereotaxically (2 µL) into the hippocampus, as described.5,10,31 Forty-eigth hours later, mice were euthanized, brains fixed, removed, and hippocampal sections (40 µm) were cryoprotected and subjected to TACS-Tdt apoptosis detection (R&D Systems, no. TA4625) with methylene green counterstain as described.38 Sections were visualized with an upright microscope (Axio-Imager; Zeiss), and images were captured digitally (12-bit camera, 25 MHz, 1388 × 1040 resolution), processed (AxioVision 4.7, Zeiss), imported to Photoshop-CS4 as 8-bit TIFF files, and scale-adjusted equally for all frames.
9. Pharmacokinetic and Lupus Disease-Free Survival Studies
Pharmacokinetic analyses of intravenous and oral dosing of reduced HIV protease inhibitor compounds dissolved in PBS were performed on male CD-1 mice of 6–10 weeks of age (n = (3/group)/ time point). Whole blood was protein precipitated with acetonitrile (50:50 v/v), and supernatant was analyzed for compound recovery by HPLC/MS (Supporting Information, Figure S2). For SLE disease free survival, female NZB/W F1 mice were used at 22 weeks of age, prior to onset of severe proteinuria. Prior to the initial dose, proteinuria scores and body weights were measured and groups were assigned such that each group had an equivalent mean proteinuria score and body weight (n = 10 mice/group). Prestudy body weight measurements were performed once weekly, and proteinuria measurements were performed once every 2 weeks beginning at 22 weeks of age.
Animals received dose administrations of test or vehicle twice daily by intraperitoneal (ip) injection beginning on day 1 (at 22 weeks of age). Doses were administered at 5.0 mL kg−1 day−1 (2.5 (mL/kg)/ injection) per animal. Urine was collected before the initial dose administration on day −1 and once every 2 weeks thereafter by either spontaneous urination or gentle pelvic compression during manual restraint of the animal. Interim blood samples were collected by either tail nick or facial vein puncture and terminal blood samples by cardiac puncture.
Whole blood samples were collected into serum separator tubes inverted gently several times and allowed to clot for at least 30 min at room temperature. Samples were then centrifuged at 9300g for 10 min at room temperature, serum was extracted by pipet and stored at −70 °C. Euthanasia (isoflurane, to effect, in conjunction with exsanguination [terminal blood collection], followed by thoracotomy) was performed in accordance with accepted American Veterinary Medical Association (AVMA) guidelines at 46 weeks of age or presentation of humane disease-specific exit criteria which were as follows: if a mouse exhibited a body weight loss of ≥20%, it was removed from study for euthanasia; if a mouse exhibited ≥3+ proteinuria, it was retested the following week and body weights were measured three times weekly; if a mouse exhibited ≥3+ proteinuria on two consecutive weeks, it was removed from study for euthanasia.
10. Kidney Histology Studies in NZB/W F1 Mice
Kidney sections from each animal were analyzed as previously described.39 The sections were evaluated by one investigator (JMC) in a blinded fashion, without knowledge of the mouse treatment. The severity of disease was scored semiquantitatively on a scale of 0 to 3+ for each of the following features: glomerular sclerosis, glomerular hypercellularity (including glomerular cell proliferation, leukocyte exudation), glomerular crescents, interstitial nephritis (infiltration of mononuclear cells), and vasculitis (perivascular) inflammatory infiltrates.
Supplementary Material
Acknowledgments
This work was partially funded by NIH Grant R01AR05708405 (B.D.), and STTR Grant R41AR060620 (T.R.C.).
ABBREVIATIONS USED
- SLE
systemic lupus erythematosus
- Da
dalton
- dsDNA
double stranded DNA
- CNS
central nervous system
- FcR
fragment crystallizable receptor
- TLR
Toll-like receptor
- NMDAR
N-methyl-d-aspartate receptor
- NR2a
N-methyl-d-aspartate receptor 2a
- NR2b
N-methyl-d-aspartate receptor 2b
- HIV
human immunodeficiency virus
- ELISA
enzyme linked immunosorbent assay
- Ab
antibody
- IC50
inhibitory concentration 50
- MS
mass spectrometry
- HPLC
high performance liquid chromatography
- NMR
nuclear magnetic resonance
- PBS
phosphate buffered saline
Footnotes
ASSOCIATED CONTENT
The authors declare the following competing financial interest(s): B.D. and Y.A.-A. are inventors on patents related to FISLE-412 and analogues.
References
- 1.Bloom O, Cheng KF, He M, Papatheodorou A, Volpe BT, Diamond B, Al-Abed Y. Generation of a unique small molecule peptidomimetic that neutralizes lupus autoantibody activity. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10255–9. doi: 10.1073/pnas.1103555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hahn BH. Antibodies to DNA. N. Engl. J. Med. 1998;338:1359–68. doi: 10.1056/NEJM199805073381906. [DOI] [PubMed] [Google Scholar]
- 3.Gaynor B, Putterman C, Valadon P, Spatz L, Scharff MD, Diamond B. Peptide inhibition of glomerular deposition of an anti- DNA antibody. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1955–60. doi: 10.1073/pnas.94.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katz JB, Limpanasithikul W, Diamond B. Mutational analysis of an autoantibody: differential binding and pathogenicity. J. Exp. Med. 1994;180:925–32. doi: 10.1084/jem.180.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeGiorgio LA, Konstantinov KN, Lee SC, Hardin JA, Volpe BT, Diamond B. A subset of lupus anti-DNA antibodies cross-reacts with the NR2 glutamate receptor in systemic lupus erythematosus. Nat. Med. 2001;7:1189–93. doi: 10.1038/nm1101-1189. [DOI] [PubMed] [Google Scholar]
- 6.Lapteva L, Nowak M, Yarboro CH, Takada K, Roebuck-Spencer T, Weickert T, Bleiberg J, Rosenstein D, Pao M, Patronas N, Steele S, Manzano M, van der Veen JW, Lipsky PE, Marenco S, Wesley R, Volpe B, Diamond B, Illei GG. Anti-N-methyl-D-aspartate receptor antibodies, cognitive dysfunction, and depression in systemic lupus erythematosus. Arthritis Rheum. 2006;54:2505–14. doi: 10.1002/art.22031. [DOI] [PubMed] [Google Scholar]
- 7.Franchin G, Son M, Kim SJ, Ben-Zvi I, Zhang J, Diamond B. Anti-DNA antibodies cross-react with C1q. J. Autoimmun. 2013;44:34–9. doi: 10.1016/j.jaut.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shefner R, Kleiner G, Turken A, Papazian L, Diamond B. A novel class of anti-DNA antibodies identified in BALB/c mice. J. Exp. Med. 1991;173:287–96. doi: 10.1084/jem.173.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen-Solal J, Diamond B. Lessons from an anti-DNA autoantibody. Mol. Immunol. 2011;48:1328–31. doi: 10.1016/j.molimm.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Jacobi AM, Wang T, Berlin R, Volpe BT, Diamond B. Polyreactive autoantibodies in systemic lupus erythematosus have pathogenic potential. J. Autoimmun. 2009;33:270–4. doi: 10.1016/j.jaut.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitka M. Treatment for lupus, first in 50 years, offers modest benefits, hope to patients. JAMA. 2011;305:1754–5. doi: 10.1001/jama.2011.548. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz N, Goilav B, Putterman C. The pathogenesis, diagnosis and treatment of lupus nephritis. Curr. Opin. Rheumatol. 2014;26:502–9. doi: 10.1097/BOR.0000000000000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seguro LP, Rosario C, Shoenfeld Y. Long-term complications of past glucocorticoid use. Autoimmun. Rev. 2013;12:629–32. doi: 10.1016/j.autrev.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Bernatsky S, Ramsey-Goldman R, Joseph L, Boivin JF, Costenbader KH, Urowitz MB, Gladman DD, Fortin PR, Nived O, Petri MA, Jacobsen S, Manzi S, Ginzler EM, Isenberg D, Rahman A, Gordon C, Ruiz-Irastorza G, Yelin E, Bae SC, Wallace DJ, Peschken CA, Dooley MA, Edworthy SM, Aranow C, Kamen DL, Romero-Diaz J, Askanase A, Witte T, Barr SG, Criswell LA, Sturfelt GK, Blanco I, Feldman CH, Dreyer L, Patel NM, St Pierre Y, Clarke AE. Lymphoma risk in systemic lupus: effects of disease activity versus treatment. Ann. Rheum. Dis. 2014;73:138–42. doi: 10.1136/annrheumdis-2012-202099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravarty EF. Viral infection and reactivation in autoimmune disease. Arthritis Rheum. 2008;58:2949–57. doi: 10.1002/art.23883. [DOI] [PubMed] [Google Scholar]
- 16.Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28:3910–20. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Yang Z, Meng EC, Pettersen EF, Huang CC, Datta RS, Sampathkumar P, Madhusudhan MS, Sjolander K, Ferrin TE, Burley SK, Sali A. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39:D465–74. doi: 10.1093/nar/gkq1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cereto-Massague A, Ojeda MJ, Valls C, Mulero M, Garcia-Vallve S, Pujadas G. Molecular fingerprint similarity search in virtual screening. Methods. 2015;71:58–63. doi: 10.1016/j.ymeth.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Roberts NA, Martin JA, Kinchington D, Broadhurst AV, Craig JC, Duncan IB, Galpin SA, Handa BK, Kay J, Krohn A, et al. Rational design of peptide-based HIV proteinase inhibitors. Science. 1990;248:358–61. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 21.Perry CM, Frampton JE, McCormack PL, Siddiqui MA, Cvetkovic RS. Nelfinavir: a review of its use in the management of HIV infection. Drugs. 2005;65:2209–44. doi: 10.2165/00003495-200565150-00015. [DOI] [PubMed] [Google Scholar]
- 22.Oldfield V, Plosker GL. Lopinavir/ritonavir: a review of its use in the management of HIV infection. Drugs. 2006;66:1275–99. doi: 10.2165/00003495-200666090-00012. [DOI] [PubMed] [Google Scholar]
- 23.Lacy MK, Abriola KP. Indinavir: a pharmacologic and clinical review of a new HIV protease inhibitor. Conn. Med. 1996;60:723–7. [PubMed] [Google Scholar]
- 24.Chan SL, Labute P. Training a scoring function for the alignment of small molecules. J. Chem. Inf. Model. 2010;50:1724–35. doi: 10.1021/ci100227h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molecular Operating Environment (MOE), version 08. Chemical Computing Group Inc; 1010 Sherbooke St. West, Suite 910, Montreal, QC, Canada, H3A 2R7: 2013. [Google Scholar]
- 26.Sundberg EJ. Structural basis of antibody-antigen interactions. Methods Mol. Biol. 2009;524:23–36. doi: 10.1007/978-1-59745-450-6_2. [DOI] [PubMed] [Google Scholar]
- 27.Grubor NM, Hayes J, Small GJ, Jankowiak R. Cross-reactivity and conformational multiplicity of an anti-polycyclic aromatic hydrocarbon mAb. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7453–8. doi: 10.1073/pnas.0502540102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haupt VJ, Daminelli S, Schroeder M. Drug Promiscuity in PDB: Protein Binding Site Similarity Is Key. PLoS One. 2013;8:e65894. doi: 10.1371/journal.pone.0065894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thielges MC, Zimmermann J, Yu W, Oda M, Romesberg FE. Exploring the energy landscape of antibody-antigen complexes: protein dynamics, flexibility, and molecular recognition. Biochemistry. 2008;47:7237–47. doi: 10.1021/bi800374q. [DOI] [PubMed] [Google Scholar]
- 30.Faust TW, Chang EH, Kowal C, Berlin R, Gazaryan IG, Bertini E, Zhang J, Sanchez-Guerrero J, Fragoso-Loyo HE, Volpe BT, Diamond B, Huerta PT. Neurotoxic lupus autoantibodies alter brain function through two distinct mechanisms. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18569–74. doi: 10.1073/pnas.1006980107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kowal C, Degiorgio LA, Lee JY, Edgar MA, Huerta PT, Volpe BT, Diamond B. Human lupus autoantibodies against NMDA receptors mediate cognitive impairment. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19854–9. doi: 10.1073/pnas.0608397104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang EH, Volpe BT, Mackay M, Aranow C, Watson P, Kowal C, Storbeck J, Mattis P, Berlin R, Chen H, Mader S, Huerta TS, Huerta PT, Diamond B. Selective Impairment of Spatial Cognition Caused by Autoantibodies to the N-Methyl-d- Aspartate Receptor. EBioMedicine. 2015;2:755–64. doi: 10.1016/j.ebiom.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowal C, DeGiorgio LA, Nakaoka T, Hetherington H, Huerta PT, Diamond B, Volpe BT. Cognition and immunity; antibody impairs memory. Immunity. 2004;21:179–88. doi: 10.1016/j.immuni.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Horowitz DM, Furie RA. Abetimus sodium: a medication for the prevention of lupus nephritis flares. Expert Opin. Pharmacother. 2009;10:1501–7. doi: 10.1517/14656560902946419. [DOI] [PubMed] [Google Scholar]
- 35.Linnik MD, Hu JZ, Heilbrunn KR, Strand V, Hurley FL, Joh T, Consortium LJPI. Relationship between anti-double-stranded DNA antibodies and exacerbation of renal disease in patients with systemic lupus erythematosus. Arthritis Rheum. 2005;52:1129–37. doi: 10.1002/art.20980. [DOI] [PubMed] [Google Scholar]
- 36.Willett P, Barnard J, Downs G. Chemical Similarity Searching. J. Chem. Inf. Comput. Sci. 1998;38:983–996. [Google Scholar]
- 37.Ballester PJ, Richards WG. Ultrafast shape recognition to search compound databases for similar molecular shapes. J. Comput. Chem. 2007;28:1711–23. doi: 10.1002/jcc.20681. [DOI] [PubMed] [Google Scholar]
- 38.Lee JY, Huerta PT, Zhang J, Kowal C, Bertini E, Volpe BT, Diamond B. Neurotoxic autoantibodies mediate congenital cortical impairment of offspring in maternal lupus. Nat. Med. 2009;15:91–6. doi: 10.1038/nm.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shlomchik MJ, Madaio MP, Ni D, Trounstein M, Huszar D. The role of B cells in lpr/lpr-induced autoimmunity. J. Exp. Med. 1994;180:1295–306. doi: 10.1084/jem.180.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.