Abstract
Purpose
Tobacco smoking is a risk factor in several cancers, yet its roles as a putative etiologic exposure or poor prognostic factor in breast cancer are less clear. Altered DNA methylation contributes to breast cancer development and may provide a mechanistic link between smoking and gene expression changes leading to cancer development or progression.
Methods
Using a cancer-focused array, we examined methylation at 933 CpGs in 517 invasive breast tumors in the Carolina Breast Cancer Study to determine whether methylation patterns differ by exposure to tobacco smoke. Multivariable generalized linear regression models were used to compare tumor methylation profiles between smokers and never smokers, overall, or stratified on hormone receptor (HR) status.
Results
Modest differences in CpG methylation were detected at p < 0.05 in breast tumors from current or ever smokers compared with never smokers. In stratified analyses, HR− tumors from smokers exhibited primarily hypomethylation compared with tumors from never smokers; hypomethylation was similarly detected within the more homogeneous basal-like subtype. Most current smoking-associated CpG loci exhibited methylation levels in former smokers that were intermediate between those in current and never smokers and exhibited progressive changes in methylation with increasing duration of smoking. Among former smokers, restoration of methylation toward baseline (never smoking) levels was observed with increasing time since quitting. Moreover, smoking-related hypermethylation was stronger in HR+ breast tumors from blacks than in whites.
Conclusions
Our results suggest that breast tumor methylation patterns differ with tobacco smoke exposure; however, additional studies are needed to confirm these findings.
Keywords: Breast cancer, Smoking, Breast tumor, Methylation, Epigenetic
Introduction
Cigarette smoking is a well-known risk factor for a number of human cancers, including cancers of the lung [1], head and neck [2], and bladder [3]. The epidemiologic data on active smoking in breast cancer has been mixed [4, 5], but more recent studies support a modest role for smoking in breast cancer development [6–8], most often among women who smoked for a long time [6, 9–12], recently quit [12, 13], or began smoking at an early age or prior to first birth [6, 7, 10, 11, 14]. Several reports have also described stronger associations of smoking with either ER+ [7, 8, 15, 16] or ER− breast cancer [17–20]. In addition to putative etiologic effects, tobacco smoking at the time of breast cancer diagnosis is associated with worse disease-specific and overall survival [21, 22]; however, the biological mechanisms through which smoking might influence the risk of developing or dying from breast cancer are not fully understood.
Epigenetic alterations, such as DNA methylation, are thought to be part of the causal pathway in the development of malignancies, including breast cancer [23, 24]. Methylation does not alter the nucleotide sequence of DNA but can influence gene expression and may provide a mechanistic link between environmental exposures, such as smoking, and altered cellular gene expression leading to the development or progression of cancer [25, 26]. Numerous epigenome-wide studies have reported methylation differences in normal peripheral blood leukocytes (PBL) between current smokers and nonsmokers [27–30], with methylation changes at many loci being reversible after cessation of smoking [28, 29, 31], highlighting the dynamic nature of DNA methylation and the potential for modulation by environmental exposures. In cancers for which smoking is a known risk factor, differences in tumor methylation profiles have been detected between smokers and nonsmokers [32–36], but the effect of tobacco smoke exposure on the epigenetic profile of breast tumors has not been examined.
Studies in the Carolina Breast Cancer Study (CBCS) have reported the associations of smoking and breast cancer mainly among long-term smokers and recent quitters, and observed stronger associations for luminal breast cancer, particularly among African Americans [9, 16]. Additionally, we observed worse disease-specific survival among breast cancer cases who smoked (unpublished observations). In an effort to better understand the genomic effects of smoking and determine whether breast tumor methylation patterns reflect, in part, this exposure, we examined methylation profiles of invasive breast tumors in the CBCS according to active smoking status using a cancer-focused methylation array.
Materials and methods
CBCS population
The CBCS is a population-based, case–control study of breast cancer that includes women aged 20–74 years residing in 24 counties of central and eastern North Carolina. Women with a first diagnosis of invasive breast cancer between 1993 and 1996 (phase 1) were identified by the North Carolina Central Cancer Registry through a rapid case ascertainment system. Women diagnosed prior to age 50 and black/African American women were over-sampled to ensure that they comprised roughly half the study sample. Race was self-reported. Additional details of the study design are described elsewhere [37], and case characteristics are provided in Table 1. All aspects of this research were approved by the UNC School of Medicine Institutional Review Board. A total of 861 breast cancer cases were eligible for and consented to participate in the CBCS during phase 1.
Table 1.
Selected breast cancer case characteristics by smoking status
| Characteristic | Current smokers (N = 125) No. (%) | Former smokers (N = 124) No. (%) | Never smokers (N = 268) No. (%) | p-value | |
|---|---|---|---|---|---|
| Case characteristics | |||||
| Age, mean (SD) | 46.9 (10.6) | 51.0 (11.3) | 50.4 (12.5) | 0.009 | |
| Age | 50+ years | 35 (28.0) | 53 (42.7) | 111 (41.4) | 0.02 |
| <50 years | 90 (72.0) | 71 (57.3) | 157 (58.6) | ||
| Race | White/Othera | 76 (60.8) | 77 (62.1) | 148 (55.2) | 0.35 |
| Black | 49 (39.2) | 47 (37.9) | 120 (44.8) | ||
| Menopausal status | Postmenopausal | 55 (44.0) | 59 (47.6) | 128 (47.8) | 0.77 |
| Premenopausal | 70 (56.0) | 65 (52.4) | 140 (52.2) | ||
| Alcohol consumption | Never | 16 (12.8) | 21 (16.9) | 113 (42.2) | < 0.0001 |
| Ever | 109 (87.2) | 103 (83.1) | 155 (57.8) | ||
| Body mass index (BMI) | <25 (normal) | 60 (48.0) | 39 (32.2) | 86 (33.0) | 0.05 |
| 25 to <30 (overweight) | 29 (23.2) | 36 (29.8) | 80 (30.7) | ||
| >30 (obese) | 36 (28.8) | 46 (38.0) | 95 (36.4) | ||
| Smoking characteristics | |||||
| Cigarettes/day | <1/2 pack | 32 (25.6) | 51 (41.5) | – | 0.02 |
| 1/2–1 pack | 49 (39.2) | 44 (35.8) | – | ||
| >1 pack | 44 (35.2) | 28 (22.8) | – | ||
| Duration of smoking (yrs) | <10 | 19 (15.2) | 59 (48.0) | – | < 0.0001 |
| 11–20 | 22 (17.6) | 29 (23.6) | – | ||
| >20 | 84 (67.2) | 35 (28.5) | – | ||
| Time since quitting (yrs) | ≤10 | – | 54 (43.6) | – | – |
| 11+ | – | 70 (56.4) | – | ||
| Age at initiation (yrs) | <15 | 31 (24.8) | 13 (10.5) | – | 0.0008 |
| 16–20 | 54 (43.2) | 81 (65.3) | – | ||
| >20 | 40 (32.0) | 30 (24.2) | – | ||
| Clinical characteristics | |||||
| Stageb | I | 45 (39.5) | 45 (39.8) | 88 (34.7) | 0.52 |
| II | 51 (44.7) | 55 (48.7) | 139 (54.7) | ||
| III | 15 (13.2) | 9 (8.0) | 21 (8.3) | ||
| IV | 3 (2.6) | 4 (3.5) | 6 (2.4) | ||
| ER/PR status | ER+/PR+ | 59 (49.6) | 69 (58.1) | 122 (46.6) | 0.38 |
| ER+/PR− | 15 (12.6) | 9 (7.6) | 24 (9.2) | ||
| ER−/PR+ | 8 (6.7) | 7 (5.9) | 24 (9.2) | ||
| ER−/PR− | 37 (31.1) | 34 (28.6) | 92 (35.1) | ||
| Intrinsic subtype | |||||
| All cases | Luminal A | 54 (54.0) | 52 (53.6) | 106 (49.1) | 0.99 |
| Luminal B | 15 (15.0) | 14 (14.4) | 36 (16.7) | ||
| Basal-like | 19 (19.0) | 20 (20.6) | 47 (21.8) | ||
| HER2+/ER− | 6 (6.0) | 5 (5.2) | 15 (6.9) | ||
| Unclassified | 6 (6.0) | 6 (6.2) | 12 (5.5) | ||
| Black | Luminal A | 19 (52.8) | 18 (47.4) | 44 (46.3) | 0.79 |
| Luminal B | 4 (11.1) | 4 (10.5) | 14 (14.7) | ||
| Basal-like | 9 (25.0) | 12 (31.6) | 25 (26.3) | ||
| HER2+/ER− | 4 (11.1) | 3 (7.9) | 6 (6.3) | ||
| Unclassified | 0 (0) | 1 (2.6) | 6 (6.3) | ||
| White/other | Luminal A | 35 (54.7) | 34 (57.6) | 62 (51.2) | 0.83 |
| Luminal B | 11 (17.2) | 10 (17.0) | 22 (18.2) | ||
| Basal-like | 10 (15.6) | 8 (13.6) | 22 (18.2) | ||
| HER2+/ER− | 2 (3.1) | 2 (3.4) | 9 (7.4) | ||
| Unclassified | 6 (9.4) | 5 (8.5) | 6 (5.0) | ||
The white/other cases included 291 Caucasians, 3 American Indians, 6 Asians, and 1 other
According to the AJCC breast tumor staging guidelines. The differences between column totals and numbers within some categories are due to missing data in some cases for some variables
Smoking history
Risk factor information, including smoking history, was obtained from questionnaires administered to participants by trained nurse interviewers. Active smokers self-reported smoking ≥100 cigarettes, while never smokers smoked <100 cigarettes over their lifetime. On average, interviews were conducted 6 months following case ascertainment. Since a breast cancer diagnosis may influence decisions concerning smoking cessation, current smokers were defined as women who self-reported active smoking at the time of interview and women who reported smoking cessation at the same age (within a year) of case selection [16]. Former smokers quit at any age prior to the age at case selection. These data were used to derive smoking duration in years. All smoking data were recorded as categorical variables.
Clinical data and tumor tissues
Clinical data and information on tumor characteristics were obtained from medical records or histopathologic review of tumor tissue. Formalin-fixed paraffin-embedded (FFPE) tumor blocks were obtained from pathology departments at participating hospitals and underwent standardized histopathologic review [38]. The pathologist also encircled the tumor on the hematoxylin and eosin (H&E)-stained slide which was then used to guide manual microdissection of tumor from surrounding non-tumor tissue. Isolated tissue was processed for DNA using a standard proteinase K-based method as previously described [39]. Intrinsic breast tumor subtypes were previously identified using a panel of immunohistochemical markers and included luminal A (ER+ and/or PR+(HR+)/HER2−), luminal B (HR+/HER2+), basal-like (ER−/PR− (HR−)/HER2−/CK5+ or CK6+, or EGFR+), HER2+/HR−, and unclassified (all markers negative) [40].
DNA methylation analysis
DNA methylation profiling was previously accomplished using the Illumina GoldenGate Cancer Panel I methylation bead array [39]. Sodium bisulfite modification of FFPE breast tumor DNA was performed using the EZ DNA Methylation Gold kit according to the manufacturer’s protocol. Bisulfite-converted tumor DNA was analyzed for methylation using the Cancer Panel I array in the Mammalian Genotyping Core facility at UNC. The Cancer Panel I array measures methylation at CpG sites in the promoter (P) or first exon (E) of cancer-related genes, including tumor suppressor genes, oncogenes, DNA repair genes, and others. Array probe names are comprised of the gene symbol followed by the position of the CpG in the P or E upstream from the transcription start site. The methylation level at each CpG site is represented by the beta (β) value, which ranged from 0 (completely unmethylated) to 1.0 (fully methylated). Probes reported to overlap a single-nucleotide polymorphism (SNP) or repeat [41] were excluded. After quality control filtering as described previously [39], methylation analysis was completed on 517 breast tumors and included 933 CpG loci in 609 genes.
Statistical analyses
Statistical analyses were carried out using R (http://www.r-project.org/) or SAS v9.3. To identify CpG loci that showed differential breast tumor methylation between cases who were active smokers versus never smokers, generalized linear regression models (GLM) were fitted to the logit transformed β-values adjusting for age (continuous), race (African American, white/other), menopausal status, stage (1, 2, 3, 4), body mass index (BMI) (<25, 25 to <30, ≥30 kg/m2), and alcohol consumption (lifetime consumption in grams/week). BMI was considered a potential confounder as BMI differed between smoking groups and obesity (BMI ≥30) was previously associated with tumor methylation differences [42]. Similarly, alcohol consumption differed between smokers and never smokers and was previously correlated with breast tumor methylation patterns [43]. GLM stratified on hormone receptor (HR) status (HR-positive; ER+ and/or PR+, or HR-negative; ER−/PR−) was also performed as tumor methylation profiles are known to differ by HR status [39]. Volcano plots were utilized to display array-wide association patterns of differential methylation between smokers and never smokers, showing the estimated coefficients from the previously mentioned GLMs plotted against the negative logarithm of the raw p-values obtained from the associated Wald significance tests. The Benjamini-Hochberg false discovery rate (FDR) was calculated to adjust for multiple comparisons, and a threshold of <0.05 was considered significant. Spearman correlation coefficients and p-values were used to test the relationship between increasing duration of smoking among ever smokers or years since quitting among former smokers and tumor methylation at smoking-associated CpG loci identified from GLM.
Gene ontology analysis
The DAVID Bioinformatics Resources 6.8 Functional Annotation Tool (https://david.ncifcrf.gov/) was used to perform gene-GO term enrichment analysis to identify the most relevant GO terms associated with the genes found to be differentially methylated in breast tumors from smokers versus never smokers. Entrez gene IDs from each gene list were compared to the background list of 609 genes evaluated from the Illumina Cancer Panel I array after filtering. Genes with more than one CpG site were listed only once in the analysis. We performed functional annotation clustering with default settings. Terms that were significantly enriched (Benjamini-Hochberg corrected p < 0.05) are listed.
Correlation between methylation and gene expression
The relationship between methylation and gene expression at CpG loci associated with current (vs. never) smoking was tested using breast tumor data in The Cancer Genome Atlas (TCGA) [44] since gene expression data were not available in phase I of the CBCS. Methylation analysis in TCGA was performed using the Illumina Infinium HumanMethylation450 K array (450 K), and gene expression data were generated using RNA sequencing. Due to the small degree of overlap of CpG probes between the 450 K array and the Cancer Panel array used in this study, we examined the relationship between methylation and gene expression only for probes that directly overlapped the two platforms [39, 42, 45]. Pearson correlation coefficients were calculated in 581 TCGA breast tumors (from 44 blacks, 431 whites, 29 Asians, 77 of unknown race/ethnicity), or within hormone receptor-positive or hormone receptor-negative subsets, based on RNAseq (Illumina) log2 RSEM gene normalized expression values with methylation beta values for 450 K CpG probes, with significance set at p <0.05.
Results
The characteristics of cases from CBCS phase 1 evaluated for tumor methylation according to smoking status are shown in Table 1. Current smokers were younger than never or former smokers (p = 0.02), smoked for longer (p < 0.0001), and began smoking at a younger age (p = 0.0008) compared with former smokers. Smokers were more likely to consume alcohol (p < 0.0001) and have lower BMI (p = 0.05) compared with never smokers. Otherwise, case subgroups defined by smoking status did not vary significantly on other demographic or clinical characteristics.
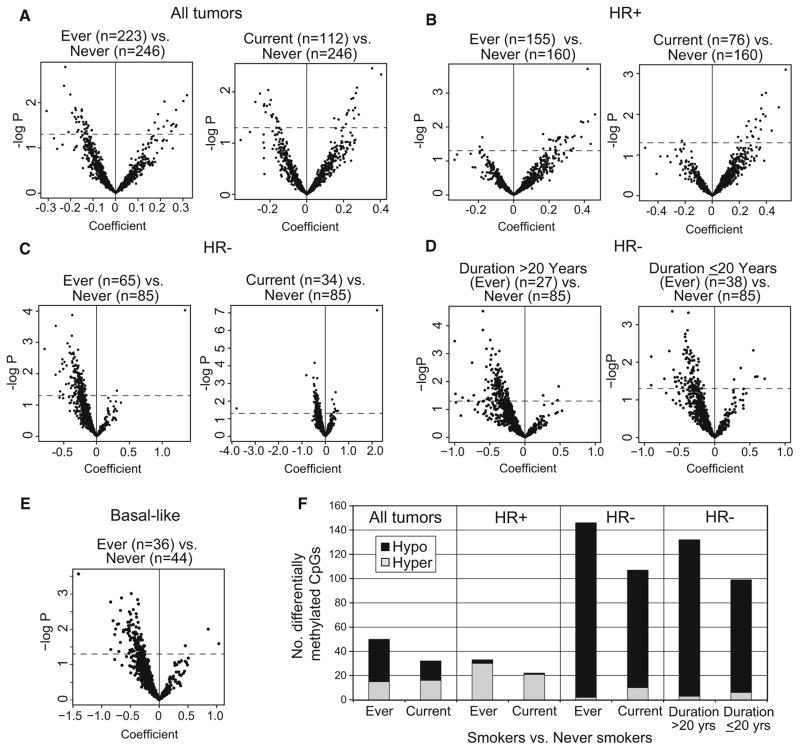
To determine whether tobacco smoke exposure was associated with differences in breast tumor epigenetic profiles, separate multivariable GLMs were used to compare methylation at 933 CpG sites between smokers and never smokers. As illustrated by volcano plots (Fig. 1a), breast tumors of current or ever smokers exhibited both increases and decreases in CpG methylation compared to never smokers at p <0.05; similar results were obtained with or without inclusion of stage in the model (Online Resource Fig. S1). We previously observed differences in DNA methylation between HR+ (ER+ and/or PR+) and HR− (ER−/PR−) breast tumors [39]; therefore, to minimize the potential contribution of HR subtype heterogeneity to methylation differences, we examined the effect of smoking stratified on HR status. The effect of smoking on methylation differed by HR status, with HR+ breast tumors from smokers exhibiting hypermethylation (Fig. 1b). In contrast, HR− breast tumors from smokers exhibited a somewhat more robust pattern of mostly CpG hypomethylation compared with tumors from never smokers (Fig. 1c) which was stronger among long-term (>20 year) smokers than in those who smoked for shorter duration (Fig. 1d). The HR− breast tumor subset included several intrinsic subtypes based on immunoprofiles (basal-like, HER2±/HR−, and unclassified), yet smoking was also associated with CpG hypomethylation in the most homogeneous subset of basal-like tumors (Fig. 1e). The divergent patterns of smoking-related differential methylation in HR− versus HR+ breast tumors seen in the volcano plots are summarized numerically in Fig. 1f. Patterns of differential hypo- or hypermethylation in former (vs. never) smokers were similar to those of current smokers (Online Resource Fig. S2).
Fig. 1.
Patterns of breast tumor differential methylation in smokers compared with never smokers. Generalized linear regression models (GLM) (logit link) adjusted for age, race, menopausal status, stage, BMI, and alcohol consumption were used to identify CpG loci differentially methylated in breast tumors in smokers versus never smokers. Volcano plots display array-wide patterns of breast tumor differential methylation in current or ever smokers among: a all cases, b HR+ cases, c HR− cases, d HR− cases according to smoking duration (long-term >20 years or shorter-term ≤20 years) among ever smokers, or e basal-like cases. Each volcano plot displays the negative log of unadjusted p-values for differences in β (proportion DNA methylated) at each probe on the y axis versus the correlation coefficient for methylation at each CpG locus on the x axis. Probes that fall above the broken line are significant at p <0.05. Probes hypomethylated in smokers have negative coefficients, while probes hypermethylated in smokers have positive coefficients. f Bar graph summarizing numbers of differentially hypomethylated or hypermethylated CpG probes at p < 0.05 in ever smokers, current smokers, long-term (>20 year), or shorter-term (<20 year) smokers versus never smokers among all cases or by HR status
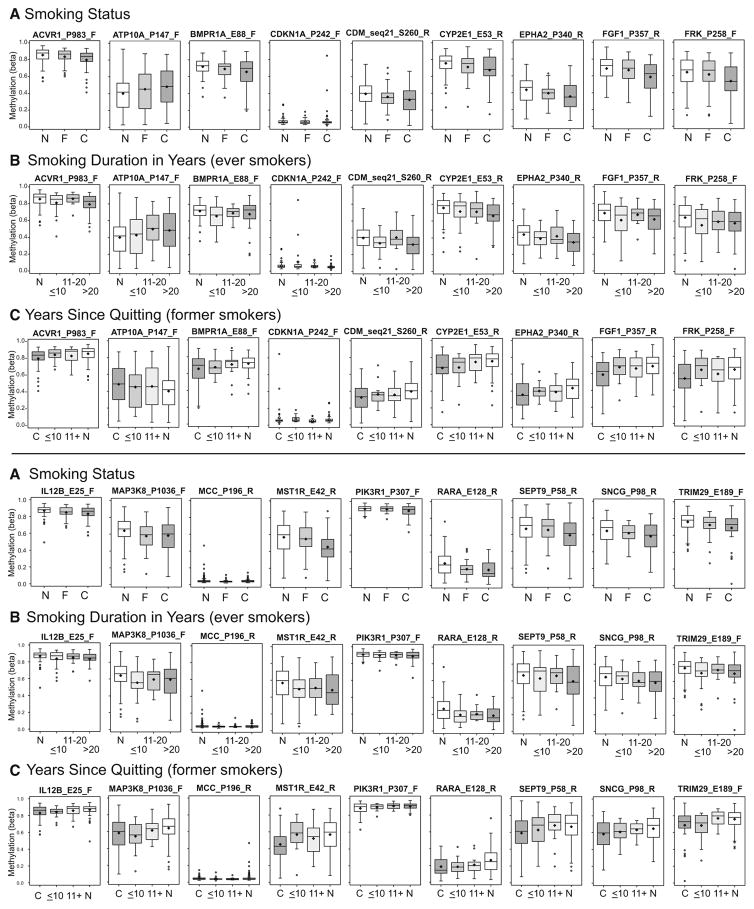
Because methylation differences in current smokers are expected to be more stable than in former smokers based on studies in peripheral blood leukocytes (PBLs), we have focused on CpG loci differing in HR+ or HR− breast tumors from current smokers. A list of loci significant at p <0.05 is given in Online Resource Table S1. The mean percent change in methylation in HR− tumors at CpG loci differing between current and never smokers was +/−11% (range 0.8–39%). Two candidate genes (CDKN1A and IL12B) showed significant current smoking-related differential methylation after FDR correction (FDR adjusted p <0.05) in HR− breast tumors. Box plots in Fig. 2 illustrate methylation levels at top CpG loci related to current smoking in HR− cases; these include several genes previously found to differ in PBLs between current smokers and nonsmokers (RARA, ACVR1, SEPT9, CDKN1A, MCC, ATP10A, and PIK3R1). Most current smoking-related CpG loci in HR− tumors exhibited methylation levels in former smokers that were intermediate between those in current and never smokers (Fig. 2a), and exhibited progressive changes in methylation with increasing duration of smoking among ever smokers (Fig. 2b; Online Resource Table S2). Among former smokers, restoration of methylation toward the baseline (never smoking) level was observed with increasing time since quitting (Fig. 2c). For a number of genes differentially methylated in current smokers (ACVR1, CYP2E1, FGF1, EPHA2, KRT5, MCF2, PTHR1, RARRES1, SEPT9, SNCG, TRIM29, VBP1 in HR− cases, and COL1A1 and TAL1 in HR+ cases), two probes independently detected similar patterns of smoking-associated differential methylation (Online Resource Table S1). Boxplots for additional current smoking-related CpGs are provided in Online Resource Fig. S3.
Fig. 2.
Boxplots illustrating methylation levels at top CpGs differing in methylation between current and never smokers with HR− breast tumors. a Methylation differences according to smoking status in current (C) (n = 37) or former (F) (n = 34) smokers compared with never smokers (N) (n = 92). b Methylation with increasing duration of smoking in years among ever smokers: N (n = 92), ≤10 (n = 25), 11–20 (n = 16), or >20 years (n = 29). c Methylation with years since quitting among former smokers: C (n = 37), ≤10 (n = 18), 11+ (n = 16), N (n = 92). Never or current smokers are included for reference in some plots as appropriate. Boxplot boundaries indicate the interquartile range, and mean and median are indicated by the black bar and diamond, respectively
Analysis of TCGA breast tumor data indicated that 39 current smoking-associated CpGs were common to the Illumina methylation platforms used in TCGA and CBCS. Approximately half of these CpGs showed significant inverse correlations between methylation and gene expression, overall or within HR-based subtypes (Online Resource Table S3), suggesting that smoking-related methylation differences may translate to alterations in tumor gene expression.
Recent studies in the CBCS indicated that smoking was more strongly associated with the development of luminal (HR+) breast cancers among blacks [16], but worse long-term survival among black smokers and smokers with ER− breast tumors (unpublished observations).
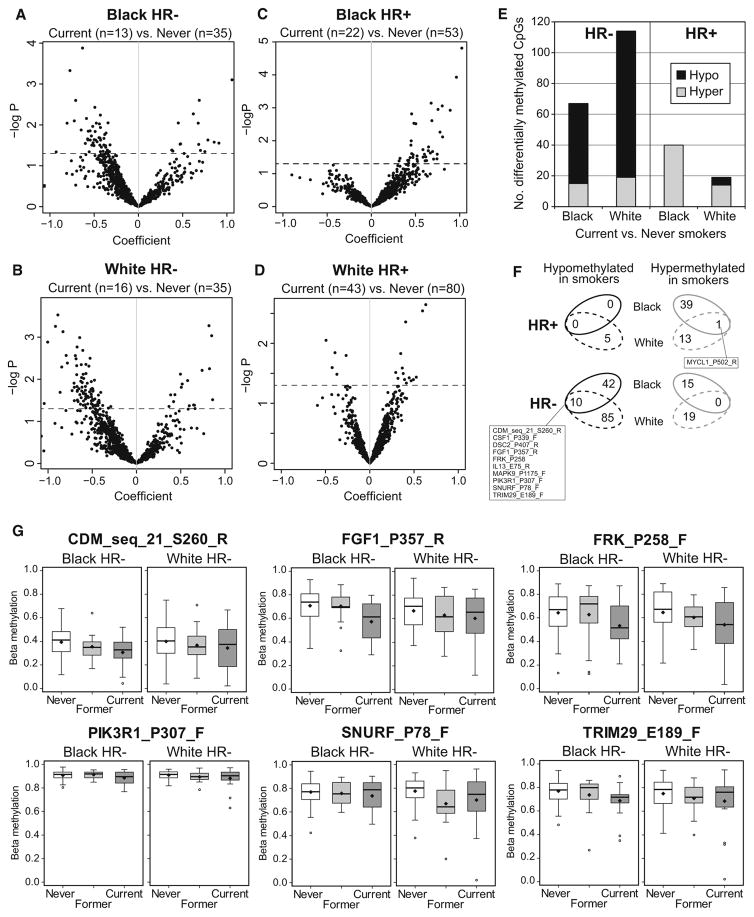
Therefore, we also examined the effect of current smoking on methylation profiles of HR+ or HR− breast tumors stratified by race. HR− breast tumors in blacks and whites similarly exhibited hypomethylation among current smokers (Fig. 3a, b), whereas among HR+ cases, black smokers exhibited a stronger pattern of hypermethylation (Fig. 3c, d); these patterns of smoking-related differential methylation are summarized in Fig. 3e. At the level of individual probes, 19% of current smoking-related CpGs in HR− tumors from black cases overlapped with those of whites (Fig. 3f; Online Resources Table S4 and Table S5), including loci in the CDM, FGF1, FRK, PIK3R1, SNURF, and TRIM29 genes, as shown in boxplots in Fig. 3g. In black smokers, three candidate CpGs in RIPK4, SMO, and MAF exhibited hypermethylation in HR+ tumors and were significant after FDR correction (Online Resource Table S5).
Fig. 3.
Patterns of breast tumor differential methylation in black or white current smokers versus never smokers with HR+ or HR− breast cancer. Generalized linear regression (GLM) models adjusted for age, race, menopausal status, stage, BMI, and alcohol consumption were used to compare breast tumor methylation beta values between current and never smokers. a Volcano plots display array-wide patterns of breast tumor differential methylation in current versus never smokers among a black HR− cases, b black HR+ cases, c white HR− cases, and d white HR+ cases. Probes that fall above the broken line are significant at p <0.05. Probes hypomethylated in smokers have negative coefficients and probes hypermethylated in smokers have positive coefficients. e Bar graph summarizing numbers of differentially hypomethylated or hypermethylated CpG probes in black and white current smokers with HR− or HR+ breast tumors. f Venn diagrams summarizing the overlap of CpG probes differentially methylated in current smokers among black and white cases with HR− or HR+ breast cancers. g Boxplots of selected smoking-related CpG loci illustrating the range of beta values in HR− breast tumors from black or white current or former smokers versus never smokers
DAVID gene ontology analysis of genes differentially methylated in current versus never smokers showed that in HR− cases, smoking was associated with significant terms such as ‘response to organic substance,’ ‘response to stress,’ and ‘response to chemical stimulus’ (Online Resource Table S6). In HR+ tumors, significant GO terms were related mainly to regulation of RNA processes or transcription.
Discussion
The results of this study suggest that exposure to tobacco smoke is associated with modest differences in breast tumor methylation patterns. Current smokers with HR− breast tumors showed primarily CpG hypomethylation compared with never smokers, whereas smokers with HR+ breast tumors showed hypermethylation, and these changes were more pronounced with longer duration of smoking but were diminshed with time since smoking cessation. Smoking-related differential methylation was not obviously explained by intrinsic subtype composition as these were very similar among the smoker and never smoker groups, and smoking-associated hypomethylation was also detected when analyses were restricted to the most homogeneous basal-like subtype. Importantly, we also adjusted for alcohol use and BMI as these differed between smokers and nonsmokers in the CBCS and previously were found to be associated with epigenetic differences in breast tumors [42, 43]. Interestingly, gene ontology analysis revealed different enrichment terms associated with smoking in HR− and HR+ tumors, with smoking-related terms in HR− tumors related to response to organic chemicals and stress, while those in HR+ tumors were associated with transcriptional regulation, suggesting that these tumor subsets may both be associated with smoking but via different pathways. Smoking-related epigenetic differences also varied somewhat by race in that the hypermethylation in HR+ breast tumors was more pronounced in black smokers, which may be consistent with epidemiologic findings from CBCS indicating that smoking was associated with the development of luminal breast cancer mainly among black women [16].
Several array-based studies have reported smoking-related methylation differences in cancers for which smoking is a major risk factor, including cancers of the lung [32, 33], bladder [34–36], and head and neck [46, 47]. In epigenome-wide studies of smoking in PBLs [27–30], hypomethylation was the predominant effect, with CpGs in the AHRR and/or F2RL3 genes showing reduced methylation in current smokers. Although these genes were not included on the Cancer Panel I array, several others previously reported to show changes in methylation in current smokers also differed in breast tumors [28–30, 48]. One recent study investigated smoking-associated differences in breast tumor promoter methylation at 13 candidate genes and found that current smoking (vs. non-current smoking) was associated with hypomethylation of DAPK [49]; however, ours is the first to report on the relationship between tobacco smoke exposure and breast tumor methylation profiles using an array-based approach.
Tobacco smoke is comprised more than 7000 chemicals, including 69 established carcinogens [50, 51], and is considered to be a complete carcinogen with the ability to act during tumor initiation, promotion, and progression [51, 52]. Nicotine, the primary addictive constituent of cigarette smoke, may have in vitro genotoxic [53], promotional [54], and anti-apoptotic effects [55] in breast tumor cells. Thus, the pro-carcinogenic effects of smoking in breast epithelium could be mediated via alterations in DNA methylation throughout the continuum of the disease process, both early in breast cancer initiation and/or later during tumor growth and progression [51, 52]. A number of mechanisms have been posited for how smoking alters DNA methylation, including induction of DNMT1 expression [56], recruitment of DNMT1 and subsequent methylation of CpGs adjacent to sites of DNA damage [57, 58], modulation of gene expression by nicotine, including of CDKN1A [59, 60], alteration of expression and DNA binding stability of transcription factors, e.g., Sp1, which can protect CpG sites from methylation [61, 62], and hypoxia, which can lead to increases in the methyl donor, S-adenosylmethionine [63]. Smoking is also associated with reduced serum folate levels [64, 65], which could result in impaired DNA methylation, especially in rapidly growing HR− tumors. Indeed, triple-negative (including basal-like) breast tumors frequently overexpress the folate receptor, FOLR1, which is thought to provide a growth advantage to tumor cells, particularly in a low-folate environment [66].
Genes found to be differentially methylated in the tumors of smokers have varied roles in breast or other cancers. CDKN1A (cyclin-dependent kinase inhibitor, p21/ WAF1) is a key negative regulator of the cell cycle and cell proliferation [67]. CDKN1A hypermethylation in breast tumors was associated with reduced mRNA and protein expression [68, 69], and in endometrial tumors, p21 protein expression was inversely associated with current smoking [70]. The pro-inflammatory cytokine, IL12, of which IL12B (p40) is a subunit, functions in cell-mediated immunity and host immune responses to tumors [71]. Genetic variants in IL12B are associated with breast cancer risk [72] and survival in chemotherapy-treated ER− breast cancer patients [73]. Other smoking-associated genes have roles in DNA repair (OGG1, BRCA1, FANCE, and TDG), signal transduction (FRK, TYRO3, EPHA2, EPHB1, MST1R, and ACVR1), cell cycle regulation (CDKN1A, MAP3K8, and MAPK9), inflammation and immune function (IL12B, IL13, IL6, and HLA-F), xenobiotic or drug metabolism (CYP2E1 and UGT1A1), or function as growth factors (FGF1, HGF, and PDGFRA) or transcription factors (TRIM29, RARA).
Strengths of this study include the large size and population-based design of the CBCS and inclusion of cases and breast tumors with relatively complete risk factor, histopathologic and clinical data. Limitations include the use of intrinsic subtyping based on a panel of IHC protein expression markers rather than gene expression subtyping (due to lack of mRNA availability from archival FFPE specimens), which may have resulted in some misclassification, mainly among luminal breast cancers [74]. However, smoking-associated differential hypomethylation observed among HR− tumors was also seen within the more accurately classified basal-like subtype [74], reducing the likelihood that the alterations in methylation in HR− tumors were due to subtype heterogeneity rather than smoking. Moreover, while cell-type heterogeneity is often an inherent problem in tissue analysis, the breast tumors in CBCS were manually dissected to enrich for tumor cells. Although the data were collected on a first generation methylation array that over-sampled genes in cancer-related pathways, many genes on the platform had strong coverage for the best-studied methylation sites in breast cancer research. Additionally, as this study focused on CpG methylation in gene promoters, the results may uniquely reflect effects of smoking in these genomic regions.
Our results indicate that tobacco smoke exposure is associated with modest differences in breast tumor epigenetic patterns and that these vary with HR status and race. Additional studies are needed to confirm our findings and to clarify the mechanisms underlying the effects of smoking on tumor methylation.
Supplementary Material
Acknowledgments
We thank the participants and staff of the CBCS for their ongoing support of the study.
Funding: This research was supported by Grants to KC from Susan G. Komen Foundation (Grant #KG081397) and from the University Cancer Research Fund of the Lineberger Comprehensive Cancer Center at the University of North Carolina at Chapel Hill. The Carolina Breast Cancer Study was also funded by the University Cancer Research Fund of North Carolina and the National Cancer Institute Specialized Program of Research Excellence (SPORE) in Breast Cancer (NIH/NCI P50-CA58223).
Footnotes
Electronic supplementary material: The online version of this article (doi:10.1007/s10549-017-4178-8) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest The authors declare they have no conflict of interest.
Research involving human subjects All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable standards.
References
- 1.Ernster VL. Female lung cancer. Ann Rev Public Health. 1996;17:97–114. doi: 10.1146/annurev.pu.17.050196.000525. [DOI] [PubMed] [Google Scholar]
- 2.Boffetta P, Mashberg A, Winkelman R, Garfinkel L. Carcinogenic effect of tobacco smoking and alcohol drinking on anatomic sites of the oral cavity and oropharynx. Int J Cancer. 1992;52:530–533. doi: 10.1002/ijc.2910520405. [DOI] [PubMed] [Google Scholar]
- 3.Castelao JE, Yuan J-M, Skipper PL, Tannenbaum SR, Gago-Dominguez M, Crowder JS, Ross RK, Yu MC. Gender-and smoking-related bladder cancer risk. J Natl Cancer Inst. 2001;93:538–545. doi: 10.1093/jnci/93.7.538. [DOI] [PubMed] [Google Scholar]
- 4.Reynolds P. Smoking and breast cancer. J Mammary Gland Biol Neoplasia. 2013;18:15–23. doi: 10.1007/s10911-012-9269-x. [DOI] [PubMed] [Google Scholar]
- 5.Macacu A, Autier P, Boniol M, Boyle P. Active and passive smoking and risk of breast cancer: a meta-analysis. Breast Cancer Res Treat. 2015;154:213–224. doi: 10.1007/s10549-015-3628-4. [DOI] [PubMed] [Google Scholar]
- 6.Dossus L, Boutron-Ruault MC, Kaaks R, Gram IT, Vilier A, Fervers B, Manjer J, Tjonneland A, Olsen A, Overvad K. Active and passive cigarette smoking and breast cancer risk: results from the EPIC cohort. Int J Cancer. 2014;134:1871–1888. doi: 10.1002/ijc.28508. [DOI] [PubMed] [Google Scholar]
- 7.Gaudet MM, Gapstur SM, Sun J, Diver WR, Hannan LM, Thun MJ. Active smoking and breast cancer risk: original cohort data and meta-analysis. J Natl Cancer Inst. 2013;105:515–525. doi: 10.1093/jnci/djt023. [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg L, Boggs DA, Bethea TN, Wise LA, Adams-Campbell LL, Palmer JR. A prospective study of smoking and breast cancer risk among African-American women. Cancer Causes Control. 2013;24:2207–2215. doi: 10.1007/s10552-013-0298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mechanic LE, Millikan RC, Player J, de Cotret AR, Winkel S, Worley K, Heard K, Heard K, Tse CK, Keku T. Polymorphisms in nucleotide excision repair genes, smoking and breast cancer in African Americans and whites: a population-based case-control study. Carcinogenesis. 2006;27:1377–1385. doi: 10.1093/carcin/bgi330. [DOI] [PubMed] [Google Scholar]
- 10.Xue F, Willett WC, Rosner BA, Hankinson SE, Michels KB. Cigarette smoking and the incidence of breast cancer. Arch Intern Med. 2011;171:125–133. doi: 10.1001/archinternmed.2010.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catsburg C, Miller AB, Rohan TE. Active cigarette smoking and risk of breast cancer. Int J Cancer. 2015;136:2204–2209. doi: 10.1002/ijc.29266. [DOI] [PubMed] [Google Scholar]
- 12.Hall IJ, Moorman PG, Millikan RC, Newman B. Comparative analysis of breast cancer risk factors among African-American women and White women. Am J Epidemiol. 2005;161:40–51. doi: 10.1093/aje/kwh331. [DOI] [PubMed] [Google Scholar]
- 13.McKenzie F, Ellison-Loschmann L, Jeffreys M, Firestone R, Pearce N, Romieu I. Cigarette smoking and risk of breast cancer in a New Zealand multi-ethnic case-control study. PLoS ONE. 2013;8(4):e63132. doi: 10.1371/journal.pone.0063132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gram IT, Park SY, Kolonel LN, Maskarinec G, Wilkens LR, Henderson BE, Le Marchand L. Smoking and risk of breast cancer in a racially/ethnically diverse population of mainly women who do not drink alcohol: the MEC study. Am J Epidemiol. 2015;182:917–925. doi: 10.1093/aje/kwv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai M, Malone KE, Tang MT, Li CI. Active smoking and the risk of estrogen receptor-positive and triple-negative breast cancer among women ages 20 to 44 years. Cancer. 2014;120:1026–1034. doi: 10.1002/cncr.28402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler EN, Tse CK, Bell ME, Conway K, Olshan AF, Troester MA. Active smoking and risk of Luminal and Basal-like breast cancer subtypes in the Carolina Breast Cancer Study. Cancer Causes Control. 2016;27:775–786. doi: 10.1007/s10552-016-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manjer J, Malina J, Berglund G, Bondeson L, Garne JP, Janzon L. Smoking associated with hormone receptor negative breast cancer. Int J Cancer. 2001;91:580–584. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1091>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 18.Nishino Y, Minami Y, Kawai M, Fukamachi K, Sato I, Ohuchi N, Kakugawa Y. Cigarette smoking and breast cancer risk in relation to joint estrogen and progesterone receptor status: a case-control study in Japan. Springerplus. 2014;3:65. doi: 10.1186/2193-1801-3-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morabia A, Bernstein M, Ruiz J, Héritier S, Diebold Berger S, Borisch B. Relation of smoking to breast cancer by estrogen receptor status. Int J Cancer. 1998;75:339–342. doi: 10.1002/(sici)1097-0215(19980130)75:3<339::aid-ijc2>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Cooper JA, Rohan TE, Cant EL, Horsfall DJ, Tilley WD. Risk factors for breast cancer by oestrogen receptor status: a population-based case-control study. Br J Cancer. 1989;59:119–125. doi: 10.1038/bjc.1989.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Passarelli MN, Newcomb PA, Hampton JM, Trentham-Dietz A, Titus LJ, Egan KM, Baron JA, Willett WC. Cigarette smoking before and after breast cancer diagnosis: mortality from breast cancer and smoking-related diseases. J Clin Oncol. 2016;34:1315–1322. doi: 10.1200/JCO.2015.63.9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braithwaite D, Izano M, Moore DH, Kwan ML, Tammemagi MC, Hiatt RA, Kerlikowske K, Kroenke CH, Sweeney C, Habel L, Castillo A, Weltzien E, Caan B. Smoking and survival after breast cancer diagnosis: a prospective observational study and systematic review. Breast Cancer Res Treat. 2012;136:521–533. doi: 10.1007/s10549-012-2276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 24.Lo PK, Sukumar S. Epigenomics and breast cancer. Pharmacogenomics. 2008;9:1879–1902. doi: 10.2217/14622416.9.12.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, Haile RW, Laird PW. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet. 2012;131:1565–1589. doi: 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KW, Pausova Z. Cigarette smoking and DNA methylation. Front Genet. 2013;4:132. doi: 10.3389/fgene.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P, Flanagan JM. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22:843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 28.Zeilinger S, Kühnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, Strauch K, Waldenberger M, Illig T. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE. 2013;8(5):e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guida F, Sandanger TM, Castagné R, Campanella G, Polidoro S, Palli D, Krogh V, Tumino R, Sacerdote C, Panico S, Severi G, Kyrtopoulos SA, Georgiadis P, Vermeulen RC, Lund E, Vineis P, Chadeau-Hyam M. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum Mol Genet. 2015;24:2349–2359. doi: 10.1093/hmg/ddu751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dogan MV, Shields B, Cutrona C, Gao L, Gibbons FX, Simons R, Monick M, Brody GH, Tan K, Beach SR, Philibert RA. The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genom. 2014;15:151. doi: 10.1186/1471-2164-15-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsaprouni LG, Yang TP, Bell J, Dick KJ, Kanoni S, Nisbet J, Viñuela A, Grundberg E, Nelson CP, Meduri E, Buil A, Cambien F, Hengstenberg C, Erdmann J, Schunkert H, Goodall AH, Ouwehand WH, Dermitzakis E, Spector TD, Samani NJ, Deloukas P. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics. 2014;9:1382–1396. doi: 10.4161/15592294.2014.969637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan Q, Wang G, Huang J, Ding Z, Luo Q, Mok T, Tao Q, Lu S. Epigenomic analysis of lung adenocarcinoma reveals novel DNA methylation patterns associated with smoking. Onco Targets Ther. 2013;6:1471–1479. doi: 10.2147/OTT.S51041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato T, Arai E, Kohno T, Takahashi Y, Miyata S, Tsuta K, Watanabe S, Soejima K, Betsuyaku T, Kanai Y. Epigenetic clustering of lung adenocarcinomas based on DNA methylation profiles in adjacent lung tissue: its correlation with smoking history and chronic obstructive pulmonary disease. Int J Cancer. 2014;135:319–334. doi: 10.1002/ijc.28684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilhelm-Benartzi CS, Koestler DC, Houseman EA, Christensen BC, Wiencke JK, Schned AR, Karagas MR, Kelsey KT, Marsit CJ. DNA methylation profiles delineate etiologic heterogeneity and clinically important subgroups of bladder cancer. Carcinogenesis. 2010;31:1972–1976. doi: 10.1093/carcin/bgq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Besaratinia A, Cockburn M, Tommasi S. Alterations of DNA methylome in human bladder cancer. Epigenetics. 2013;8:1013–1022. doi: 10.4161/epi.25927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolff EM, Chihara Y, Pan F, Weisenberger DJ, Siegmund KD, Sugano K, Kawashima K, Laird PW, Jones PA, Liang G. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. 2010;70:8169–8178. doi: 10.1158/0008-5472.CAN-10-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman B, Moorman P, Millikan R, Qaqish B, Geradts J, Aldrich T, Liu ET. The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast Cancer Res Treat. 1995;34:51–60. doi: 10.1007/BF00694745. [DOI] [PubMed] [Google Scholar]
- 38.Dressler LG, Geradts J, Burroughs M, Cowan D, Millikan RC, Newman B. Policy guidelines for the utilization of formalin-fixed, paraffin-embedded tissue sections: the UNC SPORE experience. University of North Carolina Specialized Program of Research Excellence. Breast Cancer Res Treat. 1999;58:31–39. doi: 10.1023/a:1006354627669. [DOI] [PubMed] [Google Scholar]
- 39.Conway K, Edmiston SN, May R, Kuan PF, Chu H, Bryant C, Tse CK, Swift-Scanlan T, Geradts J, Troester MA, Millikan RC. DNA methylation profiling in the Carolina Breast Cancer Study defines cancer subclasses differing in clinicopathologic characteristics and survival. Breast Cancer Res. 2014;16(5):450. doi: 10.1186/s13058-014-0450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS, Millikan RC. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 41.Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, Yang AS. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet. 2009;18:4808–4817. doi: 10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hair BY, Troester MA, Edmiston SN, Parrish EA, Robinson WR, Wu MC, Olshan AF, Swift-Scanlan T, Conway K. Body mass index is associated with gene methylation in estrogen receptor-positive breast tumors. Cancer Epidemiol Biomarkers Prev. 2015;24:580–586. doi: 10.1158/1055-9965.EPI-14-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christensen Christensen BC, Kelsey KT, Zheng S, Houseman EA, Marsit CJ, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Kushi LH, Kwan ML, Wiencke JK. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010;6(7):e1001043. doi: 10.1371/journal.pgen.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bardowell SA, Parker J, Fan C, Crandell J, Perou CM, Swift-Scanlan T. Differential methylation relative to breast cancer subtype and matched normal tissue reveals distinct patterns. Breast Cancer Res Treat. 2013;142:365–380. doi: 10.1007/s10549-013-2738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith IM, Mydlarz WK, Mithani SK, Califano JA. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer. 2007;121:1724–1728. doi: 10.1002/ijc.22889. [DOI] [PubMed] [Google Scholar]
- 47.Tessema M, Yingling CM, Liu Y, Tellez CS, Van Neste L, Baylin SS, Belinsky SA. Genome-wide unmasking of epigenetically silenced genes in lung adenocarcinoma from smokers and never smokers. Carcinogenesis. 2014;35:1248–1257. doi: 10.1093/carcin/bgt494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ambatipudi S, Cuenin C, Hernandez-Vargas H, Ghantous A, Le Calvez-Kelm F, Kaaks R, Barrdahl M, Boeing H, Aleksandrova K, Trichopoulou A, Lagiou P, Naska A, Palli D, Krogh V, Polidoro S, Tumino R, Panico S, Buenode-Mesquita B, Peeters PH, Quirós JR, Navarro C, Ardanaz E, Dorronsoro M, Key T, Vineis P, Murphy N, Riboli E, Romieu I, Herceg Z. Tobacco smoking-associated genome-wide DNA methylation changes in the EPIC study. Epigenomics. 2016;8:599–618. doi: 10.2217/epi-2016-0001. [DOI] [PubMed] [Google Scholar]
- 49.White AJ, Chen J, Teitelbaum SL, McCullough LE, Xu X, Hee Cho Y, Conway K, Beyea J, Stellman SD, Steck SE, Mordukhovich I, Eng SM, Beth Terry M, Engel LS, Hatch M, Neugut AI, Hibshoosh H, Santella RM, Gammon MD. Sources of polycyclic aromatic hydrocarbons are associated with gene-specific promoter methylation in women with breast cancer. Environ Res. 2016;145:93–100. doi: 10.1016/j.envres.2015.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.United States Department of Health and Human Services. How tobacco smoke causes disease: the biology and behavioral basis for smoking-attributable disease: a report of the surgeon general. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease, Prevention and Health Promotion, Office of Smoking and Health; Atlanta: 2010. [PubMed] [Google Scholar]
- 51.Hecht SS. Lung carcinogenesis by tobacco smoke. Int J Cancer. 2012;131:2724–2732. doi: 10.1002/ijc.27816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen RJ, Chang LW, Lin P, Wang YJ. Epigenetic effects and molecular mechanisms of tumorigenesis induced by cigarette smoke: an overview. J Oncol. 2011;2011:654931. doi: 10.1155/2011/654931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ginzkey C, Friehs G, Koehler C, Hackenberg S, Hagen R, Kleinsasser NH. Assessment of nicotine-induced DNA damage in a genotoxicological test battery. Mutat Res. 2013;751:34–39. doi: 10.1016/j.mrgentox.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Nishioka T, Kim HS, Luo LY, Huang Y, Guo J, Chen CY. Sensitization of epithelial growth factor receptors by nicotine exposure to promote breast cancer cell growth. Breast Cancer Res. 2011;13:R113. doi: 10.1186/bcr3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guha P, Bandyopadhyaya G, Polumuri SK, Chumsri S, Gade P, Kalvakolanu DV, Ahmed H. Nicotine promotes apoptosis resistance of breast cancer cells and enrichment of side population cells with cancer stem cell-like properties via a signaling cascade involving galectin-3, α9 nicotinic acetylcholine receptor and STAT3. Breast Cancer Res Treat. 2014;145:5–22. doi: 10.1007/s10549-014-2912-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin RK, Hsieh YS, Lin P, Hsu HS, Chen CY, Tang YA, Lee CF, Wang YC. The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. J Clin Invest. 2010;120:521–532. doi: 10.1172/JCI40706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee GE, Kim JH, Taylor M, Muller MT. DNA methyl-transferase 1-associated protein (DMAP1) is a co-repressor that stimulates DNA methylation globally and locally at sites of double strand break repair. J Biol Chem. 2010;285:37630–37640. doi: 10.1074/jbc.M110.148536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Satta R, Maloku E, Zhubi A, Pibiri F, Hajos M, Costa E, Guidotti A. Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc Natl Acad Sci USA. 2008;105:16356–16361. doi: 10.1073/pnas.0808699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bavarva JH, Tae H, Settlage RE, Garner HR. Characterizing the genetic basis for nicotine induced cancer development: a transcriptome sequencing study. PLoS ONE. 2013;8(6):e67252. doi: 10.1371/journal.pone.0067252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mercer BA, Wallace AM, Brinckerhoff CE, D’Armiento JM. Identification of a cigarette smoke-responsive region in the distal MMP-1 promoter. Am J Respir Cell Mol Biol. 2009;40:4–12. doi: 10.1165/rcmb.2007-0310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di YP, Zhao J, Harper R. Cigarette smoke induces MUC5AC protein expression through the activation of Sp1. J Biol Chem. 2012;287:27948–27958. doi: 10.1074/jbc.M111.334375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Q, Liu L, Zhao Y, Zhang J, Wang D, Chen J, He Y, Wu J, Zhang Z, Liu Z. Hypoxia induces genomic DNA demethylation through the activation of HIF-1α and transcriptional upregulation of MAT2A in hepatoma cells. Mol Cancer Ther. 2011;10:1113–1123. doi: 10.1158/1535-7163.MCT-10-1010. [DOI] [PubMed] [Google Scholar]
- 64.Gabriel HE, Crott JW, Ghandour H, Dallal GE, Choi SW, Keyes MK, Jang H, Liu Z, Nadeau M, Johnston A, Mager D, Mason JB. Chronic cigarette smoking is associated with diminished folate status, altered folate form distribution, and increased genetic damage in the buccal mucosa of healthy adults. Am J Clin Nutr. 2006;83:835–841. doi: 10.1093/ajcn/83.4.835. [DOI] [PubMed] [Google Scholar]
- 65.Pounis G, Di Castelnuovo AF, de Lorgeril M, Krogh V, Siani A, Arnout J, Cappuccio FP, van Dongen M, Zappacosta B, Donati MB, de Gaetano G, Iacoviello L. European Collaborative Group of the IMMIDIET Project. Folate intake and folate serum levels in men and women from two European populations: the IMMIDIET project. Nutrition. 2014;30:822–830. doi: 10.1016/j.nut.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 66.Necela BM, Crozier JA, Andorfer CA, Lewis-Tuffin L, Kachergus JM, Geiger XJ, Kalari KR, Serie DJ, Sun Z, Moreno-Aspitia A, O’Shannessy DJ, Maltzman JD, McCullough AE, Pockaj BA, Cunliffe HE, Ballman KV, Thompson EA, Perez EA. Folate receptor-α (FOLR1) expression and function in triple negative tumors. PLoS ONE. 2015;10(3):e0122209. doi: 10.1371/journal.pone.0122209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Askari M, Sobti RC, Nikbakht M, Sharma SC. Aberrant promoter hypermethylation of p21 (WAF1/CIP1) gene and its impact on expression and role of polymorphism in the risk of breast cancer. Mol Cell Biochem. 2013;382:19–26. doi: 10.1007/s11010-013-1696-5. [DOI] [PubMed] [Google Scholar]
- 69.Akhter N, Akhtar MS, Ahmad MM, Haque S, Siddiqui S, Hasan SI, Shukla NK, Husain SA. Association of mutation and hypermethylation of p21 gene with susceptibility to breast cancer: a study from north India. Mol Biol Rep. 2014;41:2999–3007. doi: 10.1007/s11033-014-3159-9. [DOI] [PubMed] [Google Scholar]
- 70.Felix AS, Sherman ME, Hewitt SM, Gunja MZ, Yang HP, Cora RL, Boudreau V, Ylaya K, Lissowska J, Brinton LA, Wentzensen N. Cell-cycle protein expression in a population-based study of ovarian and endometrial cancers. Front Oncol. 2015;5:25. doi: 10.3389/fonc.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuzhalin AE, Kutikhin AG. Interleukin-12: clinical usage and molecular markers of cancer susceptibility. Growth Factors. 2012;30:176–191. doi: 10.3109/08977194.2012.678843. [DOI] [PubMed] [Google Scholar]
- 72.Kaarvatn MH, Vrbanec J, Kulic A, Knezevic J, Petricevic B, Balen S, Vrbanec D, Dembic Z. Single nucleotide polymorphism in the interleukin 12B gene is associated with risk for breast cancer development. Scand J Immunol. 2012;76:329–335. doi: 10.1111/j.1365-3083.2012.02736.x. [DOI] [PubMed] [Google Scholar]
- 73.Lei J, Rudolph A, Moysich KB, Rafiq S, Behrens S, Goode EL, Pharoah PP, Seibold P, Fasching PA, Andrulis IL, Kristensen VN, Couch FJ, Hamann U, Hooning MJ, Nevanlinna H, Eilber U, Bolla MK, Dennis J, Wang Q, Lindblom A, Mannermaa A, Lambrechts D, García-Closas M, Hall P, Chenevix-Trench G, Shah M, Luben R, Haeberle L, Ekici AB, Beckmann MW, Knight JA, Glendon G, Tchatchou S, Alnæs GI, Borresen-Dale AL, Nord S, Olson JE, Hallberg E, Vachon C, Torres D, Ulmer HU, Rüdiger T, Jager A, van Deurzen CH, Tilanus-Linthorst MM, Muranen TA, Aittomäki K, Blomqvist C, Margolin S, Kosma VM, Hartikainen JM, Kataja V, Hatse S, Wildiers H, Smeets A, Figueroa J, Chanock SJ, Lissowska J, Li J, Humphreys K, Phillips KA, Linn S, Cornelissen S, van den Broek SA, Kang D, Choi JY, Park SK, Yoo KY, Hsiung CN, Wu PE, Hou MF, Shen CY, Teo SH, Taib NA, Yip CH, Ho GF, Matsuo K, Ito H, Iwata H, Tajima K, Dunning AM, Benitez J, Czene K, Sucheston LE, Maishman T, Tapper WJ, Eccles D, Easton DF, Schmidt MK, Chang-Claude MK kConFab Investigators. Assessment of variation in immunosuppressive pathway genes reveals TGFBR2 to be associated with prognosis of estrogen receptor-negative breast cancer after chemotherapy. Breast Cancer Res. 2015;17:18. doi: 10.1186/s13058-015-0522-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Allott EH, Cohen SM, Geradts J, Sun X, Khoury T, Bshara W, Zirpoli GR, Miller CR, Hwang H, Thorne LB, O’Connor S, Tse CK, Bell MB, Hu Z, Li Y, Kirk EL, Bethea TN, Perou CM, Palmer JR, Ambrosone CB, Olshan AF, Troester MA. Performance of three-biomarker immunohistochemistry for intrinsic breast cancer subtyping in the AMBER Consortium. Cancer Epidemiol Biomarkers Prev. 2016;25:470–478. doi: 10.1158/1055-9965.EPI-15-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.