Abstract
Analysis of the clinical characteristics of hematopoietic stem cell transplant (HSCT) donors has proven beneficial for identifying cases of heritable hematopoietic disorders. This study examines poor peripheral blood hematopoietic stem cell mobilization after granulocyte colony–stimulating factor administration among 328 donors as a potential marker for suspected familial predisposition to myeloid malignancies. Here, we present data comparing the clinical characteristics of poor-mobilizing versus nonpoor-mobilizing donors and the results of panel-based sequencing of hematopoietic genes in poor-mobilizing donors. From this analysis, we identified a novel case of a donor-derived myelodysplastic syndrome in an HSCT recipient that is consistent with clonal evolution of TET2-mutated clonal hematopoiesis of indeterminate potential (CHIP) within the donor. This study demonstrates the potential risk of using hematopoietic stem cells from a donor with CHIP and raises the question of whether there should be increased screening measures to identify such donors.
Keywords: Donor-derived leukemia, TET2 mutation, Clonal hematopoiesis of indeterminate potential
INTRODUCTION
Mobilized peripheral blood stem cells (PBSC) are the most frequent source of hematopoietic stem cells (HSC) used for allogeneic HSC transplantation (HSCT). However, genetic factors contributing to donors who mobilize PBSC poorly, and how this affects transplantation outcomes, are not well understood. Previously, our laboratory identified a case of germline predisposition to myeloid malignancies by studying related allogeneic stem cell donors who had baseline unexplained thrombocytopenia as a marker for identifying familial myelodysplastic syndrome (MDS)/acute leukemia predisposition syndromes [1]. Because poor mobilization has been observed in individuals with other heritable hematopoietic disorders [2], we hypothesized that we could identify additional individuals and families at high risk for having a germline predisposition allele by examining related allogeneic HSC donors who mobilized low numbers of PBSCs. Here, we present data of the results of panel-based sequencing of hematopoietic genes in poor-mobilizing donors.
MATERIALS AND METHODS
Subjects and Samples
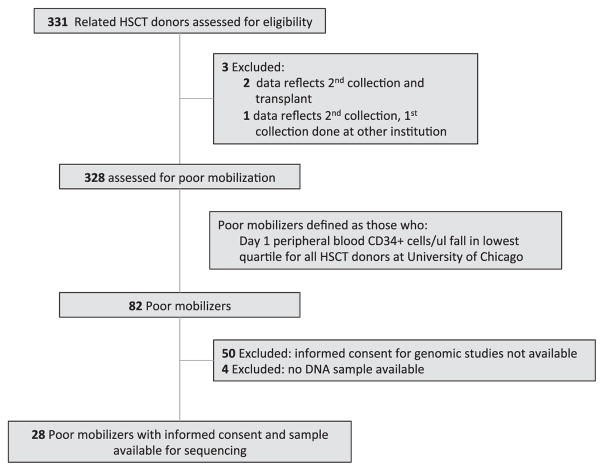
We evaluated 328 HLA-matched related HSC donors who underwent PBSC mobilization at The University of Chicago from 2001 to 2011 for transplantation into a first-degree relative with a hematopoietic malignancy. CD34+ cells in the peripheral blood (PB) were measured by flow cytometry on day 5 of mobilization after 4 to 5 days of granulocyte colony–stimulating factor (G-CSF) at 10 mcg/kg/day. We defined poor mobilizers as those whose CD34+ cell numbers fell within the lowest quartile among all donors, which included those with day 1 PB CD34+ counts ≤55.0 cells/μL). Supplemental Table S1 lists all donors who had abnormalities demonstrable in the screening complete blood cell count, regardless of mobilization parameters. Retrospective analysis of matched related transplantation data was approved by The University of Chicago institutional review board, and informed consent was obtained from 28 subjects identified as poor mobilizers who had samples available for sequencing.
Targeted Gene Panel Sequencing
Genomic DNA isolated from each donor’s mobilized PBSC product collected before transplantation was screened for mutations utilizing MarrowSeq (The University of Washington Medical Center Genetics and Solid Tumor Diagnostic Laboratory, Seattle, WA), a targeted next-generation sequencing panel (300X to 500X coverage), which includes 142 genes responsible for inherited and acquired bone marrow failure syndromes and MDS [3]. Targeted gene capture, sequencing, and analysis were performed using established protocols, and variants that were potentially damaging were confirmed by Sanger sequencing [3]. OncoHeme (The University of Chicago Department of Pathology, Chicago, IL), a second deep-sequencing next-generation sequencing panel (1000X coverage) targeting the exons of 54 genes recurrently mutated in MDS/acute leukemia, was used to identify additional acquired mutations in 1 donor/recipient pair.
RESULTS
We analyzed the clinical parameters documented on day 1 of PB collection for all matched related donors presenting for transplantation. Baseline characteristics of donors falling into the poor-mobilizer and nonpoor-mobilizer categories are given in Table 1 and Supplemental Table S2. Poor mobilizers were defined as those whose PB CD34+ cells/μL on the first day after 5 days of G-CSF administration (“day 1”) fell within the lowest quartile of the 328 donors. Among the 82 poor mobilizers, genomic DNA samples and consent were available from 28 donors (Figure 1). Poor mobilizers were significantly older (P < .001), had higher mean corpuscular volume (MCVs) (P < .001), lower total white blood cell counts (P = .03), and lower platelet counts (P = .02) than nonpoor mobilizers (Table 1).
Table 1.
Poor Mobilizer and Nonpoor Mobilizer Donor Clinical Characteristics
| Characteristic | All Donors (n = 328) | Poor Mobilizers (n = 82) | Nonpoor Mobilizers (n = 246) | P Value (Poor mobilizers versus nonpoor mobilizers) |
|---|---|---|---|---|
| Day 1 PB CD34+ counts, median, cells/μL | 86.0 | 40.7 | 107.5 | |
| range (2.0–421.9) | IQR (29.9–48.3) | IQR (75.4–141.6) | ||
| Female | 45.43% | 45.12% | 45.53% | .95 |
| Age, median, yr | 47.5 | 55 | 46 | < .001 |
| range (13–74) | IQR (43–62) | IQR (37–54) | ||
| WBC, median, K/μL | 6.6* | 6.45* | 6.7* | .03 |
| range (3.1–13.7) | IQR (5.2–7.9) | IQR (5.6–8.1) | ||
| Platelet count, median, K/μL | 250* | 240* | 255 | .02 |
| range (109–459) | IQR (201–275) | IQR (218–293) | ||
| MCV, median, fL | 89.3* | 90.8* | 88.8* | < .001 |
| range (64.7–107.6) | IQR (87.9–94.3) | IQR (85.6–91.5) |
Day 1 PB CD34+ counts were obtained after mobilization of PB HSC by G-CSF. WBC counts, platelet counts, and MCV values were taken before mobilization. IQR indicates interquartile range.
Pre-donation CBC values missing for one donor.
Figure 1.
Flow diagram of sample analysis. Flow diagram shows how related donors were deemed eligible for this study.
Among the 28 poor mobilizers sequenced using MarrowSeq (The University of Washington Medical Center Genetics and Solid Tumor Diagnostic Laboratory), clearly damaging mutations were identified in 2 individuals (7%). The first was a 63-year-old donor with mild thrombocytopenia (platelet count, 136 K/μL) and macrocytosis (MCV, 107.8 fL) whom we had reported previously with a novel deleterious germline TERT mutation (c.2908A>G; p.M970V; 48% allelic ratio), identified in the recipient’s leukemia as well [1]. Mutations in TERT are known to cause an inherited autosomal dominant telomere biology disorder [4], and we demonstrated that all lymphocyte subsets from the donor had short or very short telomeres by flow fluorescein in situ hybridization [1].
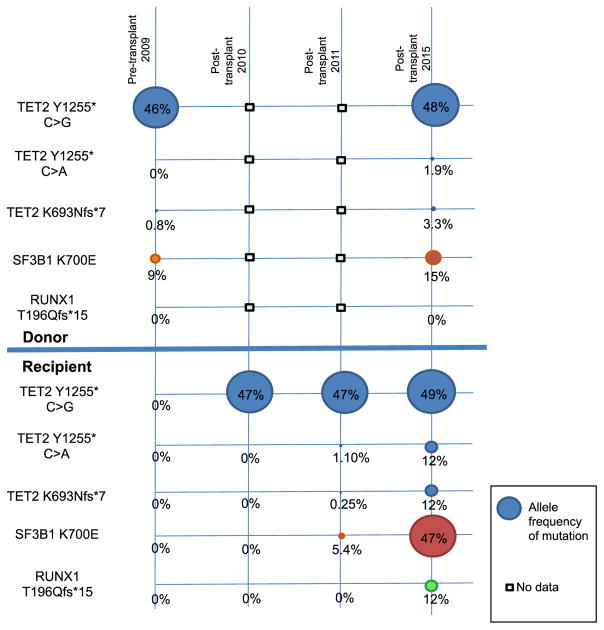
The second poor mobilizer with a deleterious mutation was a 67-year-old mildly thrombocytopenic donor (platelet count, 139 K/μL). The mobilized PBSC product contained 3 damaging mutations in TET2: c.2079del (p.K693Nfs*7; allelic ratio .8%) and c.3765C>G (p.Y1255*; allelic ratio 46%), as well as a deleterious mutation in SF3B1: c.2098A>G (p.K700E; allelic ratio 9%). Follow-up of this donor 6 years after these mutated PBSCs were collected documented persistent mild thrombocytopenia (platelet count, 121 K/μL) but otherwise normal blood counts: total white blood cell count of 5000/μL and hemoglobin of 15.6 g/dL. Molecular profiling indicated some shift of mutant allele fractions within this donor’s PB: TET2: c.2079del (p.K693Nfs*7; allelic ratio 3.3%), c.3765C>A (p.Y1255*; allelic ratio 1.9%), and c.3765C>G (p.Y1255*; allelic ratio 48%); and SF3B1: c.2098A>G (p.K700E; allelic ratio 15%).
The recipient achieved remission after reinduction chemotherapy for relapsed acute myeloid leukemia followed by matched related HSCT in 2009. Sequencing of genomic DNA from the recipient’s bone marrow 1 year after transplantation showed only the TET2 c.3765 C > G mutation (p.Y1255*; allelic ratio 47%). Two years after transplantation, however, molecular profiling of the bone marrow identified the same 3 TET2 mutations: c.3765 C > G mutation p.Y1255*; allelic ratio 47%), c.3765C>A (p.Y1255*; allelic ratio 1.1%), and c.2079del (p.K693Nfs*7; allelic ratio .25%); as well as the mutation in SF3B1 c.2098A>g (p.K700E; allelic ratio 5.4%). After HSCT, the recipient was clinically well with mild thrombocytopenia, similar to her donor, until 6 years after HSCT, when she developed transfusion-dependent anemia. A bone marrow biopsy demonstrated refractory anemia with excess blasts-2. Karyotype and microsatellite marker analysis of this bone marrow showed 95% of the cells were of male donor origin. OncoHeme sequencing of this marrow showed expansion of each of the mutated clones as well as acquisition of a new RUNX1 mutation: c.585del (p.T196Qfs*15; allelic ratio 12%) (Figure 2). Although donor germline tissue was not available, these mutations were not present in the recipient’s skin fibroblasts, making a germline hematologic malignancy syndrome due to 1 of these mutations less likely. Thus, these findings are most consistent with the presence of clonal hematopoiesis of indeterminate potential (CHIP) associated with TET2 and SF3B1 mutations in the donor [5,6], with transfer of mutation carrying cells to the recipient at the time of allogeneic stem cell transplantation.
Figure 2.
Timeline of the acquired mutations identified within the donor and recipient. Molecular profiling and allele fractions from samples at various time points are given in vertical columns. The earliest time point in the chronology is before transplantation. Data here include mobilized PBSC product from the donor and data from the recipient’s skin fibroblasts at this time frame, although they were collected after HSCT. Subsequent samples are shown in columns to the right. Allele fractions are given in circles, with the size of the circle proportional to allele fraction.
DISCUSSION
Given the age distribution of the 28 poor mobilizers whose DNA was available for sequencing (ages 60 to 69, n = 7; ages 70 to 79, n = 5; >65, n = 7), we would expect clonal hematopoiesis to be present in 5.6% of 60 to 69-year-old individuals and 9.5% of those 70 to 79 years old, or in 10% of subjects >65 years old [5,6]. We identified clonal hematopoiesis in 1 67-year-old individual in our small sample (14% of the 7 people 60 to 69 years old, or 14% of the 7 people older than 65 years old]. Thus, our results are consistent with published results [5,6].
Among the 28 sequenced poor-mobilizing donors, 3 donors had a single cytopenia [6] and no donor had more than 1 cytopenia (Supplemental Table S1). Among the 3 with a single cytopenia, a mutation in a hematopoietic gene was identified in 2 (1 with CHIP and 1 with a germline TERT mutation). Notably, all 3 with a single cytopenia had an additional complete blood count abnormality. Specifically, 2 had an elevated RDW as the single other abnormality. An elevated Red blood cell Distribution Width (RDW) was also associated with DNMT3A mutations and presence of a mutation versus absence of mutations in those with MCV >86 fL [6], suggesting that this complete blood count parameter may be a clinically useful marker of the presence of a mutation. However, given our small sample size, this should be explored further in a larger population of stem cell donors. Prior studies have shown that gross chromosomal abnormalities after G-CSF administration are not over-represented versus healthy controls [7]. Taken together, our data suggest that among poor-mobilizing donors, a single cytopenia, especially if the cytopenia is thrombocytopenia or is accompanied by an elevated RDW, warrants evaluation for hereditary or acquired mutations that increase the risk of future hematologic malignancy development. Given the curative intent of transplantation and first principle to do no harm to either recipient or donor, our data along with other published work [5,6] suggest that healthy donors with even a single unexplained cytopenia should undergo genetic evaluation.
Here, we demonstrate that both inherited and acquired genetic factors in PBSC from apparently healthy donors can contribute to poor mobilization and donor-derived malignancy after HSCT. Furthermore, to the best of our knowledge, this study presents the first example of clonal evolution of TET2- and SF3B1-mutated CHIP within a donor resulting in a donor-derived leukemia within the HSCT recipient. Replicative stress to reconstitute hematopoiesis in the recipient and/or the recipient’s damaged bone marrow microenvironment may have contributed to the development of this donor-derived leukemia [8], while the donor remains healthy despite persistent thrombocytopenia. Results of this study suggest that poor mobilizers with thrombocytopenia may carry inherited or acquired mutations in hematopoietic genes with the potential to adversely affect short-term and long-term transplantation outcomes. Genomic investigations of these donors or use of an alternative donor should be considered. Our study also raises concerns about whether we need to screen for CHIP in healthy donors with unexplained cytopenias, especially as donors of increasing age are utilized.
Supplementary Material
Acknowledgments
This research was supported by the American Society of Hematology through the ASH HONORS Award (award no. 061273 to KR), the Cancer Research Foundation (award no. 060015 to JEC and LAG), and NCI K12 CA139160/NHLBI K08 HL129088 (to JEC).
Financial disclosure: None of the authors has any conflicts of interests to disclose.
Authorship statement: K.R. conducted the experiments, compiled data, and cowrote the manuscript. E.N. conducted the experiments and compiled data. B.N. assisted with patient identification. R.M. assisted with patient identification and performing the experiments. A.W. supervised collection of hematopoietic stem cells. A.A. and K.v.B. cosupervised evaluation of HLA-matched related donors and allogeneic stem cell transplantations. R.A.L. cosupervised allogeneic stem cell transplantations. M.K.L., J.P.S., M-C.K., T.W., A.S., and S.B.K. contributed to next-generation sequencing and its analysis. J.E.C. contributed to next-generation sequencing and its analysis, analyzed data, and cowrote the manuscript. L.A.G. designed the research, supervised the conduct of the work and its analysis, and cowrote the manuscript. J.E.C. and L.A.G. contributed equally.
Footnotes
Supplementary data related to this article can be found online at doi:10.1016/j.bbmt.2016.08.002.
References
- 1.Churpek JE, Nickels E, Marquez R, et al. Identifying familial myelodysplastic/acute leukemia predisposition syndromes through hematopoietic stem cell transplantation donors with thrombocytopenia. Blood. 2012;120:5247–5249. doi: 10.1182/blood-2012-09-457945. [DOI] [PubMed] [Google Scholar]
- 2.Fogarty PF, Yamaguchi H, Wiestner A, et al. Late presentation of dyskeratosis congenita as apparently acquired aplastic anaemia due to mutations in telomerase RNA. Lancet. 2003;362:1628–1630. doi: 10.1016/S0140-6736(03)14797-6. [DOI] [PubMed] [Google Scholar]
- 3.Zhang MY, Keel SB, Walsh T, et al. Genomic analysis of bone marrow failure and myelodysplastic syndromes reveals phenotypic and diagnostic complexity. Haematologica. 2015;100:42–48. doi: 10.3324/haematol.2014.113456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vulliamy TJ, Walne A, Baskaradas A, Mason PJ, Marrone A, Dokal I. Mutations in the reverse transcriptase component of telomerase (TERT) in patients with bone marrow failure. Blood Cells Mol Dis. 2005;34:257–263. doi: 10.1016/j.bcmd.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olnes MJ, Poon A, Miranda SJ, et al. Effects of granulocyte-colony-stimulating factor on Monosomy 7 aneuploidy in healthy hematopoietic stem cell and granulocyte donors. Transfusion. 2012;52:537–541. doi: 10.1111/j.1537-2995.2011.03313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiseman DH. Donor cell leukemia: a review. Biol Blood Marrow Transplant. 2011;17:771–789. doi: 10.1016/j.bbmt.2010.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.