Abstract
Networks of redox sensor proteins within discrete microdomains regulate the flow of redox signaling. Yet, the inherent reactivity of redox signals complicates the study of specific redox events and pathways by traditional methods. Herein, we review designer chemistries capable of measuring flux and/or mimicking subcellular redox signaling at the cellular and organismal level. Such efforts have begun to decipher the logic underlying organelle-, site-, and target-specific redox signaling in vitro and in vivo. These data highlight chemical biology as a perfect gateway to interrogate how Nature choreographs subcellular redox chemistry to drive precision redox biology.
Graphical abstract
Introduction
Biological systems must respond rapidly to both external and internal stimuli to maintain homeostasis, prevent disease, and repair damage. Protein expression level changes are one way to mount dynamic responses. But to orchestrate a rapid response without relinquishing upstream control, biology often relies on posttranslational modifications (PTMs) of specific proteins to effect changes in activity, function, locale, or stability of preexisting proteins. The resulting signaling cascades can influence decision making on an organelle, cellular, and whole-organism level.
Since there are many possible sources of stress, a complex series of signaling subsystems has evolved.6 The chemical modifications that comprise these systems underpin cellular “signal exchange” (Key words) in which a specific chemical signal is transferred between different proteins or regulates PTMs on other proteins, enabling exquisite control of downstream responses. Traditional signaling events—in which hydrolytically stable adducts are formed between target proteins and the signal—are typically enzyme-catalyzed. Removal of the signal is also enzyme-catalyzed. In an emerging non-canonical signaling mechanism called “redox signaling”, reactive chemicals, including electrophilic and oxygen species (RES/ROS)—the focus of this review—serve as signaling mediators modifying discrete signaling proteins, thereby initiating signaling cascades, largely without enzyme catalysis. In contrast to canonical enzyme-catalyzed signaling, most RES/ROS are inherently reactive and diffuse rapidly, but have relatively short diffusion distances. Thus, these reactive signals likely use proximity, kinetic privilege, and privilege of occupancy (phenotypic dominance; Key words) to initiate specific downstream events with exquisite specificity.5–7 Lack of enzymatic control as well as high context dependence has made understanding redox signaling uniquely challenging.4, 8, 9 Additionally, RES/ROS show hormesis and are toxic at supraphysiological concentrations.
Key Words.
Change (of) Hands: exchange of one redox modification on a specific protein to the same redox modification on another protein/site (e.g., interconversion of disulfide bonds).
Class II Proximity Enhancement: a “virtual tethering strategy” in which a reactive ligand/signal is delivered to a target protein of interest through on-demand precision redox-signal targeting guided by proximity1 (e.g., CALI/FALI and T-REX).
Signal Exchange: conversion of one form of signaling to a gain or loss of another (e.g., a reactive oxygen species (ROS) signal regulating phosphorylation, ubiquitylation, etc.). This occurs at a signaling node.
Signaling Node: a protein at which signal exchange occurs.
Kinetic Privilege: highly reactive proteins under favorable conditions will intercept reactive species faster than less reactive proteins.
Phenotypically Dominant: a signaling event that elicits gain of function or dominant loss of function such that low-occupancy modification(s) on a single protein likely elicit(s) a phenotypic response.
Privilege of Occupancy: phenotypically-dominant modifications require intrinsically low occupancy and so will exert their influence earlier than recessive effects (such as loss of function). Redox Sensor: a protein that senses reactive electrophilic/oxygen species (RES/ROS) (e.g., Keap1,2–5 PTEN2, 5).
Redox Signal: a reactive electrophile or oxidant that can covalently modify a sensor protein eliciting downstream responses (a broad denomination of signaling).
The broader lack of enzyme regulation at the point of RES/ROS signaling initiation and target adduction means this area is ideal for study through chemical biology. This review thus focuses on non-enzymatic redox modifications as signal-initiating/transfer points. We concentrate on customized chemical biology toolsets—mostly developed over the last 5–7 years—aimed at deciphering Nature’s design principles of redox signaling subsystems in cultured mammalian cells and higher eukaryotic models. Emphasis is placed on endogenous RES/ROS. These studies show that redox-modified sensor proteins behave differently from their unmodified state and have unique functions and interactomes.
Since RES/ROS signaling is nuanced, we focus on mechanisms that operate against or exploit the heterogeneous distribution across distinct subcellular or tissue-scale compartments10, and time. Accordingly, the majority of this review is concerned with kinetically-controlled processes that help to couple RES/ROS flux to downstream signaling cascades. Given this constraint, as well as the privileged role cysteine adopts in higher organisms,11 the resurgence of cysteine-targeting covalent drugs,12 and appreciation of acquired cysteine mutations in disease,13 we limit our discussion to cysteine-specific, redox-linked modifications. Other nucleophilic residues (e.g., Histidine, Lysine) are targeted by RES/ROS,14 but to a lesser extent and are primarily detected under bolus RES/ROS treatment or after equilibration.15
Unusual suspects
Both RES and ROS are toxic and cause deleterious effects when misregulated, overproduced, or administered for a prolonged time. Only ROS are actual oxidants; RES influences redox balance through modulation of specific protein activity/function and depleting reducing agents in a redox-independent process. There are relatively few chemotypes of ROS. Endogenous ROS are mostly generated by enzymes with overlapping function/regulation,16 and classified based upon the oxidation state of molecular oxygen (Table 1). Unlike RES, ROS can form metal-bound complexes. These complexes may show different physiochemical properties to their parent compounds and can themselves form other ROS.17 These pathways have been implicated in numerous physiological processes, including a recently discovered cell-death pathway, ferroptosis.18
Table 1.
Overview of ROS
| Entry | ROS | Oxidation State | Description | Sources | Half life (s) |
|---|---|---|---|---|---|
| 119, 20 | Singlet oxygen (1O2) | 0 | Unfavored spin state | Many | 10−6 |
| 221 | Superoxide (O2•−) | −1/2 | Reactive radical | Electron transfer from NADPH to O2 by NADPH oxidases | 10−6 |
| 321 | Peroxide (H2O2) | −1 | Two-electron oxidant | Many | 10−3–10−5 |
| 421 | Hydroxyl radical (HO•) | −1 | Reactive radical | Reaction of superoxide with peroxide (Fenton reaction) | 10−9 |
| 522 | Peroxynitrite | Endogenous pseudohalogen | Reaction of superoxide with NO | 10–3 |
ROS engage in both radical and two-electron chemistry, and specific forms can interconvert. For example, 1O2 forms organic peroxides through ene/Diels–Alder chemistry with unsaturated lipids; superoxide is converted to peroxide enzymatically. Thus it can be hard to ascertain which ROS is responsible for a specific bioactivity. The half-lives of ROS also vary, although these calculations are approximations. 1O2, OH• and O2•− are universally acknowledged to be short-lived with particularly short diffusion distances; peroxides are at least an order of magnitude longer-lived than the other species discussed here (Table 1).21 However, even assuming the upper estimate of half-life for peroxide (1 ms), the resulting diffusion length for peroxide is no more than a tenth of the length of a mammalian cell,23 meaning that even peroxide is localized in cells. Many ROS forms are metabolized enzymatically through direct enzyme–ROS reaction, concurrently modifying these enzymes largely reversibly. Interestingly, these modified enzymes can orchestrate specific signaling events as we discuss in subsequent sections.
There are many types of endogenous bioactive RES. Most of these form aliphatic or aromatic adducts to nucleophilic residues that are kinetically stable. There are some RES-based acylating agents that kinetically react with cysteine, but can be transferred to other residues like lysine in a context-specific manner.15 Common examples of endogenous RES that form kinetically-stable adducts include: enals [e.g., 4-hydroxynonenal (HNE)]; enones (e.g., 15-deoxy-Δ12,14-prostaglandin-J2); α-chloro aldehydes (e.g., 2-chlorohexadecanal);24 and quinones (e.g., vitamin K). There are also dietary plant-based RES of which the most common are isothiocyanates.25 Isothiocyanates form thiocarbamylated enzyme adducts that initially form cysteine adducts that may transfer to lysine, depending on conditions/protein.15 There are also well-known synthetic RES including electrophilic drugs, such as dimethyl fumarate (Tecfidera®) approved for the treatment of multiple sclerosis, and bardoxolone methyl in Phase 2/3 trials for various conditions. These compounds likely form irreversible enzyme adducts as they are chemically similar to HNE. Both enzymatic and non-enzymatic lipid peroxidation pathways for the production of RES such as HNE from polyunsaturated fatty acids are known.26 RES are metabolized through enzymatic redox chemistry27 and by carnitine/carnosine and glutathione conjugation—the latter catalyzed by glutathione-S-transferase (GST) in cells.28, 29 Glutathione conjugation to RES does not modify GST, although cellular redox balance is affected due to glutathione depletion. The redox:glutathione adduction pathways partition roughly equally (40 % : 30 %) 2 minutes post exposure.30 Interestingly, the percentage of HNE-protein adducts formed following acute HNE exposure varies significantly depending on cell type [1% (intestinal enterocytes), 3% (hepatocytes), 8.5% (Ehrlich ascites tumor cells) of total HNE] and subcellular locale (30% HNE adducts are mitochondrial).31
General roles of RES and ROS in information transfer
Peroxide interacts with enzymes initially by forming chemically/enzymatically reversible cysteine sulfenic acids (cysteine-SOH). Subsequent oxidations are slower, producing sulfinic acid (cysteine-SO2H, mostly irreversible, although reduced by certain enzymes32) and sulfonic acid (cysteine-SO3H, irreversible).33 These modifications are redox signals.34–36 Sulfenic acid formation exerts a reverse effect on cysteine: sulfenic acids can react with nucleophiles, and are often trapped out by another thiol (signal exchange, Key words). The resulting disulfide bond can form a crosslink between two spatially distinct residues on a protein, inducing a conformational change. These disulfides can also “change hands” (Key words) and oxidize other specific protein thiols to propagate a redox signal through “disulfide relays”. Erv1/ALR and Mia40 engage in such a pathway that ultimately stimulates mitochondrial translocation of p53.37 It has recently been reported that a kinetically controlled ROS-mediated enzyme oxidation event at a signaling node (Key words) can usher an enzyme-orchestrated redox-signaling cascade through disulfide exchange.38 This shows how a “non-targeted,” initiating redox signal can be efficiently exchanged into an alternative signal that can be propagated with specificity.
RES modification can elicit protein conformational changes, presumably to a larger extent in cases in which a protein reacts with bifunctional RES such as HNE (e.g., Michael addition with cysteine and Schiff base formation with lysine6). Since many RES bind proteins irreversibly, they cannot change hands. Thus, degradation of the target protein is usually required to turn off RES signaling. A RES-modulated signaling node thus likely requires signal exchange to propagate cellular information.39 An exception is acylating RES agents, such as plant-derived isothiocyanates,25 that can themselves undergo signal exchange.15
We will discuss below many cellular pathways that are regulated by RES/ROS. Although some redox-dependent processes have not yet been studied using the chemical biology techniques discussed herein, known RES/ROS-sensitive pathways include several essential transcriptional processes mediated by the redox-sensitive transcription factors FOXO,40 NFκB41 and hypoxia inducible factor,42 among others. Numerous mRNA binding proteins have also been shown to be RES/ROS sensitive, including GAPDH.43
(A) Subcellular Redox Target Capture: Molecular Finger Print Analysis
Capture of cysteine thiols/thiolates
Functional cysteines typically have high nucleophilicity. Although an approximation, nucleophilicity generally correlates with low pKa.44 Reported pKa values for cysteines in proteins range from 2.5–12, representing at least a ±4 unit change in pKa with respect to cysteine.45 Methods to identify redox-functional cysteines typically use electrophilic probes to irreversibly modify reactive cysteines. These probes usually contain an affinity tag for enrichment of modified proteins. Early probes were acyloxymethyl- and chloromethyl-ketones. These reactive probes work well in lysates but are not readily compatible with intact cells. These reagents uncovered the activation and function of caspases during apoptosis.46,47 Iodoacetamide (1)/maleimide-functionalized affinity agents (e.g., 2–3) are also used and show some preference for labeling specific enzymes, but do not remedy compatibility issues (Figure 1).48,49, 50 o-Nitrobenzyl acetal-caged electrophiles (e.g., 4) that are compatible with live cells have been developed.51 However, the requirement for high probe concentrations and liberation of a reactive nitrosoketone side product limits this technology and its use in cells.
Figure 1.
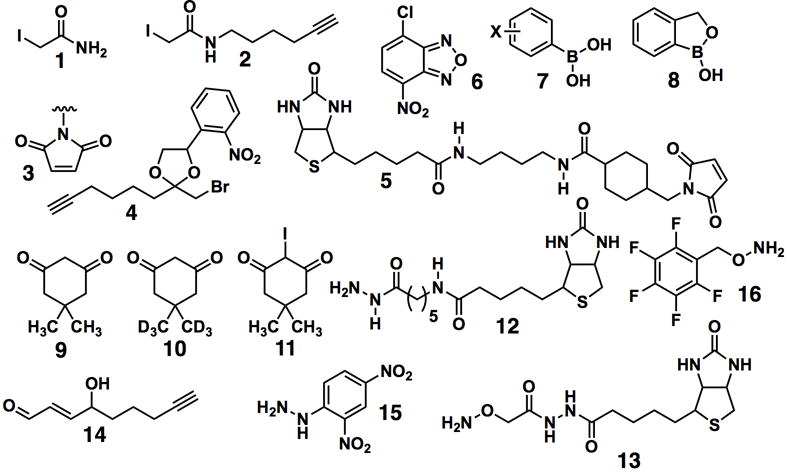
Glossary and chemical structures of representative probes highlighted in this review. (3 illustrates maleimide-functionalized affinity agents. X in 7: H, 4-NO2, 3-NO2, 3-Br, 4-Br, 4-F, 4-OCH3).
Organelle-specific variants
Some “organelle-specific” capture probes use affinity reagents tailored to a specific class of organelle-specific enzymes or reactive probes fused to a subcellular localization marker. Profiling activity changes in lysosomal cathepsins during autophagy stimulation has been achieved using epoxide-functionalized peptides that react selectively with cathepsins52. To assist targeting to the lysosome’s acidic interior, the peptides were also functionalized with a weakly basic element [3-(2,4-dinitroanilino)-3′-amino-N-methyldipropylamine]. Global profiling of mitochondrial proteins has recently been achieved using a nonspecific reactive group fused to a mitochondrial targeting sequence.53 Some (~75%) specificity for the intended organelle was achieved with simple reactive functions. The authors attempted to use photoactivatable reactive groups (phenyl azide and benzophenone) to localize the capture to the mitochondria, but both reactive groups proved unable to identify any labeled peptides by mass spectrometry (MS). Improvements will likely need photocaged electrophiles that are compatible with MS techniques.
Identification of reversible modifications to Cys
A variation on the above methods is used to identify cysteine sulfenic acids. Free thiols are blocked using an unfunctionalized alkylating agent followed by chemoselective reduction of unreactive cysteine-SOH into corresponding thiols by reducing agents (such as arsenite54). Thiols thus formed are captured with biotin-maleimide (5). MS following affinity capture, electrophoresis, and digestion allows proteins containing cysteine-SOH to be identified (Figure 2a).54 Similar methods detect cystine, cysteine-SNO and other univalent sulfur modifications.55–57 Chemical probes for cysteine-SOH harness the dual reactivity of the sulfur within sulfenic acids.58 The nucleophilicity of cysteine-SOH is utilized by probes like 4-chloro-7-nitrobenzo-2-oxa-1,3-diazole (6),59 and more recently reversible arylboronic acids (7) and benzoxaborole (8).60 The electrophilicity of cysteine-SOH has been exploited using functionalized dimedones (or analogous 1,3-dicarbonyls61) (9) that selectively displace water from sulfenic acids either in lysates or in intact cells.62–64 This approach has been converted to an MS-based method to differentiate between sulfenyl-thiol- and thiol-containing proteins by treating a lysate with D6-dimedone (10) (nucleophilic; reacts with sulfenyl-thiol) and iododimedone (11) (reacts with thiols).63, 65 These two probes produce isotopomeric products contingent only upon the oxidation state of the specific sulfur modified, assuming labeling is independent of redox state of both probe and cysteine (Figure 2b). Interestingly, bicyclo[6.1.0]nonyne traps cysteine sulfenyl acid more than 100 times faster than 1,3-dicarbonyls providing an alternative affinity probe.66 The rate of adduction is important because lysis is typically carried out under weakly reducing conditions in which cysteines are prone to oxidation. These conditions may give false positives due to competing post-lysis oxidation, especially since bolus oxidative challenges are often used to upregulate ROS in one sample relative to an unoxidized sample, rendering downstream variables hard to control. One approach to side-step many of these issues uses yeast transcription factor Yap1, which reacts selectively with cysteine-SOH to form a mixed disulfide.67 Direct MS analysis has been used in either a bottom up or top down approach.
Figure 2.
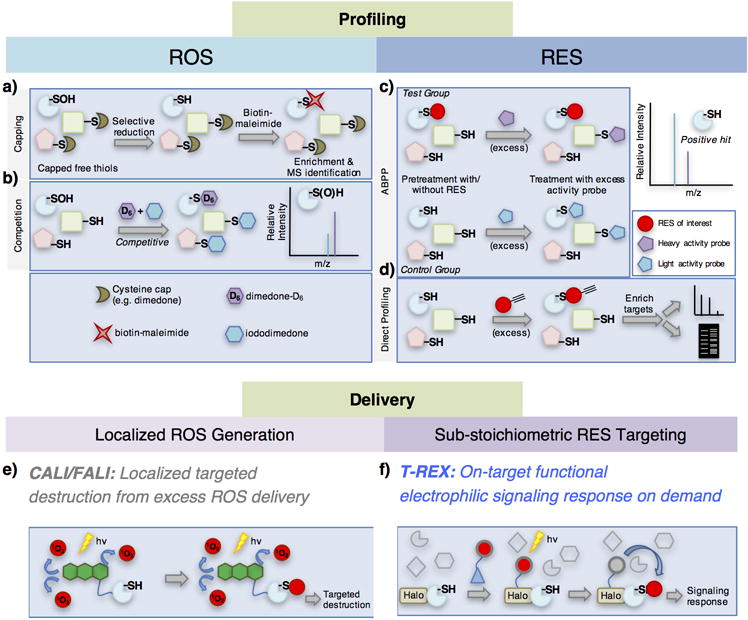
Representative ROS/RES target profiling and delivery strategies. ROS target profiling can be broken down into (a) “capping” strategies, wherein free thiols are capped before labeling of modified Cys (in this example, Cys-SOH), and (b) “chemoselective” strategies that use a chemoselective reagent to “pick out” a specific modification, such as Cys-SOH and label it selectively. Combinations of these strategies can be used in “competition,” wherein simultaneous chemoselective reactions are run on the same sample. RES target profiling can similarly be broken down into (c) activity-based proteomic profiling (ABPP) which utilizes a stepwise competition approach or (d) “direct profiling,” in which cells/lysates are treated with a functionalized probe and directly detected by MS analysis. Delivery strategies for ROS have generally been limited to (e) CALI/FALI approaches, which approximate bolus dosing, although they can be restricted to particular proteins or cellular compartments. (f) By contrast, the only available RES delivery strategy, T-REX, more closely mimics endogenous electrophile signaling and is capable of evoking a specific signaling response triggered by on-target electrophilic modifications.
Detection of RES-modified cysteine
The above strategies are not applicable to RES modifications. Activity-based protein profiling (ABPP) is most commonly used to identify RES-sensitive proteins.68 In competitive ABPP (Figure 2c), an intact cell, or more commonly a lysate, is treated with the desired RES, then lysates (either derived from cells or directly from the first step as appropriate) are treated with a cysteine-specific labeling agent, such as a functionalized iodoacetamide. A control group is prepared in parallel from lysates treated with the activity probe but with no RES pretreatment. The activity probe contains a handle that allows click attachment of biotin for enrichment and an isotopomeric barcode specific to the control group or the test sample. After enrichment of the labeled proteins using streptavidin-agarose, digestion and liberation from the beads, cysteines labeled by the reagent of choice are enriched in the sample not treated with test compound.68 This method is high-throughput and has opened a new lens on redox signaling. However, ABPP is capable of profiling only a small percentage (<5%) of cysteines within the proteome, often does not capture active-site cysteines6 and cannot prove that the specific residue profiled is modified by the ligand of interest (only that a functional impediment/loss of signal has occurred). ABPP mostly uses whole cell lysates and is prone to artifacts due to disruption of subcellular redox homeostasis post-lysis. Secondary validations for identified hits are necessary for experimental confidence. Furthermore, ABPP typically gives no information about compartmentalization, although mitochondria-specific profiling during oxidative stress using stable-isotope-labeled cells has been reported.53
Complementary alternatives to ABPP rule in specific modifications. Chemical derivatization approaches take advantage of the carbonyl group residual in many RES modifications (such as the aldehyde that is retained after Michael addition to HNE by a cysteine). Typical derivatization reagents include (a) hydrazine probe [such as dinitrophenylhydrazine (DNPH)] containing an affinity tag (e.g., 12) to create a hydrazone, and (b) tagged hydroxylamine (e.g., 13) to form an oxime. Hyrdazones require reduction by borohydride to form a stable hydrazide for identification of modified proteins following enrichment, digestion, and MS.69, 70 Oximes are sufficiently stable for direct MS analysis and the method has identified carbonyl-modified proteins in cardiac mitochondria.71 These protocols rely only on the presence of a carbonyl group residual from electrophilic modification so they detect many different electrophilic modifications. Importantly, the identity of the modification can be ascertained from MS. Antibodies detecting various lipid-derived electrophiles (LDEs) are available.72, 73 Some have affinity for particular modified residues, although several authors have questioned their specificity.73 Such antibodies are used in immunohistochemistry,74 western or dot blot, and ELISA75. Anti-HNE allowed visualization of HNE-protein adducts due to ischemia in mouse heart76 and Alzheimer’s disease.77 MS has also been used to identify directly modified proteins. Antibodies to DNPH are also available (so-called “oxy-blot” analysis) but their use is more appropriate for the analysis of non-specific carbonylation within the oxidatively-damaged proteome, as opposed to residual carbonyls from RES conjugation to cysteine or lysine.78
Modified lipid electrophiles (e.g., dye-conjugated derivatives) are available, but these reagents are non-native and results should be analyzed with caution.79 Alkyne-modified HNE (14) has also been used. The first application of this strategy involves treatment of RKO cells with 14, click chemistry with biotin-azide to append biotin to HNEylated proteins for enrichment, and MS (Figure 2d). This approach has identified many novel cellular HNE-targets.80 An extension of this method utilizes heavy- and light-isotope-labeled photo-cleavable linkers to which biotin is appended. Following treatment of cells with 14 under different conditions, heavy and light-labeled biotin reagents are conjugated to HNEylated proteins using click chemistry. Following enrichment, exposure to UV light liberates HNEylated targets tagged with a heavy or light barcode from the streptavidin beads. Subsequent MS analysis allows mapping of the HNEylated proteome under different conditions.81
Complexities
RES/ROS reactions with proteins are complex. Ignoring the many forms of RES/ROS (some of which interconvert or can convert to other RES/ROS), each RES/ROS can yield various chemical structures upon protein adduction.82 Some of these adducts may be bioactive, others potentially not. Chemical detection methods may only recognize a subset of possible protein adducts. For instance, worries have been raised that dimedone (9) may also (preferentially) probe for sulfenyl amides, the dehydration product of cysteine-SOH.83 Activity profiling (ABPP) can also lead to false positives due to functional coupling/degradation/adventitious oxidation. We thus urge researchers to validate specific modifications from proteins enriched from extracts by MS wherever possible, use a functional downstream assay under controlled conditions or use the methods below to trigger specific signals in the appropriate context.
(B) Subcellular Redox Imaging: a Biological Stakeout
Typical sensors employ a chromophore activated by a specific redox signal. On/off systems give good signal to noise, but have no internal standard, making comparison of different steady states/cells difficult. Intrinsic variation must be considered because transient expression (that gives a diverse expression profile between different cells) is often used to introduce genetically-encoded reporters (e.g., fluorescent proteins). A common strategy to sidestep problems with on/off sensors is ratiometric measurements, where the chromophore shuttles between two fluorescent states. Thus the readout is independent of expression level and can be calibrated using standards.
Genetically encoded sensors for H2O2
Development of genetic sensors typically involves mutagenesis of a known protein. Fluorescent proteins are ideal because mutants with <25% homology to wild-type (WT) can retain activity.84 The most common sensors are roGFP85 (redox-sensitive GFP) and HyPer (Figure 3a).86 These sensors couple redox-induced chemical modifications to fluorescence. However, these protein-based probes react with H2O2 and may short circuit cellular redox couples and/or misdirect signaling. The most recent probes claim to be free of this artifact, although redox balance is finely tuned and it may be hard to detect all physiologically relevant fluctuations.87 Furthermore, many sensors are not ideal; for instance both roGFP and HyPer are sensitive to pH.
Figure 3.
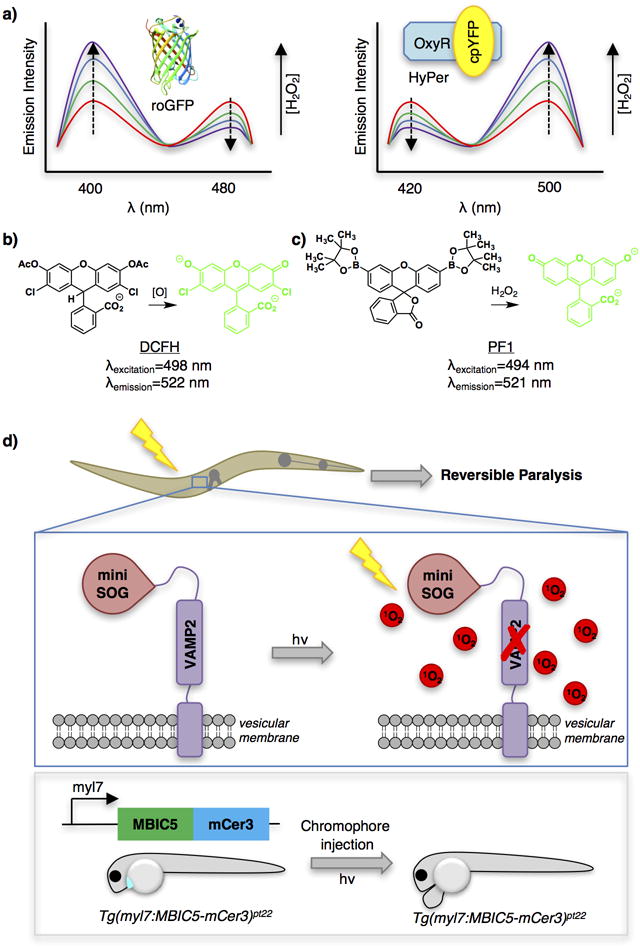
Selected examples of protein- or small-molecule-based ROS sensors (a–c) and an example application of excess ROS delivery tools in model organisms (d). (a) Fluorescent protein-based tools to measure ROS flux (left, roGFP; right, HyPer) and their respective excitation spectra across varying concentrations of H2O2. (b and c) Dichlorodihydrofluorescein (DCFH) and Peroxyfluor 1 (PF1), two turn-on small molecule probes for detecting ROS. (d) Two examples of applications in model organisms making use of chemical tools for localized generation of excess ROS. Top panel: Inhibition of synapses with CALI (InSynC) was demonstrated in C. elegans by fusing miniSOG (a genetically encodable singlet oxygen generator) with the synaptic protein VAMP-2. Worms showed reversible paralysis following light activation of miniSOG. Lower panel: A similar approach has been demonstrated in zebrafish through expression of “Fluorescence Activating Protein—Targeted and Activated Photosensitizer (FAP-TAP)” under a cardiomyocyte-specific promoter (myl7). Zebrafish exposed to light showed ablation of cardiomyocytes with prolonged damage of cardiac tissue. cpYFP, circularly-permuted yellow fluorescent protein; DCFH, dichloro-dihydro-fluorescein; mCer3, monomeric cerulean 3; myl7, myosin light chain promoter; PF1, peroxyfluor-1; roGFP, redox-sensitive GFP; SOG, singlet oxygen generator; VAMP2, vesicle-associated membrane protein 2. Also see the “Abbreviation” list at end of the manuscript. Zebrafish construct nomenclature is as follows: Tg(regulatory sequence:coding sequence) where Tg indicates transgene, regulatory sequence is a promoter (or enhancer), and coding sequence is the POI.
GFP exhibits two excitation bands, originating from neutral and anionic forms of the chromophore.88 roGFP was engineered by installation of cysteines at S147 and Q204 in p-strands 7 and 10. These two residues serve as a cysteine/disulfide redox switch to form a unique ratiometric sensor by opposing changes in the two excitation A and B bands through a single isobestic point (Figure 3a).89 roGFP2 is a variant that contains an additional S65T mutation.90 This protein has several advantages over roGFP1, including greater brightness and more favorable emission properties. roGFP2 is pH-sensitive but this affects the brightness of both emissions equally and hence only affects sensitivity.91
Next-generation sensors feature roGFP directly fused with H2O2-scavenging peroxidases like Orp1.91 Orp1 non-enzymatically reacts with H2O2 to form intramolecular disulfides that near-quantitatively change hands (Key words) with redox-sensor cysteines within fused roGFP2 in a disulfide relay process. The oxidized roGFP can be reduced in vivo affording reversibility and continual detection.92 Similarly, a rxYFP-glutaredoxin1 (Grx1) reporter measures GSH/GSSG redox couple ratio specifically.93
HyPer is an insertion of circularly permuted YFP (cpYFP) between amino acids 205 and 206 of the H2O2-sensitive regulatory domain of OxyR, a redox-sensor protein from E. coli.94 Upon exposure to H2O2, OxyR undergoes a conformational change as a result of the peroxidatic cysteine, Cys-199, undergoing a direct, non-enzymatic reaction with H2O2 followed by the resolving cysteine, Cys-208, forming a C199–C208 disulfide.95 An induced structural change around the cpYFP chromophore simultaneously reduces excitation at 420 nm (F420) and proportionally increases excitation at 500 nm (F500). The detection range of H2O2 for 1st generation HyPer was low-mid nM H2O2 and the dynamic range (F500/F420) was only 3 fold.86 Using Hyper, the intracellular tolerance for H2O2 before the onset of cell death was estimated to be less than 7.5 nM.96 The latest version, “HyPer-3,” has an improved, albeit still limited, 6-fold dynamic range.97 Redox sensitive RFPs have been developed, possibly allowing dual molecule/organelle sensing in the future.98 The emission ratio of HyPer is particularly pH-sensitive. One way to account for this variable is to express a redox-insensitive mutant or to measure pH fluctuations under the specific conditions tested.99
Organelle-specific stakeouts
By genetically targeting the redox-sensor FPs to a specific organelle, microenvironment-specific redox perturbations can be measured. These sensors should have a redox couple that equilibrates with the predominant redox-active enzyme in that compartment. For example, an ER lumen-localized roGFP probe, roGFPiE, was designed to be in equilibrium with protein disulfide isomerase (PDI), a redox-mediating chaperone in the endoplasmic reticulum (ER).100 In vivo fluorescence lifetime imaging using this probe revealed surprising redox stability during unfolded protein response (UPR), but an unexpected shift to a reducing ER in response to loss of lumen calcium.101 Another ER-specific probe led to discovery of peroxiredoxin 4 (PRDX4)-mediated restoration of the ER’s oxidative environment in endoplasmic reticulum oxidation 1 [ERO1 (an enzyme that maintains oxidizing conditions with the ER)]- deficient cells.102 Similar strategies measured mitochondria-specific redox response to TGF-β103 and hypoxia,104 as well as redox environments of endosomes, lysozomes105, and peroxisomes106. Localized H2O2 sensing was also achieved using “HyPer-Tau”—wherein HyPer is tethered to microtubules.107
Small-molecule-based H2O2 sensors
These probes tend to be on/off sensors, which is less of a problem for small-molecules because their concentration is easier to control than ectopic proteins. Common issues associated with the use of small molecules, such as variable uptake, excretion, and distribution/localization have been reported.108–110 Probes can function either by nucleophilic addition/bond migration or radical chemistry.111 2′,7′-Dichlorodihydrofluorescein (DCFH), the earliest H2O2 probe, gains fluorescence via radical chemistry (Figure 3b).112 In addition to pioneering work in mammalian cells, these probes also work in plant cells.113 Later probes built upon addition-migration chemistry use an element like boron (Figure 3c), which is a good acceptor of electrons and facilitates migration to appended weak bonds. Peroxide exposure prompts nucleophilic addition followed by bond migration, converting the non-fluorescent phenyl boronic ester to a fluorescent phenol.114, 115 These probes have seen widespread applications to detect redox flux in cells/organelles and have been expanded to ratiometric variants.116 One further stumbling point with chemical sensors is their low specificity compared to enzymatic counterparts: DCFH is activated by hydroxyl radical,117 NO,118 peroxynitrite,119 and depletion of ROS.120, 121 Boron-based probes sense HOCl and peroxynitrite >1000 times faster than peroxide.122 Another issue is irreversibility, although a reversible on/off cyanine-based sensor has been reported and was applied to H2O2 detection during L-buthionine sulphoximine (inhibitor of glutathione synthesis)-induced apoptosis of HepG2 cells and at the site of injury of zebrafish.123
Detection of endogenous RES
The first reported HNE/enal probe was DNPH (15), which forms an adduct with aldehydes with absorption at 360–380 nm.124 Similarly, the reaction between aldehydes and 1,3-cyclohexanediones produces fluorescent decahydroacridines.125 Treatment of biological samples with fluorescent variants of these probes, and subsequent HPLC analysis, demonstrated the presence of aldehyde adducts in the livers of vitamin E-deficient rats.125 A GC-MS-based method employing carbonyl-reactive pentafluorobenzyl-hydroxylamine (16) identified free HNE as a biomarker for neuronal retinal ceroidosis.126
RES versus ROS detection in terms of chemistry and biology
Probes for direct and selective in-cell detection of RES and resulting modifications are underdeveloped. There are no methods to measure free RES flux in real time, or even to measure free levels in live cells. This can be traced to: (1) the sheer number of structurally similar LDEs compared to the relatively few forms of ROS; (2) the fact that (unlike ROS) few proteins react diagnostically with RES; and (3) the fact that, even post-enzyme conjugation, many RES still contain a reactive functional group6 (e.g., a reactive carbonyl, vide supra). The remaining reactive motif likely would be detected by the RES-detection probe [whereas ROS, on the other hand, either form chemically distinct species upon reaction (e.g., disulfides) or form secondary ROS chemically distinct from the first species]. The irreversibility/long residence of RES modifications also limits the likelihood of ever being able to measure flux in real time. Unsurprisingly, many end-point RES probes are nonspecific, detecting all LDEs and their adducts. Clearly the relative merits of global versus specific profiling are context dependent, but different LDEs have different signaling properties.
(C) Uncontrolled Redox Release: Neighborhood Canvassing
Treatment of cells with H2O2 to elicit redox signaling is a widely-employed general strategy to model oxidative stress. But this approach cannot mimic endogenously-generated H2O2 in terms of pulse duration, localization, and timing.
Bolus generation of reactive oxygen species
Traditional approaches that restrict ROS generation to a specific locale include chemical tethering of a fluorophore (or genetically encoding its binding site) to a protein of interest (POI), e.g., (a) FLAsH/ReAsH;127 (b) ligand-specific protein tags such as Halotag (a protein that reacts stoichiometrically and irreversibly with chloroalkane-tethered ligands);128 (c) antibody-conjugated chromophore (e.g., malachite green)129 (d) fluorophore (e.g., fluorescein)130; (e) expression of ROS-producing enzymes; or (f) photosensitizing fluorescent proteins (FPs).131 Traditional FPs show weak photosensitization properties. KillerRed is a modified FP with a more open β-barrel structure promoting oxygen photosensitization.132 KillerRed, the subsequently developed KillerOrange,133 the monomeric miniSOG (mini singlet-oxygen generator),134 and supernova135 have been used to kill cells.131, 132, 136
Inactivation of specific proteins through spatially-restricted bolus ROS generation has also been achieved using chromophore/fluorofore-assisted light inactivation (CALI/FALI) (Figure 2e). For example, cell division arrest was achieved by chromatin-directed CALI targeting histone H2B.137 Organelle-specific ROS release has also been examined by targeting the chromophore/fluorophore to particular organelles/subcellular compartments. These experiments illustrate the complex way by which the cell responds to ROS: mitochondria-localized ROS triggers apoptosis, whereas necrosis is elicited by plasma-membrane-specific ROS.131
Uncontrolled RES generation
Bolus dosing can mimic RES-induced stress and signaling. Bolus dosing of electrophiles has given new insights into redox signaling, but reservations must be placed on the validity of bolus dosing as a physiologic redox signaling model. No RES-sensitization methods equivalent to CALI/FALI have been reported.
(D) Controlled Release: Sting Operations
Controlling ROS signaling
Methods for the controlled generation of ROS to regulate redox signaling remain limited mainly because ROS are highly diffusible, their release is hard to control, and they readily interconvert. ROS are also typically generated through sensitization of cellular oxygen (i.e., via CALI), rendering efficacy of ROS generation context dependent. The primary radius of influence of many CALI probes is small (< 5 nm), but there are examples of off-target modification/cross-linking.138 It is also hard to assess other proteins indirectly affected through reversible ROS-mediated signaling (i.e., disulfide change of hands). None-the-less, CALI-type probes have helped deconvolute redox signaling: controlled generation of superoxide/HO• through photosensitization of Pd-bacteriochlorophyll-serine changes phosphorylation of JN kinase, p38, extracellular signal-regulated kinase (ERK), and Akt.139 A photocaged polyphenol is also used for on-demand generation of H2O2 (via superoxide) in HEK293 cells. This probe elicits cofilin-actin rod formation, which also occurs upon H2O2 (50–200 μM) treatment.140 Although probe concentrations used were high (200 μM), the authors show this concentration is not toxic. Of equal importance, they show that the byproducts of photouncaging are not active. An alternative method to generate ROS that does not generate superoxide employs enzyme D-amino acid oxidase from Rhodotorula graciles, which produces H2O2 dependent upon D-amino acids.141 This strategy successfully induces antioxidant response protecting neurons from oxidative stress.142
Controlled generation of RES as a signal: T-REX
Recently, Class II proximity-enhancement through pseudo-intramolecular delivery1 has been used to modify a specific protein of interest (POI). The POI is genetically fused to Halotag143 that recognizes a photocaged precursor to a specific bioactive lipid-derived electrophile (LDE) such as HNE. The method is called targetable reactive electrophiles and oxidants (T-REX) (Figure 2f).2–5, 7 Treatment of cells with a custom-designed chloroalkane-containing photocaged LDE-precursor yields complete binding of the photocaged probe to the Halotag. Following photoactivation that rapidly (t1/2 < 1 min) releases a maximum of one equivalent of LDE, the POI fused to Halotag will be labeled with LDE, provided the POI’s reaction with LDE is rapid. Because the amount of LDE liberated is maximally stoichiometric to the POI and localized to the microenvironment of the POI, this technique mirrors “endogenous signaling” and has a proven ability to elicit downstream signaling through single-protein-target modification.3 Several proteins have proven amenable to T-REX, including Keap1,2–5 Akt37 and PTEN.2, 5 triggering downstream responses in the antioxidant response pathway and FOXO transcriptional activation, and regulation of membrane-bound PIP2/PIP3 phosphoinositides, respectively).
The key differences between RES and ROS sensitization methods are stoichiometry and specificity. One chromophore can generate many ROS, meaning that CALI/FALI etc., approximate overload approaches. Although “selective” target protein perturbation has been demonstrated, a significant percentage of ROS sensitization strategies is applied to killing/ablation/overstressing cells. There is currently no sensitization method to form RES, mainly because a chemical reaction, as opposed to photosensitization/intersystem-crossing/electron transfer, is required to convert RES precursors to active RES. T-REX is broadly analogous to CALI, but the need for a photocaged precursor limits the amount of RES produced. Thus T-REX is particularly well-suited for studying physiological RES signaling. Time will tell how useful T-REX-type probes are to study localized, non-targeted stress.
Additional considerations
Many of the methods that use target-specific redox signaling to modulate a protein’s function overexpress proteins bearing a genetically-encoded protein tag, such as Halo. This approach is useful for proof-of-concept studies, or to study phenotypically-dominant (Key words) modifications. But especially for loss of function (e.g., in CALI/FALI systems), endogenous POI can mask/complement the desired phenotype).144 Thus, knocking in tags to endogenous POI genetic loci or expression of a transgene in a knockout/null background should be used wherever possible.
(E) Redox Model Systems: Means, Motives, and Opportunities
Adapting these chemical biology techniques to whole organisms is challenging and underdeveloped. Many sensing techniques employ fluorescent sensors/photosentitization, which is challenging in many multicellular organisms because they are made up of tissues (which may be less permeant to light than cultured cells) and many are pigmented. Fortunately, there are multiple methods to surmount these obstacles, such as transparent organisms (C. elegans, Xenopus/zebrafish larvae), choosing translucent organs, using genetic backgrounds that have reduced pigmentation145 or using near-IR (tissue-penetrating) light (e.g., multiphoton) (Table 2). Some reports indicate that it is possible to use visible light-initiated photouncaging in mice xenografts.146 However, the magnitude of the resulting responses is relatively low. Furthermore, whole organisms are much less compatible with small molecules than cultured cells, and adsorption/distribution/metabolism/excretion/toxicity (ADMET) must be considered. Zebrafish and Xenopus have an almost fully-functioning set of organs 3-day post-fertilization.147 C elegans is surrounded by a cuticle that shields it from many chemicals, meaning that chemical biology applications are largely underdeveloped.148 Tissue/structural complexity can make even simple experiments highly complex and require intimate familiarity with specific anatomy/physiology. Nevertheless, we highlight some of the elegant examples where chemical biology was applied to study redox physiology in whole organisms.
Table 2.
Advances in studying ROS and RES signaling in model organisms.171
| Entry | Model Organism | Advantages | Limitations | Examples |
|---|---|---|---|---|
| 1172 | C. elegans | • Simple culture conditions/rapid life cycle • Transparent • Many transgenic and reporter lines readily available • Hermaphrodites can generate many cloned copies rapidly; but genetic crosses possible. • High-throughput compatible |
• Cuticle prevents small-molecule penetrance • Lacks many organs/systems analogous to humans. • The average protein homology to humans is 41% (20%~71%) |
• InSynC (CALI) |
| 2173 | D. rerio | • Transparent embryos • Pigmentation can be reduced using mutant lines or small-molecule treatment during development • Rapid, observable development • Relatively simple genetic manipulation • Fully functioning set of organs 3 days post-fertilization • High-throughput compatible |
• Genome duplication can make genetics challenging • Lack some organs/structures present in humans |
• T-REX • CALI |
| 3171 | X. laevis/tropacalis | • Transparent embryos • Rapid, observable development • High brood size • Functioning organs 3 days post-fertilization • X. tropicalis is diploid • Physiology similar to humans • Knockdown possible • High-throughput compatible |
• Difficult transgenesis • X. laevis is allotetraploid |
• CALI |
| 4171 | M. musculus | • Highly homologous to humans | • Low light penetrance for optical tools • High cost per embryo • Early development is not observable • Genetic manipulation difficult/development slow • Low throughput |
• CALI (in eyes) • CALI (xenograft) |
H2O2 overload strategies in whole organisms
Numerous photochemical methods to induce injury in transparent organisms are known, including enzymatic sensitization of prodrugs149 and direct laser ablation.150 Unfortunately, these strategies are technically complex, require intricate knowledge of anatomy, and cannot be done easily on a large scale. CALI/FALI approaches are useful alternatives. CALI/FALI-mediated ablation has been applied to Xenopus laevis embryos,151 C. elegans152, and tumor xenografts in mice.153 One interesting application is inhibition of synapses with CALI (InSynC) (Figure 3d).148 VAMP2 is a mammalian analog of the C. elegans neuronal protein synaptobrevin that localizes to synaptic vesicles. A miniSOG−/− citrine fusion of the human synaptobrevidin analog, VAMP2, rescues motility deficiency shown by synaptobrevin null worms, validating that VAMP2 is functional in C. elegans. Upon 480-nm light irradiation, worms were paralyzed but recovered over the next day (Figure 3d upper). Similar loss of function was observed when miniSOG-VAMP2-citrine was expressed in wild-type worms, showing that ROS inhibition of VAMP2 behaves in a dominant-negative manner. The latter effect is likely due to heterodimerization of VAMP2 with analogous C. elegans proteins.154 However, the authors also demonstrate off-target effects both within the synaptic vesicle and at the plasma membrane. Similar experiments have been performed in D rerio.155 This system employed a fluorescence activating protein—targeted and activated photosensitizer (FAP-TAP), a domain that binds a specific otherwise inefficient fluorophore, enhancing its fluorescence. The protein/fluorophore pair was optimized in vitro to produce high levels of singlet oxygen, when exposed to near-IR light. When fish expressing this FAP-TAP specifically in cardiac cells were injected with the fluorophore and exposed to tissue-penetrating near-IR radiation cardiac ablation was observed (Figure 3d lower). Müller glia cells in mice have also been used as a model system.156 The latter approach ingeniously uses the transparency of the eye to great effect.
Mitochondrial redox homeostasis in model organisms
Mitochondria are implicated in age-dependent increase in oxidative stress as well as cardiac damage in mice.157 Overexpression of mitochondria-specific peroxiredoxin-3 or small molecule antioxidants like MitoQ in mice have mitigated neuorodegenerative diseases.158 Generation of ROS by disruption of the mitochondrial electron transport chain and subsequent mitohormesis leads to lifespan expansion in mice.159 Similar lifespan expansion by mitohormesis or through mitochondria-selective antioxidant enzyme has been noted in yeast,160 C. elegans,161 and drosophila.162
Subcellular connectomics
Given the increasing appreciation for H2O2 diffusion through membranes163, redox signals may be one way to transfer information between different organelle/cellular compartments. Indeed, redox crosstalk between peroxisomes and mitochondria, for instance, has been reported.164, 165 Inhibition of peroxisomal catalase activity (increasing H2O2 cellular concentrations) enhances mitochondrial ROS presumably through transport of H2O2 from the peroxisome.166, 167 On the other hand, transient generation of H2O2 by expression of peroxisomal catalase rescues glucose metabolism in insulin-resistant cells by increasing mitochondrial respiration.168 Investigations into ER/cytosol crosstalk have used chemical probes to understand redox signaling. Dimedone treatment of lysates of oxidatively-stressed C elegans identifies IRE-1 sulfenylation as an evolutionarily-conserved mechanism to upregulate antioxidant response and promote longevity.169 This pathway is suppressed by mutating the target cysteine identified by dimedone, further documenting that redox signaling functions through specific modifications to form defined states with biological significance. Perhaps most surprisingly, mounting evidence indicates that this organelle homeostasis is regulated across tissues.170
Outlook
Reactive species are an often-overlooked component of cellular information transfer. This is not surprising given that redox messages are transient, have intrinsically low specificity and are context dependent. But since pioneers began to scrutinize these unusual signals, it has become clear that biology will harness all means at its disposal to control decision making. Classically biology has been a field that measures flux and response to specific perturbations. These measurements or perturbations were usually achieved using specific inhibitors or genetic manipulations, when studying canonical signaling pathways. The study of redox signaling likewise measures flux and employs specific perturbation, but since chemistry rather than enzymology dominates redox-signaling initiation, chemical biology is the ideal tool to use. Chemical biology has indeed risen to this challenge and many innovative tools have been developed. Using these new techniques, progress has been made in measuring flux of specific chemical signals. But there are still many challenges left. In our opinion focus should shift to the nuanced way in which biology uses compartmentalized RES/ROS signaling to influence its decisions at the organelle, cell/tissue, or whole-organism scale. To do this, it is our belief that we need to identify and investigate specific proteins whose modifications alone are sufficient to trigger signaling cascades in cells and whole animals under endogenous signaling conditions. At an organismal level, we need to gain precision control in terms of specificity and timing to successfully mimic endogenous redox signaling. We hope that this review will stimulate new interdisciplinary research and push researchers from different fields to tackle the challenging yet rewarding field of redox signaling, ideally in whole organisms.
Acknowledgments
NIH-New-Innovator (1DP2GM114850), NSF CAREER (CHE-1351400), Beckman Young Investigator, Burroughs Wellcome Fund CRTG, and the Sloan fellowship programs (to Y.A.) are acknowledged for supporting the RES signaling research program in the Aye lab. J.R.P. acknowledges the NIH CBI Training Grant (NIGMS T32GM008500).
Abbreviations
- ABPP
activity-based protein profiling
- Ac
acetyl
- ADMET
absorption, distribution, metabolism, excretion, toxicity
- CALI/FALI
chromophore/fluorophore-assisted light inactivation
- cpYFP
circularly-permuted yellow fluorescent protein
- Cys
cysteine
- DCFH
dichlorodihydrofluorescein
- DNPH
dinitrophenylhydrazine
- ER
endoplasmic reticulum
- ERK
extracellular signal regulated kinase
- FAP-TAP
fluorescence activating protein—targeted and activated photosensitizer
- FOXO3
forkhead box O3
- FP
fluorescent protein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GS-MS
gas chromatography-mass spectrometry
- His
histidine
- HNE
4-hydroxynonenal
- HPLC
high-performance liquid chromatography
- IR
infrared
- LDE
lipid-derived electrophile
- Lys
lysine
- mCer3
monomeric cerulean 3
- MitoQ
mitoquinone
- MS
mass spectrometry
- myl7
myosin light chain promoter
- NFκB
nuclear factor κB
- PIP2/3
phosphoinisitol bis/trisphosphate
- POI
protein of interest
- PTM
posttranslational modification
- RES
reactive electrophilic species
- roGFP
redox-sensitive green fluorescent protein
- ROS
reactive oxidative species
- SOG
singlet oxygen generator
- T-REX
targetable reactive electrophiles and oxidants
- UPR
unfolded protein response
- VAMP2
vesicle-associated membrane protein 2
- WT
wild-type
- Yap1
Yes-associated protein 1
Footnotes
Notes
A new article reporting a mitochondria-targeted two-photon fluorescent probe that can ratiometrically detect peroxynitrite in a mouse model of inflammation (J. Am. Chem. Soc., DOI: 10.1021/jacs.6b10508) appeared during the final stages of revising this review.
References
- 1.Long MJ, Poganik JR, Aye Y. On-Demand Targeting: Investigating Biology with Proximity-Directed Chemistry. J Am Chem Soc. 2016;138:3610–3622. doi: 10.1021/jacs.5b12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang X, Fu Y, Long MJ, Haegele JA, Ge EJ, Parvez S, Aye Y. Temporally controlled targeting of 4-hydroxynonenal to specific proteins in living cells. J Am Chem Soc. 2013;135:14496–14499. doi: 10.1021/ja405400k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parvez S, Fu Y, Li J, Long MJ, Lin HY, Lee DK, Hu GS, Aye Y. Substoichiometric hydroxynonenylation of a single protein recapitulates whole-cell-stimulated antioxidant response. J Am Chem Soc. 2015;137:10–13. doi: 10.1021/ja5084249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin HY, Haegele JA, Disare MT, Lin Q, Aye Y. A generalizable platform for interrogating target- and signal-specific consequences of electrophilic modifications in redox-dependent cell signaling. J Am Chem Soc. 2015;137:6232–6244. doi: 10.1021/ja5132648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parvez S, Long MJ, Lin HY, Zhao Y, Haegele JA, Pham VN, Lee DK, Aye Y. T-REX on-demand redox targeting in live cells. Nat Protoc. 2016;11:2328–2356. doi: 10.1038/nprot.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long MJ, Aye Y. The Die Is Cast: Precision Electrophilic Modifications Contribute to Cellular Decision Making. Chem Res Toxicol. 2016;29:1575–1582. doi: 10.1021/acs.chemrestox.6b00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long MJC, Parvez S, Zhao Y, Surya SL, Wang Y, Zhang S, Aye Y. Akt3 is a privileged first responder in isozyme-specific electrophile response. Nat Chem Biol. 2016 doi: 10.1038/nchembio.2284. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol. 2011;7:504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs AT, Marnett LJ. Systems Analysis of Protein Modification and Cellular Responses Induced by Electrophile Stress. Acc Chem Res. 2010;43:673–683. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu Rev Pharmacol Toxicol. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- 11.Miseta A, Csutora P. Relationship Between the Occurrence of Cysteine in Proteins and the Complexity of Organisms. Mol Biol Evol. 2000;17:1232–1239. doi: 10.1093/oxfordjournals.molbev.a026406. [DOI] [PubMed] [Google Scholar]
- 12.Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 13.Khan S, Vihinen M. Spectrum of disease-causing mutations in protein secondary structures. BMC Struct Biol. 2007;7:56. doi: 10.1186/1472-6807-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szweda LI, Uchida K, Tsai L, Stadtman ER. Inactivation of Glucose-6-phosphate Dehydrogenase by 4-Hydroxy-2-nonenal. J Biol Chem. 1993;268:3342–3347. [PubMed] [Google Scholar]
- 15.Nakamura T, Kawai Y, Kitamoto N, Osawa T, Kato Y. Covalent Modification of Lysine Residues by Allyl Isothiocyanate in Physiological Conditions: Plausible Transformation of Isothiocyanate from Thiol to Amine. Chem Res Toxicol. 2009;22:536–542. doi: 10.1021/tx8003906. [DOI] [PubMed] [Google Scholar]
- 16.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Byrnes RW. Association of redox-active iron bound to high molecular weight structures in nuclei with inhibition of cell growth by H2O2. Free Radic Biol Med. 1999;26:49–60. doi: 10.1016/s0891-5849(98)00165-8. [DOI] [PubMed] [Google Scholar]
- 18.Dixon SJ, Stockwell BR. The role of iron and reactive oxygen species in cell death. Nat Chem Biol. 2014;10:9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 19.Onyango AN. Endogenous Generation of Singlet Oxygen and Ozone in Human and Animal Tissues: Mechanisms, Biological Significance, and Influence of Dietary Components. Oxid Med Cell Longev. 2016;2016:2398573. doi: 10.1155/2016/2398573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skovsen E, Snyder JW, Lambert JDC, Ogilby PR. Lifetime and Diffusion of Singlet Oxygen in a Cell. J Phys Chem B. 2005;109:8570–8573. doi: 10.1021/jp051163i. [DOI] [PubMed] [Google Scholar]
- 21.Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Hydrogen peroxide: a metabolic by - product or a common mediator of ageing signals? Nat Rev Mol Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- 22.Szabo C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- 23.Lim JB, Huang BK, Deen WM, Sikes HD. Analysis of the lifetime and spatial localization of hydrogen peroxide generated in the cytosol using a reduced kinetic model. Free Radic Biol Med. 2015;89:47–53. doi: 10.1016/j.freeradbiomed.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Duerr MA, Aurora R, Ford DA. Identification of glutathione adducts of alpha-chlorofatty aldehydes produced in activated neutrophils. J Lipid Res. 2015;56:1014–1024. doi: 10.1194/jlr.M058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawson AP, Long MJC, Coffey RT, Qian Y, Weerapana E, Oualid FE, Hedstrom L. Naturally Occurring Isothiocyanates Exert Anticancer Effects by Inhibiting Deubiquitinating Enzymes. Cancer Res. 2015;75:5130–5142. doi: 10.1158/0008-5472.CAN-15-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schopfer FJ, Cipollina C, Freeman BA. Formation and signaling actions of electrophilic lipids. Chem Rev. 2011;111:5997–6021. doi: 10.1021/cr200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leffel EK, Cunningham C, Sloat BR, Hebel H, Bilby C. RLIP76 protein reduces 4-HNE generated during oxidative stress and results in protection in well characterized animal models of acute radiation syndrome. New Horiz Transl Med. 2015;2:56–57. [Google Scholar]
- 28.Xie Z, Baba SP, Sweeney BR, Barski OA. Detoxification of aldehydes by histidine-containing dipeptides: from chemistry to clinical implications. Chem Biol Interact. 2013;202:288–297. doi: 10.1016/j.cbi.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalleau S, Baradat M, Gueraud F, Huc L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013;20:1615–1630. doi: 10.1038/cdd.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siems WG, Capuozzò E, Verginelli D, Salerno C, Crifo C, Grune T. Inhibition of NADPH Oxidase-Mediated Superoxide Radical Formation in PMA-Stimulated Human Neutrophils by 4-Hydroxynonenal–Binding to -SH and -NH2 Groups. Free Radic Res. 2009;27:353–0358. doi: 10.3109/10715769709065774. [DOI] [PubMed] [Google Scholar]
- 31.Siems W, Grune T. Intracellular metabolism of 4-hydroxynonenal. Mol Aspects Med. 2003;24:167–175. doi: 10.1016/s0098-2997(03)00011-6. [DOI] [PubMed] [Google Scholar]
- 32.Lo Conte M, Carroll KS. The redox biochemistry of protein sulfenylation and sulfinylation. J Biol Chem. 2013;288:26480–26488. doi: 10.1074/jbc.R113.467738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Santamarina S, Boronat S, Hidalgo E. Reversible cysteine oxidation in hydrogen peroxide sensing and signal transduction. Biochemistry. 2014;53:2560–02580. doi: 10.1021/bi401700f. [DOI] [PubMed] [Google Scholar]
- 35.Gupta V, Carroll KS. Sulfenic acid chemistry, detection and cellular lifetime. Biochim Biophys Acta. 2014;1840:847–875. doi: 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowther WT, Haynes AC. Reduction of cysteine sulfinic acid in eukaryotic, typical 2-Cys peroxiredoxins by sulfiredoxin. Antioxid Redox Signal. 2011;15:99–109. doi: 10.1089/ars.2010.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhuang J, Wang PY, Huang X, Chen X, Kang JG, Hwang PM. Mitochondrial disulfide relay mediates translocation of p53 and partitions its subcellular activity. Proc Natl Acad Sci USA. 2013;110:17356–17361. doi: 10.1073/pnas.1310908110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobotta MC, Liou W, Stocker S, Talwar D, Oehler M, Ruppert T, Scharf AN, Dick TP. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat Chem Biol. 2015;11:64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- 39.Grune T, Davies KJA. The proteasomal system and HNE-modified proteins. Mol Aspects Med. 2003;24:195–204. doi: 10.1016/s0098-2997(03)00014-1. [DOI] [PubMed] [Google Scholar]
- 40.Klotz LO, Sanchez-Ramos C, Prieto-Arroyo I, Urbanek P, Steinbrenner H, Monsalve M. Redox regulation of FoxO transcription factors. Redox Biol. 2015;6:51–72. doi: 10.1016/j.redox.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung SN, Yang WK, Kim J, Kim HS, Kim EJ, Yun H, Park H, Kim SS, Choe W, Kang I, Ha J. Reactive oxygen species stabilize hypoxia-inducible factor-1 alpha protein and stimulate transcriptional activity via AMP-activated protein kinase in DU145 human prostate cancer cells. Carcinogenesis. 2008;29:713–721. doi: 10.1093/carcin/bgn032. [DOI] [PubMed] [Google Scholar]
- 43.Mikhed Y, Gorlach A, Knaus UG, Daiber A. Redox regulation of genome stability by effects on gene expression, epigenetic pathways and DNA damage/repair. Redox Biol. 2015;5:275–289. doi: 10.1016/j.redox.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poole LB. The basics of thiols and cysteines in redox biology and chemistry. Free Radic Biol Med. 2015;80:148–157. doi: 10.1016/j.freeradbiomed.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roos G, Foloppe N, Messens J. Understanding the pK(a) of redox cysteines: the key role of hydrogen bonding. Antioxid Redox Signal. 2013;18:94–127. doi: 10.1089/ars.2012.4521. [DOI] [PubMed] [Google Scholar]
- 46.Faleiro L, Kobayashi R, Fearnhead H, Lazebnik Y. Multiple species of CPP32 and Mch2 are the major active caspases present in apoptotic cells. EMBO J. 1997;16:2271–2281. doi: 10.1093/emboj/16.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berger AB, Witte MD, Denault JB, Sadaghiani AM, Sexton KM, Salvesen GS, Bogyo M. Identification of early intermediates of caspase activation using selective inhibitors and activity-based probes. Mol Cell. 2006;23:509–521. doi: 10.1016/j.molcel.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 48.Banerjee R, Pace NJ, Brown DR, Weerapana E. 1,3,5-Triazine as a modular scaffold for covalent inhibitors with streamlined target identification. J Am Chem Soc. 2013;135:2497–2500. doi: 10.1021/ja400427e. [DOI] [PubMed] [Google Scholar]
- 49.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong HL, Liebler DC. Mitochondrial protein targets of thiol-reactive electrophiles. Chem Res Toxicol. 2008;21:796–804. doi: 10.1021/tx700433m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abo M, Weerapana E. A Caged Electrophilic Probe for Global Analysis of Cysteine Reactivity in Living Cells. J Am Chem Soc. 2015;137:7087–7090. doi: 10.1021/jacs.5b04350. [DOI] [PubMed] [Google Scholar]
- 52.Wiedner SD, Anderson LN, Sadler NC, Chrisler WB, Kodali VK, Smith RD, Wright AT. Organelle-specific activity-based protein profiling in living cells. Angew Chem Int Ed Engl. 2014;53:2919–2922. doi: 10.1002/anie.201309135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yasueda Y, Tamura T, Fujisawa A, Kuwata K, Tsukiji S, Kiyonaka S, Hamachi I. A Set of Organelle-Localizable Reactive Molecules for Mitochondrial Chemical Proteomics in Living Cells and Brain Tissues. J Am Chem Soc. 2016;138:7592–7602. doi: 10.1021/jacs.6b02254. [DOI] [PubMed] [Google Scholar]
- 54.Saurin AT, Neubert H, Brennan JP, Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc Natl Acad Sci USA. 2004;101:17982–17987. doi: 10.1073/pnas.0404762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol. 2004;2:e333. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perez VI, Pierce A, de Waal EM, Ward WF, Bokov A, Chaudhuri A, Richardson A. Detection and quantification of protein disulfides in biological tissues a fluorescence-based proteomic approach. Methods enzymol. 2010;473:161–177. doi: 10.1016/S0076-6879(10)73008-1. [DOI] [PubMed] [Google Scholar]
- 57.Majmudar JD, Konopko AM, Labby KJ, Tom CT, Crellin JE, Prakash A, Martin BR. Harnessing Redox Cross-Reactivity To Profile Distinct Cysteine Modifications. J Am Chem Soc. 2016;138:1852–1859. doi: 10.1021/jacs.5b06806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta V, Paritala H, Carroll KS. Reactivity, Selectivity, and Stability in Sulfenic Acid Detection: A Comparative Study of Nucleophilic and Electrophilic Probes. Bioconjug Chem. 2016;27:1411–1418. doi: 10.1021/acs.bioconjchem.6b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olojo RO, Xia RH, Abramson JJ. Spectrophotometric and fluorometric assay of superoxide ion using 4-chloro-7-nitrobenzo-2-oxa-1,3-diazole. Anal biochem. 2005;339:338–344. doi: 10.1016/j.ab.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 60.Liu CT, Benkovic SJ. Capturing a sulfenic acid with arylboronic acids and benzoxaborole. J Am Chem Soc. 2013;135:14544–14547. doi: 10.1021/ja407628a. [DOI] [PubMed] [Google Scholar]
- 61.Qian J, Wani R, Klomsiri C, Poole LB, Tsang AW, Furdui CM. A simple and effective strategy for labeling cysteine sulfenic acid in proteins by utilization of beta-ketoesters as cleavable probes. Chem Commun. 2012;48:4091–4093. doi: 10.1039/c2cc17868k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, Gupta V, Tallman KA, Porter NA, Carroll KS, Liebler DC. Global, in situ, site-specific analysis of protein S-sulfenylation. Nat Protoc. 2015;10:1022–1037. doi: 10.1038/nprot.2015.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupta V, Carroll KS. Rational design of reversible and irreversible cysteine sulfenic acid-targeted linear C-nucleophiles. Chem Commun. 2016;52:3414–3417. doi: 10.1039/c6cc00228e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seo YH, Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc Natl Acad Sci USA. 2009;106:16163–16168. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seo YH, Carroll KS. Quantification of protein sulfenic acid modifications using isotope-coded dimedone and iododimedone. Angew Chem Int Ed Engl. 2011;50:1342–1345. doi: 10.1002/anie.201007175. [DOI] [PubMed] [Google Scholar]
- 66.Poole TH, Reisz JA, Zhao W, Poole LB, Furdui CM, King SB. Strained cycloalkynes as new protein sulfenic acid traps. J Am Chem Soc. 2014;136:6167–6170. doi: 10.1021/ja500364r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takanishi CL, Ma LH, Wood MJ. A Genetically Encoded Probe for Cysteine Sulfenic Acid Protein Modification in Vivo. Biochemistry. 2007;46:14725–14732. doi: 10.1021/bi701625s. [DOI] [PubMed] [Google Scholar]
- 68.Wang C, Weerapana E, Blewett MM, Cravatt BF. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat Methods. 2014;11:79–85. doi: 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Codreanu SG, Kim HY, Porter NA, Liebler DC. Biotinylated probes for the analysis of protein modification by electrophiles. Methods Mol Biol. 2012;803:77–95. doi: 10.1007/978-1-61779-364-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol Cell Proteomics. 2009;8:670–680. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chavez JD, Wu J, Bisson W, Maier CS. Site-specific proteomic analysis of lipoxidation adducts in cardiac mitochondria reveals chemical diversity of 2-alkenal adduction. J Proteomics. 2011;74:2417–2429. doi: 10.1016/j.jprot.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nagata N, Nishizaki Y, Watanabe N, Tsuda M, Matsuzaki S. An enzyme immune assay for serum anti-acetaldehyde adduct antibody using low-density lipoprotein adduct and its significance in alcoholic liver injury. Alcohol Clin Exp Res. 1998;22:150S–155S. doi: 10.1111/acer.1998.22.s3_part1.150s. [DOI] [PubMed] [Google Scholar]
- 73.Weber D, Milkovic L, Bennett SJ, Griffiths HR, Zarkovic N, Grune T. Measurement of HNE-protein adducts in human plasma and serum by ELISA-Comparison of two primary antibodies. Redox Biol. 2013;1:226–233. doi: 10.1016/j.redox.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang Z, Gan Y, Liu Q, Yin JX, Liu Q, Shi J, Shi FD. CX3CR1 deficiency suppresses activation and neurotoxicity of microglia/macrophage in experimental ischemic stroke. J Neuroinflammation. 2014;11:1–13. doi: 10.1186/1742-2094-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uchida K, Osawa T, Hiai H, Toyokuni S. 4-Hydroxy-2-nonenal-trapping ELISA: direct evidence for the release of a cytotoxic aldehyde from oxidized low density lipoproteins. Biochem Biophys Res Commun. 1995;212:1068–1073. doi: 10.1006/bbrc.1995.2078. [DOI] [PubMed] [Google Scholar]
- 76.Eaton P, Li JM, Hearse DJ, Shattock MJ. Formation of 4-hydroxy-2-nonenal-modified proteins in ischemic rat heart. Am J Physiol. 1999;276:H935–943. doi: 10.1152/ajpheart.1999.276.3.H935. [DOI] [PubMed] [Google Scholar]
- 77.Reed TT, Pierce WM, Markesbery WR, Butterfield DA. Proteomic identification of HNE-bound proteins in early Alzheimer disease: Insights into the role of lipid peroxidation in the progression of AD. Brain Res. 2009;1274:66–76. doi: 10.1016/j.brainres.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 78.Dalle-Donne I, Rossi R, Giustarini D, Milzani A, Colombo R. Protein carbonyl groups as biomarkers of oxidative stress. Clinica Chimica Acta. 2003;329:23–38. doi: 10.1016/s0009-8981(03)00003-2. [DOI] [PubMed] [Google Scholar]
- 79.Landar A, Zmijewski JW, Dickinson DA, Le Goffe C, Johnson MS, Milne GL, Zanoni G, Vidari G, Morrow JD, Darley-Usmar VM. Interaction of electrophilic lipid oxidation products with mitochondria in endothelial cells and formation of reactive oxygen species. Am J Physiol Heart Circ Physiol. 2006;290:H1777–1787. doi: 10.1152/ajpheart.01087.2005. [DOI] [PubMed] [Google Scholar]
- 80.Vila A, Tallman KA, Jacobs AT, Liebler DC, Porter NA, Marnett LJ. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem Res Toxicol. 2008;21:432–444. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang J, Tallman KA, Porter NA, Liebler DC. Quantitative chemoproteomics for site-specific analysis of protein alkylation by 4-hydroxy-2-nonenal in cells. Anal Chem. 2015;87:2535–2541. doi: 10.1021/ac504685y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein Adducts Generated from Products of Lipid Oxidation: Focus on HNE and ONE. Drug Metab Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 83.Forman HJ, Fukuto JM, Miller T, Zhang H, Rinna A, Levy S. The chemistry of cell signaling by reactive oxygen and nitrogen species and 4-hydroxynonenal. Arch Biochem Biophys. 2008;477:183–195. doi: 10.1016/j.abb.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stepanenko OV, Verkhusha VV, Kuznetsova IM, Uversky VN, Turoverov KK. Fluorescent Proteins as Biomarkers and Biosensors: Throwing Color Lights on Molecular and Cellular Processes. Curr Protein Pept Sci. 2008;9:338–369. doi: 10.2174/138920308785132668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barata AG, Dick TP. In vivo imaging of H2O2 production in Drosophila. Methods Enzymol. 2013;526:61–82. doi: 10.1016/B978-0-12-405883-5.00004-1. [DOI] [PubMed] [Google Scholar]
- 86.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 87.Morgan B, Van Laer K, Owusu TN, Ezerina D, Pastor-Flores D, Amponsah PS, Tursch A, Dick TP. Real-time monitoring of basal H2O2 levels with peroxiredoxin-based probes. Nat Chem Biol. 2016;12:437–443. doi: 10.1038/nchembio.2067. [DOI] [PubMed] [Google Scholar]
- 88.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 89.Cannon MB, Remington SJ. Redox-sensitive green fluorescent protein: probes for dynamic intracellular redox responses. A review, Methods Mol Biol. 2008;476:51–65. doi: 10.1007/978-1-59745-129-1_4. [DOI] [PubMed] [Google Scholar]
- 90.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 91.Lukyanov KA, Belousov VV. Genetically encoded fluorescent redox sensors. Biochim Biophys Acta. 2014;1840:745–756. doi: 10.1016/j.bbagen.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 92.Gutscher M, Sobotta MC, Wabnitz GH, Ballikaya S, Meyer AJ, Samstag Y, Dick TP. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J Biol Chem. 2009;284:31532–31540. doi: 10.1074/jbc.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schwarzlander M, Dick TP, Meyer AJ, Morgan B. Dissecting Redox Biology Using Fluorescent Protein Sensors. Antioxid Redox Signal. 2016;24:680–712. doi: 10.1089/ars.2015.6266. [DOI] [PubMed] [Google Scholar]
- 94.Christman MF, Storz G, Ames BN. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci USA. 1989;86:3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 96.Huang BK, Sikes HD. Quantifying intracellular hydrogen peroxide perturbations in terms of concentration. Redox Biol. 2014;2:955–962. doi: 10.1016/j.redox.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bilan DS, Pase L, Joosen L, Gorokhovatsky AY, Ermakova YG, Gadella TW, Grabher C, Schultz C, Lukyanov S, Belousov VV. HyPer-3: a genetically encoded H2O2 probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem Biol. 2013;8:535–542. doi: 10.1021/cb300625g. [DOI] [PubMed] [Google Scholar]
- 98.Fan Y, Chen Z, Ai HW. Monitoring redox dynamics in living cells with a redox-sensitive red fluorescent protein. Anal Chem. 2015;87:2802–2810. doi: 10.1021/ac5041988. [DOI] [PubMed] [Google Scholar]
- 99.Poburko D, Santo-Domingo J, Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J Biol Chem. 2011;286:11672–11684. doi: 10.1074/jbc.M110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laboissière MCA, Sturley SL, Raines RT. The Essential Function of Protein-disulfide Isomerase Is to Unscramble Non-native Disulfide Bonds. J Biol Chem. 1995;270:28006–28009. doi: 10.1074/jbc.270.47.28006. [DOI] [PubMed] [Google Scholar]
- 101.Avezov E, Cross BC, Kaminski Schierle GS, Winters M, Harding HP, Melo EP, Kaminski CF, Ron D. Lifetime imaging of a fluorescent protein sensor reveals surprising stability of ER thiol redox. J Cell Biol. 2013;201:337–349. doi: 10.1083/jcb.201211155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zito E, Melo EP, Yang Y, Wahlander A, Neubert TA, Ron D. Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol Cell. 2010;40:787–797. doi: 10.1016/j.molcel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jain M, Rivera S, Monclus EA, Synenki L, Zirk A, Eisenbart J, Feghali-Bostwick C, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial reactive oxygen species regulate transforming growth factor-beta signaling. J Biol Chem. 2013;288:770–777. doi: 10.1074/jbc.M112.431973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Al-Mehdi AB, Pastukh VM, Swiger BM, Reed DJ, Patel MR, Bardwell GC, Pastukh VV, Alexeyev MF, Gillespie MN. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci Signal. 2012;5:ra47. doi: 10.1126/scisignal.2002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Austin CD, Wen X, Gazzard L, Nelson C, Scheller RH, Scales SJ. Oxidizing potential of endosomes and lysosomes limits intracellular cleavage of disulfide-based antibody-drug conjugates. Proc Natl Acad Sci USA. 2005;102:17987–17992. doi: 10.1073/pnas.0509035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ivashchenko O, Van Veldhoven PP, Brees C, Ho YS, Terlecky SR, Fransen M. Intraperoxisomal redox balance in mammalian cells: oxidative stress and interorganellar cross-talk. Mol Biol Cell. 2011;22:1440–1451. doi: 10.1091/mbc.E10-11-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Warren EA, Netterfield TS, Sarkar S, Kemp ML, Payne CK. Spatially-resolved intracellular sensing of hydrogen peroxide in living cells. Sci Rep. 2015;5:16929. doi: 10.1038/srep16929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fujikawa Y, Morgan B, Dick TP. Fluorescent imaging of redox species in multicellular organisms. In: Jakob U, Reichmann D, editors. Oxidative Stress and Redox Regulation. Springer; Dordrecht: 2013. pp. 119–155. [Google Scholar]
- 109.Ludescher C, Thaler J, Drach D, Drach J, Spitaler M, Gattringer C, Huber H, Hofmann J. Detection of activity of P-glycoprotein in human tumour samples using rhodamine 123. Br J Haematol. 1992;82:161–168. doi: 10.1111/j.1365-2141.1992.tb04608.x. [DOI] [PubMed] [Google Scholar]
- 110.Huai-Yun H, Secrest DT, Mark KS, Carney D, Brandquist C, Elmquist E, Miller DW. Expression of multidrug resistance-associated protein (MRP) in brain microvessel endothelial cells. Biochem Biophys Res Commun. 1998;243:816–820. doi: 10.1006/bbrc.1997.8132. [DOI] [PubMed] [Google Scholar]
- 111.Fina NJ, Edwards JO. The alpha effect. A review, Int J Chem Kinet. 2016;5:1–26. [Google Scholar]
- 112.LeBel CP, Ischiropoulos H, Bondy SC. Evaluation of the probe 2’,7’-dichlorofluorescin as an indicator of reactive oxygen species formation and oxidative stress. Chem Res Toxicol. 1992;5:227–231. doi: 10.1021/tx00026a012. [DOI] [PubMed] [Google Scholar]
- 113.Watkins JM, Hechler PJ, Muday GK. Ethylene-induced flavonol accumulation in guard cells suppresses reactive oxygen species and moderates stomatal aperture. Plant Physiol. 2014;164:1707–1717. doi: 10.1104/pp.113.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lippert AR, Van de Bittner GC, Chang CJ. Boronate oxidation as a bioorthogonal reaction approach for studying the chemistry of hydrogen peroxide in living systems. Acc Chem Res. 2011;44:793–804. doi: 10.1021/ar200126t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lo LC, Chu CY. Development of highly selective and sensitive probes for hydrogen. Chem Commun. 2003:2728–2729. doi: 10.1039/b309393j. [DOI] [PubMed] [Google Scholar]
- 116.Lin VS, Dickinson BC, Chang CJ. Boronate-based fluorescent probes: imaging hydrogen peroxide in living systems. Methods Enzymol. 2013;526:19–43. doi: 10.1016/B978-0-12-405883-5.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wrona M, Patel K, Wardman P. Reactivity of 2′,7′-dichlorodihydrofluorescein and dihydrorhodamine 123 and their oxidized forms toward carbonate, nitrogen dioxide, and hydroxyl radicals. Free Radic Biol Med. 2005;38:262–270. doi: 10.1016/j.freeradbiomed.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 118.Gunasekar PG, Kanthasamy AG, Borowitz JL, Isom GE. Monitoring intracellular nitric oxide formation by dichlorofluorescin in neuronal cells. J Neurosci Methods. 1995;61:15–21. doi: 10.1016/0165-0270(95)00018-p. [DOI] [PubMed] [Google Scholar]
- 119.Possel H, Noack H, Augustin W, Keilhoff G, Wolf G. 2,7-Dihydrodichlorofluorescein diacetate as a fluorescent marker for peroxynitrite formation. FEBS Lett. 1997;416:175–178. doi: 10.1016/s0014-5793(97)01197-6. [DOI] [PubMed] [Google Scholar]
- 120.Jakubowski W, Bartosz G. 2,7-dichlorofluorescin oxidation and reactive oxygen species: what does it measure? Cell Biol Int. 2000;24:757–760. doi: 10.1006/cbir.2000.0556. [DOI] [PubMed] [Google Scholar]
- 121.Chen X, Zhong Z, Xu Z, Chen L, Wang Y. 2′,7′-Dichlorodihydrofluorescein as a fluorescent probe for reactive oxygen species measurement: Forty years of application and controversy. Free Radic Res. 2010;44:587–604. doi: 10.3109/10715761003709802. [DOI] [PubMed] [Google Scholar]
- 122.Sikora A, Zielonka J, Lopez M, Joseph J, Kalyanaraman B. Direct oxidation of boronates by peroxynitrite: mechanism and implications in fluorescence imaging of peroxynitrite. Free Radic Biol Med. 2009;47:1401–1407. doi: 10.1016/j.freeradbiomed.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu K, Qiang M, Gao W, Su R, Li N, Gao Y, Xie Y, Kong F, Tang B. A near-infrared reversible fluorescent probe for real-time imaging of redox status changes in vivo. Chem Sci. 2013;4:1079. [Google Scholar]
- 124.Benedetti A, Comporti M, Esterbauer H. Identification of 4-hydroxynonenal as a cytotoxic product originating from the peroxidation of liver microsomal lipids. Biochim Biophys Acta. 1980;620:281–296. doi: 10.1016/0005-2760(80)90209-x. [DOI] [PubMed] [Google Scholar]
- 125.Yoshino K, Sano M, Fujita M, Tomita I. Formation of aliphatic aldehydes in rat plasma and liver due to vitamin E deficiency. Chem Pharm Bull (Tokyo) 1986;34:5184–5187. [PubMed] [Google Scholar]
- 126.Siakotos AN, Bray R, Dratz E, van Kuijk F, Sevanian A, Koppang N. 4-Hydroxynonenal: a specific indicator for canine neuronal-retinal ceroidosis. Am J Med Genet Suppl. 1988;5:171–181. doi: 10.1002/ajmg.1320310620. [DOI] [PubMed] [Google Scholar]
- 127.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 128.Takemoto K, Matsuda T, McDougall M, Klaubert DH, Hasegawa A, Los GV, Wood KV, Miyawaki A, Nagai T. Chromophore-assisted light inactivation of HaloTag fusion proteins labeled with eosin in living cells. ACS Chem Biol. 2011;6:401–406. doi: 10.1021/cb100431e. [DOI] [PubMed] [Google Scholar]
- 129.Jay DG. Selective destruction of protein function by chromophore-assisted laser inactivation. Proc Natl Acad Sci USA. 1988;85:5454–5458. doi: 10.1073/pnas.85.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Beck S, Sakurai T, Eustace BK, Beste G, Schier R, Rudert F, Jay DG. Fluorophore-assisted light inactivation: a high-throughput tool for direct target validation of proteins. Proteomics. 2002;2:247–255. doi: 10.1002/1615-9861(200203)2:3<247::aid-prot247>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 131.Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. A genetically encoded photosensitizer. Nat Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 132.Carpentier P, Violot S, Blanchoin L, Bourgeois D. Structural basis for the phototoxicity of the fluorescent protein KillerRed. FEBS Lett. 2009;583:2839–2842. doi: 10.1016/j.febslet.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 133.Sarkisyan KS, Zlobovskaya OA, Gorbachev DA, Bozhanova NG, Sharonov GV, Staroverov DB, Egorov ES, Ryabova AV, Solntsev KM, Mishin AS, Lukyanov KA. KillerOrange, a Genetically Encoded Photosensitizer Activated by Blue and Green Light. PLoS One. 2015;10:e0145287. doi: 10.1371/journal.pone.0145287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Takemoto K, Matsuda T, Sakai N, Fu D, Noda M, Uchiyama S, Kotera I, Arai Y, Horiuchi M, Fukui K, Ayabe T, Inagaki F, Suzuki H, Nagai T. SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci Rep. 2013;3:2629. doi: 10.1038/srep02629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pletnev S, Gurskaya NG, Pletneva NV, Lukyanov KA, Chudakov DM, Martynov VI, Popov VO, Kovalchuk MV, Wlodawer A, Dauter Z, Pletnev V. Structural basis for phototoxicity of the genetically encoded photosensitizer KillerRed. J Biol Chem. 2009;284:32028–32039. doi: 10.1074/jbc.M109.054973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Serebrovskaya EO, Gorodnicheva TV, Ermakova GV, Solovieva EA, Sharonov GV, Zagaynova EV, Chudakov DM, Lukyanov S, Zaraisky AG, Lukyanov KA. Light-induced blockage of cell division with a chromatin-targeted phototoxic fluorescent protein. Biochem J. 2011;435:65–71. doi: 10.1042/BJ20101217. [DOI] [PubMed] [Google Scholar]
- 138.Rahmanzadeh R, Hüttmann G, Gerdes J, Scholzen T. Chromophore-assisted light inactivation of pKi-67 leads to inhibition of ribosomal RNA synthesis. Cell Prolif. 2007;40:422–430. doi: 10.1111/j.1365-2184.2007.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Posen Y, Kalchenko V, Seger R, Brandis A, Scherz A, Salomon Y. Manipulation of redox signaling in mammalian cells enabled by controlled photogeneration of reactive oxygen species. J Cell Sci. 2005;118:1957–1969. doi: 10.1242/jcs.02323. [DOI] [PubMed] [Google Scholar]
- 140.Miller EW, Taulet N, Onak CS, New EJ, Lanselle JK, Smelick GS, Chang CJ. Light-activated regulation of cofilin dynamics using a photocaged hydrogen peroxide generator. J Am Chem Soc. 2010;132:17071–17073. doi: 10.1021/ja107783j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Alim I, Haskew-Layton RE, Aleyasin H, Guo H, Ratan RR. Spatial, temporal, and quantitative manipulation of intracellular hydrogen peroxide in cultured cells. Methods Enzymol. 2014;547:251–273. doi: 10.1016/B978-0-12-801415-8.00014-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Haskew-Layton RE, Payappilly JB, Smirnova NA, Ma TC, Chan KK, Murphy TH, Guo H, Langley B, Sultana R, Butterfield DA, Santagata S, Alldred MJ, Gazaryan IG, Bell GW, Ginsberg SD, Ratan RR. Controlled enzymatic production of astrocytic hydrogen peroxide protects neurons from oxidative stress via an Nrf2-independent pathway. Proc Natl Acad Sci USA. 2010;107:17385–17390. doi: 10.1073/pnas.1003996107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 144.Vitriol EA, Uetrecht AC, Shen F, Jacobson K, Bear JE. Enhanced EGFP-chromophore-assisted laser inactivation using deficient cells rescued with functional EGFP-fusion proteins. Proc Natl Acad Sci USA. 2007;104:6702–6707. doi: 10.1073/pnas.0701801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wenner M. The most transparent research. Nat Med. 2009;15:1106–1109. doi: 10.1038/nm1009-1106. [DOI] [PubMed] [Google Scholar]
- 146.Shirmanova MV, Serebrovskaya EO, Lukyanov KA, Snopova LB, Sirotkina MA, Prodanetz NN, Bugrova ML, Minakova EA, Turchin IV, Kamensky VA, Lukyanov SA, Zagaynova EV. Phototoxic effects of fluorescent protein KillerRed on tumor cells in mice. J Biophotonics. 2013;6:283–290. doi: 10.1002/jbio.201200056. [DOI] [PubMed] [Google Scholar]
- 147.Barros TP, Alderton WK, Reynolds HM, Roach AG, Berghmans S. Zebrafish: an emerging technology for in vivo pharmacological assessment to identify potential safety liabilities in early drug discovery. Br J Pharmacol. 2008;154:1400–1413. doi: 10.1038/bjp.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lin JY, Sann SB, Zhou K, Nabavi S, Proulx CD, Malinow R, Jin Y, Tsien RY. Optogenetic inhibition of synaptic release with chromophore-assisted light inactivation (CALI) Neuron. 2013;79:241–253. doi: 10.1016/j.neuron.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Curado S, Stainier DY, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3:948–954. doi: 10.1038/nprot.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fang-Yen C, Gabel CV, Samuel ADT, Bargmann CI, Avery L. Laser Microsurgery in Caenorhabditis elegans. In: Wilson L, Tran P, editors. Methods Cell Biol. Elsevier; 2012. pp. 177–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jewhurst K, Levin M, McLaughlin KA. Optogenetic Control of Apoptosis in Targeted Tissues of Xenopus laevis Embryos. J Cell Death. 2014;7:25–31. doi: 10.4137/JCD.S18368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Qi YB, Garren EJ, Shu X, Tsien RY, Jin Y. Photo-inducible cell ablation in Caenorhabditis elegans using the genetically encoded singlet oxygen generating protein miniSOG. Proc Natl Acad Sci USA. 2012;109:7499–7504. doi: 10.1073/pnas.1204096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shirmanova M, Yuzhakova D, Snopova L, Perelman G, Serebrovskaya E, Lukyanov K, Turchin I, Subochev P, Lukyanov S, Kamensky V, Zagaynova E. Towards PDT with Genetically Encoded Photosensitizer KillerRed: A Comparison of Continuous and Pulsed Laser Regimens in an Animal Tumor Model. PLoS One. 2015;10:e0144617. doi: 10.1371/journal.pone.0144617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fdez E, Martinez-Salvador M, Beard M, Woodman P, Hilfiker S. Transmembrane-domain determinants for SNARE-mediated membrane fusion. J Cell Sci. 2010;123:2473–2480. doi: 10.1242/jcs.061325. [DOI] [PubMed] [Google Scholar]
- 155.He J, Wang Y, Missinato MA, Onuoha E, Perkins LA, Watkins SC, St Croix CM, Tsang M, Bruchez MP. A genetically targetable near-infrared photosensitizer. Nat Methods. 2016;13:263–268. doi: 10.1038/nmeth.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Byrne LC, Khalid F, Lee T, Zin EA, Greenberg KP, Visel M, Schaffer DV, Flannery JG. AAV-mediated, optogenetic ablation of Muller Glia leads to structural and functional changes in the mouse retina. PLoS One. 2013;8:e76075. doi: 10.1371/journal.pone.0076075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Logan A, Shabalina IG, Prime TA, Rogatti S, Kalinovich AV, Hartley RC, Budd RC, Cannon B, Murphy MP. In vivo levels of mitochondrial hydrogen peroxide increase with age in mtDNA mutator mice. Aging Cell. 2014;13:765–768. doi: 10.1111/acel.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Santini E, Turner KL, Ramaraj AB, Murphy MP, Klann E, Kaphzan H. Mitochondrial Superoxide Contributes to Hippocampal Synaptic Dysfunction and Memory Deficits in Angelman Syndrome Model Mice. J Neurosci. 2015;35:16213–16220. doi: 10.1523/JNEUROSCI.2246-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lapointe J, Stepanyan Z, Bigras E, Hekimi S. Reversal of the mitochondrial phenotype and slow development of oxidative biomarkers of aging in long-lived Mclk1+/− mice. J Biol Chem. 2009;284:20364–20374. doi: 10.1074/jbc.M109.006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Scialo F, Sriram A, Fernandez-Ayala D, Gubina N, Lohmus M, Nelson G, Logan A, Cooper HM, Navas P, Enriquez JA, Murphy MP, Sanz A. Mitochondrial ROS Produced via Reverse Electron Transport Extend Animal Lifespan. Cell Metab. 2016;23:725–734. doi: 10.1016/j.cmet.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Appenzeller-Herzog C, Banhegyi G, Bogeski I, Davies KJ, Delaunay-Moisan A, Forman HJ, Gorlach A, Kietzmann T, Laurindo F, Margittai E, Meyer AJ, Riemer J, Rutzler M, Simmen T, Sitia R, Toledano MB, Touw IP. Transit of H2O2 across the endoplasmic reticulum membrane is not sluggish. Free Radic Biol Med. 2016;94:157–160. doi: 10.1016/j.freeradbiomed.2016.02.030. [DOI] [PubMed] [Google Scholar]
- 164.Schrader M, Grille S, Fahimi HD, Islinger M. Peroxisome interactions and cross-talk with other subcellular compartments in animal cells. Subcell Biochem. 2013;69:1–22. doi: 10.1007/978-94-007-6889-5_1. [DOI] [PubMed] [Google Scholar]
- 165.Lismont C, Nordgren M, Van Veldhoven PP, Fransen M. Redox interplay between mitochondria and peroxisomes. Front Cell Dev Biol. 2015;3:35. doi: 10.3389/fcell.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Walton PA, Pizzitelli M. Effects of peroxisomal catalase inhibition on mitochondrial function. Front Physiol. 2012;3:108. doi: 10.3389/fphys.2012.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mueller S, Weber A, Fritz R, Mutze S, Rost D, Walczak H, Volkl A, Stremmel W. Sensitive and real-time determination of H2O2 release from intact peroxisomes. Biochem J. 2002;363:483–491. doi: 10.1042/0264-6021:3630483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Barbosa MR, Sampaio IH, Teodoro BG, Sousa TA, Zoppi CC, Queiroz AL, Passos MA, Alberici LC, Teixeira FR, Manfiolli AO, Batista TM, Cappelli AP, Reis RI, Frasson D, Kettelhut IC, Parreiras-e-Silva LT, Costa-Neto CM, Carneiro EM, Curi R, Silveira LR. Hydrogen peroxide production regulates the mitochondrial function in insulin resistant muscle cells: effect of catalase overexpression. Biochim Biophys Acta. 2013;1832:1591–1604. doi: 10.1016/j.bbadis.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 169.Kim DH. IRE1 Sulfenylation by Reactive Oxygen Species Coordinates Cellular Stress Signaling. Mol Cell. 2016;63:541–542. doi: 10.1016/j.molcel.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 170.Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wheeler GN, Brandli AW. Simple vertebrate models for chemical genetics and drug discovery screens: lessons from zebrafish and Xenopus. Dev Dyn. 2009;238:1287–1308. doi: 10.1002/dvdy.21967. [DOI] [PubMed] [Google Scholar]
- 172.Kaletta T, Hengartner MO. Finding function in novel targets: C.elegans as a model organism. Nat Rev Drug Discov. 2006;5:387–398. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- 173.Basu S, Sachidanandan C. Zebrafish: a multifaceted tool for chemical biologists. Chem Rev. 2013;113:7952–7980. doi: 10.1021/cr4000013. [DOI] [PubMed] [Google Scholar]


