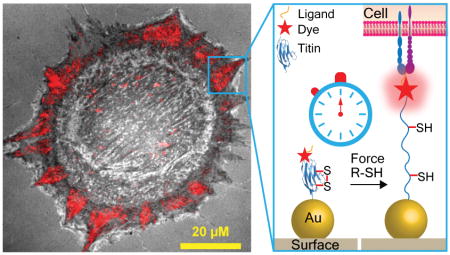
Abstract
Mechanical forces transmitted through integrin transmembrane receptors play important roles in a variety of cellular processes ranging from cell development to tumorigenesis. Despite the importance of mechanics in integrin function, the magnitude of integrin forces within adhesions remains unclear. Literature suggests a range from 1 to 50 pN, but the upper limit of integrin forces remains unknown. Herein we challenge integrins with the most mechanically stable molecular tension probe, which is comprised of the immunoglobulin 27th (I27) domain of cardiac titin flanked with a fluorophore and gold nanoparticle. Cell experiments show that integrin forces unfold the I27 domain, suggesting that integrin forces exceed 80 pN. The introduction of a disulfide bridge within I27 “clamps” the probe and resists mechanical unfolding. Importantly, the addition of a reducing agent initiates SH exchange, thus unclamping I27 at a rate that is dependent on the applied force. By recording the rate of S-S reduction in clamped I27 we infer that integrins apply 110+/−15 pN within focal adhesions. The rates of S-S exchange are heterogeneous and integrin subtype-dependent. Nanoparticle titin tension sensors along with kinetic analysis of unfolding demonstrate that a subset of integrins apply tension many fold greater than previously reported.
Keywords: integrins, mechanotransduction, biophysics, focal adhesion
Graphical Abstract
Cells adhere to the extracellular matrix (ECM) through transmembrane integrin receptors consisting of an α and β subunit.1 For example, fibronectin (FN) ECM molecules are primarily recognized by α5β1 and αvβ3 integrins that bind the Arg-Gly-Asp (RGD) and Pro-His-Ser-Arg-Asn (PHSRN) motifs.2 Integrin sub-type expression levels are cell type-dependent and dynamic, responding to the chemical and mechanical properties of the ECM. Therefore, determining the magnitude of forces experienced by individual integrins is important in better understanding the mechanisms of mechanotransduction processes in cell adhesion.
Integrin activation typically triggers the formation of supramolecular protein assemblies, focal adhesions (FAs), that are comprised of hundred of different structural and signaling molecules.3 Collectively, the FA functions to physically and chemically link the cytoskeleton to the ECM.4 FA assembly/disassembly is highly orchestrated in space and time to accommodate cell polarization and migration activities.5 While the chemical composition of FAs and their dynamics are currently under investigation, there is limited knowledge about the mechanical forces experienced and transmitted throughout these structures and especially the primary load-bearing molecules within FAs, the integrin receptors.
The most common technique to characterize cell adhesion forces is traction force microscopy (TFM), which indicates that forces for adhered fibroblasts range from 1 to 2 nN/μm2.6 By estimating the number of bound integrins per unit area (1000 molecules/ μm2 7), the force per receptor was also estimated at 1–2 pN per integrin.6a However, the challenge with TFM pertains to its limited spatial resolution (~ 1 μm) and force sensitivity (~nN) because of the crosslinked nature of the polymer. In addition, the deformable TFM substrates influence the cell biology, further convoluting the results.8
To address these issues, we pioneered the development of molecular tension fluorescence microscopy (MTFM)9, which allows for optical imaging of pN-level receptor-mediated forces with high spatial and temporal resolution. MTFM probes consist of an extendable macromolecule such as polyethylene glycol (PEG) or oligonucleotides that are flanked by a fluorophore-quencher pair undergoing resonance energy transfer (RET). MTFM probes are immobilized onto a substrate and modified with a ligand, such that receptor applied tension extends the linker and dequenches the fluorophore, generating a significant increase in donor emission intensity.
To tailor MTFM probes for integrins, we previously modified PEG24 polymer with the cyclic RGDfK peptide and anchored the probe using biotin-streptavidin association.9f Surprisingly, fibroblasts cultured on this probe for 45 min caused biotin-streptavidin dissociation. Nanoparticle-based MTFM probes immobilized using thiol-gold binding, were more mechanically robust and showed that integrin forces exceed 10–15 pN in magnitude.9d, 9e Supporting this result, we and others observed that DNA hairpin tension probes are denatured due to integrin mediated forces, indicating forces in excess of 16 pN.9c, 10 Likewise, tension gauge tether probes, consisting of DNA duplexes decorated with the cRGDfK peptide indicate that integrin activation is associated with peak forces up to ~50 pN in magnitude.11 In contrast, Dunn and colleagues determined that integrin-mediated forces are in the range of 1–5 pN based on single molecule imaging of a spidersilk elastin-based tension probe.12 Therefore, there is discrepancy in the reported integrin forces within FAs.
In this report, we investigate the magnitude of integrin tension within mature FAs by engineering the 27th immunoglobulin domain of cardiac titin (I27) which is structurally and mechanically well studied both experimentally13 and through theoretical modeling14 and displays highly reversible force-extension curves.15 Based on single molecule force spectroscopy, I27 unfolding requires significantly greater forces (~80–200 pN)16 when compared to DNA denaturation (~4–50 pN), PEG extension (1–15 pN), and spidersilk elastin extension (~2–6 pN), and therefore investigating integrin forces at ranges that are greater than values studied thus far. Moreover, by using the kinetics of titin unfolding rather than the equilibrium distribution of unfolded domains, we more precisely analyze high-magnitude integrin forces within mature FAs and determine the relative contribution of the α5β1 and αvβ3 integrins. In principle, this kinetic approach can be used to determine absolute integrin forces at any time point during FA maturation.
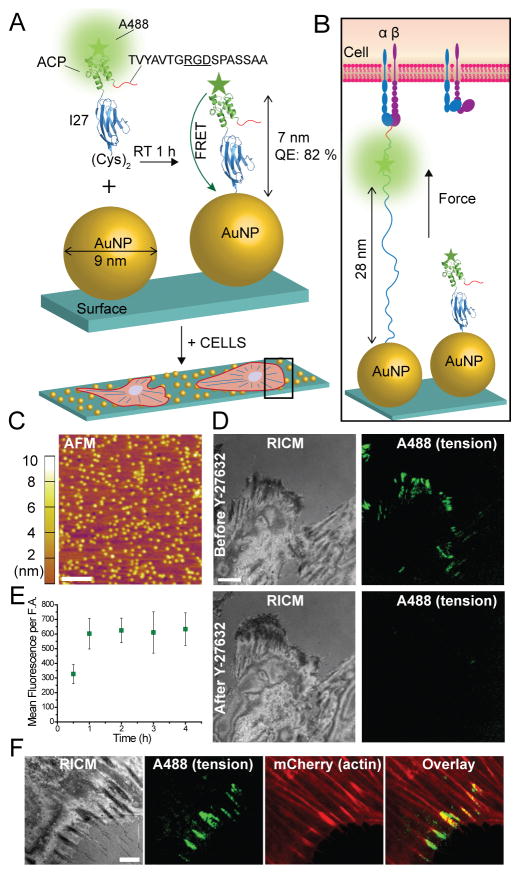
To label the I27 domain with a fluorophore, we initially introduced the acyl carrier protein (ACP)-tag near the N-terminus which can be enzymatically modified at its serine residue with a fluorophore with over 90% yield (Figure 1A).17 The probe displayed the “GRGDS” motif, specific for integrins, at the N-terminus (Figure 1B). At the C-terminus, two cysteine residues were introduced for immobilization onto a 9 nm gold nanoparticle (AuNP, see AFM in Figure 1C); which serves as an efficient quencher through nanometal surface energy transfer (NSET).9d The I27 probes were conjugated to Alexa488 and then characterized using UV-vis, SDS-PAGE, fluorescence spectrometry, and CD (Figure S1 A–C). We initially asked whether integrin-mediated forces would unfold the RGD-ACP(A488)-I27 probe (Figure 1B). Protein incubation conditions were optimized to ensure that probes were specifically bound through the thiol-Au association with minimal non-specific adsorption.18 This required incubating the AuNP surface with a binary mixture of passivating mPEG, SH(CH2-CH2-O)8COOH, along with the RGD-ACP(A488)-I27 probe at 1:12 molar stoichiometry for 1 hr at RT. By releasing the sensor from the AuNP we determined that each 9 nm AuNP presented an average of 5.2 sensors (Figure S1D).19 Also note that passivation was important to maintain proper orientation and function of the probe, since cells failed to engage surfaces lacking passivation with the mPEG (data not shown). With these conditions, the quenching efficiency of the ACP-tagged probes was measured at 82%, which was consistent with the dimensions of the ACP and I27 structures, and suggesting a ~7 nm fluorophore-AuNP distance.
Figure 1.
I27-based MTFM probes reveal integrin forces within FAs. (A–B) Schematic illustration showing the RGD-ACP(A488)-I27 probe and the mechanism of sensing (PDB ID: TIT1 and 2492 for I27 and ACP, respectively). The probe is modified at the N terminus with FN binding motif TVYAVTGRGDSPASSAA. The C terminus included two cysteine residues for binding to the AuNP. (C) AFM image of 9 nm AuNPs immobilized on the glass coverslip. Scale bar, 200 nm. (D) Representative RICM and fluorescence images of cells incubated on the tension probe surface before and after treatment with a myosin inhibitor Y-27632 (40 μM) for 30 min. Scale bar, 10 μm. (E) Plot of the average fluorescence tension signal per FA as a function of time. The error bars represent the SEM from n = 5 cells at each time point.
When rat embryonic fibroblasts (REFs) were incubated on the RGD-ACP(A488)-I27 probe surface for 1 hr, we observed significant signal in total internal reflection fluorescence (TIRF) that coincided with the cell edge as indicated by reflection interference contrast microscopy (RICM) (Figure 1D). The increase in fluorescence suggested that integrin-mediated forces unfolded the I27, the ACP domain, or both I27 and ACP domains, thus separating the Alexa488 reporter from the AuNP surface. To confirm that I27/ACP unfolding is driven by the cytoskeleton and specifically myosin II, we treated cells with the Rho-associated protein kinase (ROCK) inhibitor, Y-27632, for 30 min and re-imaged the sample. Following ROCK inhibition, the fluorescence signal was diminished (Figure 1D) indicating that signal is reversible and mediated by myosin contractility. Further controls show colocalization of RGD-ACP(A488)-I27 signal with the termini of F-actin stress fibers (Figure S1E). Time lapse analysis of the tension probe signal indicated that fluorescence increased over a period of ~1 hr and then held steady for at least 4 hrs (Figure 1E). Beyond 5 hrs, we typically observe a slight decrease in tension signal, likely due to a combination of thiol displacement and protease activity. The results obtained with the RGD-ACP(A488)-I27 nanoparticle probe suggest that this protein engineering strategy is suitable for investigating integrin forces, but the poorly characterized mechanical stability of ACP made it difficult to ascertain which domains mechanically unfolded.
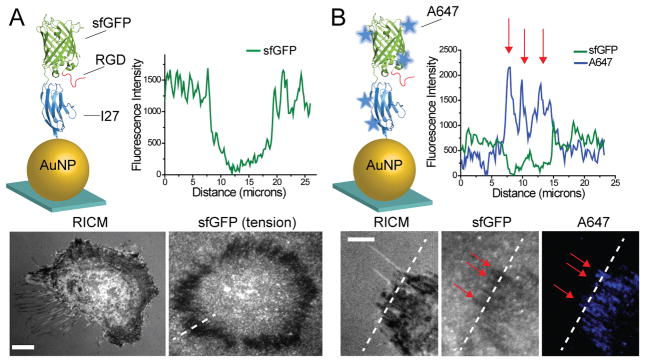
We next incorporated superfolder green fluorescent protein (sfGFP) within the I27 MTFM probe, replacing the ACP tag. sfGFP was selected due to its mechanical stability (~100 pN unfolding force20), increased brightness when compared to other GFP reporters21 and enhanced folding when fused with other proteins. The RGD-sfGFP-I27 probe was characterized using SDS PAGE, fluorescence, CD and absorption spectrometry (Figure S2 A–C) and immobilized onto AuNPs as described above (Figure 2A). Surprisingly, when REF cells were cultured onto the RGD-sfGFP-I27 tension probe surface for 3 hrs, sfGFP emission was quenched at the perimeter of the cell (Figure 2A). Given that mechanical unfolding of GFP quenches fluorescence, we suspected that integrin-mediated forces were sufficient to unfold the β-barrel of GFP. To verify the source of sfGFP quenching, we labeled free lysine residues of the sfGFP-I27 probe with an amine-reactive Alexa647 (Figure 2B). In this case, REF cells showed an increase in Alexa647 signal that coincided with regions of sfGFP quenching. Moreover, treating cells with ROCK inhibitor did not reverse the quenched sfGFP, but did lead to a decrease in Alexa647 emission (Figure S2D. These experiments demonstrate that integrin-mediated forces denature and irreversibly quench sfGFP, suggesting significant tension, but it remained unclear whether I27 was stable toward integrin tension.
Figure 2.
Integrin forces irreversibly quench and unfold sfGFP. (A) Schematic illustrating the RGD-sfGFP-I27 probe functionalized onto immobilized AuNPs (sfGFP PDB ID: 2B3P). Representative fluorescence and RICM images of REF cells cultured onto the RGD-sfGFP-I27 sensor surface for 3 hrs. Line scan analysis shows a loss in fluorescence at the cell edge, coinciding with the expected regions of mature FAs. (B) The RGD-sfGFP-I27 probe was non-specifically labeled with Alexa 647, purified and immobilized onto the AuNP surface. Representative RICM, GFP fluorescence and Alexa 647 fluorescence images of cells incubated on the probe surface for 3 hrs. Line scan (dashed white line) analysis through the stress fibers (red arrows) shows anti-correlation between Alexa 647 and sfGFP emission. Scale bar, 10 μm.
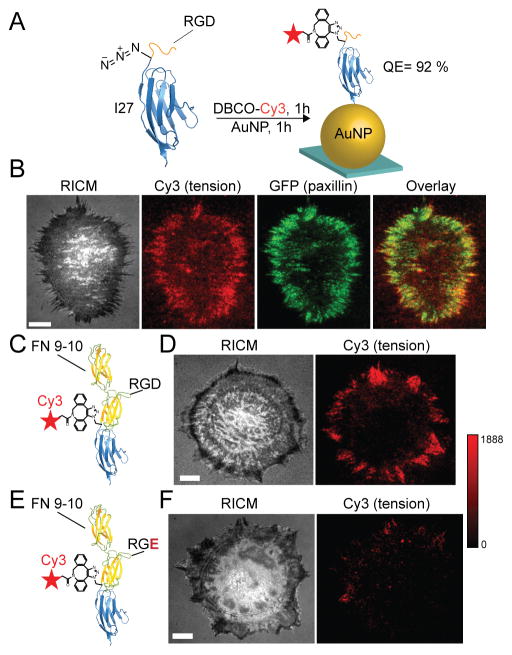
Unnatural amino acid incorporation was next used to express an I27 MTFM probe labeled with the p-azidophenylalanine amino acid. This azide-modified protein was then modified with the Cy3 dye using Cu-free click cycloaddition coupling (Figure 3A). The RGD-Cy3-I27 probe was characterized by SDS-PAGE, ESI-MS, CD, and fluorescence spectrometry (Figure S3 A–E), and showed 92% quenching upon AuNP immobilization. This quenching efficiency roughly agreed with the predicted distance between the dye and the AuNP surface (~4 nm). Within 1 hr of incubating REFs on this MTFM probe surface, we observed Cy3 fluorescence signal at the edges of the cell, suggesting mechanically-driven I27 unfolding at these sites. Following ROCK inhibition with 40 μM of Y-27632 for 30 min, the fluorescence signal was diminished (Figure S3F) showing that signal is reversible and mediated by myosin contractility. To confirm that tension signal is associated with FAs, we transfected the REFs with GFP-paxillin, a widely accepted FA marker, and found colocalization with Cy3 tension signal (Figure 3A). Importantly, this result unambiguously shows that the I27 domain is mechanically unfolded due to integrin forces within 1 hr of cell adhesion.
Figure 3.
Integrin forces unfold I27 through RGD and FN. (A) Illustration of the RGD-Cy3-I27 MTFM sensor containing the p-azidophenylalanine amino acid. The dye was conjugated with Cu-free cycloaddition and was subsequently immobilized onto the AuNP surface. The quenching efficiency was 92% (top). Representative RICM, GFP-paxillin, Cy3 tension and overlay of Cy3/GFP shows colocalization of FAs with tension. (B) FN (9–10) sensor was labeled with the Alexa 647 and co-incubated on the AuNP surface with RGD-I27-Cy3 for 1 hr (FN 9–10 PDB ID: 1FNF). Representative RICM and fluorescence images of a REF cell cultured on a binary tension probe surface for 1 hr. The RGD-Cy3-I27 and FN-A647-I27 fluorescence signals are displayed from the highlighted region of interest. Overlay of these channels and line scan analysis shows colocalization of both sensors. Scale bar, 10 μm.
The RGD sequence derived from FN is widely used in model cell adhesion studies.22 However, one persistent question is whether the linear RGD sequence recapitulates the properties of more physiologically relevant ECM proteins such as domains of FN or full length FN. We rationalized that integrin-ligand binding and mechanics may be enhanced for FN domains compared to the RGD peptide fragment. Importantly, recombinant protein expression allows for facile engineering of virtually any expressible protein ligand within these I27 MTFM probes. Therefore, the sensor was redesigned to present FN domains 9 and 10 at the N-terminus, providing both the synergy (PHSRN) and GRGDS motifs (Figure S4 A and B). Initial experiments using FN-Cy3-I27 probe showed similar spatial and temporal patterns of fluorescence as the RGD-Cy3-I27 probe (Figure S4C). To better understand the specificity of force transmission to the FN tension probe, we generated a control probe presenting RGE rather than the RGD sequence within the FN 10 domain. In this case, we found that REF cells spread on the sensor surface but failed to generate a fluorescence response. Thus suggesting that the RGD sequence within FN is required for triggering a chemo-mechanical response that generates sufficient tension to unfold I27 (Figure S4 D–F).
To quantitatively compare the differences in tension between RGD and FN, we encoded each probe with a different fluorophore and randomly immobilized an equimolar binary mixture of RGD-Cy3-I27 and FN-A647-I27 conjugates onto AuNPs, as described above (Figure 3B). REFs were then plated on the binary mixture of spectrally encoded MTFM probes for 1 hr and imaged using dual channel TIRF. No observable difference in the spatial distribution of tension signal was noted when both channels were compared at 1 hr (see line scan in Figure 3B). This finding suggests that the spatial distribution of mechanical tension (sufficient for I27 unfolding) is similar for the GRGDS and FN III 9–10 ligands, and that the synergy site does not significantly alter cell mechanics characterized by I27 unfolding.
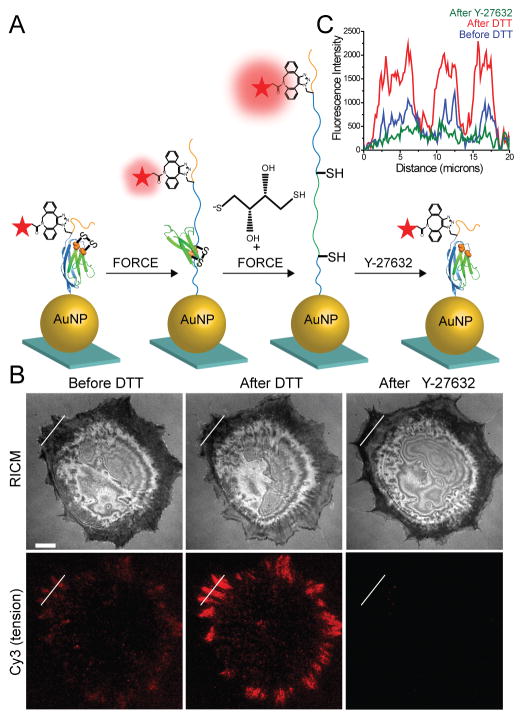
Given that I27 unfolds within 1 hr of cell plating and the probe does not dissociate from the surface, we infer that integrin forces range from ~80 pN up to ~1000 pN with the precise value depending on the loading rate. However, if the integrin receptor is challenged with an MTFM probes that does not mechanically unfold, then this probe will more precisely define the upper limit of integrin tension within FAs. Therefore, we introduced a buried disulfide bridge within I27 between the 32nd and 75th residues as previously reported.23 Using this reengineered I27, we generated disulfide “clamped” RGD-Cy3-I27 MTFM probe (Figure S5 A and B). As illustrated in Figure 4A, the disulfide bond traps 43 amino acids (green) and leaves the other amino acids unsequestered (blue) under mechanical load. Mechanical extension of this clamped I27 stops at the disulfide bond, placing the dye at a distance of 12 nm, which is predicted to produce a QE of ~80%. Addition of dithiothreitol (DTT) reduction agent leads to disulfide reduction and opening of the clamp. This allows for extension of the dye away from the AuNP surface to fully dequench the fluorophore. Moreover, inhibition of cell-applied force should lead to reversible refolding of I27.
Figure 4.
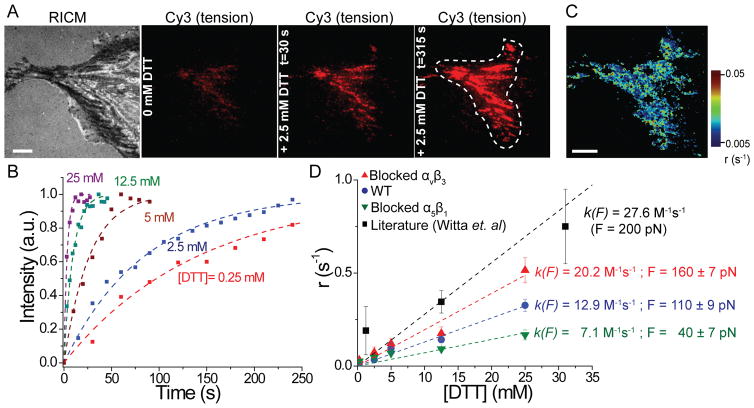
Covalently locked I27 resists complete mechanical unfolding by integrin forces. (A) Schematic illustration of disulfide clamped I27 tension sensor. The disulfide bridge is indicated by orange spheres. The probe was labeled with Cy3 dye and functionalized onto a AuNP surface (QE ~ 92%). When cells apply tension to the clamped MTFM sensor, I27 is stretched to the position of the disulfide clamp, resulting in solvent exposure of the disulfide. I27 can be further mechanically extended only in the presence of reducing agent, such as DTT. Refolding of the sensor is expected after ROCK kinase inhibitor (Y-27632) treatment. (B) Representative RICM and clamped I27 tension signal for REF cells incubated onto the sensor surface for 2 hrs before and after treatment with 0.25 mM DTT for 10 min and then after treatment with Y-27632 (40 μM) for 30 min. Scale bar, 10 μm. (C) Line scan analysis of the region of interest in (B) shows the fluorescence intensity through a FA before and after DTT addition and after Y-27632 treatment.
In a typical experiment testing the clamped RGD-Cy3-I27, REFs were plated on the probe surface for 2 hrs, resulting in a weak fluorescence signal at regions that coincided with FAs (Figure 4B). This signal is likely associated with partial unfolding of I27 up to the covalent “lock”. Upon addition of 0.25 mM DTT, time lapse movies showed that the signal rapidly increased, suggesting reduction of the disulfide bond and complete mechanical unfolding of the clamped probe (Figure 4B, 5A, Movie 1). To confirm that opening of the clamped sensor is myosin driven and the signal is reversible, we treated cells with the ROCK inhibitor (Y-27632), and found that the signal was diminished below the original values, prior to DTT incubation (Figure 4B and C). Control experiments indicated that pretreatment of the clamped RGD-Cy3-I27 probe with 1 mM DTT for 10 min, prior to cell plating, did not lead to increase in fluorescence and did not alter probe response following cell plating (Figure S5D). Treating 50 nM of clamped RFD-Cy3-I27 probe with 10 mM DTT in solution displayed ~10% reduction in emission (Figure S5D) whereas probe that was immobilized to the AuNPs displayed an average of ~2% decrease in emission upon treatment with DTT. These results confirm that the buried disulfide bridge locks the I27 probe preventing mechanical unfolding, and the reduction of the disulfide bond is dependent on both the cell-applied force, and reducing agent.
Figure 5.
Kinetic measurements of S-S reduction to determine integrin forces. (A) Representative RICM and fluorescence images of clamped I27 sensor before and after DTT addition at t = 30 and 315 sec. White dashed line shows the region of interest that was analyzed. Scale bar, 10 μm. (B) Heat map of disulfide reduction rates generated from the fluorescence video of the cell depicted in (A). Scale bar, 10 μm. (C) Representative kinetic plots showing an increase in fluorescence tension signal at different DTT concentrations. Dashed lines represent single-exponential fits used to determine τr. (D) Plot of the rate of disulfide reduction (r = 1/τr) as function of [DTT] for REF cells (blue), REF cells blocked with αvβ3 (red) and α5β1 antibodies (green). k(F) and F were derived from the linear fit of each data set. Black squares and fit (dashed line) are from Witta et al. PNAS, 2006 where clamped I27 was subjected to a constant force of 200 pN, which generated a slope of 27.6 M−1s−1. This previous work included DTT concentration up to 125 mM, but the additional data points were omitted for clarity. Error bars represent the S.E.M. of r obtained from n = 5 cells at each concentration of DTT (0.5 mM, 2.5 mM, 5 mM, 12.5 mM and 25 mM)..
Through a series of single molecule experiments23–24, Fernandez and coworkers found that the rate of disulfide reduction, r, for the clamped I27 was exponentially dependent on force, in accordance with the Bell model, and they showed that r was also linearly dependent on [DTT]. These relations are: r=k(F)[DTT] and k(F) = A exp((FΔxr-Ea)/kBT), where A is a pre-exponential factor, Δxr is the distance to the transition state of the reaction, and Ea is activation energy for the disulfide bond reduction. By varying the force and [DTT], and measuring r, Fernandez and coworkers determined Δxr, A and Ea for mechanically catalyzed disulfide reduction in I27. By using these validated constants for I27, we next inferred the magnitude of force experienced by the I27 probe by recording the rate of disulfide reduction as determined from the rate of fluorescence increase (Figure S6). Although Δxr, A and Ea were determined in three reports using different mechanical loading conditions ( See SI Table)24a, 24d, 25, we used fitting values of A = 1.3 × 1012 M −1s−1, Δxr = 0.34 Å and Ea = 65 kJ from the most relevant study where k(F) was determined by varying DTT concentration.24a
We recorded the rate of fluorescence increase for cells treated with DTT concentration ranging from 0.25 mM up to 25 mM (Figure 5C; n = 25 cells total). By fitting the increase of fluorescence versus time, we measured the time constant of disulfide reduction, τr, and thus determined the rate of disulfide reduction (r = 1/τr) from cell experiments. DTT contractions greater than 25 mM induced a reduction in cell adhesion, suggesting cell toxicity. In contrast, treating REFs expressing GFP-β3 with 2.5 mM DTT for 10 min did not lead to any observable change in cell morphology or GFP signal (Figure S5E). To show the variation in r values in a single cell, we generated a heat map of rates (ranging from 0.005 to 0.05 s−1) using the programming software (IDL). We next plotted the average r for different DTT concentrations (Figure 5D) and then determined k(F) from the slope of this plot (Figure S6). Assuming that the integrin force is constant within the time window of observation (<4 min) with minimal receptor turnover, we estimate that unfolded tension probes experience an average of 110 +/−15 pN (Figure S6). This value suggests that a subset of integrins experience forces that have been significantly underestimated.
To investigate force dependence on integrin subtype, we treated the cells with blocking antibodies against αvβ3 and α5β1 and repeated the kinetic measurements described above. The efficiency of antibody blocking was confirmed by using GFP-β3 expressing cells (Figure S7A). Blocking with both antibodies abrogated cell adhesion, indicating that these two integrins are the primary receptors mediating cell adhesion (Figure S7B). Cells primarily using αvβ3 integrins displayed slowed rates of disulfide reduction indicating a tension of 40 +/− 10 pN. This is in contrast to cells employing α5β1 integrins, which showed accelerated rates of disulfide reduction with average forces of 160 +/− 25 pN. These values are consistent with precedent indicating greater rupture forces for α5β1 compared to αvβ3.26
There is disagreement in the literature regarding the magnitude of integrin-ligand tension within FAs. Dunn and colleagues suggest an integrin-ligand force of 1–5 pN in human foreskin fibroblasts by using single molecule FRET.12a In agreement with this estimate, Schwartz et al. calculate cell traction forces of a few pN per integrin receptor in mouse embryo fibroblasts by averaging cell stress over the average number of integrins per unit area.6a In contrast, we previously inferred greater integrin forces > 20 pN per receptor based on the observation of mechanically induced biotin-streptavidin dissociation in many cell types.9f The tension gauge tether (TGT) technique also suggests greater integrin forces and a peak force of 56 pN for integrin activation.11
The I27 tension probes are more mechanically robust than any other sensor tested to date, thus investigating a greater range of integrin tension values. Except for the disulfide clamped I27, all probes (RGD-ACP(A488)-I27, RGD-sfGFP-I27, RGD-Cy3-I27, FN-A647-I27) were mechanically unfolded by integrin forces displaying a fluorescence response. The sfGFP probe showed mechanically-induced quenching which has been investigated by theory and experiments20a, 27 and is thought to require ~100–200 pN. This surprising result indicates that genetically encoded tension probes for cell adhesion proteins are susceptible to mechanical quenching. For example, genetically encoding an integrin tension probe akin to the vinculin tension sensor28 is not likely to succeed. Although I27 is a highly reversible folder, sfGFP does not recover its full fluorescence after refolding, which may explain why treating cells with the ROCK inhibitor did not lead to full recovery of sfGFP fluorescence.
Importantly, using equilibrium distribution of fold to unfolded tension probes to estimate integrin forces is limited for the following reasons: a) mechanical unfolding is highly dependent on the duration of force, which is unknown because FA maturation is heterogeneous, b) mechanical unfolding is dependent on the loading rate, which is also unknown during FA maturation, and c) once the applied forces exceed a threshold causing unfolding, this defines a minimum tension but not the average tension. Using single molecule techniques can, in principle, overcome some of these limitations but produces a sparse density of reporters. We overcome these limitations by recording the rate of disulfide reduction. Note that a fundamental assumption for force estimates determined by using the kinetics of disulfide reduction are the reaction parameters measured by Fernandez and colleagues. Any significant errors in those measurements will lead to significant deviations in our reported values. Nonetheless, these parameters have been validated in a number of papers investigating different proteins.23–24
By blocking REFs with monoclonal antibodies against αvβ3 and α5β1 we showed that the cell-generated forces are integrin-subtype dependent. The rate of I27 unfolding by αvβ3 integrins is three fold smaller than that for the α5β1 integrins. The mechanism behind differential force levels for different integrin sub-types is likely due to the different functions of each integrins within cell-ECM adhesions. During cell migration, force sensing at the cell edge requires constant recycling of αvβ3 integrins that promotes mechanotransduction and reinforcement of other integrins and proteins to the sites of adhesions.29 However, cells dampen migration by engaging more α5β1 integrins and by generating stable adhesion sites that require stronger integrin-ligand bonds to resist high forces. Sheetz et al. found that clustering of α5β1 integrins enhances rupture forces by six-fold compared to the αvβ3 integrins.26 This is consistent with our results.
High magnitude forces reported through the I27 probe likely pertain to a small subset of force bearing receptors. Indeed, the average integrin tension within FAs is 2–4 pN as determined by TFM.6a Also DNA- and PEG-based MTFM probes sensitive to ~1–20 pN forces generate signal much more rapidly following cell plating (~10 min compared to ~30 min for I27). Therefore, the results reveal the existence of “hot” receptors that disproportionately carry a greater fraction of cell traction forces compared to the vast majority of integrins recruited to FAs.7 High magnitude integrin forces exist with FN and RGD ligands as both types of I27 probes display similar signal. Future experiments with multiplexed tension probes will generate a more comprehensive histogram of integrin force distributions within FAs.
In summary, we report a new class of I27-based tension probes that provides many advantages over the previously reported MTFM probes. First, I27 tension probes are mechanically responsive to a greater dynamic range of forces, and, in principle, the response threshold can range from ~40 pN to ~100 pN by inserting point mutations within the domain. This feature significantly expands the current range of forces detected by MTFM probes. Second, the disulfide bridge clamped sensor circumvents caveats inherent to the transient nature of focal adhesion maturation and the loading rate dependence of mechanical extension, which is a long-standing issue with FRET-based tension sensors. Last, I27-based protein sensors can be easily engineered to incorporate virtually any recombinant protein ligand. This enables the study of mechanotransduction in an expanded number of membrane receptors.
Supplementary Material
Acknowledgments
We thank Dr. Oskar Laur from Custom Cloning Core Facility Yerkes-Microbiology at Emory University for sequencing all the plasmids. We thank Leann Quertinmont (Emory University) for DH5-alpha and BL21(DE3) competent E. coli cells. We thank Dr. Benjamin Geiger (Department of Molecular Cell Biology at Weizmann Institute) for REF cells stably expressing GFP-tagged β3-integrins. We also thank Dr. Ada Cavalcanti-Adam (Max Planck Institute for Intelligent Systems, Stuttgart, Germany) for plasmid encoding mCherry-lifeact. K.S. acknowledges support from the NIH (R01-GM097399), the Alfred P. Sloan Research Fellowship, and the NSF CAREER award.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Materials and methods, quantification of integrin tension for REF cells and additional figures are included. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–87. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Redick SD, Settles DL, Briscoe G, Erickson HP. Defining fibronectin’s cell adhesion synergy site by site-directed mutagenesis. The Journal of cell biology. 2000;149(2):521–7. doi: 10.1083/jcb.149.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humphrey JD, Dufresne ER, Schwartz MA. Mechanotransduction and extracellular matrix homeostasis. Nature reviews. Molecular cell biology. 2014;15(12):802–12. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468(7323):580–4. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nature reviews. Molecular cell biology. 2010;11(9):633–43. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Sabass B, Gardel ML, Waterman CM, Schwarz US. High resolution traction force microscopy based on experimental and computational advances. Biophysical journal. 2008;94(1):207–20. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(4):1484–9. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiseman PW, Brown CM, Webb DJ, Hebert B, Johnson NL, Squier JA, Ellisman MH, Horwitz AF. Spatial mapping of integrin interactions and dynamics during cell migration by image correlation microscopy. J Cell Sci. 2004;117(Pt 23):5521–34. doi: 10.1242/jcs.01416. [DOI] [PubMed] [Google Scholar]
- 8.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 9.(a) Stabley DR, Jurchenko C, Marshall SS, Salaita KS. Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nat Methods. 2012;9(1):64–7. doi: 10.1038/nmeth.1747. [DOI] [PubMed] [Google Scholar]; (b) Jurchenko C, Salaita KS. Lighting up the Force: Investigating Mechanisms of Mechanotransduction Using Fluorescent Tension Probes. Mol Cell Biol. 2015 doi: 10.1128/MCB.00195-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zhang Y, Ge C, Zhu C, Salaita K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nature communications. 2014;5:5167. doi: 10.1038/ncomms6167. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Liu Y, Medda R, Liu Z, Galior K, Yehl K, Spatz JP, Cavalcanti-Adam EA, Salaita K. Nanoparticle tension probes patterned at the nanoscale: impact of integrin clustering on force transmission. Nano Lett. 2014;14(10):5539–46. doi: 10.1021/nl501912g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Liu Y, Yehl K, Narui Y, Salaita K. Tension sensing nanoparticles for mechano-imaging at the living/nonliving interface. Journal of the American Chemical Society. 2013;135(14):5320–3. doi: 10.1021/ja401494e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Jurchenko C, Chang Y, Narui Y, Zhang Y, Salaita KS. Integrin-generated forces lead to streptavidin-biotin unbinding in cellular adhesions. Biophysical journal. 2014;106(7):1436–46. doi: 10.1016/j.bpj.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blakely BL, Dumelin CE, Trappmann B, McGregor LM, Choi CK, Anthony PC, Duesterberg VK, Baker BM, Block SM, Liu DR, Chen CS. A DNA-based molecular probe for optically reporting cellular traction forces. Nat Methods. 2014;11(12):1229–32. doi: 10.1038/nmeth.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340(6135):991–4. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 2013;13(9):3985–9. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Morimatsu M, Mekhdjian AH, Chang AC, Tan SJ, Dunn AR. Visualizing the interior architecture of focal adhesions with high-resolution traction maps. Nano Lett. 2015;15(4):2220–8. doi: 10.1021/nl5047335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Botello E, Harris NC, Sargent J, Chen WH, Lin KJ, Kiang CH. Temperature and chemical denaturant dependence of forced unfolding of titin I27. The journal of physical chemistry B. 2009;113(31):10845–8. doi: 10.1021/jp9002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu H, Isralewitz B, Krammer A, Vogel V, Schulten K. Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophysical journal. 1998;75(2):662–71. doi: 10.1016/S0006-3495(98)77556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276(5315):1109–12. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM. Point mutations alter the mechanical stability of immunoglobulin modules. Nature structural biology. 2000;7(12):1117–20. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z, Cironi P, Lin AJ, Xu Y, Hrvatin S, Golan DE, Silver PA, Walsh CT, Yin J. Genetically encoded short peptide tags for orthogonal protein labeling by Sfp and AcpS phosphopantetheinyl transferases. ACS Chem Biol. 2007;2(5):337–46. doi: 10.1021/cb700054k. [DOI] [PubMed] [Google Scholar]
- 18.Aubin-Tam ME. Conjugation of nanoparticles to proteins. Methods Mol Biol. 2013;1025:19–27. doi: 10.1007/978-1-62703-462-3_3. [DOI] [PubMed] [Google Scholar]
- 19.Yehl K, Joshi JP, Greene BL, Dyer RB, Nahta R, Salaita K. Catalytic deoxyribozyme-modified nanoparticles for RNAi-independent gene regulation. ACS nano. 2012;6(10):9150–7. doi: 10.1021/nn3034265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(a) Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(46):16192–7. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Otten M, Ott W, Jobst MA, Milles LF, Verdorfer T, Pippig DA, Nash MA, Gaub HE. From genes to protein mechanics on a chip. Nat Methods. 2014;11(11):1127–30. doi: 10.1038/nmeth.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24(1):79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 22.Pankov R, Yamada KM. Fibronectin at a glance. Journal of cell science. 2002;115(Pt 20):3861–3. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 23.Ainavarapu SR, Brujic J, Huang HH, Wiita AP, Lu H, Li L, Walther KA, Carrion-Vazquez M, Li H, Fernandez JM. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophysical journal. 2007;92(1):225–33. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.(a) Wiita AP, Ainavarapu SR, Huang HH, Fernandez JM. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(19):7222–7. doi: 10.1073/pnas.0511035103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ainavarapu SR, Wiita AP, Huang HH, Fernandez JM. A single-molecule assay to directly identify solvent-accessible disulfide bonds and probe their effect on protein folding. Journal of the American Chemical Society. 2008;130(2):436–7. doi: 10.1021/ja077851s. [DOI] [PubMed] [Google Scholar]; (c) Wiita AP, Perez-Jimenez R, Walther KA, Grater F, Berne BJ, Holmgren A, Sanchez-Ruiz JM, Fernandez JM. Probing the chemistry of thioredoxin catalysis with force. Nature. 2007;450(7166):124–7. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Liang J, Fernandez JM. Kinetic measurements on single-molecule disulfide bond cleavage. Journal of the American Chemical Society. 2011;133(10):3528–34. doi: 10.1021/ja109684q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koti Ainavarapu SR, Wiita AP, Dougan L, Uggerud E, Fernandez JM. Single-molecule force spectroscopy measurements of bond elongation during a bimolecular reaction. Journal of the American Chemical Society. 2008;130(20):6479–87. doi: 10.1021/ja800180u. [DOI] [PubMed] [Google Scholar]
- 26.Roca-Cusachs P, Gauthier NC, Del Rio A, Sheetz MP. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(38):16245–50. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saeger J, Hytonen VP, Klotzsch E, Vogel V. GFP’s mechanical intermediate states. PLoS One. 2012;7(10):e46962. doi: 10.1371/journal.pone.0046962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466(7303):263–6. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White DP, Caswell PT, Norman JC. alpha v beta3 and alpha5beta1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. The Journal of cell biology. 2007;177(3):515–25. doi: 10.1083/jcb.200609004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.