ABSTRACT
The developmental accumulation of proliferative germ cells in the C. elegans hermaphrodite is sensitive to the organismal environment. Previously, we found that the TGFβ signaling pathway links the environment and proliferative germ cell accumulation. Neuronal DAF-7/TGFβ causes a DAF-1/TGFβR signaling cascade in the gonadal distal tip cell (DTC), the germline stem cell niche, where it negatively regulates a DAF-3 SMAD and DAF-5 Sno-Ski. LAG-2, a founding DSL ligand family member, is produced in the DTC and activates the GLP-1/Notch receptor on adjacent germ cells to maintain germline stem cell fate. Here, we show that DAF-7/TGFβ signaling promotes expression of lag-2 in the DTC in a daf-3-dependent manner. Using ChIP and one-hybrid assays, we find evidence for direct interaction between DAF-3 and the lag-2 promoter. We further identify a 25 bp DAF-3 binding element required for the DTC lag-2 reporter response to the environment and to DAF-7/TGFβ signaling. Our results implicate DAF-3 repressor complex activity as a key molecular mechanism whereby the environment influences DSL ligand expression in the niche to modulate developmental expansion of the germline stem cell pool.
KEY WORDS: GLP-1/Notch, SMAD, DAF-3, C. elegans, Sno-Ski
Summary: In the C. elegans germline, a DAF-3 binding element is required for the distal tip cell Notch pathway response to the environment and to DAF-7/TGFβ signaling, modulating developmental expansion of the stem cell pool.
INTRODUCTION
To maintain tissues, stem cells and their dividing progeny are tightly regulated. In addition to direct signals from the stem cell niche, global signals reporting environmental and physiological conditions can influence the outcome of stem cell decisions (Drummond-Barbosa, 2008; Hubbard et al., 2012; Laws and Drummond-Barbosa, 2017). However, our understanding of the mechanisms underlying this regulation remains incomplete.
The C. elegans germ line is a powerful model for exploring how stem cell behavior is influenced by the combined action of signaling from the local niche and by organismal cues that report physiologically relevant conditions. In hermaphrodites, a single distal tip cell (DTC) caps each of the two gonad arms and functions as the germline stem cell niche. DTC-to-germline signaling via the Notch pathway and gap junctions governs stem cell fate and proliferation (Kimble and White, 1981; Austin and Kimble, 1987; Starich et al., 2014). At least two of the 10 DSL family ligands in the C. elegans genome (Chen and Greenwald, 2004), LAG-2 and APX-1, are produced by the DTC and activate the GLP-1/Notch receptor on the surface of germ cells to maintain germline stem cell fate (Henderson et al., 1994; Nadarajan et al., 2009). The DAF-7/TGFβ signaling pathway was identified for its role in dauer formation (Larsen et al., 1995). If animals meet unfavorable conditions as early larvae, the resulting decrease in DAF-7/TGFβ signaling promotes formation of the stress-resistant, non-aging dauer larva. In this role, DAF-7 signals through the DAF-1 type I and DAF-4 type II receptors and DAF-8 and DAF-14 R-Smads to negatively regulate a DAF-3 SMAD/DAF-5 Sno-Ski transcriptional repressor complex (Gumienny, 2013). We identified the DAF-7/TGFβ signaling pathway in a genetic screen for genes that modulate the accumulation of proliferative germ cells in later larval stages, after the time that the dauer decision is made (Dalfó et al., 2012). In this role, it uses the same components and regulatory logic as in dauer formation (Fig. 1A), but the DAF-1/TGFβR complex and downstream transcriptional regulators act in the DTC (Dalfó et al., 2012). Unlike the DAF-2/insulin-IGF-like signaling pathway that also influences larval germline progenitor cell accumulation (Hubbard et al., 2012; Michaelson et al., 2010), DAF-7/TGFβ signaling influences germline stem cell fate and not the rate of germ cell cycle progression. Finally, in at least one genetic scenario DAF-7/TGFβ can act independently of the GLP-1/Notch receptor.
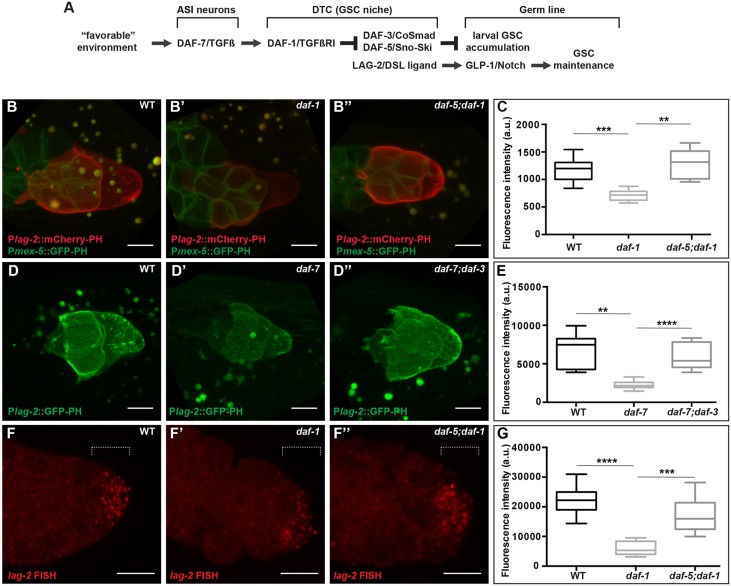
Fig. 1.
DAF-7/TGFβ signaling promotes lag-2 expression in the DTC. (A) Summary of the influences of larval DAF-7/TGFβ and GLP-1/Notch signaling reported prior to this work. (B-E) Expression of lag-2 (3 kb upstream) reporters. naIs37: membrane-bound mCherry (mCherry-PH) low-copy reporter in the distal gonad arm of (B) wild-type, (B′) daf-1 and (B″) daf-5;daf-1 animals. naSi1 [Pmex-5::GFP-PH] marks germ cell membranes. See Fig. S1 for additional genotypes. naSi8: membrane-bound GFP (GFP-PH) single-copy reporter in (D) wild-type, (D′) daf-7 and (D″) daf-7; daf-3 animals. (C,E) Quantification of mCherry and GFP signals, respectively. (F-G) In situ hybridization with lag-2 probes of dissected gonads from (F) wild-type, (F′) daf-1 and (F″) daf-5; daf-1 animals (magnified relative to other panels). (C,E,G) Values correspond to mean pixel intensity (C,E) and sum pixel intensity (G) in arbitrary units, measured in the DTC as described in the Materials and Methods. The boxes indicate the minimum-maximum range of samples quantified. Dotted lines mark the area (distal 5 μm) where the signal was quantified in G. This signal is in the DTC, not in the germ line. Scale bars: 5 μm. Mutant alleles: daf-1(m40), daf-5(e1386), daf-7(e1372) and daf-3(e1376). **P<0.01, ***P<0.001, ****P<0.0001, two-tailed Student's t-test. n≥15 animals, one DTC scored per animal. Error bars represent s.e.m. In all cases, P>0.05 for wild type versus double mutants.
Here, we report that TGFβ receptor signaling promotes the expression of lag-2 in the late larval DTC, defining a direct mechanistic link between TGFβ and Notch signaling. We also extend previous findings on the transcriptional regulation of lag-2. We find that DTC expression of lag-2 is reduced when TGFβ signaling is low but is restored in the absence of daf-3 or daf-5. Similarly, lag-2 reporter expression is reduced in unfavorable environments, in a manner dependent on daf-3. Using one-hybrid and ChIP assays, we find evidence for direct interaction between DAF-3 and the lag-2 promoter. Using transcriptional reporters, we define a TGFβ response element in the lag-2 promoter and show that eliminating one of two potential DAF-3-binding sites abrogates the response to TGFβ receptor signaling and to low food, suggesting that it may serve as a direct target for the TGFβ pathway within the lag-2 promoter. We propose a working model and discuss our findings in the context of previous work, of TGFβ-Notch and environment-Notch interactions in general.
RESULTS
DAF-7/TGFβ signaling promotes lag-2 expression
We sought to determine the mechanistic link between TGFβR signaling in the DTC and germline stem cell maintenance. Previously, we showed that TGFβR signaling in the DTC promotes germ cell accumulation during late larval stages by preventing germ cell differentiation. Our previous work suggested that the TGFβR can act in parallel to GLP-1/Notch (Dalfó et al., 2012). This conclusion was based on: (1) the observation that reducing TGFβ signaling in a glp-1(null) mutant, which was also mutant for gld-1 and gld-2, to permit production and maintenance of proliferative germ cells, reduced the number of germ cells in the resulting germline tumors; and (2) our inability to detect consistent changes in then-existing lag-2 reporter expression upon manipulation of the TGFβ pathway (Dalfó et al., 2012; E.J.A.H., unpublished). Although the residual effects of TGFβ signaling in the glp-1(null) mutant (albeit tumorous) background still argue for a partially GLP-1-independent role for TGFβ signaling (see Discussion), results presented here suggest that TGFβ signaling also influences GLP-1 activity by modulating expression of a DSL ligand, LAG-2, in the DTC.
Given our provisional conclusion that GLP-1/Notch was not acting downstream of TGFβ, we set out to investigate the possibility that the DTC-germline interface might be disrupted when TGFβ signaling was low. We built DTC membrane-bound reporters (using GFP or mCherry fusions to the PH domain of the rat PLC1δ1) driven by the 3 kb lag-2 upstream region used by others (Henderson et al., 1994). We introduced the new reporter transgenes into the worm genome using microparticle bombardment, a technique that results in fewer copies than traditional transgenes borne on extrachromosomal arrays. We examined the DTC in the fourth larval stage (L4) as DAF-7/TGFβ signaling affects proliferative germ cell accumulation before and during this stage (Dalfó et al., 2012). We observed that the reporters were expressed at an overall lower level in wild type than reporters we had examined previously.
To our surprise, the new lower-copy reporters were expressed at significantly lower levels in daf-7 or daf-1 mutant backgrounds than in the wild type, and their expression was restored to wild-type levels in daf-7 or daf-1 double mutants with daf-3 or daf-5 (Fig. 1B,C, Fig. S1A-C). The restoration of expression in these double mutants is consistent with the logic of the canonical DAF-7/TGFβ pathway that we previously implicated in the regulation of the germline proliferative pool (Fig. 1A). Similar effects were observed with a single-copy reporter introduced by MosSCI (Fig. 1D,E) and with either mCherry or GFP reporters. A different non-lag-2 DTC-expressed reporter showed no such regulation (Fig. S1D,E). Therefore, we conclude that the lower copy number and expression levels of the newer reporters reveals modulation by the DAF-7/TGFβ pathway that was not detectable with high-copy reporters.
To determine whether the reporters were reflecting changes in endogenous lag-2 mRNA in the DTC, we performed fluorescence in situ hybridization experiments on dissected gonad preparations. Our results were consistent with the reporter analysis (Fig. 1F,G), suggesting that lag-2 expression in the DTC is regulated positively by DAF-7/TGFβ signaling and negatively by DAF-3 and DAF-5.
Environmental conditions alter lag-2 expression, dependent on the DAF-7/TGFβ signaling pathway
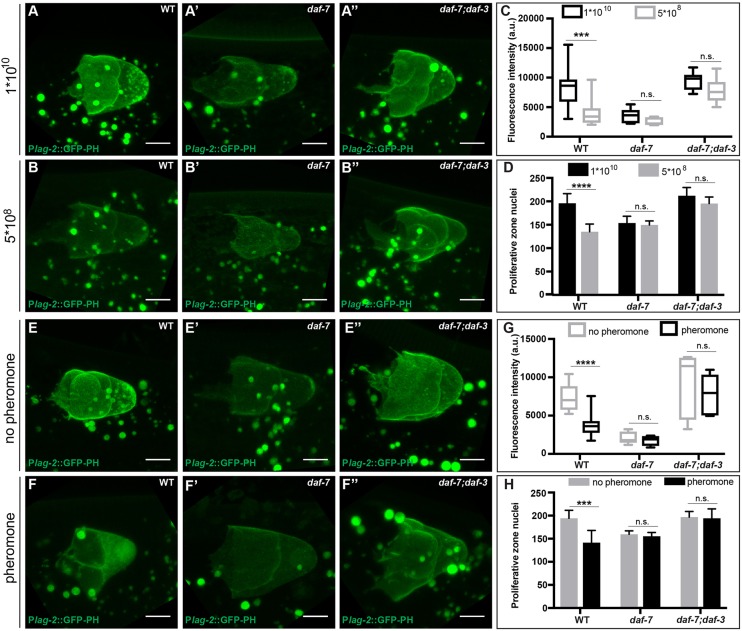
Previously, we showed that DAF-7/TGFβ signaling regulates the accumulation of proliferative zone cells in the larval germ line in response to low food or high dauer pheromone, two conditions that reduce daf-7 expression in ASI neurons (Ren et al., 1996; Schackwitz et al., 1996). To determine whether lag-2 expression is similarly modulated, we measured GFP expression from a single-copy lag-2 reporter under favorable (high food or low pheromone) and unfavorable (low food or high pheromone) conditions (Fig. 2). We found that lag-2 reporter expression was diminished in either unfavorable condition relative to the favorable condition in the wild type (Fig. 2B,F). The daf-7 mutant displayed lower expression that was not further reduced by unfavorable conditions (Fig. 2B′,F′). In the daf-7; daf-3 double mutant, lag-2 reporter expression is restored to that of favorable conditions in the wild type, even under unfavorable conditions (Fig. 2B″,F″). These results suggest that environmental regulation of lag-2 expression occurs through the canonical DAF-7/TGFβ pathway. Consistent with previous results (Dalfó et al., 2012), germ cell accumulation in all these environmental and genetic combinations parallels observed changes in lag-2 reporter expression (Fig. 2).
Fig. 2.
Reduced lag-2 DTC expression under adverse environmental conditions depends on daf-3. (A-B″) Expression levels of the lag-2 single-copy reporter naSi8 in the DTC of late L4 animals reared from early L3 on a high (1×1010) or low (5×108) concentration of OP50 bacteria in (A,B) wild-type, (A′,B′) daf-7 and (A″,B″) daf-7; daf-3 animals. (C) Quantification of the GFP signal in the DTC in animals exposed to high or low bacterial concentrations. (D) Number of proliferative zone nuclei per gonad arm in early adults (collected from the same plates as in previous panels) reared from early L3 under high or low bacterial concentrations. (E-F″) Expression levels of lag-2 single-copy reporter (naSi8) in the DTC of late L4 animals in the absence or presence of exogenous dauer pheromone introduced in the early L3 in (E,F) wild-type, (E′,F′) daf-7 and (E″,F″) daf-7; daf-3 animals. A slight enrichment of nuclear GFP was observed under high-pheromone conditions (but not under low-food conditions) in a majority of animals, but its significance was not further investigated. (G) Quantification of the GFP signal in the DTC in animals exposed or not exposed to exogenous dauer pheromone. (C,G) Mean pixel intensity (arbitrary units), measured in the DTC as described in the Materials and Methods. The boxes indicate the minimum-maximum range of samples quantified. (H) Number of proliferative zone nuclei of early adults (collected from the same plates as in previous panels) exposed or not exposed to exogenous dauer pheromone introduced in the early L3 stage. Scale bars: 5 μm. Mutant alleles were daf-7(e1372) and daf-3(e1376). n.s. indicates P>0.05, ***P<0.001, ****P<0.0001, two-tailed Student's t-test. n≥15 animals, one DTC scored per animal (GFP quantifications) or one gonadal arm per animal (proliferative zone quantifications). Error bars represent s.e.m. Additional statistical analysis in GFP quantifications: P>0.05 for the wild type under high-food or no pheromone conditions versus daf-7; daf-3 double mutant under low-food or with pheromone conditions, respectively.
Canonical DAF-3 SMAD-binding sites in the lag-2 promoter do not mediate DAF-7/TGFβ signaling in the DTC
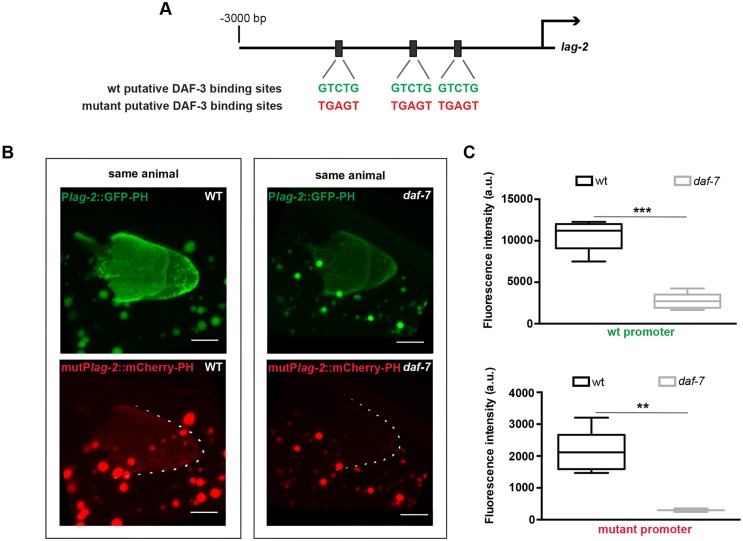
We hypothesized that lag-2 expression may be negatively regulated by the DAF-3 repressor complex through direct interaction with lag-2 regulatory sequences. Within the 3 kb region upstream of lag-2, we found three instances of the 5 bp DAF-3 binding motif (Fig. 3A) previously defined in the myo-2 promoter (Thatcher et al., 1999), and that are also present within a DAF-3-bound region in the promoters of daf-7 and daf-8, as determined by whole-animal ChIP (Park et al., 2010). If these sites mediate DAF-3 repressor activity, mutating them should render lag-2 expression insensitive to the loss of daf-7 or daf-1. We generated two independent lines, one bearing a promoter with all three sites mutated that drives mCherry and the other bearing the wild-type promoter that drives GFP (Fig. 3A). We crossed them to generate a strain bearing both reporters and assessed their expression in the wild type and daf-7 mutant. As expected, we observed lower wild-type promoter (‘wt lag-2 promoter’, green in Fig. 3B) expression in the daf-7 mutant background. However, we also observed lower expression from the mutant promoter in daf-7 relative to wild type (Fig. 3B,C). The differences in mutant promoter-driven expression were also apparent in strains without the wild-type GFP reporter (data not shown). These results suggest that the predicted canonical DAF-3-binding sites do not confer regulation of lag-2 expression by the DAF-7/TGFβ pathway in this context.
Fig. 3.
Canonical DAF-3-binding sites are not required for the effect of DAF-7/TGFβ on lag-2 DTC expression. (A) Positions of three canonical DAF-3-binding sites (Thatcher et al., 1999) within 3 kb upstream of the lag-2 ATG. (B) Distal gonad arms from two animals, each expressing two reporters: the wild-type single-copy naSi8 GFP-PH reporter; and naIs96, an integrated reporter in which mCherry-PH is driven by a lag-2 promoter, in which three canonical DAF-3-binding sites were mutated as in A. The expression of each reporter was measured in individual wild-type (left) and daf-7(e1372) (right) animals. Scale bars: 5 μm. (C) Quantification of GFP (top) and mCherry (bottom) in the DTC in the different genetic backgrounds. Mean pixel intensity (arbitrary units), measured in the DTC as described in the Materials and Methods. The boxes indicate the minimum-maximum range of samples quantified. **P<0.01, ***P<0.001, two-tailed Student's t-test. n≥15 animals, one DTC scored per animal. Error bars represent s.e.m.
An ∼100 bp sequence upstream of lag-2 is required for the response to DAF-7/TGFβ signaling
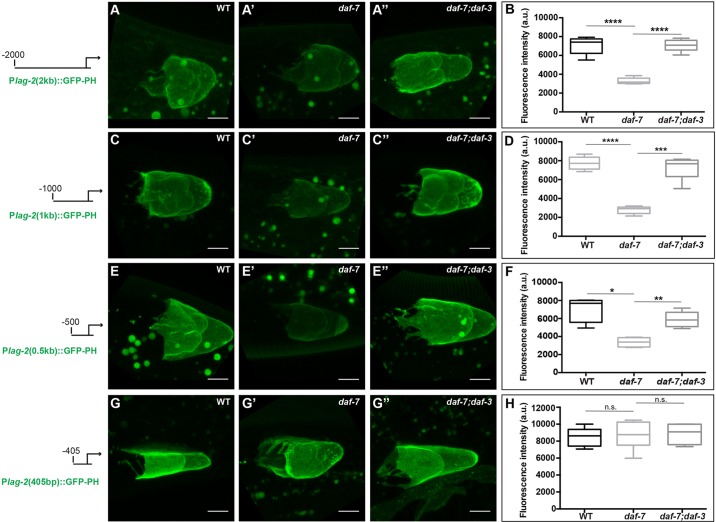
To identify the TGFβ-responsive region of the lag-2 promoter, we performed promoter deletion analysis. We generated strains expressing reporters of 2.0, 1.0 and 0.5 kb upstream of the ATG. We then crossed the transgenes into daf-7 and daf-7; daf-3 mutant backgrounds and compared their expression (Fig. 4). Consistent with previous reports of lag-2 expression in head neurons (Ouellet et al., 2008; Singh et al., 2011), our 1.0 kb promoter drove reporter expression in head neurons, as well as in the DTC. This expression in neurons was not grossly affected by DAF-7/TGFβ pathway signaling and served as an internal control (Fig. S2). Each of the three promoter regions tested showed reduced expression in the daf-7 single mutant that was restored to wild-type levels in daf-7; daf-3 double mutants (Fig. 4A-F). We next assessed a promoter truncated at −405 bp. This reporter no longer responded to the loss of daf-7: expression was similar in wild type, daf-7 and daf-7; daf-3. Taken together, these results define a key TGFβ-responsive element (TRE) near or within the −405 to −500 region upstream of lag-2 (Fig. 4G,H).
Fig. 4.
A region between −500 bp and −405 bp in the lag-2 promoter is crucial for the DTC response to DAF-7/TGFβ. Expression levels of lag-2 low-copy GFP-PH reporters driven by (A-B) 2 kb, (C-D) 1 kb, (E-F) 500 bp and (G-H) 405 bp upstream in the DTC of wild-type, daf-7(e1372) and daf-7(e1372); daf-3(e1376) animals. Transgene alleles are naIs81, naIs84, naIs87 and naIs98, respectively. (B,D,F,H) Mean pixel intensity (arbitrary units), measured in the DTC as described in the Materials and Methods. The boxes indicate the minimum-maximum range of samples quantified. Scale bars: 5 μm. n.s. indicates P>0.05, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, two-tailed Student's t-test. n≥15 animals, one DTC scored per animal. Error bars represent s.e.m. In all cases, P>0.05 for wild type versus double mutants.
A 25 bp sequence mediates the lag-2 promoter response to TGFβ signaling and the environment
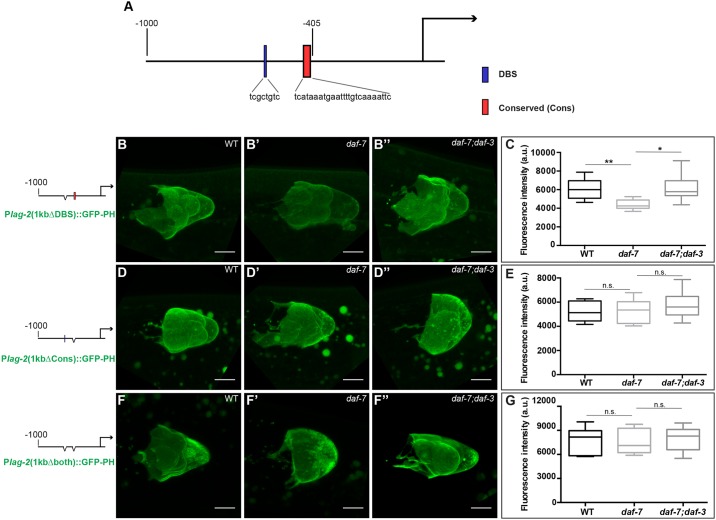
Closer inspection of the TRE upstream of lag-2 revealed homology to a cis-acting regulatory sequence named the PD motif, which was characterized in the osm-9 promoter in the context of continuous versus postdauer (PD) development (Sims et al., 2016). The osm-9 PD motif consists of two sequence elements: a DAF-3-binding site (osm-9p-DBS) and a conserved sequence (osm-9p-Cons) that is found in the upstream regulatory regions of ∼1000 genes of the C. elegans genome (Sims et al., 2016). At the lag-2 locus, we identified an 8 bp sequence with homology to the osm-9 promoter that partially overlaps with the osm-9p-DBS at −577 to −570 upstream of the lag-2 ATG (lag-2p-DBS). Whereas the osm-9p-DBS contains a canonical SMAD-binding sequence (GTCT), the lag-2p-DBS does not (Fig. S3). In addition, we identified a 25 bp loosely conserved sequence from −432 to −407 (lag-2p-Cons) that, although not identical to the osm-9p-Cons, satisfies much of the conserved (Cons) sequence ‘rule’ (Fig. S3). The lag-2p-DBS sequence is upstream of the TRE, and the lag-2p-Cons is within the TRE that we defined by promoter deletion analysis (Fig. 5A). We generated reporter strains with each site deleted individually or together in the context of the 1 kb lag-2 upstream fragment (Fig. 5B-G). Expression of the reporter lacking the lag-2p-DBS sequence was still reduced in daf-7 relative to wild type, albeit not as much as when the sequence was intact (Fig. 5B,C). By contrast, the reporter lacking the lag-2p-Cons motif was expressed at the same level in the wild type and in the daf-7 mutant (Fig. 5D,E). These results indicate that the 25 bp lag-2p-Cons sequence is required for the TGFβ response in the L4 DTC.
Fig. 5.
The conserved (Cons) sequence is required for the response of lag-2 to DAF-7/TGFβ signaling in the DTC. (A) Positions of the DAF-3-binding site (DBS) and conserved (Cons) sequences. (B-G) Expression of lag-2 (1 kb, low-copy) GFP-PH reporters in the DTC of wild-type, daf-7(e1372) and daf-7(e1372);daf-3(e1376) animals. Reporters are lacking (B-C) DBS sequence (1kbΔDBS), naIs100; (D-E) conserved (Cons) sequence (1kbΔCons), naIs102; or both (1kbΔboth, F-G), naIs106. (C,E,G) Quantification of GFP expression; mean pixel intensity (arbitrary units), measured in the DTC as described in Materials and Methods. The boxes indicate the minimum-maximum range of samples quantified. Scale bars: 5 µm. n.s. indicates P>0.05, *P<0.05, **P<0.01, two-tailed Student's t-test. n≥15 animals, one DTC scored per animal. Error bars represent s.e.m. In all cases, P>0.05 for wild type versus double mutants.
To determine whether lag-2 reporter expression in response to the environment (Fig. 2) is similarly regulated by the Cons sequence, we repeated the low-food experiment in strains bearing the 1 kb lag-2 upstream fragment with and without the Cons sequence. We measured expression of GFP in the DTC of L4 animals raised on low or high bacterial concentration and, as a positive control for the effects of low food, measured the number of proliferative germ cells. We observed that in the absence of the Cons sequence, the reporter was no longer sensitive to the food level (Fig. S4), suggesting that the Cons sequence mediates the daf-3-dependent effect of low food on lag-2 expression.
DAF-3 binds upstream of lag-2
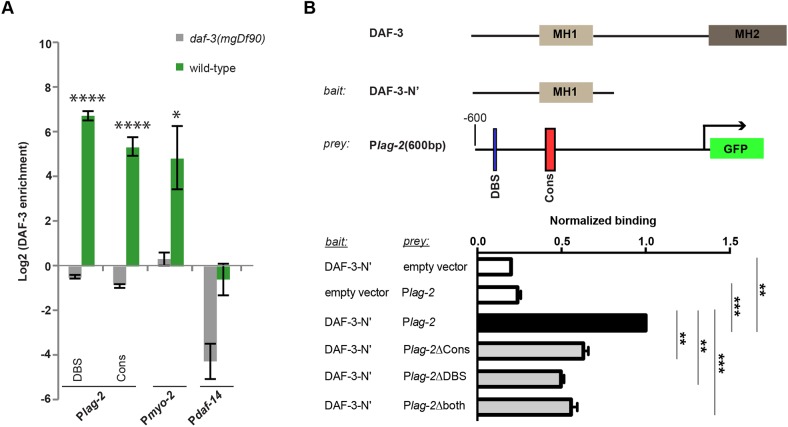
In the osm-9 promoter, DAF-3 SMAD is enriched at the osm-9p-DBS within the PD motif to downregulate osm-9 in ADL neurons of animals that passed through the dauer stage (Sims et al., 2016). To determine whether DAF-3 is enriched at the PD motif elements upstream of lag-2, we performed DAF-3 immunoprecipitation on whole worms using a commercially available antibody followed by quantitative PCR of the lag-2 upstream sequences in the wild type and the daf-3(mgDf90) mutant. Our results indicate that DAF-3 is significantly enriched at the lag-2p-DBS in wild-type L4 animals compared with background levels of enrichment in the daf-3(mgDf90) strain (Fig. 6A). As expected, DAF-3 was not enriched at the osm-9p-DBS in larval L4 animals that experienced continuous (non-dauer) development (Sims et al., 2016). Interestingly, we found that DAF-3 was enriched at the Cons sequences in both lag-2 (lag-2p-Cons) and osm-9 (osm-9p-Cons), suggesting that binding may occur at multiple sites within the PD motif (Fig. S5A). As positive controls, we verified DAF-3 enrichment at its characterized binding sites in the myo-2, daf-7 and daf-8 promoters (Park et al., 2010; Thatcher et al., 1999). Furthermore, DAF-3 was not enriched in the lag-2-coding region, similar to our negative control, daf-14 (Fig. S5A) (Park et al., 2010). Together, these results indicate that the lag-2 upstream regulatory region contains the lag-2p-DBS and lag-2p-Cons sequence components of the PD motif, both of which have enriched DAF-3 binding in the ChIP assay.
Fig. 6.
DAF-3 can bind the lag-2 promoter. (A) Log2 normalized enrichment of DAF-3 SMAD binding to lag-2 DBS and conserved (Cons) elements in the wild type and in daf-3(mgDf90) mutants. Upstream regulatory regions of myo-2 and daf-14 serve as positive and negative controls, respectively. Bar graph represents IP-qPCR data, normalized to DAF-3 enrichment at the actin act-2 promoter (Park et al., 2010). n≥2 biologically independent trials. Significant enrichment in wild type relative to daf-3(mgDf90) is indicated by *P<0.05, ****P<0.0001, two-tailed Student's t-test. Data are mean±s.e.m. (B) Bacterial one-hybrid assay. The DAF-3 N-terminal fragment (up to amino acid 250; DAF-3-N’) was used as bait and 600 bp upstream of the lag-2 ATG [Plag-2(600 bp)] was used as prey. Bar graph represents FACS analysis data (mean GFP fluorescence) of different baits and preys, normalized to the signal from DAF-3-N’ and Plag-2. Empty prey and bait vectors served as negative controls. Binding of DAF-3-N’ to 600 bp of the lag-2 promoter lacking Cons (Plag-2ΔCons), DBS (Plag-2ΔDBS) or both (Plag-2Δboth). **P<0.01, ***P<0.001, two-tailed Student's t-test. Data are mean±s.e.m.
One-hybrid studies support direct binding of DAF-3 to both PD motif elements within the lag-2 upstream region
We sought independent evidence for binding of DAF-3 to the lag-2 promoter. We turned to a bacterial one-hybrid approach that has advantages of high sensitivity and quantifiable results (Noyes, 2012). We found that the N-terminal DNA-binding region of DAF-3 showed greater binding in the presence of the 600 bp sequence upstream of the lag-2 ATG (Fig. 6B, Fig. S5B). To assess the relevance of the PD motif in this binding assay, we deleted or scrambled the lag-2p-DBS and lag-2p-Cons sequences individually and in tandem. We found that deleting either site lowered binding (Fig. 6B). Although scrambling only one sequence had no effect, scrambling both significantly reduced binding (Fig. S5B). We conclude that, although the Cons site alone is required to regulate DTC reporter expression, binding can occur at either site, consistent with the whole-worm ChIP-qPCR analysis.
DISCUSSION
Our results indicate that the environment impacts the expression of a DSL ligand, lag-2, in the germline stem cell niche through TGFβ signaling. When conditions are poor (low food or high pheromone), lag-2 reporter expression is reduced, and this reduction is dependent on DAF-3 SMAD activity. Furthermore, this reduction is dependent on a 25 bp conserved (Cons) region of the lag-2 promoter (Fig. S4). Together with our previous studies (Dalfó et al., 2012), this work suggests a simple working model (Fig. 7) in which proliferative germ cells accumulate in favorable conditions in response to DAF-7/TGFβ signaling that ensures high levels of LAG-2 in the DTC, that then signals to GLP-1/Notch in the germ line. That DAF-7/TGFβ signaling may modulate Notch activity is consistent with previous results showing that low daf-1 activity enhances the phenotype of a glp-1 reduction-of-function mutant and that, similar to glp-1, daf-1 does not affect the mitotic index of the proliferative pool (Dalfó et al., 2012).
Fig. 7.
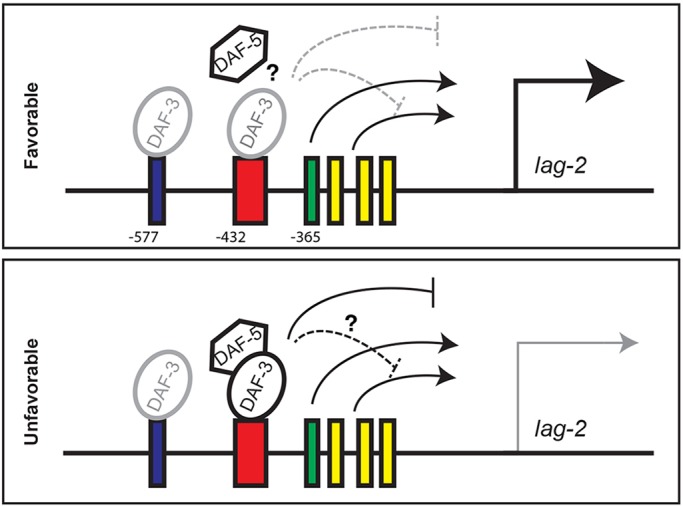
A working model for the regulation of late larval DTC-expressed lag-2. (Top) Under favorable conditions, DAF-7/TGFβ pathway activity inhibits repressor activity of the DAF-3/DAF-5 complex, possibly by interfering with complex formation or stability. lag-2 expression is driven by positive regulators, including HLH-2, which binds the E-box (green), and unknown regulators that bind other conserved sequences (yellow) shown in Fig. S7. DAF-3 may bind both the DBS (blue) and the Cons (red) sequences under favorable conditions, but it is not active. (Bottom) Under unfavorable conditions, the DAF-3/DAF-5 complex is active and represses lag-2 expression, possibly by interfering with non-HLH-2 activators. See Discussion for details.
Our current results show that DAF-3 SMAD can bind both the DBS and the Cons sequences in the lag-2 promoter in the L4 (in whole-worm ChIP), even when conditions are favorable and animals have experienced continuous development. Work by others has shown that DAF-3 can also bind to mdl-1, daf-7 and daf-8 promoters in non-dauer animals (Deplancke et al., 2006; Park et al., 2010), and Sims et al. (2016) demonstrated that DAF-3 bound the osm-9 promoter in continuously developing adult animals, which was dependent upon functional ZFP-1/AF10 protein and endogenous RNAi pathways. Spacing may also be important for DAF-3 binding of DBS and Cons, as binding in the bacterial one-hybrid assay was maintained when the sites were individually scrambled (rather than deleted) but was lost when both were scrambled (Fig. S5B). Subcellular localization of DAF-3 is not grossly altered by changes in TGFβ signaling (Patterson et al., 1997). Thus, DAF-3 activity as a transcriptional repressor may not correlate solely with its ability to bind DNA. For example, it could depend on post-translational modifications or additional DNA- binding partners. In unfavorable conditions when TGFβ receptor signaling is low, we speculate that the DAF-3 repressor complex at the Cons sequence becomes active in the DTC (Fig. 7).
How does the DAF-3 repressor complex repress transcription? Among many possible mechanisms, the active repressor complex may interfere with activators at nearby sites. The helix-loop-helix (HLH) transcription factor HLH-2/Da activates lag-2 via enhancer box (E-box) sequences (Krause et al., 1997) that are close to the Cons sequence. However, we found that the active DAF-3 complex is unlikely to interfere with HLH-2, because depleting hlh-2 by RNAi reduced lag-2 reporter expression in wild-type, daf-7 and daf-7; daf-3 (Fig. S6 and supplementary Materials and Methods for RNAi methods). Interfering with hlh-2 or E-boxes reduces but does not eliminate DTC lag-2 expression, suggesting that other transcription factors contribute (Chesney et al., 2009; Karp, 2003; Karp and Greenwald, 2004). We found highly conserved sequences within 500 bp upstream of the ATG: a GC-box, a C/EBP box and an uncharacterized sequence (Fig. S7). Deletion of any one of these sites abrogated reporter expression (data not shown). Therefore, the DAF-3 repressor complex may interfere with activators that bind these sites.
If modulation of DTC lag-2 expression and its subsequent effects on germline GLP-1/Notch receptor activity were the only mechanism by which the larval germ line responds to DAF-7/TGFβ, the germline progenitor pool should be insensitive to TGFβ in the absence of glp-1. However, we previously showed that fewer germline progenitor cells accumulate when DAF-7/TGFβ signaling is low, even in the absence of the GLP-1/Notch receptor, albeit in a tumorous germline context (Dalfó et al., 2012). These results suggest a glp-1-independent role for TGFβ. One possibility is that the other Notch receptor in the C. elegans genome, LIN-12, has a partially redundant germline-autonomous role in maintaining germline stem cells. Null mutations in lin-12 cause sterility, likely secondary to its roles in the somatic gonad (Greenwald et al., 1983; Seydoux et al., 1990), and no obvious larval germline lin-12 expression is detected by in situ hybridization (NEXTDB; Shin-i and Kohara, 1999). We found that lin-12 RNAi directed primarily to the germ line (in rrf-1; Sijen et al., 2001) did not impact the size of the proliferative pool in glp-1(+) or glp-1(rf), a highly sensitized background for germline stem cell loss (Fig. S8; see supplementary Materials and Methods for RNAi methods). Additional models include a Notch signaling-independent role for DTC lag-2 or a lag-2-independent role for TGFβ. For example, TGFβ signaling may influence DTC-germline gap junctions that promote proliferation (Starich et al., 2014). Unlike the well-characterized role for TGFβ in the non-tumorous scenario, it is unknown whether the glp-1-independent effect of TGFβ on tumor cell number is DTC autonomous or daf-3 dependent. In any case, we propose that TGFβ regulation of DTC lag-2 expression reported here accounts for much of the germ cell fate regulation by TGFβ in the presence of glp-1.
In apparent conflict with our results, and as noted by Dalfó et al., (2012), Park et al. (2010) reported that daf-8/R-Smad negatively regulates lag-2 high-copy reporter expression in the DTC and germline proliferative zone size, independently of daf-3. Dalfó et al. (2012) observed positive regulation (i.e. less accumulation of proliferative germ cells in daf-7, daf-1, daf-8 and daf-14 mutants), dependent on daf-3 and daf-5. Here, consistent with Dalfó et al., (2012) we show that DAF-7/TGFβ signaling promotes late larval lag-2 expression and proliferative germ cell accumulation in a daf-3- and daf-5-dependent manner. A major difference in the two studies is that Park et al. (2010) focused on adults, whereas we focused on synchronized late larval stages. It remains possible that DAF-8 plays opposite roles in DTC lag-2 regulation at different life stages. We also observed that the DBS and Cons sites may mediate activation (rather than repression) of a lag-2 reporter in neurons, indicating cell-specific regulatory differences (data not shown). Consistent with Park et al. (2010), we found enrichment of DAF-3 at the daf-7 and daf-8 promoters using whole-worm ChIP (Fig. S5A). A more complete understanding awaits additional comparative studies of all pathway components in specific cell types at different stages in tightly synchronized animals reared in well-controlled environments.
TGFβ-mediated regulation of DSL ligands and Notch signaling in other systems
Examples of DSL ligand regulation by TGFβ superfamily members exist in several systems. This may reflect an ancient relationship between these signaling pathways that, together with Wnt, existed in the earliest metazoans (Richards and Degnan, 2010). Examples of positive regulation include TGFβ promoting Jag1 expression or activity in the epithelial-to-mesenchymal transition in mammalian cell lines (Zavadil et al., 2004), muscle differentiation of mesenchymal stem cells (Kurpinski et al., 2010), and an endothelial-to-hematopoietic transition in zebrafish (Monteiro et al., 2016). Positive regulation also occurs in endothelial cell lines where a Smad1/5-binding motif was identified upstream of Jag1 by ChIP-seq (Morikawa et al., 2011). Nodal also positively regulates Ci-Delta2 transcription in notochord specification in Ciona (Hudson and Yasuo, 2005, 2006). It will be of interest to determine how positive regulation by TGFβ occurs through cis-acting sequences upstream of DSL ligand genes in vivo in other systems.
Environmental regulation of Notch signaling
Examples of environmental regulation of canonical Notch signaling in response to DSL ligand activity are emerging. In C. elegans, LAG-2 is expressed in many different cell types and stages. Expression of lag-2 in IL2 neurons is dauer specific. This regulation depends on putative forkhead-binding sites 1.3 kb upstream of the start site (Ouellet et al., 2008). In the background of daf-7 mutants that form dauers constitutively but are still sensitive to dauer recovery cues, Ouellet et al. (2008) found that loss of lag-2 or glp-1, or ablation of IL2, interfered with dauer maintenance. In light of our results, it will be interesting to determine whether the sequences implicated in lag-2 regulation in the DTC also mediate DAF-3 repression of lag-2 expression in specific neurons that regulate dauer entry, maintenance and recovery. It is noteworthy that the Cons-binding site we define as crucial for modulation of lag-2 in the DTC was discovered for its role in the regulation of osm-9 in postdauer ADL neurons via DAF-3 binding (Sims et al., 2016).
The dauer decision also impacts lag-2 expression in the context of vulval precursor cell (VPC) fate specification. Here, the DAF-2 pathway, rather than the TGFβ pathway, transduces environmental signals via inhibition of DAF-16 that acts autonomously to prevent lag-2 expression in P6.p during dauer and thereby forestalls fate specification (Karp and Greenwald, 2013). Regulation of a 1 kb lag-2 reporter in VPCs (but not in the DTC or in neurons) is also sensitive to pha-4 RNAi (Chen and Riddle, 2008). Because PHA-4 FOXA acts in both transcriptional regulation and environmental signaling, this regulation may also contribute to the lag-2 response to environmental cues. It will be interesting to determine whether other (non-LAG-2) DSL ligands in C. elegans in other cellular contexts are regulated by the environment.
In Drosophila, regulation of Notch signaling by the environment or physiology has been documented in at least two contexts: the ovary and the adult brain. In the ovary, Notch activity is required to maintain somatic cap cells that, in turn, maintain germline stem cells (GSCs) (Song et al., 2007). Under poor nutrient conditions, low insulin signaling allows FOXO to promote expression of the glycosyltransferase Fringe, which, in turn, negatively regulates Notch receptor activity. Insulin signaling in this context also acts in a Notch-independent manner to promote adhesion between the cap cells and the GSCs (Hsu and Drummond-Barbosa, 2009, 2011; Yang et al., 2013). Delta-dependent modulation of Notch activity in the adult Drosophila brain occurs in response to several stimuli, including odorants (Lieber et al., 2011), ultimately contributing to changes in the volume of glomeruli and in activity of specific neurons (Kidd and Lieber, 2016; Kidd et al., 2015).
It remains to be determined whether environmental or physiological regulation of Notch signaling is widespread among animals, whether it occurs primarily via modulation of ligand expression and whether direct regulation of ligand expression by TGFβ is a common mechanism. Environmental regulation of Notch signaling could conceivably contribute to disease states such as developmental defects and cancer, in which aberrant Notch signaling has been implicated (Aster, 2014). Modulation of Notch signaling by the environment or animal physiology may have fewer consequences in relatively fast or highly robust Notch-mediated cell fate decisions. It may be more consequential in circumstances where Notch activity is required over time (e.g. maintenance of developmental states such as dauer, maintenance of stem cells or response to long-term olfactory cues) or in relatively plastic developmental or physiological processes that are important for survival or reproduction and could thereby confer evolutionary advantage.
MATERIALS AND METHODS
Strains and plasmids
Strains were derived from N2 wild type (Bristol) and handled using standard methods (Brenner, 1974). Synchronization was performed by 2 h hatch-off as described previously (Pepper et al., 2003). Unless otherwise indicated, worms were grown on OP50 at 20°C. Transgene alleles are noted in figure legends; corresponding plasmids are in Table S1, together with full genotypes of all strains. Plasmids generated for this study were: pGC457, pGC630, pGC642, pGC643, pGC644, pGC680, pGC681, pGC682, pGC683 and pGC684; see Table S1 for plasmid construction details and primer sequences. Transgenic strains were integrated by microparticle bombardment (Praitis et al., 2001) into DP38 unc-119(ed3), except for MosSCI-generated naSi8 (Frøkjær-Jensen et al., 2008). To compare transgene expression in different mutant backgrounds, each transgene insertion was generated in an otherwise wild-type (non-TGFβ-pathway mutant) background and subsequently crossed into the different TGFβ pathway gene mutants.
Microscopy and image analysis
For fluorescence images, live animals were immobilized with 0.2 mM levamisole in M9 on a 4% agarose pad and imaged at 63×/1.20 objective on Leica SP5 confocal microscope. For each experiment, control and experimental animals (e.g. wild type, daf-7 and daf-7;daf-3) were imaged in a single session. All microscope (e.g. laser power, gain, pin hole) and camera settings (e.g. exposure times) were established and held constant for imaging sessions for each transgene. Exposure times and gain were adjusted to insure sub-saturating levels. Approximately 25 images (z-stacks) were captured at intervals of 0.46 µm, and were analyzed using ImageJ (http://rsb.info.nih.gov/ij). Figures show z-stacks as maximum intensity projections.
Quantification of lag-2 reporter expression
After summing the intensity for the entire projection, bandpass filtering and thresholding were performed to help distinguish between DTC and gut granule signals. The ‘analyze particles’ function was applied and mean pixel intensity was measured in all particles that were manually designated as DTC (i.e. omitting gut granule signals that appear as large bright spots in some figure panels, depending on the orientation of the animal). At least 15 animals were analyzed per genotype.
Quantification of lag-2 in situ hybridization
The ImageJ ‘object counter 3D’ plug-in was used. The threshold was set such that pixel intensity was measured only from the in situ signal, and the pixel intensity from all the detected dots in all stacks in each image was summed. At least 15 animals were analyzed per genotype.
Germ nuclei counts
Whole-worm fixation, staining, microscopy and proliferative zone determination were performed according to Michaelson et al. (2010). Germ nuclei counts used a semi-automated ImageJ plug-in described previously (Korta et al. 2012).
In situ hybridization
Custom Stellaris fluorescent in situ hybridization probes were designed against the cDNA of lag-2 using the Stellaris FISH Probe Designer (https://www.biosearchtech.com/support/tools/design-software/stellaris-probe-designer) and 48 probes labeled with Quasar 670 were obtained from Biosearch Technologies. L4 animals were dissected and subjected to a procedure similar to Biosearch recommendations and to that described by Lee et al. (2016) with modifications as follows: after washing twice with M9 buffer, gonads were fixed (3.7% formaldehyde in 1×PBS RNase free) for 45 min at room temperature. Following two washes with 1×PBS, gonads were resuspended in 70% ethanol for 30 min. Following ethanol removal, samples were washed in wash buffer (2×SSC, 10% deionized formamide) for 5 min. After wash buffer removal, samples were placed in hybridization buffer (228 mM dextran sulfate, 2×SSC, 10% deionized formamide) mixed with lag-2 probe at 0.25 µm final concentration at 37°C overnight in the dark. After hybridization solution removal, samples were washed with wash buffer for 30 min at 37°C in the dark. Samples were then washed in 2×SSC. Following 2×SSC removal, samples were mounted using Antifade Prolong Gold mounting medium (Life Technologies). All solutions were made with nuclease-free water.
Bacterial reduction and dauer pheromone assays
Both pheromone and food (OP50) assays were performed as described by Dalfó et al. (2012). Synchronized early L3 worms were washed and distributed on plates at 25°C and analyzed in the L4 for lag-2 DTC expression and as early adults for number of nuclei in the proliferative zone. Pairs of conditions (high/low food and with/without pheromone) were carried out in parallel in at least three independent experiments and, in each case, L4 and adult animals were taken from the same plates to ensure they experienced the same conditions. Bacterial concentrations were established by serial dilutions as described previously (Korta et al., 2012). Dauer pheromone preparation was carried out as described previously (Zhang et al., 2013). In this case, 66 μl of crude pheromone was added to 1 ml of NGM agar, the amount that caused 100% of animals to enter dauer in a wild-type population at 25°C.
Chromatin immunoprecipitation
DAF-3 chromatin immunoprecipitation (ChIP) was performed as described previously (Sims et al., 2016) using Novus Biologicals NB100-1924 Lot A1, and packed worm pellets (∼500 µl) of N2 L4 larvae. The negative control, which is particularly important for this potentially cross-reactive antibody, was a packed pellet (∼500 µl) of mixed population GR1311 daf-3(mgDf90).
Real-time PCR
Quantitative real-time PCR was carried out using 1 µl of the DAF-3 ChIP using the iTaq Universal SYBR Green Supermix (BioRad). Primers used were: MO2389 and MO2390 for the region in the lag-2 promoter homologous to an osm-9 promoter region with a potential DAF-3 binding site (DBS); MO2391 and MO2388 for the conserved PD motif in the lag-2 promoter; MO2339 and MO2340 for the DBS in the osm-9 promoter; MO2392 and MO2393 for the conserved sequence in the PD motif in the osm-9 promoter; MO2262 and MO2263 for detection of the C sub-element in the myo-2 promoter; MO2398 and MO2399 for a region in exon 1 of lag-2; and MO2402 and MO2403 for a region in exon 3 of lag-2 (see Table S1). Regions in the promoters of daf-7 and daf-8 were used as positive controls and daf-14 as a negative control (Park et al., 2010). Ct values were normalized using act-2 (Park et al., 2010).
Bacterial one-hybrid assay
Mutant and wild-type versions the lag-2 upstream sequence (600 bp) were cloned into MRB1H-reporter vector (also known as ‘GHUC’, see Table S1) (Oakes et al., 2016) between NotI and EcoRI upstream of the HIS3-GFP cassette, generating GHUC-1, GHUC-2, GHUC-3, GHUC-4, GHUC-5, GHUC-6 and GHUC-7. DNA encoding amino acids 1-250 of DAF-3 (DAF-3-N’) was cloned into the pB1Hw2-omega vector (Noyes et al., 2008) between KpnI and XbaI, creating an omega-DAF-3-N’ fusion in pB1Hw2-Daf3250 (see Table S1). Combinations of the DAF-3-N’ pB1Hw2-omega vector or pB1Hw2-Daf3250 (bait) and lag-2 promoter MRB1H-reporter vectors or GHUC-1 to GHUC-7 (prey) were transformed into USO Δomega cells (Noyes et al., 2008) and selected on Kan/Amp to recover cells with both plasmids. Three replicate colonies from each bait-prey combination were inoculated into 5 ml rich media with Kan/Amp, and incubated at 37°C for ∼8 h (OD600=0.5). From each culture, 2.5 µl was used to inoculate 5 ml supplemented minimal NM media containing histidine, uracil, IPTG, Kan/Amp (Noyes et al., 2008) and grown at 37°C overnight (OD600∼1.0-2.0). On ice, 1 ml from each sample was pelleted, washed once with PBS and re-pelleted. PBS was removed and the pellet resuspended in 1 ml PBS+1% FBS. Resuspended cells (100 µl) were added to 1.5 ml PBS+1% FBS in a FACS tube. The mean GFP fluorescence of lag-2 mutant samples and controls and lag-2 scrambled samples and controls were measured with a Sony SH800 cell sorter and a BD LSRII HTS sorter, respectively. Mean fluorescence values were determined from at least 10,000 cells. Sequences for scrambled lag-2 DBS and Cons sequences were: 5′-ctatcact-3′ and 5′-ctgcgggcaggccccactggggcct-3′, respectively.
Acknowledgements
We thank Yossi Capua and Jesus Martinez-Gomez for technical assistance, Chris Rushlow for advice, and members of the Nance and Hubbard labs for advice, discussions and comments on the manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We also thank WormBase and the NYU School of Medicine's Microscopy Core, which is partially supported by a Cancer Center Support Grant (P30CA016087), for the use of confocal microscope, which is partially supported by NCRR S10 RR024708.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: O.P., M.B.N., S.E.H., E.J.A.H.; Methodology: O.P., M.B.N., S.E.H., E.J.A.H.; Validation: O.P., M.C.O., M.B.N., S.E.H., E.J.A.H.; Formal analysis: O.P., M.C.O., M.B.N., S.E.H., E.J.A.H.; Investigation: O.P., M.C.O., K.Y.H.; Resources: O.P., M.C.O., M.B.N., S.E.H., E.J.A.H.; Data curation: O.P., M.B.N., S.E.H., E.J.A.H.; Writing - original draft: O.P., M.B.N., S.E.H., E.J.A.H.; Writing - review & editing: O.P., M.C.O., M.B.N., S.E.H., E.J.A.H.; Visualization: O.P., M.C.O., S.E.H.; Supervision: M.B.N., S.E.H., E.J.A.H.; Project administration: M.B.N., S.E.H., E.J.A.H.; Funding acquisition: E.J.A.H., S.E.H.
Funding
This work was supported by the National Institutes of Health (R01GM102254 and R01GM061706 to E.J.A.H.; R15GM111094 to S.E.H.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.147660.supplemental
References
- Aster J. C. (2014). In brief: Notch signalling in health and disease. J. Pathol. 232, 1-3. 10.1002/path.4291 [DOI] [PubMed] [Google Scholar]
- Austin J. and Kimble J. (1987). glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51, 589-599. 10.1016/0092-8674(87)90128-0 [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. and Greenwald I. (2004). The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell 6, 183-192. 10.1016/S1534-5807(04)00021-8 [DOI] [PubMed] [Google Scholar]
- Chen D. and Riddle D. L. (2008). Function of the PHA-4/FOXA transcription factor during C. elegans post-embryonic development. BMC Dev. Biol. 8, 26 10.1186/1471-213X-8-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesney M. A., Lam N., Morgan D. E., Phillips B. T. and Kimble J. (2009). C. elegans HLH-2/E/Daughterless controls key regulatory cells during gonadogenesis. Dev. Biol. 331, 14-25. 10.1016/j.ydbio.2009.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalfó D., Michaelson D. and Hubbard E. J. A. (2012). Sensory regulation of the C. elegans germline through TGF-β-dependent signaling in the niche. Curr. Biol. 22, 712-719. 10.1016/j.cub.2012.02.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B., Mukhopadhyay A., Ao W., Elewa A. M., Grove C. A., Martinez N. J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J. S., Hope I. A. et al. (2006). A gene-centered C. elegans protein-DNA interaction network. Cell 125, 1193-1205. 10.1016/j.cell.2006.04.038 [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa D. (2008). Stem cells, their niches and the systemic environment: an aging network. Genetics 180, 1787-1797. 10.1534/genetics.108.098244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Wayne Davis M., Hopkins C. E., Newman B. J., Thummel J. M., Olesen S.-P., Grunnet M. and Jorgensen E. M. (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375-1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I. S., Sternberg P. W. and Horvitz H. R. (1983). The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34, 435-444. 10.1016/0092-8674(83)90377-X [DOI] [PubMed] [Google Scholar]
- Gumienny T. L. (2013). TGF-β signaling in C. elegans. In WormBook (ed. The C. elegans Research Community). WormBook, 10.1895/wormbook.1.145.1http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. T., Gao D., Lambie E. J. and Kimble J. (1994). lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120, 2913-2924. [DOI] [PubMed] [Google Scholar]
- Hsu H.-J. and Drummond-Barbosa D. (2009). Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc. Natl Acad. Sci. USA 106, 1117-1121. 10.1073/pnas.0809144106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.-J. and Drummond-Barbosa D. (2011). Insulin signals control the competence of the Drosophila female germline stem cell niche to respond to Notch ligands. Dev. Biol. 350, 290-300. 10.1016/j.ydbio.2010.11.032 [DOI] [PubMed] [Google Scholar]
- Hubbard E. J. A., Korta D. Z. and Dalfó D. (2012). Physiological control of germline development. In Germ Cell Development in C. elegans, pp. 101-131. New York, NY: Springer. [Google Scholar]
- Hudson C. and Yasuo H. (2005). Patterning across the ascidian neural plate by lateral Nodal signalling sources. Development 132, 1199-1210. 10.1242/dev.01688 [DOI] [PubMed] [Google Scholar]
- Hudson C. and Yasuo H. (2006). A signalling relay involving Nodal and Delta ligands acts during secondary notochord induction in Ciona embryos. Development 133, 2855-2864. 10.1242/dev.02466 [DOI] [PubMed] [Google Scholar]
- Karp X. (2003). Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. 17, 3100-3111. 10.1101/gad.1160803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X. and Greenwald I. (2004). Multiple roles for the E/Daughterless ortholog HLH-2 during C. elegans gonadogenesis. Dev. Biol. 272, 460-469. 10.1016/j.ydbio.2004.05.015 [DOI] [PubMed] [Google Scholar]
- Karp X. and Greenwald I. (2013). Control of cell-fate plasticity and maintenance of multipotency by DAF-16/FoxO in quiescent Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 110, 2181-2186. 10.1073/pnas.1222377110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S. and Lieber T. (2016). Mechanism of Notch pathway activation and its role in the regulation of olfactory plasticity in Drosophila melanogaster. PLoS ONE 11, e0151279 10.1371/journal.pone.0151279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S., Struhl G. and Lieber T. (2015). Notch is required in adult Drosophila sensory neurons for morphological and functional plasticity of the olfactory circuit. PLoS Genet. 11, e1005244 10.1371/journal.pgen.1005244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J. E. and White J. G. (1981). On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81, 208-219. 10.1016/0012-1606(81)90284-0 [DOI] [PubMed] [Google Scholar]
- Korta D. Z., Tuck S. and Hubbard E. J. A. (2012). S6K links cell fate, cell cycle and nutrient response in C. elegans germline stem/progenitor cells. Development 139, 859-870. 10.1242/dev.074047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Park M., Zhang J. M., Yuan J., Harfe B., Xu S. Q., Greenwald I., Cole M., Paterson B. and Fire A. (1997). A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development 124, 2179-2189. [DOI] [PubMed] [Google Scholar]
- Kurpinski K., Lam H., Chu J., Wang A., Kim A., Tsay E., Agrawal S., Schaffer D. V. and Li S. (2010). Transforming growth factor-β and notch signaling mediate stem cell differentiation into smooth muscle cells. Stem Cells 28, 734-742. 10.1002/stem.319 [DOI] [PubMed] [Google Scholar]
- Larsen P. L., Albert P. S. and Riddle D. L. (1995). Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139, 1567-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws K. M. and Drummond-Barbosa D. (2017). Control of germline stem cell lineages by diet and physiology. Results Probl. Cell Differ. 59, 67-99. 10.1007/978-3-319-44820-6_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Sorensen E. B., Lynch T. R. and Kimble J. (2016). C. elegans GLP-1/Notch activates transcription in a probability gradient across the germline stem cell pool. Elife 5, e18370 10.7554/eLife.18370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber T., Kidd S. and Struhl G. (2011). DSL-Notch signaling in the Drosophila brain in response to olfactory stimulation. Neuron 69, 468-481. 10.1016/j.neuron.2010.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D., Korta D. Z., Capua Y. and Hubbard E. J. A. (2010). Insulin signaling promotes germline proliferation in C. elegans. Development 137, 671-680. 10.1242/dev.042523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro R., Pinheiro P., Joseph N., Peterkin T., Koth J., Repapi E., Bonkhofer F., Kirmizitas A. and Patient R. (2016). Transforming growth factor beta drives hemogenic endothelium programming and the transition to hematopoietic stem cells. Dev. Cell 38, 358-370. 10.1016/j.devcel.2016.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M., Koinuma D., Tsutsumi S., Vasilaki E., Kanki Y., Heldin C.-H., Aburatani H. and Miyazono K. (2011). ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res. 39, 8712-8727. 10.1093/nar/gkr572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S., Govindan J. A., McGovern M., Hubbard E. J. A. and Greenstein D. (2009). MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development 136, 2223-2234. 10.1242/dev.034603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes M. B. (2012). Analysis of specific protein-DNA interactions by bacterial one-hybrid assay. Methods Mol. Biol. 786, 79-95. 10.1007/978-1-61779-292-2_5 [DOI] [PubMed] [Google Scholar]
- Noyes M. B., Meng X., Wakabayashi A., Sinha S., Brodsky M. H. and Wolfe S. A. (2008). A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 36, 2547-2560. 10.1093/nar/gkn048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes B. L., Xia D. F., Rowland E. F., Xu D. J., Ankoudinova I., Borchardt J. S., Zhang L., Li P., Miller J. C., Rebar E. J. et al. (2016). Multi-reporter selection for the design of active and more specific zinc-finger nucleases for genome editing. Nat. Commun. 7, 10194 10.1038/ncomms10194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet J., Li S. and Roy R. (2008). Notch signalling is required for both dauer maintenance and recovery in C. elegans. Development 135, 2583-2592. 10.1242/dev.012435 [DOI] [PubMed] [Google Scholar]
- Park D., Estevez A. and Riddle D. L. (2010). Antagonistic Smad transcription factors control the dauer/non-dauer switch in C. elegans. Development 137, 477-485. 10.1242/dev.043752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G. I., Koweek A., Wong A., Liu Y. and Ruvkun G. (1997). The DAF-3 Smad protein antagonizes TGF-beta-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes Dev. 11, 2679-2690. 10.1101/gad.11.20.2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A. S.-R., Killian D. J. and Hubbard E. J. A. (2003). Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163, 115-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D. and Austin J. (2001). Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P., Lim C. S., Johnsen R., Albert P. S., Pilgrim D. and Riddle D. L. (1996). Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274, 1389-1391. 10.1126/science.274.5291.1389 [DOI] [PubMed] [Google Scholar]
- Richards G. S. and Degnan B. M. (2010). The dawn of developmental signaling in the Metazoa. Cold Spring Harb. Symp. Quant. Biol. 74, 81-90. 10.1101/sqb.2009.74.028 [DOI] [PubMed] [Google Scholar]
- Schackwitz W. S., Inoue T. and Thomas J. H. (1996). Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17, 719-728. 10.1016/S0896-6273(00)80203-2 [DOI] [PubMed] [Google Scholar]
- Seydoux G., Schedl T. and Greenwald I. (1990). Cell-cell interactions prevent a potential inductive interaction between soma and germline in C. elegans. Cell 61, 939-951. 10.1016/0092-8674(90)90060-R [DOI] [PubMed] [Google Scholar]
- Shin-i T. and Kohara Y. (1999). NEXTDB: the expression pattern map database for C. elegans. Genome Inform. 10, 213-214. [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K. L., Parrish S., Timmons L., Plasterk R. H. and Fire A. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465-476. 10.1016/S0092-8674(01)00576-1 [DOI] [PubMed] [Google Scholar]
- Sims J. R., Ow M. C., Nishiguchi M. A., Kim K., Sengupta P. and Hall S. E. (2016). Developmental programming modulates olfactory behavior in C. elegans via endogenous RNAi pathways. Elife 5, e11642 10.7554/eLife.11642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Chao M. Y., Somers G. A., Komatsu H., Corkins M. E., Larkins-Ford J., Tucey T., Dionne H. M., Walsh M. B., Beaumont E. K. et al. (2011). C. elegans notch signaling regulates adult chemosensory response and larval molting quiescence. Curr. Biol. 21, 825-834. 10.1016/j.cub.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Call G. B., Kirilly D. and Xie T. (2007). Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development 134, 1071-1080. 10.1242/dev.003392 [DOI] [PubMed] [Google Scholar]
- Starich T. A., Hall D. H. and Greenstein D. (2014). Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in Caenorhabditis elegans. Genetics 198, 1127-1153. 10.1534/genetics.114.168815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher J. D., Haun C. and Okkema P. G. (1999). The DAF-3 Smad binds DNA and represses gene expression in the Caenorhabditis elegans pharynx. Development 126, 97-107. [DOI] [PubMed] [Google Scholar]
- Yang S.-A., Wang W.-D., Chen C.-T., Tseng C.-Y., Chen Y.-N. and Hsu H.-J. (2013). FOXO/Fringe is necessary for maintenance of the germline stem cell niche in response to insulin insufficiency. Dev. Biol. 382, 124-135. 10.1016/j.ydbio.2013.07.018 [DOI] [PubMed] [Google Scholar]
- Zavadil J., Cermak L., Soto-Nieves N. and Böttinger E. P. (2004). Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 23, 1155-1165. 10.1038/sj.emboj.7600069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Noguez J. H., Zhou Y. and Butcher R. A. (2013). Analysis of ascarosides from Caenorhabditis elegans using mass spectrometry and NMR spectroscopy. Methods Mol. Biol. 1068, 71-92. 10.1007/978-1-62703-619-1_6 [DOI] [PMC free article] [PubMed] [Google Scholar]