ABSTRACT
Patterning of the facial skeleton involves the precise deployment of thousands of genes in distinct regions of the pharyngeal arches. Despite the significance for craniofacial development, how genetic programs drive this regionalization remains incompletely understood. Here we use combinatorial labeling of zebrafish cranial neural crest-derived cells (CNCCs) to define global gene expression along the dorsoventral axis of the developing arches. Intersection of region-specific transcriptomes with expression changes in response to signaling perturbations demonstrates complex roles for Endothelin 1 (Edn1) signaling in the intermediate joint-forming region, yet a surprisingly minor role in ventralmost regions. Analysis of co-variance across multiple sequencing experiments further reveals clusters of co-regulated genes, with in situ hybridization confirming the domain-specific expression of novel genes. We then created loss-of-function alleles for 12 genes and uncovered antagonistic functions of two new Edn1 targets, follistatin a (fsta) and emx2, in regulating cartilaginous joints in the hyoid arch. Our unbiased discovery and functional analysis of genes with regional expression in zebrafish arch CNCCs reveals complex regulation by Edn1 and points to novel candidates for craniofacial disorders.
KEY WORDS: Craniofacial, Zebrafish, Jaw, Skeleton, Cartilage, Bone, Cranial neural crest, Dorsoventral patterning, Endothelin, Edn1, Jagged-Notch, Dlx5a, Hand2, Emx2, Fsta, Mrrf
Summary: Dissecting the genetic programs in distinct regions of the pharyngeal arches illuminates the roles of Edn1 and Jagged-Notch signaling and identifies novel domain-specific genes in craniofacial patterning.
INTRODUCTION
The vertebrate facial skeleton is generated from cranial neural crest-derived cells (CNCCs) that populate a series of pharyngeal arches (Platt, 1893; Schilling and Kimmel, 1994). Signaling from endodermal and ectodermal epithelia, as well as from CNCCs themselves, establishes nested patterns of gene expression in arch CNCCs, in particular along the dorsoventral axis (Medeiros and Crump, 2012; Mork and Crump, 2015). CNCCs then progressively adopt a number of fates, including cartilage, bone, and ligament (Bronner and LeDouarin, 2012), with a subset of cells remaining as progenitors for later differentiation and possibly adult repair (Paul et al., 2016). The shapes and functions of distinct facial regions are inextricably tied to the selection of these cell fates and the subsequent growth and rearrangements of skeletal cells (Kimmel et al., 1998). The earliest fate adopted by arch CNCCs is cartilage, which occurs first in ventral-intermediate arch regions and then spreads to ventral and dorsal poles (Barske et al., 2016). Domain-specific differences in cartilage versus bone fates are likely to contribute to region-specific skeletal morphologies. In dorsal and intermediate domains, early cartilage differentiation must be actively suppressed to ensure proper formation of joints and later-forming intramembranous bones (Askary et al., 2015; Nichols et al., 2016). Identifying the molecular differences that prefigure regional cell fate choices and behaviors is therefore key to unraveling how the facial skeleton is assembled.
Candidate-based approaches, as well as forward genetic screens in zebrafish (Piotrowski et al., 1996; Schilling et al., 1996), have identified key members of craniofacial signaling pathways and their downstream targets (Minoux and Rijli, 2010). Edn1 signaling is required for gene expression and subsequent skeletal patterning in the intermediate and ventral-intermediate regions of the arches, including the joint-forming domain (Kurihara et al., 1994; Clouthier et al., 1998; Miller et al., 2000; Ozeki et al., 2004; Sato et al., 2008; Gordon et al., 2013). The Bmp pathway has an overlapping function in patterning the lower face (Tucker et al., 1998; Bonilla-Claudio et al., 2012), although it appears to be preferentially required for gene expression in ventralmost arch regions (Alexander et al., 2011; Zuniga et al., 2011). By contrast, Jagged-Notch signaling is required to pattern dorsal arch CNCCs, at least in the hyoid and posterior mandibular arches of zebrafish (Zuniga et al., 2010; Barske et al., 2016). Downstream targets have also been identified, including Edn1 activation and Jagged-Notch inhibition of the Dlx3/4/5/6 family in ventral-intermediate CNCCs (Beverdam et al., 2002; Depew et al., 2002; Talbot et al., 2010; Zuniga et al., 2010), and Edn1 and Bmp regulation of Hand2 in ventral CNCCs (Thomas et al., 1998; Miller et al., 2003; Yanagisawa et al., 2003; Zuniga et al., 2011; Bonilla-Claudio et al., 2012). However, the extent to which Edn1 and Notch globally regulate dorsoventral gene expression remains incompletely understood.
Recently, genome-wide expression profiling in mice has identified stage-specific expression signatures of craniofacial compartments, such as the mandibular, maxillary and frontonasal prominences (Feng et al., 2009; Fujita et al., 2013; Brunskill et al., 2014; Hooper et al., 2017). Similar studies have revealed genes regulated by Bmp4 (Bonilla-Claudio et al., 2012) and Dlx5/6 (Jeong et al., 2008). These studies largely relied on dissection of facial prominences rather than purification of the arch CNCCs that generate the facial skeleton. As the arches consist of not only CNCCs, but also endodermal and ectodermal epithelia and mesodermal cores, whether the identified genes were expressed in CNCCs was not always clear.
In the current study, we use the nested expression of hand2:GFP and dlx5a:GFP transgenes along the dorsoventral axis to identify genes with domain-specific expression in CNCCs of the zebrafish mandibular and hyoid arches. By combining this domain-specific profiling with effects of altered signaling on arch CNCCs (Barske et al., 2016), we demonstrate global roles for Edn1 and Jagged-Notch signaling in establishing intermediate/ventral-intermediate and dorsal arch gene expression, respectively, yet only a minor role for Edn1 in the ventralmost arches. We then used gene editing to test the requirements for 12 previously uncharacterized domain-specific genes and found opposing requirements for two new Edn1 targets, fsta and emx2, in coordinating skeletal development in the intermediate hyoid arch. Whereas fsta inhibits cartilage differentiation in the developing hyoid joint, emx2 promotes cartilage differentiation at the connection points between individual hyoid cartilages. Thus, in addition to providing a global description of dorsoventral gene expression in arch CNCCs, these findings uncover a complex role for Edn1 in balancing skeletal differentiation in the intermediate arches.
RESULTS
Generation of domain-specific arch transcriptomes by combinatorial transgene labeling
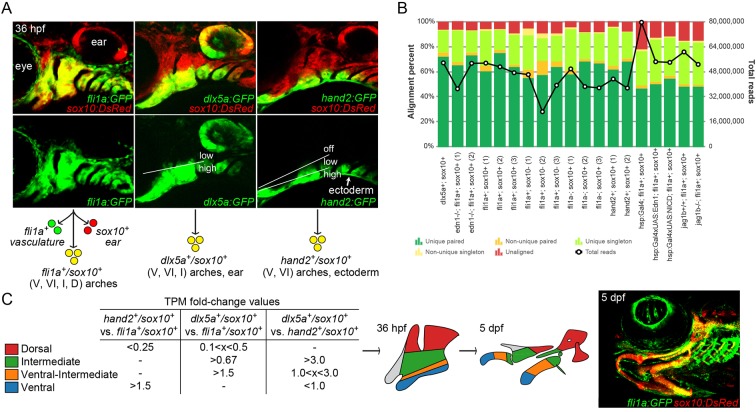
We have previously reported using dual labeling by sox10:DsRed and fli1a:GFP transgenes to purify all arch CNCCs from embryos at 20, 28, and 36 h post-fertilization (hpf), followed by mRNA isolation, cDNA library construction and deep sequencing (Barske et al., 2016). Here, we performed two additional replicates at 36 hpf to better define a minimum set of 472 arch CNCC-enriched genes. Next, we took advantage of the nested patterns of hand2:GFP and dlx5a:GFP transgenes to isolate distinct subsets of arch CNCCs along the dorsoventral axis at 36 hpf, followed by RNA sequencing (RNAseq) (Fig. 1A). Whereas fluorescence activated cell sorting (FACS) of hand2:GFP+; sox10:DsRed+ cells enriches for the ventralmost CNCCs of the arches, FACS of dlx5a:GFP+; sox10:DsRed+ cells enriches for a broader domain of ventral to intermediate arch CNCCs (and the otic vesicle).
Fig. 1.
Isolation of arch CNCCs for RNAseq. (A) Cells were sorted using transgenic lines that label different populations of arch CNCCs. fli1a:GFP and sox10:DsRed overlap throughout arch CNCCs (yellow); dlx5a:GFP and sox10:DsRed in CNCCs of ventral (V) to intermediate (I) domains, as well as some dorsal (D) domain CNCCs and the otic vesicle; and hand2:GFP and sox10:DsRed in ventral and more weakly in ventral-intermediate (VI) CNCCs [modified from Barske et al. (2016) and Medeiros and Crump (2012)]. (B) Breakdown of alignment results for each RNAseq experiment, showing the percentage of reads aligned to a unique site in the genome and whether both paired-end reads were aligned. The line graph shows the total number of reads acquired for each sample. (C) Dorsal, intermediate, ventral-intermediate, and ventral domains were defined by their relative enrichment levels (i.e. fold-change of TPM values) among hand2:GFP; sox10:DsRed, dlx5a:GFP; sox10:DsRed, and fli1a:GFP; sox10:DsRed cells. These domains correspond to specific parts of the arches at 36 hpf that give rise to the larval craniofacial skeleton at 5 dpf (shown for context in a fli1a:GFP; sox10:DsRed fish).
We then compared how RNAseq data from different experiments align to the zebrafish genome. In our analysis, we included new hand2:GFP and dlx5a:GFP samples, as well as the previously described sox10:DsRed+; fli1a:GFP+ cells from wild types, edn1 mutants, jag1b mutants, and embryos with elevated Edn1 (hsp70l:Gal4; UAS:Edn1) and Notch (hsp70l:Gal4; UAS:Notch1a-ICD) signaling. For each experiment, the majority of reads (∼80-90%) could be aligned to a unique position in the zebrafish genome, and the percentage of uniquely aligned reads was independent of the total number of reads for each particular sample (Fig. 1B). The uniformly high percentage of uniquely aligned reads is a positive indication of the quality of the RNAseq data.
Transcriptomic analysis reveals genes differentially enriched in hand2:GFP and dlx5a:GFP domains
In order to compare gene expression levels between samples, read counts were normalized to yield transcript per million values (TPMs). We identified the set of 472 genes enriched in arch CNCCs by filtering for genes with average TPMs greater than 2 across the three wild-type fli1a:GFP+; sox10:DsRed+ samples at 36 hpf, as well as for those enriched 1.5-fold or higher in double-positive versus single-positive cells (i.e. non-CNCC) (Tables S4 and S5). We then took advantage of the relative levels of hand2:GFP and dlx5a:GFP to subdivide arch CNCCs into four dorsoventral domains (Fig. 1C). As previously described, hand2:GFP displays graded expression from strong in ventralmost domains to weaker in more ventral-intermediate regions, and dlx5a:GFP transitions from strong in ventral and intermediate regions to weaker in dorsal domains (Medeiros and Crump, 2012). We therefore binned genes into clusters by comparing their relative expression in cells sorted with different transgenes: hand2:GFPhigh; dlx5a:GFPhigh (ʻventral', 15 genes), hand2:GFPlow; dlx5a:GFPhigh (ʻventral-intermediate', 22 genes), hand2:GFP−; dlx5a:GFPhigh (ʻintermediate', 16 genes), and hand2:GFP−; dlx5a:GFPlow (ʻdorsal', 30 genes) (see Fig. 1C, Table 1 for cut-off values). Confirming the validity of this filtering strategy, each dorsoventral cluster includes several genes with known expression in that domain (Table S1). For example, the ventral cluster includes endogenous hand2 (Miller et al., 2003) and homologs of genes known to be expressed in the ventral/distal domains of the murine arches (foxf1 and foxf2a) (Jeong et al., 2004); the ventral-intermediate cluster includes endogenous dlx5a, as well as dlx3b, dlx4b and msxe (msx1a) (Miller et al., 2000); the intermediate cluster includes grem2b (Zuniga et al., 2011) and genes required for joints such as irx7 (Askary et al., 2015) and nkx3.2 (Miller et al., 2003); and the dorsal cluster includes jag1b (Zuniga et al., 2010) and homologs of murine genes with dorsal arch expression (pou3f3a and pou3f3b) (Jeong et al., 2008).
Table 1.
Predicted genes with dorsoventral-restricted arch expression

In situ validation of novel domain-specific gene expression in the arches
Given the inclusion of known genes with correctly predicted dorsoventral expression, we sought to validate the expression of uncharacterized genes in each domain-specific list. To do so, we conducted fluorescent in situ hybridization in 36 hpf embryos, along with sox10:GFPCAAX (membrane GFP) in a second color to highlight all arch CNCCs.
Ventral genes
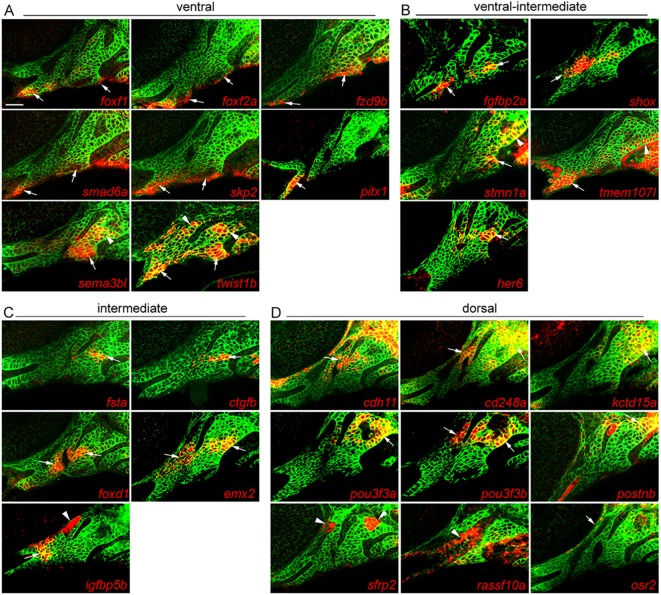
Of the eight predicted ventral genes tested, all showed some expression in the ventral mandibular or hyoid arches, yet their expression patterns were distinct (Fig. 2A). Similar to hand2 (Miller et al., 2000), we observed expression of foxf1, foxf2a, fzd9b, smad6a and skp2 in the ventralmost CNCCs of both arches, with pitx1 showing more limited ventral expression in only the mandibular arch. By contrast, sema3bl and twist1b were expressed in the ventral domain and also, to a lesser degree, in subsets of dorsal arch CNCCs, reminiscent of the published expression pattern of barx1 (Nichols et al., 2013; Barske et al., 2016), another gene on our ventral list. The weaker expression of these genes in dorsal relative to ventral arch CNCCs, together with their lower levels in the intermediate domain, is likely to explain why they were enriched in the hand2:GFP dataset and classified as ventral genes by our filtering scheme.
Fig. 2.
Arch expression of predicted domain-specific genes. Whole-mount fluorescent in situ hybridizations for select genes were performed in sox10:GFPCAAX embryos at 36 hpf, with anti-GFP staining (green) showing CNCCs of the mandibular and hyoid arches. (A) foxf1, foxf2a, fzd9b, smad6a and skp2 are expressed in ventral domains of both arches, pitx1 only in the ventral mandibular arch, and sema3bl and twist1b in ventral and dorsal CNCCs. (B) fgfbp2a and shox are expressed in ventral-intermediate CNCCs, stmn1a in ventral-intermediate and dorsal CNCCs, tmem107I in ventral mandibular and posterior dorsal hyoid arches, and her6 in a more dorsal domain. (C) fsta, ctgfb, foxd1, emx2 and igfbp5b show specific intermediate domain expression; igfbp5b is also expressed in arch mesoderm. (D) cdh11, cd248a, kctd15a, pou3f3a, pou3f3b and postnb are expressed in dorsal CNCCs, sfrp2 in dorsal arch mesoderm, rassf10a in epithelia, and osr2 between the dorsal first arch and eye. Arrows point to expression in predicted domains, and arrowheads to other arch domains. Single optical sections are shown. Scale bar: 20 μm.
Ventral-intermediate genes
Among the predicted ventral-intermediate genes, two out of five tested showed expression within the ventral-intermediate domain (fgfbp2a and shox), one displayed both ventral-intermediate and some dorsal expression (stmn1a), one was expressed in the ventral mandibular arch and dorsal posterior hyoid arch cells (tmem107l), and one in intermediate regions (her6) (Fig. 2B). The ventral-intermediate list also contains several known genes with apparent arch-wide expression [hoxa2b and hoxb2a (Hunter and Prince, 2002) and dlx1a (Sperber et al., 2008)] or ventral and dorsal expression domains (gsc) (Miller et al., 2000), although a closer examination of these previous reports suggests higher ventral-intermediate expression for these genes at 36 hpf.
Intermediate genes
Of the four previously characterized genes on this list, three are expressed in the intermediate joint-forming region and required for joints (irx7, grem2b and nkx3.2) (Miller et al., 2003; Zuniga et al., 2011; Askary et al., 2015). The inclusion of dlx4a might reflect the broader expression of Dlx3-6 genes in both ventral-intermediate and intermediate domains (Talbot et al., 2010), although other members of this family were filtered into the ventral-intermediate category. All five newly tested genes showed highly specific expression in the intermediate domain, including fsta, igfbp5b, ctgfb and homologs of mouse genes expressed in intermediate arch regions – emx2 (Compagnucci et al., 2013) and foxd1 (Jeong et al., 2004) (Fig. 2C). Interestingly, fsta and ctgfb were largely restricted to the hyoid arch and igfbp5b to the mandibular arch.
Dorsal genes
We found six of nine predicted dorsal genes to be enriched in the dorsal mandibular and hyoid arches: cadherin 11 (cdh11), pou3f3a and pou3f3b [homologs of mouse Pou3f3 with dorsal expression (Jeong et al., 2008)], cd248a, kctd15a (see also Gharbi et al., 2012) and postnb (Fig. 2D). The other three genes were excluded from the dlx5a:GFP and hand2:GFP expression domains, as predicted, but they did not present typical ʻdorsal' expression patterns: sfrp2 showed dorsal-specific expression but in mesoderm, rassf10a was largely confined to the surface ectoderm rather than arch CNCCs, and osr2 was expressed in a few cells between the mandibular arch and the eye (Swartz et al., 2011). Although not tested here, previous reports also suggest dorsal-enriched expression of ednraa from 28-36 hpf (Nair et al., 2007; Zuniga et al., 2010) and expression of gata3 in the more anterior maxillary prominence (Sheehan-Rooney et al., 2013b).
Distinct regulation of domain-specific genes by Edn1 and Jagged-Notch signaling
Given their major roles in dorsoventral arch patterning, we next tested how Edn1 and Jagged-Notch signaling regulate bulk expression of genes in the dorsoventral domains defined by our RNAseq analysis. We intersected our previously published RNAseq data of gain or loss of Edn1 or Jagged-Notch signaling in arch CNCCs (Barske et al., 2016) with domain-specific genes identified based on enrichment in sorted hand2:GFP and dlx5a:GFP cells. We found that the expression of intermediate and ventral-intermediate genes, but not ventral and dorsal genes, was significantly reduced in edn1−/− embryos (Fig. 3A) and increased upon Edn1 misexpression (Fig. 3B). In particular, intermediate genes were most affected by perturbation of Edn1 signaling, consistent with the sensitivity of intermediate skeletal elements (joints, symplectic, palatoquadrate) to partial reduction of Edn1 signaling in zebrafish (Miller and Kimmel, 2001). By contrast, loss of Jagged-Notch signaling in jag1b−/− embryos resulted in a downregulation of only dorsal genes (Fig. 3C), and gain of Notch signaling resulted in a downregulation of ventral, ventral-intermediate and intermediate genes (Fig. 3D). Gain of Notch signaling also showed a trend towards increasing the expression of dorsal genes (Bonferroni corrected P=0.18).
Fig. 3.
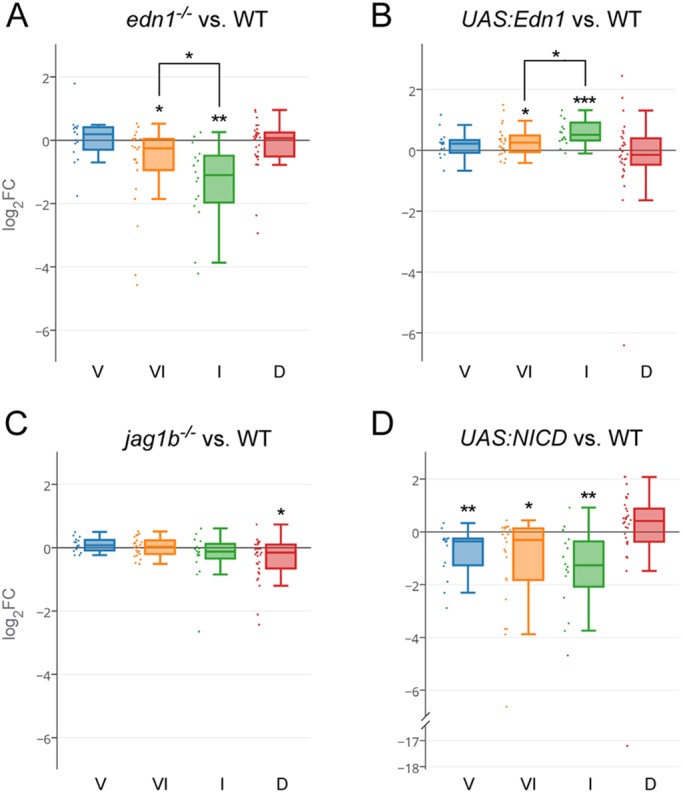
Domain regulation by Edn1 and Jagged-Notch signaling. (A) In edn1−/− mutants, intermediate (I) domain genes are those most strongly downregulated, followed by ventral-intermediate (VI) genes. Ventral (V) and dorsal (D) genes are, on average, unaffected. (B) Edn1 overexpression results in greater upregulation of intermediate than ventral genes. (C) Dorsal genes are downregulated in jag1b−/− mutants. (D) Overexpression of the Notch intracellular domain (NICD) downregulates ventral, ventral-intermediate, and intermediate genes. See the Materials and Methods for details of statistical analysis. *P<0.05, **P<0.01, ***P<0.001.
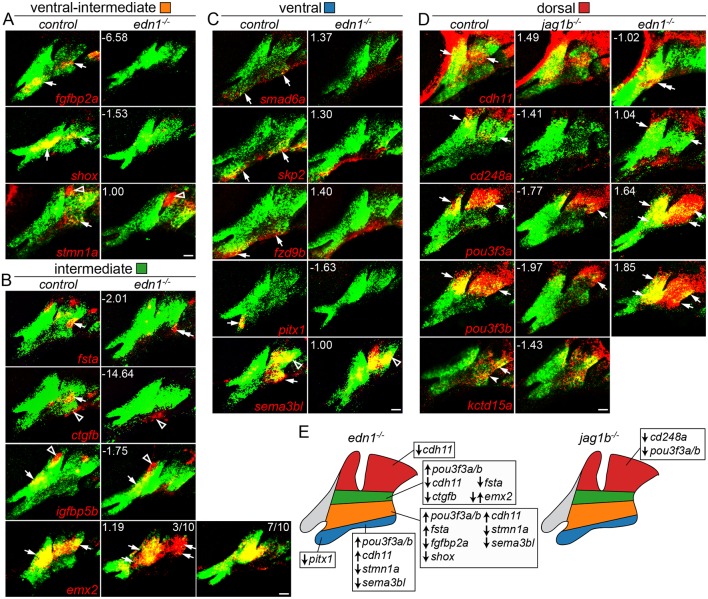
We next used in situ hybridization to confirm the predicted regulation of a subset of genes by Edn1 and Jagged-Notch signaling (Fig. 4). Ventral-intermediate genes fgfbp2a, shox and stmn1a, and intermediate genes fsta and ctgfb, were reduced in edn1 mutants. However, the intermediate gene igfbp5b was unaffected and the intermediate gene emx2 was variably upregulated or downregulated in edn1 mutants. Consistent with RNAseq data, ventral genes smad6a, skp2 and fzd9b were unaffected, and the ventral but not dorsal expression of sema3b1 and the mandibular-specific ventral expression of pitx1 were lost in edn1 mutants. Of the dorsal genes examined, pou3f3a and pou3f3b were ventrally expanded in edn1 mutants, cdh11 expression shifted ventrally, and cd248a was largely unaffected. Reciprocally, pou3f3a, pou3f3b and cd248a were reduced in jag1b mutants, with cdh11 and kctd15a expression unaffected. In summary, we find intermediate and ventral-intermediate genes, but only a subset of ventral and dorsal genes, to be regulated by Edn1 signaling, and a subset of dorsal genes to be regulated by Jagged-Notch signaling.
Fig. 4.
Changes in domain-specific gene expression in edn1 and jag1b mutants. Two-color fluorescent in situ hybridizations were performed for genes of interest (red) and dlx2a (green) to label arch CNCCs at 36 hpf. Numbers indicate the gene expression fold-change in mutant versus wild-type fli1a:GFP+; sox10:dsRed+ CNCCs as determined by RNAseq. Maximum intensity projections show the mandibular (left) and hyoid (right) arches. (A) The ventral-intermediate expression of fgfbp2a, shox and stmn1a is lost in edn1 mutants, yet pouch expression of stmn1a is unaffected. (B) In edn1 mutants, intermediate expression of fsta and ctgfb is lost, emx2 is variably upregulated or downregulated, and CNCC and mesoderm expression of igfbp5b is unaltered. We also note some ectopic ventral hyoid fsta expression in edn1 mutants. (C) Ventral expression of smad6a, skp2 and fzd9b is normal in edn1 mutants, yet ventral expression of pitx1 and sema3bl is lost. Note that dorsal expression of sema3bl is unaffected. (D) In jag1b mutants, dorsal expression of cd248a is lost, pou3f3a and pou3f3b are reduced, and cdh11 and kctd15a are unaffected. In edn1 mutants, cdh11 expression shifts to ventral CNCCs, pou3f3a and pou3f3b are ectopically expressed in ventral CNCCs, and cd248a is largely unaffected. Arrows indicate expression in predicted arch domains, open arrowheads indicate additional expression domains, and double arrows show expansion into other CNCC domains in mutants. Unless stated otherwise, consistent expression patterns were seen in a minimum of three wild types and three mutants for each experiment. Scale bars: 20 μm. (E) Summary of verified gene expression changes in edn1 and jag1b mutants. Unaffected genes are not listed.
Co-expression network analysis of pharyngeal arch genes
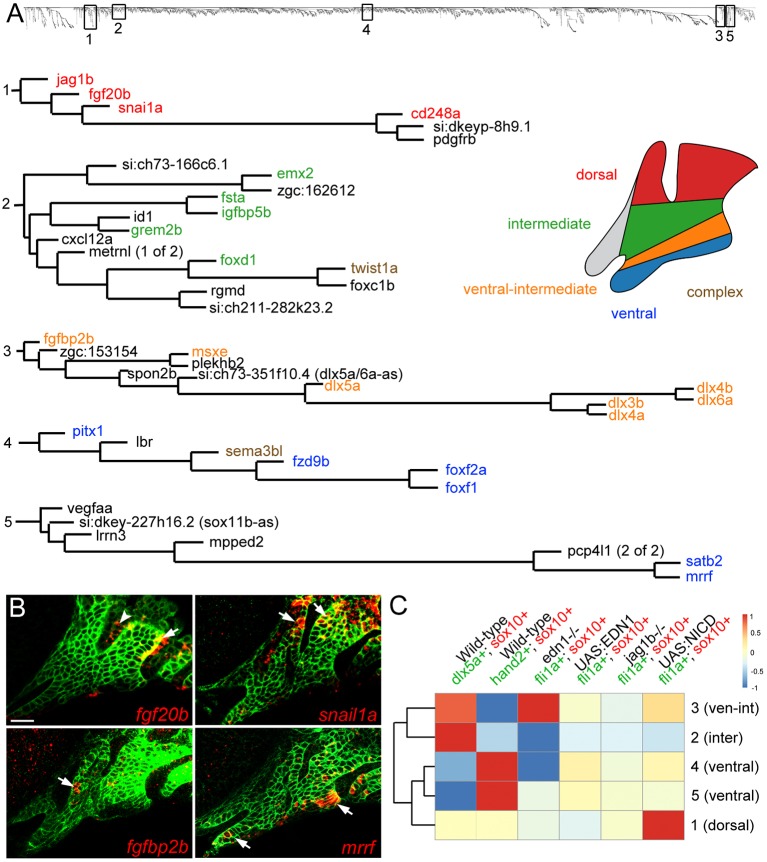
As an independent strategy to uncover genes co-expressed in arch domains, we performed a weighted gene co-expression network analysis (WGCNA) (Zhang and Horvath, 2005) across 19 of our RNAseq datasets (see Fig. 1B). We limited this analysis to the 6000 genes exhibiting the greatest variance across all datasets and showing an expression level above 2 TPM in at least one experiment. A searchable dendrogram (Fig. S5) reflects the topological overlap metric (TOM), which is a measure of the correspondence in expression between genes across samples. In order to determine the utility of TOM in uncovering novel genes within known networks, we examined five representative branches containing genes with validated dorsoventral-restricted expression (Fig. 5A). Cluster 1 is composed of six genes, including the known dorsal gene jag1b (Zuniga et al., 2010). Of these, cd248a, fgf20b and snail1a (snai1a) were detected in dorsal arch CNCCs (Fig. 2D, Fig. 5B). Cluster 2 contains 14 genes, including a known intermediate gene (grem2b) required for joint formation in zebrafish (Zuniga et al., 2011), as well as four newly validated intermediate genes (emx2, fsta, igfbp5b, foxd1) (Fig. 2C). This cluster also contains twist1a, which displays more complex expression in dorsal and ventral arch domains (Germanguz et al., 2007). Cluster 3 contains 11 genes, including five tightly clustered Dlx genes (dlx3b, dlx4a, dlx4b, dlx5a, dlx6a) known to be co-expressed in the ventral-intermediate domain (Talbot et al., 2010), as well as another known ventral-intermediate gene (msxe) (Miller et al., 2000) and a non-coding RNA (si:ch673-351f10.4) in an analogous position to the mouse Evf2 (Dlx6os1) gene, an antisense transcript that promotes the expression of the Dlx5-6 locus (Feng et al., 2006). This cluster also contains an uncharacterized gene, fgfbp2b, which we find to be expressed in a subset of ventral-intermediate first arch CNCCs (Fig. 5B). We also examined two distinct branches containing ventral-restricted genes. We verified five out of six genes in cluster 4 as being restricted to the ventralmost arches (pitx1, fzd9b, foxf1, foxf2a) or expressed more strongly in the ventral arches (sema3bl) (Fig. 2A). Cluster 5 contains a known ventral-restricted gene (satb2) (Sheehan-Rooney et al., 2013a) that tightly co-varies with mrrf, which we find to have similar ventral-restricted expression (Fig. 5B).
Fig. 5.
Co-variance network analysis reveals cohorts of similarly regulated arch genes. (A) Five representative clusters (1-5) were chosen from the dendrogram (top) generated by co-variance analysis. Gene names are color-coded based on expression patterns that are published or verified in this study. ‘Complex’ refers to genes with broader expression in multiple domains. (B) Four genes discovered by co-variance analysis were confirmed by in situ hybridization (red) of sox10:GFPCAAX embryos at 36 hpf; anti-GFP staining (green) marks CNCCs of the mandibular and hyoid arches. Arrows indicate CNCC expression and the arrowhead indicates fgf20b expression in the first pharyngeal pouch. Scale bar: 20 μm. (C) TOM driver array analysis (see Materials and Methods) shows experiments that drove clustering (red) or disrupted clustering (blue). The dendrogram on the left shows the relatedness of clusters based on which datasets drove their clustering.
We next asked which RNAseq experiments informed gene co-regulation by iteratively computing the average TOM disruption caused by removing experimental groups, thus producing a TOM driver score for each experiment (Fig. 5C). Expression in dlx5a:GFP+ cells was the strongest driver for cluster 2, containing known and validated intermediate genes, and a strong driver for cluster 3, containing ventral-intermediate-restricted genes. By contrast, expression in hand2:GFP+ cells was the strongest driver for clusters 4 and 5, consistent with these clusters containing known and newly validated ventral-restricted genes. Consistently, the ventral-intermediate (3) and intermediate (2) clusters and the two ventral clusters (4 and 5) formed separate subgroups when compared across all experiments. By contrast, the dorsal cluster (1) was driven by gain-of-function Notch signaling and not relative enrichment in dlx5a:GFP+ and hand2:GFP+ cells. Interestingly, expression in edn1 mutants was a strong driver for the ventral-intermediate cluster 3, yet disrupted intermediate cluster 2 and ventral cluster 4. This finding is consistent with our in situ validation showing opposite Edn1 regulation of intermediate genes emx2 and fsta in cluster 2, and regulation of sema3bl and pitx1 but not fzd9b in cluster 4. TOM driver analysis thus represents a powerful method for understanding how sets of genes share common expression domains and/or regulation by distinct signaling pathways in the developing face.
We also applied WGCNA to our filtered dorsoventral lists to help resolve false positives (Fig. S1). For genes whose expression patterns were validated by in situ hybridization (Fig. 2), 8/8 ventral, 3/3 ventral-intermediate, 4/5 intermediate and 6/6 dorsal genes clustered together on distinct branches. The one outlier was ctgfb, although repeating WGCNA without Edn1-related RNAseq datasets revealed that this was due in part to stronger regulation by Edn1 signaling. This analysis also revealed two classes of ventral-intermediate genes: those with more restricted ventral-intermediate expression (similar to the Dlx3-6 class) and those with dual ventral-intermediate and dorsal domains (e.g. sema3bl and twist1b). We also found that 4/5 genes identified as false positives (her6, osr2, tmem107l, rassf10a) did not cluster with other genes in their predicted dorsoventral domains. The sole exception was sfrp2, which clustered with dorsal genes despite in situ validation revealing expression in dorsal mesoderm and not CNCCs. Thus, combining dorsoventral filtering and WGCNA analysis decreased the false positive rate and uncovered distinct classes of arch expression patterns.
Absence of larval skeletal defects in loss-of-function mutants for many domain-specific genes
We next sought to uncover potential requirements for novel domain-specific genes in zebrafish craniofacial development. We used TALEN and CRISPR technologies to introduce early frameshift mutations in 12 genes (cd248a, ctgfa, ctgfb, cdh11, emx2, fsta, fstb, her6, mrrf, sfrp2, osr1, osr2; see Table S2 for details) and analyzed homozygous mutant embryos for cartilage and bone defects at 5 days post-fertilization (dpf). For all mutants except fsta and emx2, no craniofacial skeletal defects were observed (data not shown). ctgfa; ctgfb and osr1; osr2 double mutants also failed to display obvious craniofacial skeletal defects. Although not displaying larval craniofacial defects, mrrf mutants grew more slowly than wild-type siblings, rarely survived past 1 month and, even before general growth defects were apparent, were unable to regenerate their tail fins (Fig. S2).
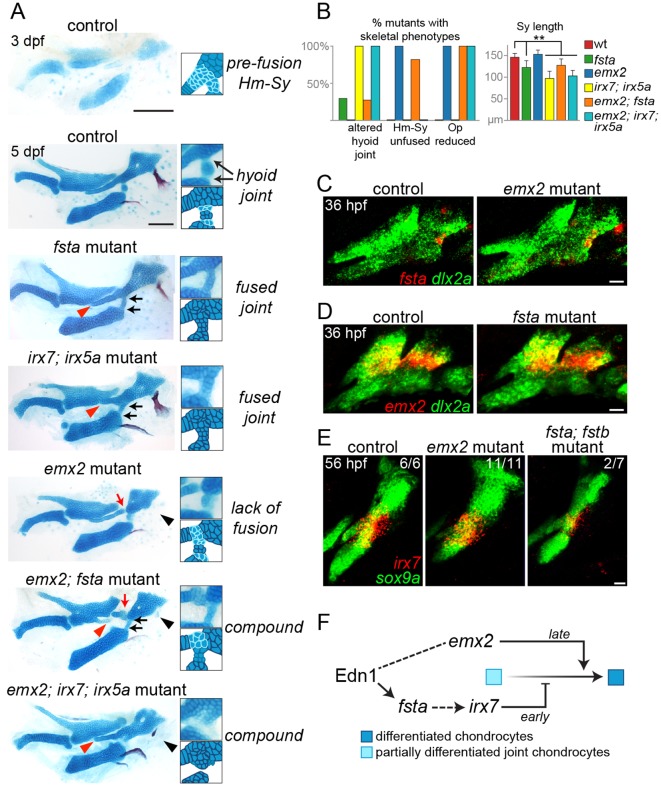
Opposite requirements for fsta and emx2 in hyoid cartilage development
Homozygous mutants for fsta and emx2, two new Edn1 targets expressed in the intermediate domain, displayed defects in the hyoid arch skeleton (Fig. 6A,B). In fsta mutants, we detected variable alterations of the hyoid joint, a compound joint in which a small interhyal cartilage makes connections to the hyomandibular and ceratohyal cartilages on either side. The interhyal cartilage was reduced and made abnormal cartilaginous connections with adjacent cartilages, the symplectic cartilage was reduced in length, and the connection between the hyomandibular and symplectic cartilages was thickened. fsta; fstb double mutants displayed a subtle enhancement of craniofacial defects compared with fsta single mutants (Fig. S3). These joint and symplectic phenotypes are similar to those reported for irx7; irx5a mutants (Fig. 6A) (Askary et al., 2015), and we correspondingly observed a reduction in arch irx7 expression in 2/7 fsta; fstb mutants (Fig. 6E).
Fig. 6.
Distinct roles of Fsta and Emx2 in hyoid skeletal development. (A) Dissected mandibular and hyoid skeletons stained with Alcian Blue (cartilage) and Alizarin Red (bone). At 3 dpf, cells between the hyomandibular (Hm) and symplectic (Sy) cartilages and at the forming hyoid joint region stain weakly with Alcian Blue, reflecting their chondrogenic immaturity. By 5 dpf, rearrangements among cells at the Hm-Sy junction result in elongation of Sy, and maturation of cells at the junction fuses Hm and Sy. Cells in the hyoid joint remain immature and weakly Alcian Blue positive. In fsta mutants, the Sy is shortened (red arrowhead), with a build up of chondrocytes in the Hm-Sy junction, and hyoid joint cells inappropriately mature into chondrocytes strongly stained by Alcian Blue, fusing the joint (black arrows). These phenotypes are similar to those of irx7; irx5a mutants. In emx2 mutants, the Hm and Sy cartilages fail to fuse completely (red arrow), and the opercle bone is lost (black arrowhead). emx2; fsta mutants show a compound phenotype, including hyoid joint fusion and reduction of Sy. Hm and Sy are disconnected in emx2; fsta but not emx2; irx7; irx5a mutants. Scale bars: 100 μm. (B) Penetrance of skeletal phenotypes in each genotype: n (sides)=57 (fsta−/−), 42 (emx2−/−), 5 (irx7−/−; irx5a−/−), 11 (emx2−/−; fsta−/−) and 7 (emx2−/−; irx7−/−; irx5a−/−). **P<0.01 versus wild type. (C,D) Two-color in situ hybridization at 36 hpf shows that fsta expression (red) is normal in emx2 mutants, and emx2 expression (red) is normal in fsta mutants. (E) Hyoid joint expression of irx7 at 56 hpf is unaffected in emx2 mutants and reduced in 2/7 fsta; fstb mutants. In green, dlx2a labels arch CNCCs and sox9a labels chondrocytes. Scale bars: 20 μm in C-E. (F) Model for Emx2 and Fsta function in the hyoid arch. At early stages, Fsta promotes irx7 expression, preventing differentiation and cartilage matrix accumulation between Hm and Sy and allowing chondroprogenitors to rearrange into the single stack of Sy chondrocytes. Following rearrangements, Emx2 promotes chondrogenic differentiation to fuse Hm and Sy into a seamless cartilage. By contrast, there is a continuous requirement for Fsta function at the nearby hyoid joint to maintain its patency through active inhibition of chondrogenesis.
By contrast, emx2 mutants had separated symplectic and hyomandibular cartilages, with weakly Alcian Blue-positive cells evident at the interface. As these two elements start out separate in wild types at 3 dpf (Fig. 6A), we interpret the emx2 phenotype as a failure of later cartilage fusion. These mutants also have a near complete loss of the opercle bone (a hyoid arch derivative) and abnormalities in the palatoquadrate cartilage (a mandibular arch derivative). The expression of fsta and irx7 is unaffected in emx2 mutants, and emx2 expression is unaffected in fsta mutants (Fig. 6C-E). Further, loss of emx2 did not restore normal hyoid joint formation to fsta or irx7; irx5a mutants, and, conversely, loss of fsta or irx7 and irx5a failed to rescue the opercle bone loss of emx2 mutants (Fig. 6A,B). fsta and emx2, two intermediate domain genes regulated in distinct ways by Edn1, therefore act in parallel pathways, with fsta acting upstream of irx7 to promote early joint and symplectic formation and emx2 promoting later cartilage fusion and bone development (Fig. 6F).
DISCUSSION
Our global gene expression analysis of zebrafish pharyngeal arch CNCCs revealed general principles of arch patterning and novel expression patterns and functions of genes not previously implicated in craniofacial development. The intersection of domain-specific gene expression with changes upon signaling perturbation uncovered distinct roles for Edn1 signaling along the dorsoventral axis that might help explain the complex phenotypes of edn1 mutants. In particular, we identified new roles for two distinctly regulated Edn1 target genes, fsta and emx2, in coordinating joint, cartilage, and bone morphogenesis in the intermediate regions of the developing arches.
Identification of novel domain-specific arch genes
We used two complementary methods to identify co-expressed modules of genes in mandibular and hyoid arch CNCCs. The first approach took advantage of the graded expression of hand2:GFP and dlx5a:GFP transgenes along the dorsoventral axis to group genes into four compartments, and the second approach mined co-variation across 19 RNAseq datasets to identify genes with similar expression patterns and/or regulation.
A limitation of the first strategy is that filtering thresholds are empirically determined, and that genes must pass all thresholds to be included (which is likely to account for some known genes, such as dlx6a, being excluded). Empirical shifting of thresholds, guided in part by anchoring well-characterized genes in each cluster, led to a balance between the number of false positives (genes not expressed in the predicted domains) and false negatives (genes with known domain-specific expression not being included). An advantage of the second, co-variance strategy is that it is unbiased, although both the relative enrichment in hand2:GFP+ and dlx5a:GFP+ domains and expression changes in response to signaling perturbation drive clustering. Nonetheless, considerable concordance between the approaches points to the validity of each. For example, four ventral genes (pitx1, fzd9b, foxf1, foxf2a), four ventral-intermediate genes (dlx3b, dlx4b, dlx5a, msxe) and five intermediate genes (emx2, fsta, igfbp5b, grem2b, foxd1) were similarly identified by hand2:GFP/dlx5a:GFP filtering and co-variance analysis.
Using these types of analyses, we uncovered a number of new genes with validated domain-specific expression. In the ventral domain, these included S-phase kinase-associated protein 2 (skp2) and mitochondrial ribosome recycling factor (mrrf), perhaps reflecting distal growth of this domain to elongate the lower jaw (Bonilla-Claudio et al., 2012; Medeiros and Crump, 2012). In the ventral-intermediate domain we uncovered specific expression of Fgf-binding proteins (fgfbp2a and fgfbp2b), suggesting fine regulation of Fgf signaling in this domain. In the intermediate domain, we discovered two putative Bmp inhibitors (fsta and ctgfb) with tightly restricted expression near the developing hyoid joint, consistent with prior data showing that complex regulation of Bmp signaling is important for joint specification (Salazar et al., 2016; Smeeton et al., 2017). In the dorsal domain, we uncovered selective expression of genes previously implicated in earlier neural crest and ectomesenchyme development, including a Snail transcription factor (snai1a) (LaBonne and Bronner-Fraser, 2000), cadherin 11 (cdh11) (McLennan et al., 2015), potassium channel tetramerization domain-containing 15a (kctd15a), which interacts with tfap2a (Zarelli and Dawid, 2013), and endosialin (cd248a) (Das and Crump, 2012); this signature is consistent with our previous findings that dorsal arch CNCCs differentiate later than other arch CNCCs (Barske et al., 2016). Of the 31 genes tested by in situ hybridization, 26 showed expression in the predicted domain, three showed CNCC-specific expression outside the predicted domain, and two were expressed in non-CNCC arch tissues. Further, applying WGCNA to the dorsoventral gene lists correctly predicted 4/5 false positives while only excluding 1/26 true positives. The one exception was secreted frizzled-related protein 2 (sfrp2), which showed dorsal-specific expression in arch mesoderm but not CNCCs. Its mouse homolog has also been reported to be upregulated in Dlx5/6 mutants, consistent with dorsal enrichment (Jeong et al., 2008). We do not know whether the inclusion of sfrp2 in our CNCC datasets reflects expression in CNCCs below the level of detection by in situ hybridization or contamination of our FACS-sorted populations by a few non-CNCC arch cells. Nonetheless, our analysis pipeline accurately predicted domain-specific expression for a high proportion of genes, including nine not previously implicated in craniofacial development.
Region-specific roles of Edn1 and Jagged-Notch signaling in arch patterning
Previous work had suggested greater roles for Edn1 signaling in intermediate versus more ventral domains (Alexander et al., 2011; Zuniga et al., 2011), and for Jagged-Notch signaling in the dorsal arches (Zuniga et al., 2010). By analyzing how gene modules of distinct arch domains are affected by signaling perturbations, we confirm this on a genomic scale (as summarized in Fig. 4E). A more prominent role for Edn1 in controlling gene expression in intermediate mandibular and hyoid arch domains, which generate joints and the palatoquadrate and symplectic cartilages, helps explain why these skeletal elements are most sensitive to partial reduction of Edn1 function (Miller and Kimmel, 2001) and mutation of its downstream effectors Plcb3 and Mef2ca (Walker et al., 2006, 2007). Conversely, the ventralmost elements of the mandibular and hyoid arches, such as the basihyal, are spared in severe edn1 mutants (Miller et al., 2000), consistent with the expression of most ventral genes in this study (smad6a, skp2, fzd9b) and in previous reports (satb2) (Sheehan-Rooney et al., 2013a) being unaffected by Edn1 perturbations. However, some ventral genes, such as hand2 and pitx1, are lost in edn1 mutants, although regulation of hand2 may be indirect through the Edn1 targets Dlx5/6 (Miller et al., 2003; Yanagisawa et al., 2003). Edn1-independent ventral genes might instead depend on Bmp signaling. Smad6 and Satb2 were identified as direct targets of Bmp-dependent pSmads in mice (Bonilla-Claudio et al., 2012), and satb2 is a target of Bmp signaling in zebrafish (Sheehan-Rooney et al., 2013a).
In the dorsal domain, only a subset of genes are regulated by Jagged-Notch signaling (e.g. cd248a, pou3f3a and pou3f3b, but not cdh11 and kctd15a), consistent with the relatively mild dorsal phenotypes of jag1b and notch2; notch3 mutants (Zuniga et al., 2010; Barske et al., 2016) and suggesting Notch-independent regulation of some aspects of dorsal identity.
Whereas we found generally good correspondence between changes in RNAseq values and in situ validation in edn1 and jag1b mutants, in situ validation but not RNAseq revealed differences in stmn1a, sema3bl and cdh11 expression in edn1 mutants. As stmn1a and sema3bl show broad arch expression, profiling all arch CNCCs is likely to dilute the effect of selective loss of their ventral expression domains in mutants. Likewise, a shift of cdh11 expression from dorsal to ventral domains in mutants would not necessarily result in a total expression difference throughout arch CNCCs. These findings suggest that examining expression changes in CNCCs sorted from distinct arch domains in animals with signaling perturbations might be a better way to detect how signaling affects expression patterns.
A lack of obvious craniofacial phenotypes in mutants for many arch-specific genes
The ease of genetic manipulation makes zebrafish an attractive system for performing reverse genetic analysis of craniofacial development. However, homozygous loss-of-function mutants for only two of the 12 domain-specific genes tested showed clear facial cartilage and/or bone phenotypes in larvae. There are several possible explanations for the lack of observable phenotypes. First, although we selected mutations causing premature translational termination before crucial conserved domains, it remains possible that some mutations do not create true nulls. Second, some mutants might have craniofacial defects that we failed to appreciate, for example in other arch derivatives such as ligaments or long-lived progenitors. Third, maternal contribution of mRNA and/or protein could compensate for zygotic loss-of-function. In some cases (e.g. ctgfa), maternal-zygotic null mutants did not display larval craniofacial defects. The growth delay and tail fin regeneration defects of mrrf mutants could be explained by depletion of remaining maternal stores, similar to previous reports for other mutants in mitochondrial proteins (Rahn et al., 2015). It thus remains possible that mrrf expression in the ventralmost arches reflects rapid growth and/or metabolism of this domain. Fourth, there might be genetic compensation (Rossi et al., 2015). Large-scale mutational screens in zebrafish have found a surprisingly small number of genes required for larval viability (∼6%), suggesting a high degree of genetic redundancy in zebrafish (Kettleborough et al., 2013). In addition, the identification of multiple alleles for craniofacial mutants suggests that previous screens are approaching saturation for obvious larval skeletal defects (Neuhauss et al., 1996; Piotrowski et al., 1996; Schilling et al., 1996; Nissen et al., 2006). Our findings therefore indicate that many of the single gene mutants with obvious craniofacial patterning defects in zebrafish might have already been found.
Complex regulation by Edn1 coordinates intermediate arch morphogenesis
A curious feature of edn1 mutants, as well as mutants for its effector mef2ca, is the phenotypic variability of intermediate domain-derived skeletal elements, including joints and the opercle bone (Kimmel et al., 2003; DeLaurier et al., 2014). Our analysis of two newly identified Edn1 target genes, emx2 and fsta, may shed some light on this variability. For example, the gain or loss of the opercle in edn1 mutants might reflect the observed variability in emx2 regulation, given that loss of emx2 suppressed the opercle expansion seen in some edn1 mutants without rescuing ventral cartilage loss (Fig. S4). The loss of the hyoid joint and the reduction in symplectic cartilage in fsta mutants are also similar to what is seen in hypomorphic Edn1 pathway mutants (Miller et al., 2000; Walker et al., 2006, 2007; DeLaurier et al., 2014), consistent with our finding that intermediate domain fsta expression is lost in edn1 mutants. Our previous analysis of similar phenotypes in irx7; irx5a compound mutants showed that inappropriate chondrogenic differentiation at the junction between the nascent symplectic and hyomandibular cartilages prevents these cells from rearranging and thus lengthening the symplectic (Askary et al., 2015). By contrast, the hyomandibular and symplectic cartilages fail to connect in emx2 mutants. Temporal regulation by Edn1 in the intermediate domain may therefore result in Fsta blocking cartilage differentiation at early stages to allow symplectic elongation, with Emx2 promoting cartilage differentiation at later stages to fuse the symplectic and hyomandibular into a seamless cartilage (Fig. 6F). As Bmp activity inhibits irx7 expression (Askary et al., 2015), Fsta might promote irx7 at the hyoid joint by limiting Bmp signaling. The low penetrance of irx7 loss and joint fusion in fsta/b mutants might be due to functional redundancy with other putative Bmp inhibitors expressed at the joint, including ctgfb (this study), grem2b (Zuniga et al., 2011) and chordin (Miller et al., 2003). Interestingly, Emx2 mutant mice lack the incus cartilage of the middle ear (Rhodes et al., 2003), which is homologous to the palatoquadrate affected in fish emx2 mutants. Part of the arch patterning function of Emx2 might thus be conserved from fish to mammals.
Our transcriptome-driven analysis of arch regionalization has therefore provided new insights into how Edn1 signaling regulates a delicate balance of cartilage differentiation to fine-tune skeletal shape. In the future, the analysis pipeline presented here should help to reveal regulatory changes in additional mutants that disrupt facial patterning.
MATERIALS AND METHODS
Zebrafish lines
The University of Southern California Institutional Animal Care and Use Committee approved all experiments on zebrafish (Danio rerio). Published lines include Tg(hand2:eGFP) (Kikuchi et al., 2011), dlx5aj1073Et (referred to here as dlx5a:GFP) (Talbot et al., 2010), Tg(fli1a:eGFP)y1 (Lawson and Weinstein, 2002), Tg(sox10:DsRed-Express)el10 (Das and Crump, 2012), Tg(sox10:GFPCAAX), irx7el538, irx5el574 (Askary et al., 2015), sucker/edn1tf216 (Miller et al., 2000) and jag1bb1105 (Zuniga et al., 2010).
FACS and RNAseq
fli1a:GFP; sox10:DsRed and hand2:GFP; sox10:DsRed fish were incrossed to generate embryos, and dlx5a:GFP; sox10:DsRed fish were outcrossed to avoid homozygosity of the dlx5aj1073Et insertional allele. Embryos were dissociated as previously described (Covassin et al., 2006), with minor modifications (Barske et al., 2016). Cells were sorted based on GFP and DsRed expression on a MoFlo Astrios instrument (Beckman-Coulter) into RLT lysis buffer (Qiagen), and total RNA was extracted using the RNeasy Micro Kit (Qiagen). RNA integrity was assessed on Bioanalyzer Pico RNA chips (Agilent), cDNA synthesized with the SMART-Seq Ultra Low Input RNA Kit (Clontech), and libraries generated with the Kapa Hyper Prep Kit (Kapa Biosystems) and NextFlex adapters (Bioo Scientific). 75 bp paired-end sequencing was performed on a NextSeq 500 machine (Illumina).
RNAseq data analysis and statistical tests
After trimming using Partek Flow default criteria, sequencing reads were aligned to zebrafish GRCz10 (Ensembl_v80) using TopHat 2 (https://ccb.jhu.edu/software/tophat/index.shtml). Aligned reads were quantified using Partek E/M and normalized to yield TPM values, controlling for sequencing depth disparities across samples (Wagner et al., 2012). Data are accessible through GEO series accession GSE95812. To test whether log2 fold-change values for each group of genes were significantly different to zero (Fig. 3), we used the Shapiro-Wilk test for normality to determine whether a one-sample t-test or a Wilcoxon signed-rank test was appropriate. The Bonferroni correction was then applied on the resulting P-values of one-tailed tests to account for multiple comparisons. The Mann–Whitney U-test for two independent samples was performed in Excel 2016 (Microsoft) using Real Statistics Resource Pack software (release 4.9; www.real-statistics.com) to compare effects of Edn1 and Notch signaling, as the data from at least one group were not distributed normally.
Co-variance analysis
A weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008) was run on m genes exhibiting the highest variance across n samples (m=6000, n=19), yielding an m×m topological overlap matrix (TOM) that links genes by correspondence of correlated genes. The exponent β was selected to yield scale-free topology as defined by minimum power required to output maximal R2. We computed the TOM driver array (TDA) by taking the average TOM value across genes of interest, then deriving the deviation in TOM when each sample was removed:
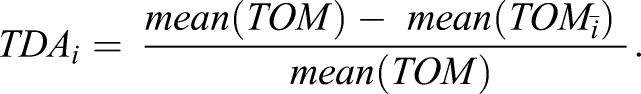 |
(1) |
These values are normalized to produce nTDA, which spans [−1,1]:
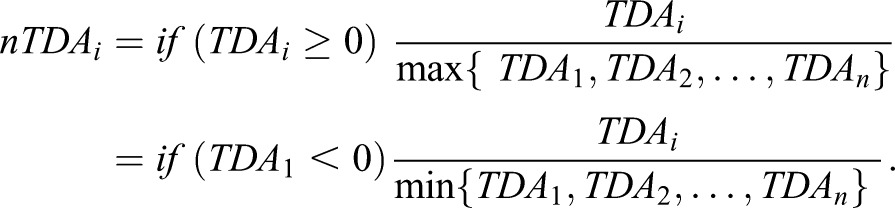 |
(2) |
Samples with a positive TDA drive clustering, whereas samples with negative TDA disrupt clustering.
In situ hybridization and immunohistochemistry
Partial cDNAs were PCR amplified with Herculase II Fusion Polymerase (Agilent), cloned into pCR_Blunt_II_Topo (ThermoFisher Scientific), linearized, and synthesized with SP6 or T7 RNA polymerase (Roche Life Sciences) as specified (Table S3). In situ hybridization was performed as described (Zuniga et al., 2010), co-staining for dlx2a (Akimenko et al., 1994), sox9a, or with rabbit anti-GFP antibody (Torrey Pines Biolabs, TP401; 1:1000) to highlight arch CNCCs or early cartilages. Imaging was performed with a Zeiss LSM800 confocal microscope and presented as optical sections or maximum intensity projections as specified. Typically, six to ten controls and three to seven mutants were imaged for each probe.
Mutant generation and skeletal staining
Twelve mutant lines were created via TALEN (Sanjana et al., 2012) or CRISPR/Cas9 (Jao et al., 2013) mutagenesis as described (Barske et al., 2016). Germline founders were detected by screening their F1 progeny by restriction digestion of PCR products, followed by sequencing to identify frameshift indels (Table S2). Alcian Blue and Alizarin Red staining of cartilage and bone were performed as described (Walker and Kimmel, 2007). Symplectic cartilage length was measured with ImageJ (NIH) and compared using unpaired t-tests.
Acknowledgements
We thank Megan Matsutani and Jennifer DeKoeyer Crump for fish care, Lora Barsky and Jeffrey Boyd at the USC Stem Cell Flow Cytometry Core Facility for FACS, Charles Nicolet at the USC Norris Cancer Center Molecular Genomics Core for sequencing, and Yibu Chen for sequencing analysis.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.A., P.X., L.B., B.B., J.G.C.; Methodology: A.A., P.X., L.B., P.B., B.B.; Software: A.A., M.B.; Validation: A.A., L.B.; Formal analysis: A.A., P.X., L.B., J.G.C.; Investigation: A.A., P.X., L.B., P.B.; Resources: A.A., J.G.C.; Data curation: A.A., P.X., L.B., M.B.; Writing - original draft: A.A., P.X., L.B., M.B., J.G.C.; Writing - review & editing: A.A., P.X., L.B., M.A.B., J.G.C.; Supervision: M.A.B., J.G.C.; Project administration: J.G.C.; Funding acquisition: M.A.B., J.G.C.
Funding
Research was funded by grants from the National Institute of Dental and Craniofacial Research (NIDCR) (DE018405) to J.G.C., National Institute of Neurological Disorders and Stroke (R00NS080913) to M.A.B., A.P. Giannini Foundation and NIDCR (K99DE026239) to L.B., and National Institute on Deafness and Other Communication Disorders (training grant T32 DC009975) to A.A. Deposited in PMC for release after 12 months.
Data availability
RNAseq data are available at Gene Expression Omnibus through series accession number GSE95812.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.151712.supplemental
References
- Akimenko M. A., Ekker M., Wegner J., Lin W. and Westerfield M. (1994). Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci. 14, 3475-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C., Zuniga E., Blitz I. L., Wada N., Le Pabic P., Javidan Y., Zhang T., Cho K. W., Crump J. G. and Schilling T. F. (2011). Combinatorial roles for BMPs and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development 138, 5135-5146. 10.1242/dev.067801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askary A., Mork L., Paul S., He X., Izuhara A. K., Gopalakrishnan S., Ichida J. K., McMahon A. P., Dabizljevic S., Dale R. et al. (2015). Iroquois proteins promote skeletal joint formation by maintaining chondrocytes in an immature state. Dev. Cell 35, 358-365. 10.1016/j.devcel.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barske L., Askary A., Zuniga E., Balczerski B., Bump P., Nichols J. T. and Crump J. G. (2016). Competition between Jagged-Notch and endothelin1 signaling selectively restricts cartilage formation in the zebrafish upper face. PLoS Genet. 12, e1005967 10.1371/journal.pgen.1005967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverdam A., Merlo G. R., Paleari L., Mantero S., Genova F., Barbieri O., Janvier P. and Levi G. (2002). Jaw transformation with gain of symmetry after Dlx5/Dlx6 inactivation: mirror of the past? Genesis 34, 221-227. 10.1002/gene.10156 [DOI] [PubMed] [Google Scholar]
- Bonilla-Claudio M., Wang J., Bai Y., Klysik E., Selever J. and Martin J. F. (2012). Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development 139, 709-719. 10.1242/dev.073197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner M. E. and LeDouarin N. M. (2012). Development and evolution of the neural crest: an overview. Dev. Biol. 366, 2-9. 10.1016/j.ydbio.2011.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill E. W., Potter A. S., Distasio A., Dexheimer P., Plassard A., Aronow B. J. and Potter S. S. (2014). A gene expression atlas of early craniofacial development. Dev. Biol. 391, 133-146. 10.1016/j.ydbio.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier D. E., Hosoda K., Richardson J. A., Williams S. C., Yanagisawa H., Kuwaki T., Kumada M., Hammer R. E. and Yanagisawa M. (1998). Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development 125, 813-824. [DOI] [PubMed] [Google Scholar]
- Compagnucci C., Debiais-Thibaud M., Coolen M., Fish J., Griffin J. N., Bertocchini F., Minoux M., Rijli F. M., Borday-Birraux V., Casane D. et al. (2013). Pattern and polarity in the development and evolution of the gnathostome jaw: both conservation and heterotopy in the branchial arches of the shark, Scyliorhinus canicula. Dev. Biol. 377, 428-448. 10.1016/j.ydbio.2013.02.022 [DOI] [PubMed] [Google Scholar]
- Covassin L., Amigo J. D., Suzuki K., Teplyuk V., Straubhaar J. and Lawson N. D. (2006). Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish. Dev. Biol. 299, 551-562. 10.1016/j.ydbio.2006.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. and Crump J. G. (2012). Bmps and id2a act upstream of Twist1 to restrict ectomesenchyme potential of the cranial neural crest. PLoS Genet. 8, e1002710 10.1371/journal.pgen.1002710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaurier A., Huycke T. R., Nichols J. T., Swartz M. E., Larsen A., Walker C., Dowd J., Pan L., Moens C. B. and Kimmel C. B. (2014). Role of mef2ca in developmental buffering of the zebrafish larval hyoid dermal skeleton. Dev. Biol. 385, 189-199. 10.1016/j.ydbio.2013.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew M. J., Lufkin T. and Rubenstein J. L. (2002). Specification of jaw subdivisions by Dlx genes. Science 298, 381-385. 10.1126/science.1075703 [DOI] [PubMed] [Google Scholar]
- Feng J., Bi C., Clark B. S., Mady R., Shah P. and Kohtz J. D. (2006). The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470-1484. 10.1101/gad.1416106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Leach S. M., Tipney H., Phang T., Geraci M., Spritz R. A., Hunter L. E. and Williams T. (2009). Spatial and temporal analysis of gene expression during growth and fusion of the mouse facial prominences. PLoS ONE 4, e8066 10.1371/journal.pone.0008066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K., Taya Y., Shimazu Y., Aoba T. and Soeno Y. (2013). Molecular signaling at the fusion stage of the mouse mandibular arch: involvement of insulin-like growth factor family. Int. J. Dev. Biol. 57, 399-406. 10.1387/ijdb.120110ys [DOI] [PubMed] [Google Scholar]
- Germanguz I., Lev D., Waisman T., Kim C. H. and Gitelman I. (2007). Four twist genes in zebrafish, four expression patterns. Dev. Dyn. 236, 2615-2626. 10.1002/dvdy.21267 [DOI] [PubMed] [Google Scholar]
- Gharbi N., Zhao X.-F., Ellingsen S. and Fjose A. (2012). Zebrafish enhancer trap line showing maternal and neural expression of kctd15a. Dev. Growth Differ. 54, 241-252. 10.1111/j.1440-169X.2011.01326.x [DOI] [PubMed] [Google Scholar]
- Gordon C. T., Petit F., Kroisel P. M., Jakobsen L., Zechi-Ceide R. M., Oufadem M., Bole-Feysot C., Pruvost S., Masson C., Tores F. et al. (2013). Mutations in endothelin 1 cause recessive auriculocondylar syndrome and dominant isolated question-mark ears. Am. J. Hum. Genet. 93, 1118-1125. 10.1016/j.ajhg.2013.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper J. E., Feng W., Li H., Leach S. M., Phang T., Siska C., Jones K. L., Spritz R. A., Hunter L. E. and Williams T. (2017). Systems biology of facial development: contributions of ectoderm and mesenchyme. Dev. Biol. 426, 97-114. 10.1016/j.ydbio.2017.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter M. P. and Prince V. E. (2002). Zebrafish hox paralogue group 2 genes function redundantly as selector genes to pattern the second pharyngeal arch. Dev. Biol. 247, 367-389. 10.1006/dbio.2002.0701 [DOI] [PubMed] [Google Scholar]
- Jao L.-E., Wente S. R. and Chen W. (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 110, 13904-13909. 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Mao J., Tenzen T., Kottmann A. H. and McMahon A. P. (2004). Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 18, 937-951. 10.1101/gad.1190304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Li X., McEvilly R. J., Rosenfeld M. G., Lufkin T. and Rubenstein J. L. R. (2008). Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development 135, 2905-2916. 10.1242/dev.019778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettleborough R. N. W., Busch-Nentwich E. M., Harvey S. A., Dooley C. M., de Bruijn E., van Eeden F., Sealy I., White R. J., Herd C., Nijman I. J. et al. (2013). A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496, 494-497. 10.1038/nature11992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Major R. J., Blum N., Dahn R. D., Begemann G. and Poss K. D. (2011). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397-404. 10.1016/j.devcel.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Miller C. T., Kruze G., Ullmann B., BreMiller R. A., Larison K. D. and Snyder H. C. (1998). The shaping of pharyngeal cartilages during early development of the zebrafish. Dev. Biol. 203, 245-263. 10.1006/dbio.1998.9016 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ullmann B., Walker M., Miller C. T. and Crump J. G. (2003). Endothelin 1-mediated regulation of pharyngeal bone development in zebrafish. Development 130, 1339-1351. 10.1242/dev.00338 [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Kurihara H., Suzuki H., Kodama T., Maemura K., Nagai R., Oda H., Kuwaki T., Cao W.-H., Kamada N. et al. (1994). Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature 368, 703-710. 10.1038/368703a0 [DOI] [PubMed] [Google Scholar]
- LaBonne C. and Bronner-Fraser M. (2000). Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev. Biol. 221, 195-205. 10.1006/dbio.2000.9609 [DOI] [PubMed] [Google Scholar]
- Langfelder P. and Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N. D. and Weinstein B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307-318. 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- McLennan R., Schumacher L. J., Morrison J. A., Teddy J. M., Ridenour D. A., Box A. C., Semerad C. L., Li H., McDowell W., Kay D. et al. (2015). Neural crest migration is driven by a few trailblazer cells with a unique molecular signature narrowly confined to the invasive front. Development 142, 2014-2025. 10.1242/dev.117507 [DOI] [PubMed] [Google Scholar]
- Medeiros D. M. and Crump J. G. (2012). New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev. Biol. 371, 121-135. 10.1016/j.ydbio.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. T. and Kimmel C. B. (2001). Morpholino phenocopies of endothelin 1 (sucker) and other anterior arch class mutations. Genesis 30, 186-187. 10.1002/gene.1061 [DOI] [PubMed] [Google Scholar]
- Miller C. T., Schilling T. F., Lee K., Parker J. and Kimmel C. B. (2000). sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development 127, 3815-3828. [DOI] [PubMed] [Google Scholar]
- Miller C. T., Yelon D., Stainier D. Y. R. and Kimmel C. B. (2003). Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development 130, 1353-1365. 10.1242/dev.00339 [DOI] [PubMed] [Google Scholar]
- Minoux M. and Rijli F. M. (2010). Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development 137, 2605-2621. 10.1242/dev.040048 [DOI] [PubMed] [Google Scholar]
- Mork L. and Crump G. (2015). Zebrafish craniofacial development: a window into early patterning. Curr. Top. Dev. Biol. 115, 235-269. 10.1016/bs.ctdb.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S., Li W., Cornell R. and Schilling T. F. (2007). Requirements for Endothelin type-A receptors and Endothelin-1 signaling in the facial ectoderm for the patterning of skeletogenic neural crest cells in zebrafish. Development 134, 335-345. 10.1242/dev.02704 [DOI] [PubMed] [Google Scholar]
- Neuhauss S. C., Solnica-Krezel L., Schier A. F., Zwartkruis F., Stemple D. L., Malicki J., Abdelilah S., Stainier D. Y. and Driever W. (1996). Mutations affecting craniofacial development in zebrafish. Development 123, 357-367. [DOI] [PubMed] [Google Scholar]
- Nichols J. T., Pan L., Moens C. B. and Kimmel C. B. (2013). barx1 represses joints and promotes cartilage in the craniofacial skeleton. Development 140, 2765-2775. 10.1242/dev.090639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J. T., Blanco-Sánchez B., Brooks E. P., Parthasarathy R., Dowd J., Subramanian A., Nachtrab G., Poss K. D., Schilling T. F. and Kimmel C. B. (2016). Ligament versus bone cell identity in the zebrafish hyoid skeleton is regulated by mef2ca. Development 143, 4430-4440. 10.1242/dev.141036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen R. M., Amsterdam A. and Hopkins N. (2006). A zebrafish screen for craniofacial mutants identifies wdr68 as a highly conserved gene required for endothelin-1 expression. BMC Dev. Biol. 6, 28 10.1186/1471-213X-6-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki H., Kurihara Y., Tonami K., Watatani S. and Kurihara H. (2004). Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mech. Dev. 121, 387-395. 10.1016/j.mod.2004.02.002 [DOI] [PubMed] [Google Scholar]
- Paul S., Schindler S., Giovannone D., de Millo Terrazzani A., Mariani F. V. and Crump J. G. (2016). Ihha induces hybrid cartilage-bone cells during zebrafish jawbone regeneration. Development 143, 2066-2076. 10.1242/dev.131292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski T., Schilling T. F., Brand M., Jiang Y. J., Heisenberg C. P., Beuchle D., Grandel H., van Eeden F. J., Furutani-Seiki M., Granato M. et al. (1996). Jaw and branchial arch mutants in zebrafish II: anterior arches and cartilage differentiation. Development 123, 345-356. [DOI] [PubMed] [Google Scholar]
- Platt J. B. (1893). Ectodermic origin of the cartilages of the head. Anat. Anz. 8, 506-509. [Google Scholar]
- Rahn J. J., Bestman J. E., Stackley K. D. and Chan S. S. L. (2015). Zebrafish lacking functional DNA polymerase gamma survive to juvenile stage, despite rapid and sustained mitochondrial DNA depletion, altered energetics and growth. Nucleic Acids Res. 43, 10338-10352. 10.1093/nar/gkv1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes C. R., Parkinson N., Tsai H., Brooker D., Mansell S., Spurr N., Hunter A. J., Steel K. P. and Brown S. D. M. (2003). The homeobox gene Emx2 underlies middle ear and inner ear defects in the deaf mouse mutant pardon. J. Neurocytol. 32, 1143-1154. 10.1023/B:NEUR.0000021908.98337.91 [DOI] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M. and Stainier D. Y. R. (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230-233. 10.1038/nature14580 [DOI] [PubMed] [Google Scholar]
- Salazar V. S., Gamer L. W. and Rosen V. (2016). BMP signalling in skeletal development, disease and repair. Nat. Rev. Endocrinol. 12, 203-221. 10.1038/nrendo.2016.12 [DOI] [PubMed] [Google Scholar]
- Sanjana N. E., Cong L., Zhou Y., Cunniff M. M., Feng G. and Zhang F. (2012). A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 7, 171-192. 10.1038/nprot.2011.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Kurihara Y., Asai R., Kawamura Y., Tonami K., Uchijima Y., Heude E., Ekker M., Levi G. and Kurihara H. (2008). An endothelin-1 switch specifies maxillomandibular identity. Proc. Natl. Acad. Sci. USA 105, 18806-18811. 10.1073/pnas.0807345105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling T. F. and Kimmel C. B. (1994). Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development 120, 483-494. [DOI] [PubMed] [Google Scholar]
- Schilling T. F., Piotrowski T., Grandel H., Brand M., Heisenberg C. P., Jiang Y. J., Beuchle D., Hammerschmidt M., Kane D. A., Mullins M. C. et al. (1996). Jaw and branchial arch mutants in zebrafish I: branchial arches. Development 123, 329-344. [DOI] [PubMed] [Google Scholar]
- Sheehan-Rooney K., Swartz M. E., Lovely C. B., Dixon M. J. and Eberhart J. K. (2013a). Bmp and Shh signaling mediate the expression of satb2 in the pharyngeal arches. PLoS ONE 8, e59533 10.1371/journal.pone.0059533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan-Rooney K., Swartz M. E., Zhao F., Liu D. and Eberhart J. K. (2013b). Ahsa1 and Hsp90 activity confers more severe craniofacial phenotypes in a zebrafish model of hypoparathyroidism, sensorineural deafness and renal dysplasia (HDR). Dis. Model. Mech. 6, 1285-1291. 10.1242/dmm.011965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeeton J., Askary A. and Crump J. G. (2017). Building and maintaining joints by exquisite local control of cell fate. Wiley Interdiscip. Rev. Dev. Biol. 6, doi: 10.1002/wdev.245 10.1002/wdev.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber S. M., Saxena V., Hatch G. and Ekker M. (2008). Zebrafish dlx2a contributes to hindbrain neural crest survival, is necessary for differentiation of sensory ganglia and functions with dlx1a in maturation of the arch cartilage elements. Dev. Biol. 314, 59-70. 10.1016/j.ydbio.2007.11.005 [DOI] [PubMed] [Google Scholar]
- Swartz M. E., Sheehan-Rooney K., Dixon M. J. and Eberhart J. K. (2011). Examination of a palatogenic gene program in zebrafish. Dev. Dyn. 240, 2204-2220. 10.1002/dvdy.22713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot J. C., Johnson S. L. and Kimmel C. B. (2010). hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development 137, 2507-2517. 10.1242/dev.049700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., Kurihara H., Yamagishi H., Kurihara Y., Yazaki Y., Olson E. N. and Srivastava D. (1998). A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development 125, 3005-3014. [DOI] [PubMed] [Google Scholar]
- Tucker A. S., Matthews K. L. and Sharpe P. T. (1998). Transformation of tooth type induced by inhibition of BMP signaling. Science 282, 1136-1138. 10.1126/science.282.5391.1136 [DOI] [PubMed] [Google Scholar]
- Wagner G. P., Kin K. and Lynch V. J. (2012). Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281-285. 10.1007/s12064-012-0162-3 [DOI] [PubMed] [Google Scholar]
- Walker M. B. and Kimmel C. B. (2007). A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82, 23-28. 10.1080/10520290701333558 [DOI] [PubMed] [Google Scholar]
- Walker M. B., Miller C. T., Coffin Talbot J., Stock D. W. and Kimmel C. B. (2006). Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev. Biol. 295, 194-205. 10.1016/j.ydbio.2006.03.028 [DOI] [PubMed] [Google Scholar]
- Walker M. B., Miller C. T., Swartz M. E., Eberhart J. K. and Kimmel C. B. (2007). phospholipase C, beta 3 is required for Endothelin1 regulation of pharyngeal arch patterning in zebrafish. Dev. Biol. 304, 194-207. 10.1016/j.ydbio.2006.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H., Clouthier D. E., Richardson J. A., Charite J. and Olson E. N. (2003). Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development 130, 1069-1078. 10.1242/dev.00337 [DOI] [PubMed] [Google Scholar]
- Zarelli V. E. and Dawid I. B. (2013). Inhibition of neural crest formation by Kctd15 involves regulation of transcription factor AP-2. Proc. Natl. Acad. Sci. USA 110, 2870-2875. 10.1073/pnas.1300203110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. and Horvath S. (2005). A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
- Zuniga E., Stellabotte F. and Crump J. G. (2010). Jagged-Notch signaling ensures dorsal skeletal identity in the vertebrate face. Development 137, 1843-1852. 10.1242/dev.049056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E., Rippen M., Alexander C., Schilling T. F. and Crump J. G. (2011). Gremlin 2 regulates distinct roles of BMP and Endothelin 1 signaling in dorsoventral patterning of the facial skeleton. Development 138, 5147-5156. 10.1242/dev.067785 [DOI] [PMC free article] [PubMed] [Google Scholar]