ABSTRACT
The blastema is a mass of progenitor cells responsible for regeneration of amputated salamander limbs and fish fins. Previous studies have indicated that resident cell sources producing the blastema contribute lineage-restricted progeny to regenerating tissue. However, these studies have labeled general cell types rather than granular cell subpopulations, and they do not explain the developmental transitions that must occur for distal structures to arise from cells with proximal identities in the appendage stump. Here, we find that regulatory sequences of tph1b, which encodes an enzyme that synthesizes serotonin, mark a subpopulation of fibroblast-like cells restricted to the joints of uninjured adult zebrafish fins. Amputation stimulates serotonin production in regenerating fin fibroblasts, yet targeted tph1b mutations abrogating this response do not disrupt fin regeneration. In uninjured animals, tph1b-expressing cells contribute fibroblast progeny that remain restricted to joints throughout life. By contrast, upon amputation, tph1b+ joint cells give rise to fibroblasts that distribute across the entire lengths of regenerating fin rays. Our experiments visualize and quantify how incorporation into an appendage blastema broadens the progeny contributions of a cellular subpopulation that normally has proximodistal restrictions.
KEY WORDS: Clonal analysis, Fin regeneration, Regeneration, Zebrafish
Summary: In zebrafish, a tph1b-expressing population of fibroblast-like cells is restricted to the joints of uninjured fins but contributes more broadly during regeneration.
INTRODUCTION
Zebrafish vigorously regenerate fins after an amputation injury, an analogous event to limb regeneration in salamanders (Tornini and Poss, 2014). Following fin amputation, cells in the tissue stump reorganize, migrate or are displaced, and proliferate, forming a structure called the blastema. Blastemal cells are progenitors for the predominant cells in the mesenchymal compartment of fin rays: osteoblasts and fibroblasts (Knopf et al., 2011; Singh et al., 2012; Tornini et al., 2016; Tu and Johnson, 2011). Previous reports indicated that formation of an appendage blastema and subsequent regeneration in fish fins, salamander limbs and murine digit tips do not involve major changes in the lineage potentials of resident cells (Knopf et al., 2011; Kragl et al., 2009; Lehoczky et al., 2011; Rinkevich et al., 2011; Singh et al., 2012; Stewart and Stankunas, 2012; Tornini et al., 2016; Tu and Johnson, 2011).
Although understanding of the blastema is deepening, cells in a restricted region of an appendage must somehow acquire the potential to contribute to new structures over a wide proximodistal (PD) range. How a cellular population can alter contributions during growth and homeostasis to those during aggressive regeneration has not, to our knowledge, been examined in a regenerating appendage. Here, we compare such contributions in zebrafish fins and reveal how a normally restricted cellular subpopulation, fibroblasts that we identify as being associated with joints and expressing the marker tph1b, can broaden its PD contribution domains during appendage regeneration.
RESULTS AND DISCUSSION
Fibroblasts adjacent to fin joints express tph1b
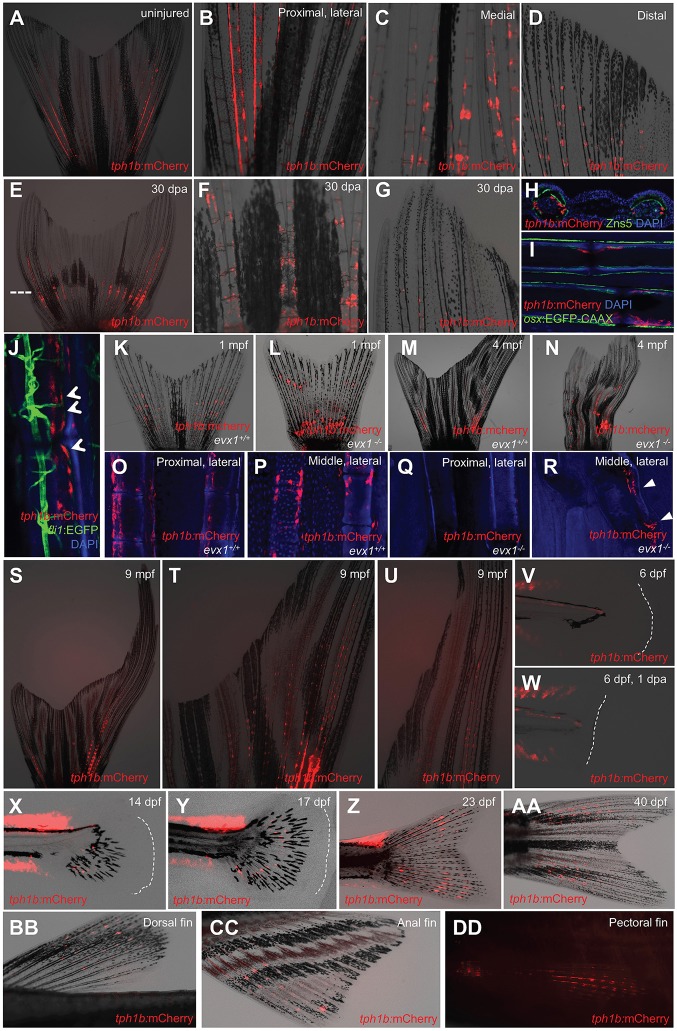
Recently, we found that tph1b is induced in medially located blastemal cells after zebrafish fin amputation (Tornini et al., 2016). Upon close inspection, we observed that tph1b:mCherry is also detectable in uninjured adult fins at much lower levels than during regeneration. Expression was localized near joints, with higher expression in more proximal joints and in lateral rays (Fig. 1A). Stronger tph1b:mCherry reporter fluorescence was detected on the outer boundaries of lateral hemirays in proximal segments, possibly as a locally induced response to higher levels of mechanical stress (Fig. 1B), and lower expression was detected near joints in medial rays (Fig. 1C). No tph1b+ cells were observed within two or three segments from the distal tip in any ray of the caudal fin (Fig. 1D). After 30 days of regeneration, expression driven by tph1b regulatory sequences again becomes restricted to joint regions and absent from distal ray segments (Fig. 1E-G). tph1b-driven fluorescence did not colocalize with osteoblast markers, but was limited to fibroblasts directly adjacent to osteoblasts in the intersegmental zones near joints (Fig. 1H,I). We did not observe colocalization of tph1b:mCherry with fluorescence from the endothelial cell reporter fli1:EGFP (Lawson and Weinstein, 2002), although tph1b+ cells were often adjacent to fli1+ endothelial cells (Fig. 1J).
Fig. 1.
tph1b+ fibroblasts localize to adult zebrafish fin joints. (A-D) tph1b:mCherry expression in uninjured adult zebrafish caudal fins. (A-D) Whole fin (A), and magnified proximal dorsal rays (B), medial rays (C) and distal mediolateral rays (D). tph1b:mCherry is expressed near fin joints. (E-G) tph1b:mCherry expression in whole (E), medial (F) or distal (G) caudal fins at 30 days post-amputation (dpa). Dashed line indicates the amputation plane. (H) Transverse section of uninjured tph1b:mCherry rays, co-stained with DsRed (red, tph1b:mCherry), ZNS-5 (osteoblasts, green) and DAPI (nuclei, blue). (I,J) Optical section through uninjured tph1b:mCherry; osx:EGFP-CAAX (I) or tph1b:mCherry; fli1:EGFP (J) fin ray, stained with DAPI (nuclei, blue). tph1b:mCherry-expressing cells do not colocalize with osteoblast markers or fli1:EGFP-expressing cells (arrowheads, J). (K-N) Wholemounted tph1b:mCherry caudal fins in wild-type (K,M) or jointless evx1−/− (L,N) background at 1 (K,L) or 4 (M,N) months post-fertilization (mpf). (O-R) Optical sections of tph1b:mCherry fin rays in a wild-type (O,P) or evx1−/− (Q,R) background, showing proximal (O,Q) or middle (P,R) lateral rays stained with DAPI (nuclei, blue). tph1b:mCherry localizes to proximal lateral rays and middle lateral rays near mature joints, but is absent in jointless fins, except for injury-induced expression (R, arrowheads). (S-U) tph1b:mCherry expression in the longfin (lof) background, shown at 9 mpf in lateral (T) and distal (U) ray tissues. (V,W) tph1b:mCherry expression at 6 dpf (V) or at 6 dpf, 1 dpa (W). Expression is not induced in larval finfold tissue after injury. Dashed lines indicate the tailfin boundary. (X-AA) tph1b:mCherry expression during caudal fin development at 14 (X), 17 (Y), 23 (Z) and 40 (AA) dpf. Dashed lines indicate fin boundary. (BB-DD) tph1b:mCherry expression in adult dorsal (BB), anal (CC) and pectoral (DD) fins, excluding distal segments.
evx1−/− mutants lack fin joints, yet their fin rays develop breaks in the absence of experimental injury, likely due to stress (Schulte et al., 2011). evx1−/− caudal fins express tph1b:mCherry near fin ray breaks (Fig. 1K-N), approximating the regions where joints would normally form (Fig. 1L,N). No tph1b:mCherry expression was detected in evx1−/− caudal fin rays other than at these breaks, confirming tph1b:mCherry as a marker of joint-associated cells in the absence of injury (Fig. 1O-R). These results suggest that tph1b regulatory elements are moderately activated in a small cellular population by mechanical stress at joints, and strongly activated across broad tissue regions after major injury. A different mutant, long fin (lof), adds segments to the distal tips at a rate disproportionate to animal growth (Iovine and Johnson, 2000). tph1b:mCherry reporter expression was similar in lof and wild-type caudal fins, including exclusion from distal ray segments (Fig. 1S-U).
We did not observe expression in developing larval fin folds (Fig. 1V), first detecting expression around 14 days post-fertilization (dpf) near developing joints (Fig. 1X-AA). We also did not detect tph1b:mCherry induction after resection of the larval fin fold at 5 dpf (Fig. 1W), indicating that tph1b regulatory elements are preferentially active in joints or regenerating tissue of rayed fin structures. Other adult fins also showed expression near joints, again excluding the distal-most segments (Fig. 1BB-DD). These findings identify an apparent subpopulation of joint-associated tph1b+ fibroblasts that localize near osteoblasts and vasculature.
Tph1b synthesizes serotonin in blastemal cells and fibroblasts but is dispensable for fin regeneration
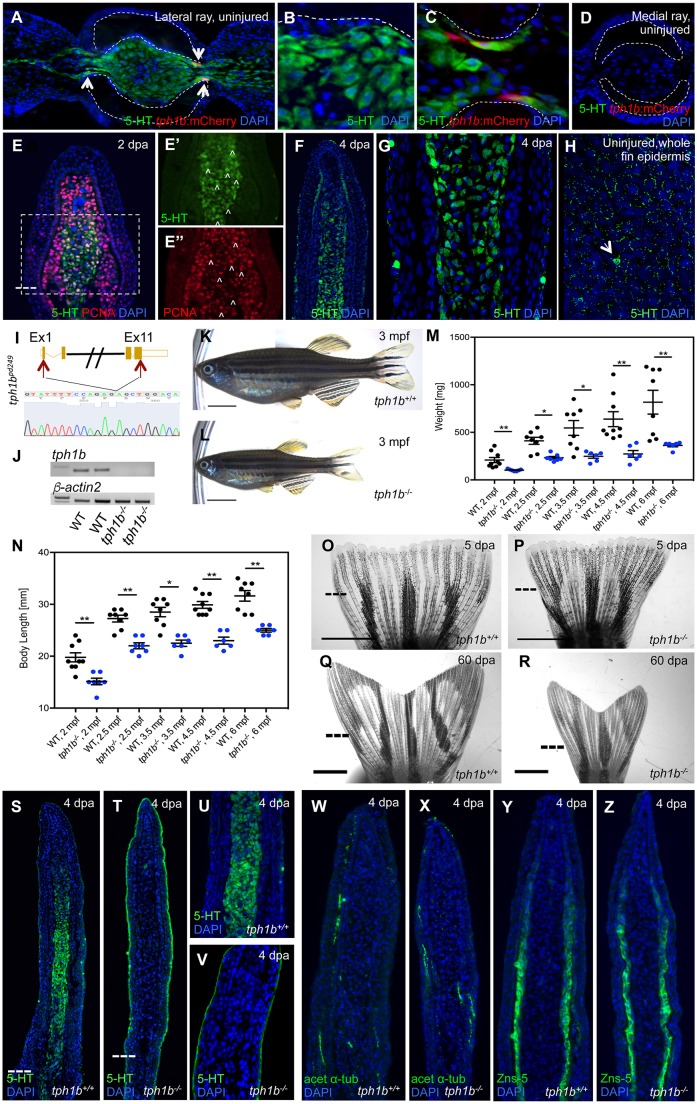
tph1b encodes a tryptophan hydroxylase, the rate-limiting enzyme in peripheral serotonin synthesis. Serotonin signaling is evolutionarily conserved and performs numerous roles across the animal kingdom (Berger et al., 2009; Curran and Chalasani, 2012). It has also been reported to modulate tissue regeneration in vertebrate contexts (Lesurtel et al., 2006; Barreiro-Iglesias et al., 2015). In uninjured caudal fins, serotonin was present in mesenchymal cells of lateral but not medial rays (Fig. 2A-D). During fin regeneration, intracellular serotonin was conspicuous in proliferating and non-proliferating blastema cells at 2 dpa and fibroblasts of 4 dpa regenerating rays (Fig. 2E,F), and occasionally observed in distal basal epithelial cells. Vesicular serotonin was observed in superficial epithelial cells of uninjured and regenerating fin epidermis (Fig. 2F-H). These data indicate that serotonin production is a stress-induced signal in zebrafish fin fibroblasts.
Fig. 2.
Tph1b is dispensable for fin regeneration but important for organismal growth. (A-D) Transverse section of lateral (A-C) or medial (D) rays of uninjured tph1b:mCherry caudal fins, stained for serotonin (5-HT, green) and nuclei (DAPI, blue). (B) Intracellular serotonin expression in fibroblasts. (C) Hemiray region shows tph1b+ cells (arrows) occasionally colocalized with serotonin staining. (D) Weak tph1b:mCherry expression, and no serotonin expression, in medial rays. Dashed lines outline the hemiray. (E) Longitudinal section of a 2 dpa blastema, stained for serotonin (5-HT, green), proliferating cells (PCNA, red) and nuclei (DAPI, blue). Serotonin is present in blastemal progenitors. Dashed line indicates the amputation plane. Separate channels for 5-HT (E′, green) and PCNA (E″, red) in the boxed area in E. Arrowheads indicate cells co-labeling for 5-HT and PCNA. Dashed line indicates the amputation plane. (F,G) Longitudinal sections of a 4 dpa blastema, stained for serotonin (5-HT, green) and nuclei (DAPI, blue). (G) Intracellular serotonin synthesis in fibroblasts and vesicular serotonin in some superficial epidermal cells. (F) 5-HT staining was occasionally detected in distal basal epithelial cells. (H) Optical section of uninjured whole caudal fin epidermis, stained for serotonin (5-HT, green) and nuclei (DAPI, blue). Stronger expression (arrow) in putative gland cells. (I) Schematic of the tph1b gene sequence, sgRNA target sites (red arrows, top) and chromatogram verification of tph1b sequence deletion (bottom). (J) RT-PCR for tph1b or β-actin2 on 3 dpa fins from tph1b−/− or wild-type siblings. tph1b mRNA (exons 6-7) is absent in tph1b−/− mutant fins. n=3 for each group. (K,L) Three mpf wild-type (J) or tph1b−/− (K) zebrafish. tph1b−/− fish are significantly smaller than wild-type siblings. At least four separate clutches were analyzed. (M,N) Mass (mg) (M) or body length (mm) (N) of wild type (black) or tph1b mutants (tph1b−/−, blue) (data are mean±s.e.m.). *P<0.01, **P<0.001; Mann–Whitney two-tailed test, n=6-9 per group. (O-R) Bright-field images of fins at 5 (O,P) or 60 dpa (Q,R). Wild-type (O,Q) and tph1b−/− (P,R) fins regenerate normally, despite differences in body size. Dashed lines indicate the amputation plane. (S-V) Longitudinal sections of wild-type (S) or tph1b−/− (T) fins at 4 dpa, stained for serotonin (5-HT, green) and nuclei (DAPI, blue). Higher magnification views are shown in U (tph1b+/+) and V (tph1b−/−). Serotonin synthesis is abrogated in tph1b−/− regenerating fins; epidermal 5-HT is still detected. Dashed lines indicate the amputation plane. (W-Z) Longitudinal sections of wild-type (V,Y) or tph1b−/− (W,Z) fin rays at 4 dpa, stained for osteoblasts (ZNS-5, green) (W,X) or nerves (acetylated α-tubulin, green) (Y,Z) and nuclei (DAPI, blue). tph1b−/− fins display normal osteoblast and nerve marker expression. Scale bars: 5 mm in K,L; 2 mm in O-R.
To determine functions of Tph1b during fin regeneration, we generated a tph1b mutant allele (tph1bpd249) that lacks 8.3 kb of the tph1b genomic sequence and all coding sequence, and is a presumed null mutant (Fig. 2I). RT-PCR analysis detected no tph1b mRNA for exons 6-7 in mutant fin regenerates (Fig. 2J). tph1b−/− mutants developed normal fins but were on average 20% shorter and weighed 55% less than wild-type siblings (Fig. 2K-N). Size differences were evident at juvenile stages and persisted through adulthood (Fig. 2M,N). These results are consistent with the reported effects of serotonin deficiency on growth in fruitflies (Neckameyer et al., 2007), nematodes (Loer and Kenyon, 1993; Srinivasan et al., 2008; Sze et al., 2000; Waggoner et al., 1998) and mice (Cote et al., 2003). tph1b−/− fins regenerated grossly normally compared with wild-type siblings when assessed at 5 and 60 dpa (Fig. 2O-R). The mesenchymal compartment of regenerating tph1b−/− fins lacked a detectable serotonin synthesis response (Fig. 2S-V), although epidermal serotonin remained detectable. Mutant regenerates displayed grossly normal osteoblast alignment and innervation (Fig. 2W-Z).
Taken together, these results indicate that Tph1b locally synthesizes serotonin in blastemal fibroblast progenitors, but that tph1b and its serotonin production is dispensable for normal fin regeneration. Growth stunting in tph1b mutants indicate that this source of serotonin has other essential physiological roles.
Expansion of contributions by tph1b+ joint fibroblasts following amputation injury
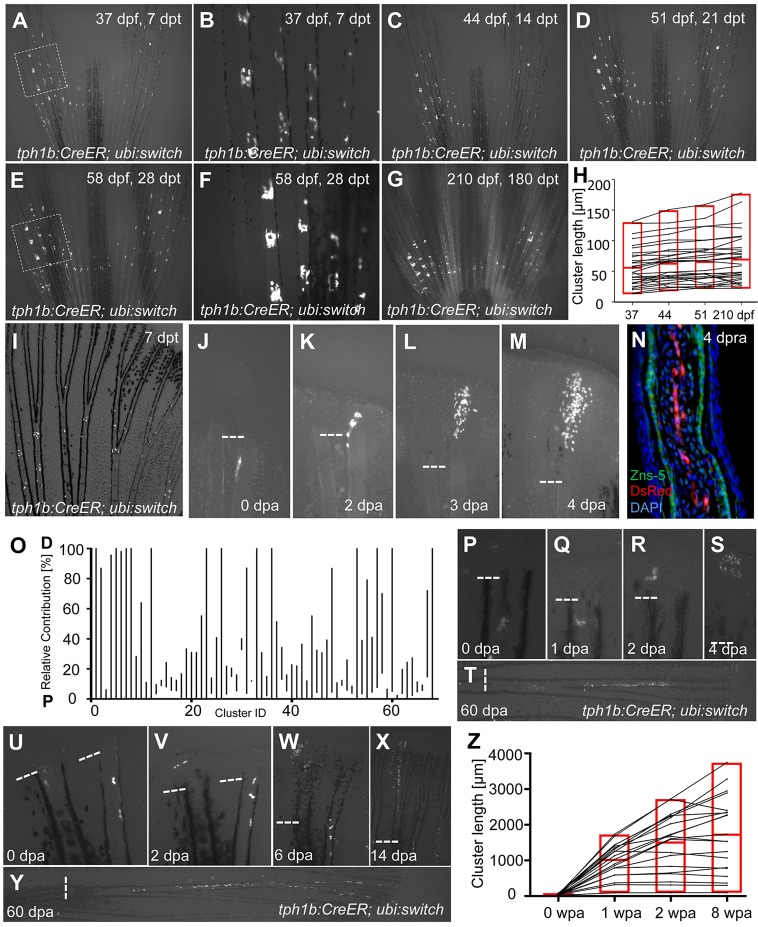
To assess developmental contributions by joint-associated tph1b+ cells to growth and tissue homeostasis, we crossed tph1b:CreER animals (Tornini et al., 2016) with a ubi:switch reporter line that, without Cre-mediated recombination, reports EGFP in most cells (Mosimann et al., 2011). No leaky recombination was detected during development or ontogeny (Tornini et al., 2016). To examine contributions of tph1b-expressing cells in growing juvenile animals, we induced recombination tph1b:CreER; ubi:switch fish at 30 dpf, labeling cells near developing joints by 7 days post-tamoxifen treatment (dpt) (Fig. 3A,B). These cells remained labeled through development and into adulthood, even at 6 months post-labeling (Fig. 3C-G). During this period, we did not detect contributions to more distal segments or beyond regions of tissue between joints (Fig. 3H). These data indicate that joint-associated tph1b+ fibroblasts contribute specifically to peri-joint tissue during normal growth and homeostatic conditions. We speculate that these cells may provide structural support to fins and joints, as they have low proliferative activity, remain in the same location for 6 months or more and are not detectable from jointless fins, which do not have structural support such as ligaments.
Fig. 3.
tph1b+ fibroblasts contribute to peri-joint regions in the absence of injury but broaden contributions during regeneration. (A-G) tph1b:CreER; ubi:switch fish were treated with 4 µM tamoxifen at 30 dpf, and labeled cells in caudal fins were tracked at 7 (A,B), 14 (C), 21 (D), 28 (E,F) and 180 (G) days post-tamoxifen treatment (dpt). The boxed areas in A and E are magnified in B and F, respectively, showing limited contributions of tph1b+ cells and progeny to local joint tissue throughout adulthood. (H) Plot of tph1b+ proximodistal (PD) cluster lengths (in µm) during normal growth measured at 37, 44, 51 and 210 dpf. tph1b:CreER; ubi:switch fish were treated as in A. n=31 clusters. Labeled cells remained closely associated to joints. Red box indicates minimum/maximum values, with the average value in the middle. (I) An uninjured adult tph1b:CreER; ubi:switch caudal fin, treated with 4 µM tamoxifen and imaged at 7 dpt. tph1b+ cells near joints are labeled. (J-M) Contribution of a labeled tph1b+ cell in tph1b:CreER; ubi:switch fish, shown at 0 (J), 2 (K), 3 (L) and 4 (M) dpa. Dashed line indicates amputation plane. (N) Longitudinal section of a tph1b:CreER; ubi:switch fin regenerate at 4 days post-re-amputation (dpra), stained for osteoblasts (ZNS-5, green), mCherry (DsRed, red) and nuclei (DAPI, blue). We saw no evidence for contributions by tph1b+ joint-associated cells to osteoblasts during regeneration. (O) Plot of final relative position along the PD axis in which tph1b+ cell progeny are distributed, normalized for regenerated ray length as a percentage (0, proximal; 100, distal). Fins were amputated near labeled tph1b+ cells and progeny were tracked through 60 dpa. n=68. P, proximal; D, distal. (P-T) Contribution of a tph1b+ labeled cell in tph1b:CreER; ubi:switch fish, shown at 0 (P), 1 (Q), 2 (R), 4 (S) and 60 (T) dpa. Joint-associated tph1b+ cells can contribute progeny to regenerated tissue well beyond joint areas. (U-Y) Contribution of a tph1b+ labeled cell in tph1b:CreER; ubi:switch fish (left ray), shown at 0 (U), 2 (V), 6 (W), 14 (X) and 60 (Y) dpa. Joint-associated tph1b+ cells can contribute progeny across the entire PD axis of the regenerate. (Z) Plot of tph1b+ progeny lengths (in µm) during fin regeneration. Adult tph1b:CreER; ubi:switch fish were treated as in I. At 7 dpt, fins were amputated distal to labeled cells, and progeny lengths were measured at 0, 1, 2 and 8 weeks post-amputation (wpa). n=20. Labeled cells significantly expanded their contributions.
To test how an amputation injury affects this restricted cell type, we first induced recombination in uninjured tph1b:CreER; ubi:switch transgenic fish at adult stages, labeling peri-joint cells as expected (Fig. 3I). At 7 dpt, we amputated tph1b:CreER; ubi:switch zebrafish fins just distal to the labeled cells. Labeled progeny were then tracked and imaged daily from 0 to 4 dpa, then at 6, 8, 10, 14 and 60 dpa. We longitudinally tracked 68 progeny clusters.
Following amputation, tph1b+ joint fibroblasts were recruited to help establish the blastema. Recruitment occurred if cells were within one segment of the amputation plane (∼250 μm), and was observed in 68 out of 85 (or 80%) amputated rays with labeled cells within the recruitment zone. Labeled cells were present in all regions of the 2 dpa blastema and the early (4 dpa) regenerate (Fig. 3J-M). We saw no evidence that tph1b+ joint cells contributed osteoblasts to regenerated structures (Fig. 3N). We calculated labeled cell cluster lengths and relative proximodistal (PD) contributions during regeneration at 0, 1, 2 and 8 weeks post-amputation (wpa), respectively (Fig. 3Z; compare to Fig. 3H). Whereas progeny were restricted in both localization and expansion during growth and homeostasis, the ultimate relative PD contributions of tph1b+ cells during regeneration were profoundly heterogeneous (Fig. 3O), with tph1b+ fibroblasts contributing prominently to inter-joint regions (Fig. 3P-T) and along the entire PD axis of the regenerating fin ray, including distal segments (Fig. 3U-Y). Strikingly, whereas labeled tph1b+ cell clusters expanded by an average of only 1.3 times in the PD axis between 37 dpf to 7 mpf, cell clusters in fin regenerates expanded by an average of 50.8 times (Fig. 3H,Z). Thus, upon amputation, tph1b+ cells broaden their range of contributions from their restricted localization near joints to the entire length of the regenerated fin.
To assess potentials of tph1b+ cells associated with spontaneous injury in the evx1−/− background, we permanently labeled a small number of cells in these areas using tph1b:CreER; ubi:switch transgenes and tamoxifen treatment (Fig. S1). Upon amputation, these labeled cells showed a similar ability to joint-associated and blastemal tph1b+ cells to contribute to regenerating tissue (Fig. S1B,C). Finally, to assess a requirement for tph1b+ joint cells, we generated transgenic tph1b:mCherry-Nitroreductase (tph1b:mCherry-NTRpd275) zebrafish that enable metronidazole (Mtz)-induced genetic cell ablation. Upon Mtz treatment for 3 days, mCherry fluorescence intensity was reduced by an average of 87%, indicating strong ablation of tph1b+ joint-associated cells (Fig. S1D,E). We amputated fins at 3 days post-treatment (dpt) and tracked regeneration for 7 days, noting no gross difference in regeneration between vehicle- and Mtz-treated fins (Fig. S1F,G). This suggests that tph1b+ joint cell progeny, like the tph1b gene product, are dispensable for normal fin regeneration and that plasticity in the system compensates for their lost contributions.
Conclusions
Here, we have identified a fibroblast subpopulation in zebrafish fins that is tightly associated with proximal fin joints throughout life. Upon injury, tph1b+ fibroblasts associated with joints help assemble the regeneration blastema, after which their developmental potentials expand to include tph1b− inter-joint fibroblasts along the entire PD axis. Our results do not dispute the dogma of lineage restriction as the primary mechanism of cellular contributions to fin regeneration; i.e. fibroblasts do not appear to create osteoblasts, or vice versa. However, they argue that developmental potentials of contributing cells can change at a granular level. Previous studies in zebrafish fins have either permanently labeled cells randomly during embryonic development (Tu and Johnson, 2011) or labeled marker-specific populations of cells that are spatially unrestricted (Knopf et al., 2011; Singh et al., 2012), masking the contributions of spatially restricted subpopulations. The results we describe identify and characterize a regional fibroblast population that is co-opted to help reconstitute the broader fibroblast population. How the blastema alters the developmental paths of resident cells through molecular signaling remains a principal outstanding question for the field.
MATERIALS AND METHODS
Zebrafish maintenance and procedures
Wild-type, evx1i232 mutant (Schulte et al., 2011), long fin (lof)d2 mutant (Iovine and Johnson, 2000), tph1bpd249 mutant, tph1b:mCherryens700 (Tornini et al., 2016), ubi:switchcz1701 (Mosimann et al., 2011), tph1b:CreERpd250 (Tornini et al., 2016) or tph1b:mCherry-NTRpd275 (J.D.T. and K.D.P., unpublished) transgenic zebrafish of the outbred Ekkwill (EK) strain of both sexes were used for all experiments, ranging in age from 5 days to 12 months (as specified). Water temperature for animals was maintained at 26°C. Zebrafish were anaesthetized with phenoxyethanol, and caudal fins were amputated using razor blades. Body-length measurements were acquired from the tip of the jaw to the caudal peduncle. Except where noted, at least five fish were used per experiment. Experiments with zebrafish were approved by the Institutional Animal Care and Use Committee at Duke University, NC, USA.
Generation of tph1b mutant zebrafish
tph1bpd249 mutants were generated using CRISPR/Cas9 technology. Briefly, sgRNA was transcribed in vitro from a template using the MEGAscript T7 Transcription Kit (Thermo Scientific). The 5′ non-coding sequence was targeted using oligo 5′-gcgTAATACGACTCACTATAGGGGGAGGAGCACGCGCCTCGTTTTAGAGCTAGAAATAGC-3′ (target sequence underlined). Exon 11 was targeted using oligo 5′-gcgTAATACGACTCACTATAGGAGGAGCTCAGACACGAGCGTTTTAGAGCTAGAAATAGC-3′ (target sequence underlined). One-cell stage embryos were co-injected with sgRNAs (25 ng/μl) and Cas9 protein (0.5 μg/μl). Deletion lines were screened from dorsal fin DNA for presence of a ∼850 bp band using primers: tph1b_del_F, 5′-AATGTACAGGCCCTCAAACA-3′; and tph1b_del_R, 5′-AAGACAATCTCTAGACAAGG-3′. Homozygous mutants were screened for absence of a 455 bp amplicon of exon 2 using primers: tph1b_ex2_F, 5′-TCCAACTTTTTCCAATGGATG-3′; and tph1b_ex2_R, 5′-TCAAAATACAAGTAACCGCAACA-3′. RT-PCR for tph1b was performed on cDNA from 3 dpa tph1b mutant or wild-type sibling fins (n=3) using primers: tph1b_F, 5′-TACCTGCAGAACCTGCCTCT-3′; and tph1b_R, 5′-GCCAAAAAGTCTCTGGGTGA-3′ to yield an expected 172 bp amplicon. Controls primers used for β-actin2 are: Bact_F, 5′-TGATGAGGCTCAGAGCAAGA-3′; and Bact_R, 5′-CACAATCCACACTTCCATGC-3′.
Histology
Immunohistochemistry on cryosections of 4% paraformaldehyde-fixed fins was performed as previously described (Nachtrab et al., 2013), using an antibody against DsRed (rabbit, 632496, Clontech, 1:500), 5-HT (rabbit, ImmunoStar, 1:5000) or ZNS-5 (mouse, ZIRC, 1:200). Staining for serotonin with an anti-5-HT antibody was performed with 0.3% Triton X-100 in 1× PBS at 4°C overnight. Imaging of 12 μm tissue sections was carried out using a Leica DM6000B compound microscope or Zeiss LSM 700 confocal microscope.
Fate mapping
Adult tph1b:CreER (Tornini et al., 2016) were crossed to ubi:switch fish (Mosimann et al., 2011) and were screened for ubiquitous EGFP fluorescence as larvae and for strong EGFP+ lens fluorescence (for the tph1b:CreER transgene) as juveniles. To permanently label tph1b+ cells and their progeny, fish were treated with tamoxifen at specified concentrations and timepoints for 6-12 h. Caudal fins were amputated near labeled cells (evidenced as small clusters of about one to four cells), typically removing 50-60% of the tissue. Imaging was performed as described using a Zeiss AxioZoom microscope. No animal or sample was excluded unless the animal died during experiments, transgenic expression within a cluster was silenced or additional recombination events were evident. Clutchmates were randomized into different groups of animals and maintained at a constant aquarium density of 12-15 fish per 3 l for experiments, except for experiments under optimal conditions (6-9 fish per 3 l). All raw images were processed using FIJI software, and images were organized by individual fish identifiers to track respective cell clusters over time. Raw images were segmented for cluster position, amputation plane and distal plane, and measurements were extracted from raw data in FIJI software. Plots were generated using GraphPad Prism software.
Acknowledgements
We thank M. Kathryn Iovine for sharing evx1−/− mutant fish; Marika Kapsimali for sharing tph1b:mCherry transgenic fish; Alberto Puliafito for help with quantitative analyses; Junsu Kang for comments on the manuscript; and Jim Burris, Shane Miller, Brandylyn Thomas, Nicolette Mariano, Tess Thoren and Torin Laffredo for fish care.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.D.P., V.A.T.; Methodology: V.A.T., J.D.T.; Formal analysis: V.A.T., J.D.T., R.L.A.; Investigation: V.A.T., J.D.T., R.L.A.; Resources: K.D.P.; Writing - original draft: V.A.T.; Writing - review & editing: K.D.P., V.A.T.; Supervision: K.D.P.; Project administration: K.D.P.; Funding acquisition: K.D.P.
Funding
This work was supported by Graduate Research Fellowships from the National Science Foundation (DGF 1106401 to V.A.T. and R.L.A.) and by a grant from the National Institutes of Health (R01 GM074057 to K.D.P.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.155655.supplemental
References
- Barreiro-Iglesias A., Mysiak K. S., Scott A. L., Reimer M. M., Yang Y., Becker C. G. and Becker T. (2015). Serotonin promotes development and regeneration of spinal motor neurons in zebrafish. Cell Rep. 13, 924-932. 10.1016/j.celrep.2015.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M., Gray J. A. and Roth B. L. (2009). The expanded biology of serotonin. Annu. Rev. Med. 60, 355-366. 10.1146/annurev.med.60.042307.110802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote F., Thevenot E., Fligny C., Fromes Y., Darmon M., Ripoche M.-A., Bayard E., Hanoun N., Saurini F., Lechat P. et al. (2003). Disruption of the nonneuronal tph1 gene demonstrates the importance of peripheral serotonin in cardiac function. Proc. Natl. Acad. Sci. USA 100, 13525-13530. 10.1073/pnas.2233056100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran K. P. and Chalasani S. H. (2012). Serotonin circuits and anxiety: what can invertebrates teach us? Invert. Neurosci. 12, 81-92. 10.1007/s10158-012-0140-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine M. K. and Johnson S. L. (2000). Genetic analysis of isometric growth control mechanisms in the zebrafish caudal Fin. Genetics 155, 1321-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf F., Hammond C., Chekuru A., Kurth T., Hans S., Weber C. W., Mahatma G., Fisher S., Brand M., Schulte-Merker S. et al. (2011). Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 20, 713-724. 10.1016/j.devcel.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Kragl M., Knapp D., Nacu E., Khattak S., Maden M., Epperlein H. H. and Tanaka E. M. (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60-65. 10.1038/nature08152 [DOI] [PubMed] [Google Scholar]
- Lawson N. D. and Weinstein B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307-318. 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- Lehoczky J. A., Robert B. and Tabin C. J. (2011). Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc. Natl. Acad. Sci. USA 108, 20609-20614. 10.1073/pnas.1118017108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesurtel M., Graf R., Aleil B., Walther D. J., Tian Y., Jochum W., Gachet C., Bader M. and Clavien P. A. (2006). Platelet-derived serotonin mediates liver regeneration. Science 312, 104-107. 10.1126/science.1123842 [DOI] [PubMed] [Google Scholar]
- Loer C. M. and Kenyon C. J. (1993). Serotonin-deficient mutants and male mating behavior in the nematode Caenorhabditis elegans. J. Neurosci. 13, 5407-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C., Kaufman C. K., Li P., Pugach E. K., Tamplin O. J. and Zon L. I. (2011). Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169-177. 10.1242/dev.059345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtrab G., Kikuchi K., Tornini V. A. and Poss K. D. (2013). Transcriptional components of anteroposterior positional information during zebrafish fin regeneration. Development 140, 3754-3764. 10.1242/dev.098798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckameyer W. S., Coleman C. M., Eadie S. and Goodwin S. F. (2007). Compartmentalization of neuronal and peripheral serotonin synthesis in Drosophila melanogaster. Genes Brain Behav. 6, 756-769. 10.1111/j.1601-183X.2007.00307.x [DOI] [PubMed] [Google Scholar]
- Rinkevich Y., Lindau P., Ueno H., Longaker M. T. and Weissman I. L. (2011). Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature 476, 409-413. 10.1038/nature10346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte C. J., Allen C., England S. J., Juárez-Morales J. L. and Lewis K. E. (2011). Evx1 is required for joint formation in zebrafish fin dermoskeleton. Dev. Dyn. 240, 1240-1248. 10.1002/dvdy.22534 [DOI] [PubMed] [Google Scholar]
- Singh S. P., Holdway J. E. and Poss K. D. (2012). Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev. Cell 22, 879-886. 10.1016/j.devcel.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Sadegh L., Elle I. C., Christensen A. G. L., Faergeman N. J. and Ashrafi K. (2008). Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab. 7, 533-544. 10.1016/j.cmet.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S. and Stankunas K. (2012). Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev. Biol. 365, 339-349. 10.1016/j.ydbio.2012.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze J. Y., Victor M., Loer C., Shi Y. and Ruvkun G. (2000). Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403, 560-564. 10.1038/35000609 [DOI] [PubMed] [Google Scholar]
- Tornini V. A. and Poss K. D. (2014). Keeping at arm's length during regeneration. Dev. Cell 29, 139-145. 10.1016/j.devcel.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornini V. A., Puliafito A., Slota L. A., Thompson J. D., Nachtrab G., Kaushik A.-L., Kapsimali M., Primo L., Di Talia S. and Poss K. D. (2016). Live monitoring of blastemal cell contributions during appendage regeneration. Curr. Biol. 26, 2981-2991. 10.1016/j.cub.2016.08.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S. and Johnson S. L. (2011). Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell 20, 725-732. 10.1016/j.devcel.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner L. E., Zhou G. T., Schafer R. W. and Schafer W. R. (1998). Control of alternative behavioral states by serotonin in Caenorhabditis elegans. Neuron 21, 203-214. 10.1016/S0896-6273(00)80527-9 [DOI] [PubMed] [Google Scholar]