Abstract
Coronary Artery Disease (CAD) is not only the most common form of heart disease, but also the leading cause of death in both men and women[1]. We present a system that is able to automatically predict whether patients develop coronary artery disease based on their narrative medical histories, i.e., clinical free text. Although the free text in medical records has been used in several studies for identifying risk factors of coronary artery disease, to the best of our knowledge our work marks the first attempt at automatically predicting development of CAD. We tackle this task on a small corpus of diabetic patients. The size of this corpus makes it important to limit the number of features in order to avoid overfitting. We propose an ontology-guided approach to feature extraction, and compare it with two classic feature selection techniques. Our system achieves state-of-the-art performance of 77.4% F1 score.
Graphical abstract
1. Introduction
Coronary Artery Disease (CAD) is not only the most common form of heart disease, but also the leading cause of death in both men and women[1]. The second track of the 2014 i2b2/UTHealth challenge targeted the automatic identification of risk factors for CAD: a complex clinical NLP task which could benefit from concept extraction, assertion classification, diagnosis extraction, medication extraction, smoking history, and family history[2].
Free text is considered to be a rich source of information for purposes of health care operations and research[3]. Furthermore, there have been several recent studies demonstrating the effectiveness of natural language processing (NLP) and machine learning methods for disease detection using clinical free text, which are discussed as related works in this paper. Thus, our aim is to develop a model for automatically predicting development of CAD from clinical free text.
We study the 2014 i2b2 Heart Disease Risk Factors Challenge Data[24] from a different perspective. Our purpose is to develop a system that automatically predicts patients who develop CAD based on their narrative medical histories before a diagnosis of CAD. For this purpose, we examine common risk factors for CAD—these risk factors consist of many of the same known risk factors for type-2 diabetes. They include high cholesterol, high-blood pressure, obesity, lack of physical activity, unhealthy diet, and stress[25]. As all patients in the corpus have diabetes, they are all at high risk for CAD and carry a lot of the same overall risk factors. This makes it challenging to separate the patients who actually develop the disease from those who do not. Additionally, solving this task on a small corpus requires special attention to overfitting. Our hypothesis is that it is possible to predict whether patients will develop CAD using a domain ontology to reduce the high dimensional nature of free text medical records.
Our approach to CAD prediction is unique in that we examine unstructured data (i.e., clinical free text in patients’ electronic medical records (EMRs)) to predict which patients will develop a CAD diagnosis in the future. This is a natural language processing (NLP) and machine learning task. We believe that our system can complement, supplement and even provide a second perspective to existing CAD models that use only non-textual, structured data for predicting the disease[23]. As part of the original 2014 i2b2/UTHealth challenge, several teams developed systems with the goal of identifying risk factors for heart disease[4–22]. However, to the best of our knowledge our work marks the first attempt at automatically predicting development of CAD using free text in medical records.
We approach the CAD prediction task as a document classification problem. This means that we treat each record as one sample, independent of any previous or future sample (i.e., we disregard the longitudinal nature of the data). We simply classify if given one patient record at a discrete point in time that patient will eventually develop (or not develop) a CAD diagnosis. To improve classifier performance, we propose an ontology-guided approach to feature extraction and compare this with two standard feature selection techniques. Specifically, our novel feature extraction technique automatically filters out features based on domain knowledge in the Unified Medical Language System (UMLS).
The clinical application of our model is in classification of patients who will develop CAD in difficult-to-discriminate situations; e.g., when patients are all at high risk for CAD and carry many of the risk factors.
Related works
There is a well established volume of research outlining natural language processing and machine learning methods for disease classification in clinical free text. Pineda et al. applied the pipeline-based NLP tool Topaz to extract 31 UMLS concept unique identifier (CUI) features for classification of influenza in emergency department free-text reports[26]. The team compared seven different classifiers to an expert-built Bayesian classifier and achieved a 93% F1 score.
Similar methods have been applied to detect thromboembolic disease in free-text radiology reports[27]. Specifically, Pham et al. developed a system that pre-processed documents using a simple sentence segmenter and tokenizer. They created a lexicon to define concept types, which were incorporated into the feature space along with filtered unigrams and bigrams. They then experimented with Weka to train Support Vector Machine (SVM) and Maximum Entropy (MaxEnt) classifiers, of which the MaxEnt classifier achieved the highest F1 score of 98%.
Furthermore, Redd et al. developed a set of retrieval criteria for identifying patients at risk for scleroderma renal crisis in electronic medical records[28]. The team developed their NLP system using data from the Veterans Informatics and Computing Infrastructure (VINCI). Their concept extraction criteria included specific disease and symptom mentions related to systemic sclerosis (SSc). The group then trained an SVM classifier to detect documents that indicated a diagnosis of SSc and reported an F1 score of 87.3%.
Several teams have experimented with domain ontologies to guide feature extraction for text classification. Wang et al., e.g., established a concept hierarchy by mapping raw terms to medical concepts using the UMLS, which they then searched to obtain the optimal concept set[29]. This feature selection technique improved the overall accuracy of their text classification system as compared with Principal Component Analysis (PCA) for dimensionality reduction.
Additionally, Garla and Brand exploited the UMLS ontology during feature engineering to improve the performance of machine-learning-based classifiers trained on the 2008 i2b2 Obesity Challenge Data Set[30]. This data set includes 15 diseases, including CAD, and its classification based on one narrative record per patient. To enhance feature ranking for this task, Garla and Brand propagated contingency tables of concepts in UMLS to their hypernyms, which they refer to as the propagated information gain. They then assigned each concept the highest propagated information of any hypernym. The use of this technique yielded the greatest performance improvement for their system, however, it did not improve performance on the classification of CAD.
For predicting CAD before it develops, we experimented with Naive Bayes, SVM, and MaxEnt classifiers and tested dimensionality reduction techniques including PCA, mutual information, and domain ontology-guided feature extraction. Our hypothesis is that the medical concepts most relevant to predicting CAD have formally defined relationships as such in the UMLS Semantic Network that can be exploited to automatically predict the disease. By engineering features around these concepts, as opposed to constructing features for every possible concept in our documents, we focus our feature space on information that really matters for our task. The reduction in the feature, in turn, results in simpler and more robust models, that run without any significant loss of performance.
2. Data
The 2014 i2b2 Heart Disease Risk Factors Challenge data set consists of 1,304 longitudinal records of a total of 296 diabetic patients. Each patient in the corpus belongs to one of three cohorts:
patients who had a CAD diagnosis in the first record of their patient profile
patients who developed a CAD diagnosis sometime later in their patient profile
patients who did not develop a CAD diagnosis
The criteria for classifying CAD and no-CAD patients in our study has been defined and validated in two earlier studies[31],[3]. To create the corpus for the 2014 i2b2 Heart Disease Risk Factors Challenge, an expert cardiologist developed the definition for CAD. Specifically, the following search criteria were used against Partners HealthCare Electronic Medical Records (EMR)[31]:
at least 3 CAD codes or 1 procedure code for a coronary revascularization
at least 4 codified mentions of beta-adrenergic inhibitor medications
at least 4 codified mentions of anti-platelet agents (such as aspirin)
at least 4 codified mentions of statins (cholesterol lowering drugs)
For the purposes of our study, we focused on prediction of CAD before the patients were officially diagnosed, i.e., they had an annotated CAD diagnosis in the i2b2/UTHealth data. We therefore discarded from the data any records with a CAD diagnosis. This removed all patients who were diagnosed with CAD at the onset of their patient profile. Additionally, for patients who later received a CAD diagnosis, records were discarded beginning with the one in which the patient received the diagnosis. This left us with the records of the patients who did not develop CAD (referred to as no-CAD patients), and the records from those who do develop CAD before their diagnosis (referred to as CAD patients).
After discarding all records with diagnosis of CAD, we checked our CAD and no-CAD patients with respect to their level of sickness. To achieve this, we calculated normalized frequencies of the number of symptoms, diseases and medications extracted by cTAKES in each record and divided by the length of the document (i.e., the number of word tokens per record), as shown in Equation 1. Intuitively, this calculation measures the disease density of the record.
We found that the no-CAD patients were depicted as being sicker in their records than the CAD patients as described by their pre-CAD diagnosis records. We decided to control this factor so that we could make the machine learning models agnostic with respect to the level of sickness in the two populations. Thus, we matched a random subsample of records in the no-CAD patient population to levels of sickness in CAD patients (See Appendix A Figure A.1).
Using this experimental setup for the CAD prediction task resulted in 215 total patients and 516 total patient records (See Table 1) from the training and test data of the 2014 i2b2/UTHealth challenge. An analysis of the resulting corpus is provided in Appendix A Table A.1. Note that given our intent to solve a different task than the original 2014 i2b2/UTHealth shared task, the training and test data of the shared task can be merged and methods can be cross-validated.
Table 1.
Analysis of numbers of patients and number of records in the corpus.
| Patients | Records | ||||
|---|---|---|---|---|---|
| # of patients in corpus | CAD | No CAD | # of records in corpus | CAD | No CAD |
| Total | 183 | 113 | Total | 813 | 491 |
| Unused | *72 | **9 | Unused | *555 | **233 |
| Used | 111 | 104 | Used | 258 | 258 |
| Total patients used | Total records used | ||||
| 215 | 516 | ||||
Unused patients and records contain CAD diagnoses, which must be omitted for the disease prediction task.
Unused patients and records not selected during random assignment, controlled by level of patient sickness.
In our subsampling, we also analyzed coverage (i.e., the duration of longitudinal patient records) to avoid bias. For CAD patients, the average longitudinal patient history covers 12.9 months (i.e, 1.075 years), while the longest duration between first and last record for any CAD patient is 76 months (i.e., 6.3 years). The average longitudinal patient history for no-CAD patients is 18.3 months (i.e., 1.525 years), while the longest duration between first and last record for any no-CAD patient is 91 months (i.e, 7.6 years). We concluded that coverage was comparable in the two populations using the z-test (α0.05).
3. Methods
Our system processes records using Apache cTAKES, an NLP system for extracting information from clinical free-text[32]. We compared a feature extraction filter to dimensionality reduction techniques including PCA[33] and mutual information[34] using three different classifiers: (1) Naive Bayes, (2) MaxEnt, and (3) SVM. To compare performances between models, we used approximate randomization testing[35,36,37]. We tested significance over micro-average precision, recall and F1, with N= 9,999 and α0.1.
3.1. Feature Extraction
cTAKES (with the Clinical Documents pipeline[38]) performs sentence detection, part-of-speech (POS) tagging, chunking, named entity recognition, context detection, and negation detection. We use it to extract tokens, POS tags, and medical concepts. Rather than using the original tokens as they appear in the text, we use lemmas. We remove English stop words before constructing bigrams using the Natural Language Toolkit (NLTK)[39].
We extract Concept Unique Identifiers (CUIs) and Type Unique Identifiers (TUIs) from cTAKES output. CUIs are codes assigned by the UMLS Metathesaurus to specific biomedical and health related concepts, which include anatomical, symptom, procedure, medication and disease-related information[40]. TUIs represent the semantic type for each CUI. For example, the CUI associated with “pain” is c0030193, and a common semantic type for pain is “finding,” represented by the TUI t184.
All features are then normalized using frequencies, as follows:
where the numerator ni,j is the number of occurrences of the i-th term in j-th document, and the denominator is the sum of all terms in the j-th document[41].
3.2. Feature Space
We trained a model using information extracted from cTAKES. As mentioned, all features represented are normalized by frequency. We tested lexical features that include unigrams and bigrams, as well as unigrams that are concatenated with their corresponding POS tags. Inclusion of bigrams introduced noise into the feature space, which hindered system performance. Our semantic features were engineered around UMLS concepts. For example, we evaluated positive CUIs (i.e., explicit mentions of medical concepts that are present in the patient, e.g., patient has asthma), as well as negated CUIs (i.e., explicit mentions of medical concepts that are absent in the patient, e.g., patient does not have asthma). We also added features that captured patient history attributes related to specific concepts. For example, if cTAKES extracted a history of hypertensive disease in a patient record, we accounted for this in the feature space (i.e., C0020538_history+1 = [normalized_frequency]). Accordingly, if cTAKES extracted a negated history of hypertensive disease in a record, we represented this negated concept in our feature space as well (i.e., C0020538_history−1 = [normalized_frequency]). In order to capture the proper UMLS semantic category for each medical concept, we merged CUI and TUI in one feature. We additionally tested isolated TUIs, but these broad categories had little variance throughout the corpus and merely added noise, which was detrimental to system performance. Our final feature space consisted of 45,695 total features (See Appendix A Table A.2 for a breakdown of the features). Given the size of the feature space and noise throughout, we evaluated dimensionality reduction techniques.
3.3. Dimensionality reduction and classification
We performed feature selection, dimensionality reduction and classification using 10-fold cross-validation to prevent overfitting and ensure a fair estimation of the models’ quality.
In our first run, we tested three separate classifiers that have proven effective in similar tasks[24,25]: (1) Gaussian Naïve Bayes; (2) MaxEnt; and (3) SVM with both linear and Radial Basis Function (RBF) kernels. For both types of SVM, we optimized the parameters. In the case of the linear kernel, which is based on the LIBLINEAR library, parameter optimization was built-into the classifier[41]. For the RBF kernel, we used a grid search optimization for γ and C parameters on the training portion of each fold during classification[42].
In our second run, we performed dimensionality reduction using PCA, a statistical technique that provides a lower dimensional projection of uncorrelated components from the most informative viewpoint of higher dimensional data by maximizing variance[43]. We tested different thresholds for the percentage of variance explained by the number of selected components produced during PCA, and tested these components with the same classifiers described in the first run.
Next, we employed logarithmic cuts using mutual information as a feature selection technique to trim the feature space for optimal performance in our third run. Mutual information measures the contribution of the presence or absence of a feature relative to the correct classification[44]. We chose to retain the top ranked features at different percentages using logarithmic cuts as opposed to cuts at linear intervals because we observed higher classifier performances at dramatic reductions to the feature space.
We further experimented with selective feature extraction using domain knowledge from the UMLS ontology to filter attributes. In the fourth run, we automatically searched UMLS using the UMLS REST API to gather attributes in the Semantic Network[45]. Specifically, we traversed the network to collect the attributes of all relationships, including siblings and children, related to “cardiovascular system drugs” and “cardiovascular diseases”. We include cardiovascular system drugs and cardiovascular diseases not just as direct evidence of CAD, but also as risk factors that indicate a future CAD diagnosis. This helps the system retain features commonly correlated with a CAD diagnosis, including features related to the common risk factors for CAD mentioned previously (e.g., hypertension, hypercholesterolemia, etc.). It also filters out UMLS features that are unlikely related to a CAD diagnosis, e.g., features related to “reproductive diseases”. We used the results of these queries to compile a filter of 838 CUIs attributed to different symptoms, diseases, medications, procedures and anatomical mentions associated with cardiovascular system drugs and cardiovascular diseases. During feature extraction of cTAKES output, we only included a UMLS feature if its corresponding CUI existed in the filter. This ontology-guided feature extraction step cut down total feature space by 29.6% (from 45,695 features to 32,180).
We subsequently applied PCA and mutual information feature selection to the feature space produced using the ontology-guided feature extraction filter. See Table 2 for an analysis of the number of features produced using the different dimensionality reduction techniques described in this section.
Table 2.
Number of features produced after applying different dimensionality reduction techniques.
| PCA | POV* | 99.0 | 95.0 | 90.0 | 85.0 | 80.0 | 75.0 | 70.0 | 65.0 | 60.0 |
| # of components | 426 | 333 | 259 | 205 | 163 | 130 | 109 | 102 | 62 | |
| % full feature space† | 0.932 | 0.723 | 0.567 | 0.449 | 0.357 | 0.828 | 0.239 | 0.223 | 0.136 | |
| MI | % feature space | 75.00 | 50.00 | 25.00 | 10.00 | 5.00 | 1.00 | 0.50 | 0.10 | 0.05 |
| # of features | 34,271 | 22,847 | 11,423 | 4,569 | 2,284 | 1,142 | 456 | 228 | 45 | |
| Ontology + PCA | POV* | 99.0 | 95.0 | 90.0 | 85.0 | 80.0 | 75.0 | 70.0 | 65.0 | 60.0 |
| # of components | 405 | 289 | 210 | 158 | 121 | 93 | 71 | 54 | 41 | |
| % full feature space† | 0.886 | 0.632 | 0.460 | 0.346 | 0.265 | 0.204 | 0.155 | 0.118 | 0.090 | |
| Ontology + MI† | % ontology feature space | 75.00 | 50.00 | 25.00 | 10.00 | 5.00 | 1.00 | 0.50 | 0.10 | 0.05 |
| % full feature space | 47.2 | 35.1 | 17.6 | 7.02 | 3.51 | 0.702 | 0.350 | 0.70 | 0.035 | |
| # of features | 24,135 | 16,054 | 8,027 | 3,210 | 1,605 | 321 | 160 | 32 | 16 |
Proportion of variance.
These percentages are less than 1.0% of the full feature space.
Number of Ontology features (i.e., Ontology without PCA or MI) is 32,180 features.
Lastly, we investigated ensemble classification through a weighted majority vote using the optimal settings for each dimensionality reduction technique that we evaluated.
4. Results and discussion
We used approximate randomization to test significance over micro-average precision, recall and F1, with N= 9,999 and α0.1[35,36,37]. We considered the best model to be the simplest (e.g., fewest number of features) among the highest performing runs with statistically significant increments.
Using the full feature space (i.e., 45,695 features), the Naïve Bayes classifier achieved an F1 score of 68.8%. Implementations of PCA, MI, and the ontology-guided feature extraction filter with PCA reduced dimensionality of the overall feature space, but did not significantly improve performance of the Naïve Bayes classifier. Performance did significantly improve, however, after implementing just the ontology filter during feature extraction (i.e., without PCA). This model achieved an F1 score of 76.6%, and reduced the feature space by 29.6% (i.e., 13,515 features). Applying MI to this ontology-guided feature extraction filter further reduced the feature space (with a total reduction of 52.8%) while maintaining high performance (i.e., 77.1% F1 score). Thus, we considered this model to be optimal for Naïve Bayes (See Table 3 for top classifier results by dimensionality reduction technique; See Figures 1–4 for graphs of classifiers’ results by dimensionality reduction technique; See Appendix A Tables A.3–A.8 for complete tables of classifiers’ results by dimensionality reduction technique).
Table 3.
Number of features [# f.], precision [P.], recall [R.], F1 score [F1] of classifier top performances by dimensionality reduction technique.
| Naïve Bayes | SVM (linear kernel) | SVM (RBF kernel) | MaxEnt | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # f. | P. | R. | F1 | # f. | P. | R. | F1 | # f. | P. | R. | F1 | # f. | P. | R. | F1 | |
| Full feature space | 45,695 | 0.761 | 0.635 | 0.688 | 45,695 | 0.744 | 0.802 | 0.769 | 45,695 | 0.714 | 0.829 | 0.765 | 45,695 | 0.740 | 0.790 | 0.762 |
| PCA | 333 | 0.541 | 0.957 | 0.691 | 426 | 0.739 | 0.798 | 0.765 | 426 | 0.709 | 0.825 | 0.760 | 426 | 0.737 | 0.790 | 0.760 |
| MI | 228 | 0.628 | 0.821 | 0.710 | 2,284 | 0.746 | 0.799 | 0.770 | 22,847 | 0.728 | 0.825 | 0.772 | 11,423 | 0.753 | 0.775 | 0.761 |
| Ontology | 32,180 | 0.872 | 0.689 | 0.766 | 32,180 | 0.762 | 0.795 | 0.774 | 32,180 | 0.747 | 0.760 | 0.747 | 32,180 | 0.733 | 0.806 | 0.764 |
| Ontology PCA | 210 | 0.538 | 0.942 | 0.684 | 405 | 0.748 | 0.779 | 0.758 | 405 | 0.718 | 0.775 | 0.741 | 405 | 0.726 | 0.810 | 0.763 |
| Ontology MI | 24,135 | 0.879 | 0.693 | 0.771 | 1,605 | 0.747 | 0.798 | 0.769 | 1,605 | 0.715 | 0.795 | 0.747 | 8,027 | 0.723 | 0.790 | 0.752 |
Note: Performance of shaded region is the simplest model that is not significantly different from the top overall model (i.e., F1 of 77.4%).
Figure 1.
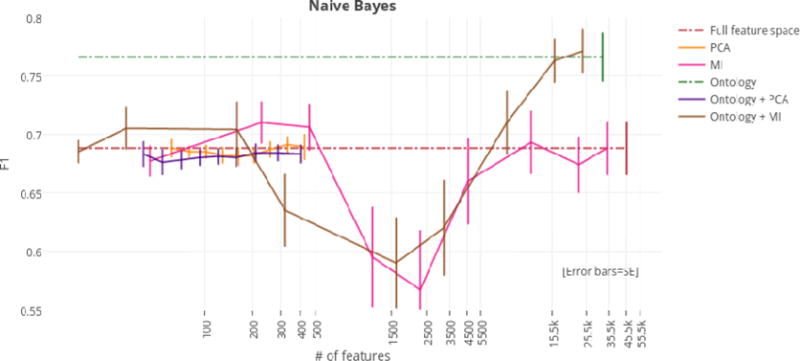
F1 scores by dimensionality reduction technique for the Naïve Bayes classifier.
Figure 4.
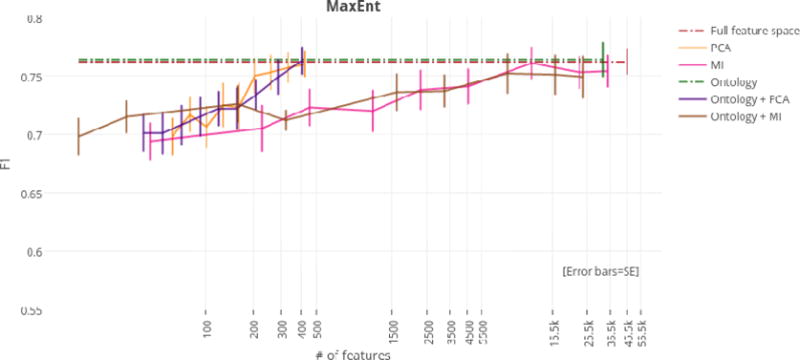
F1 scores by dimensionality reduction technique for the MaxEnt classifier.
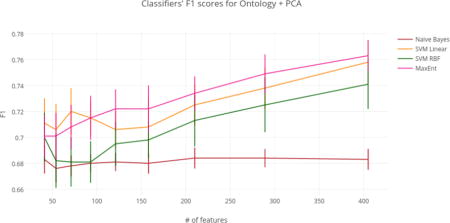
Using just the ontology filter during feature extraction (i.e., without MI or PCA), SVM linear achieved the highest overall F1 score of any classifier at 77.4%. However, regardless of dimensionality reduction technique, top performances for the SVM linear, SVM RBF, and MaxEnt classifiers were not significantly different from the highest overall F1 score of 77.4%. For this reason, models that achieved top performance using the fewest numbers of features were considered optimal. At only 405 features (a reduction of 99.1%), the ontology-guided feature extraction filter with PCA produced the most compressed feature space. This dimensionality reduction technique resulted in performances of 75.8%, 74.1%, and 76.3% F1 score for the SVM linear, SVM RBF, and MaxEnt classifiers respectively.
Our approach of ensemble classification through a weighted majority vote did not significantly improve system performance (See Appendix A Table A.9 for ensemble classification results).
Based on these results, we propose that the best dimensionality reduction technique is PCA with an ontology-guided feature extraction filter (See Figure 5 for of a graph of classifier results using this approach).
Figure 5.
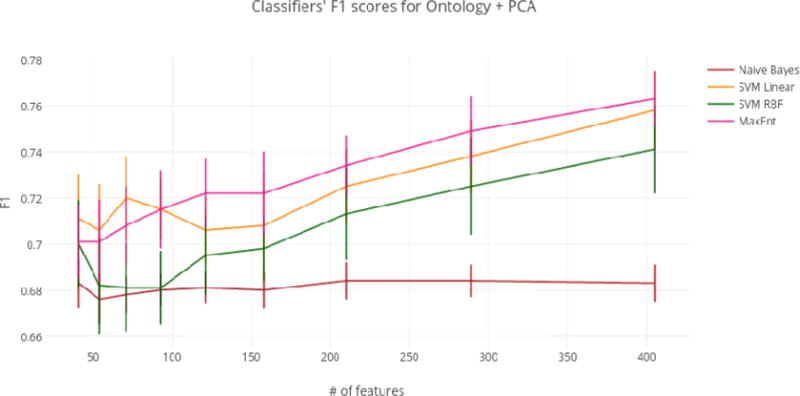
Classifiers’ F1 scores for Ontology + PCA.
An analysis of the feature selection results provided insights into the characteristics that differentiate the CAD and no-CAD patient populations in the data set. In general, several of the top 100 ranked features belonged to semantic categories important to predicting CAD. These semantic categories include medications (e.g., Simvastatin, Lipitor, Metoprolol, Atenolol, Metformin, Novolog, Trazodone, and Penicillin); symptoms and signs for diseases (e.g., illness, tobacco and history of pain); diseases (e.g., stroke, Hairy Cell Leukemia); procedures (e.g., cardiac catheterization, creatinine blood test, white blood cell count and appendectomy); and anatomical sites (e.g., artery, gastrointestinal, cerebellar and neurological). Of CUIs extracted for both CAD and no-CAD patient populations, there was only a 1.0% difference in positive mentions for congestive heart failure (CHF). Additionally, every patient record contained at least one CUI in the feature extraction filter, which is consistent with the original design of the corpus. Of the highest ranked features, the most frequent CUIs extracted were all symptom, diseases, or medications (See Appendix A – Table A.10).
We expect that as the number of available training samples increases, the selective feature extraction method becomes less necessary. This is because our model better learns which features to select for classification as training samples increase. However, in the presence of noisy data in which the number of features greatly outnumbers the number of samples, exploiting domain knowledge to automatically filter features is an effective method of dimensionality reduction.
The results show that no-CAD patients were more hypertensive (by 9.69%) and experienced a much higher incidence of stroke (by 17.05%). These patients were prescribed medications to treat hypertension and prevent further strokes at a higher rate. These medications include statins (e.g., Simvastatin), ACE inhibitors (e.g., Lisinopril) and beta blockers (e.g., metoprolol and atenolol). Importantly, these are the same treatments prescribed to prevent the development of CAD[44]. The features selected to predict patients who will develop CAD seem to suggest an important outcome in this sample population: patients who suffer from hypertension and/or stroke are treated with medications that prevent the development of CAD.
These results further explain several of our system errors. For example, 55 of 80 false negative classifications (i.e., 68.6%) discuss hypertensive patients. Also, 7 false negatives contain a cerebrovascular accident, because there were only 20 cerebrovascular accidents for the entire CAD patient population (i.e., 258 records) vs. 65 in the no-CAD population. Furthermore, of the 26 false positive records, there were considerably fewer mentions of drugs that help to prevent stroke. For example, only ten records referenced beta blockers; only five records contained ACE inhibitors; and only one record mentioned a statin.
Given the average of patients who do not develop CAD (i.e., 65 years old), it is possible that our system classifies patients who have not yet been diagnosed with CAD, but who will be diagnosed with the disease in a future visit that is not covered by our dataset. For this reason, we believe that one application of our system is to automatically classify records for patients who have gone undiagnosed, or who have been misdiagnosed, with respect to CAD.
Furthermore, our system can process any number of patient records to predict CAD with relatively little overhead—especially in comparison with manual methods. This is of great use to clinical researchers for building datasets to better understand CAD manifestation. One extension of this research would be to collect more data to examine why patients go undiagnosed or are misdiagnosed for CAD even though they present with several common risk factors.
4.1. Limitations
One limitation of our experiment is embedded in the selection of patients who do not develop CAD. The average age of this population is 65 (standard deviation = 12.47; Q1 = 54.00; Q3 = 73.63)[29]. Thus, it is possible that some of these patients eventually developed CAD after record collection in the creation of the corpus. Our prediction is limited to the text available. More specifically, our system reasonably predicts CAD for a patient that—at the time the system is run, and according to the data available—has not yet been diagnosed with the disease. Evaluating additional retrospective data would increase the reliability of our results.
Another limitation of our study is embedded in the subsample of patients who develop CAD. To avoid any bias introduced by removing records that contained ground truth labels (i.e., CAD diagnoses), we controlled for sickness in both CAD and no-CAD patient populations. In doing so, we sacrificed greater longitudinal coverage for CAD patients, as we were left with 12.9 months of coverage for patients who eventually develop a CAD diagnosis. We view the 12.9 months of coverage before manifestation of CAD as a necessary tradeoff to ensure the validity of our study. However, our system is not restricted to classifying records within a certain window of time (e.g., one-year prior to CAD manifestation). Rather, the average 12.9 months of coverage for CAD patients is a summary statistic of our subsample. Nonetheless, it would be ideal to assess risk for developing CAD further in advance as it would give more time to prevent development of the disease. Evaluating more data would also help to mitigate this limitation.
In general, the relatively small size of the dataset is a limitation of our study. Although we believe that the dataset adequately represents a very specific patient population that we are interested in better understanding, it does not cover all types of patients that clinicians would encounter in the wild.
Furthermore, we discovered during error analysis that smoking histories are not extracted by the cTAKES pipeline used in our system[33[]. For example, in one patient record the doctor explicitly states, “[the patient] does not smoke,” but no CUI is extracted related to smoking (e.g., a non-smoker CUI, a negated smoker CUI, etc.). Smoking is a common risk-factor for CAD, and it is well-represented with several concepts in UMLS4.*
The importance of extracting a patient’s smoking history accurately was confirmed in the mutual information feature selection results, as the unigram “smoking” was among the most important features extracted (i.e., it was in the top 1.0% of features selected). A separate smoking status pipeline, originally developed for the 2006 i2b2 Deidentification and Smoking Challenge[47,48], was integrated into cTAKES, but assessing its benefits would require further testing.
5. Conclusion
Among diabetic patients who share similar risk factors for CAD, it is possible to reasonably predict which patients will develop the disease. We focus on predicting the development of CAD. In this study, we show that domain ontology-guided feature extraction can reduce the high dimensional nature of free text medical records to improve the performance of machine learning methods for classification. Furthermore, we demonstrate that classic dimensionality reduction techniques complement this approach. Finally, we explain that the features selected to predict patients who will develop CAD seem to suggest an important outcome in this sample population: patients who suffer from hypertension and/or stroke are treated with medications that prevent the development of CAD. We conclude that the clinical application of our model is in classification of patients who will develop CAD in difficult to discriminate situations, such as when patients are all at high risk for CAD and carry many of the risk factors.
Figure 2.
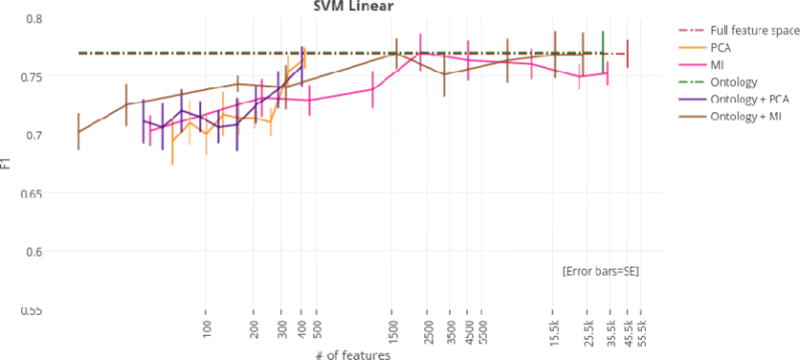
F1 scores by dimensionality reduction technique for the SVM linear classifier.
Figure 3.
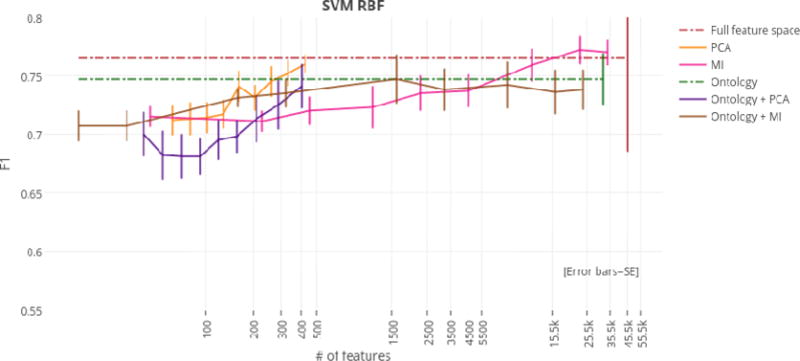
F1 scores by dimensionality reduction technique for the SVM RBF classifier.
Highlights.
A system to automatically predict coronary artery disease (CAD) from clinical narratives is proposed.
The system relies on an ontology-guided approach to feature extraction, which is compared to two classic feature selection techniques.
The system achieves state-of-the art performance of 77.4% F1-score.
Acknowledgments
We would like to thank the organizers of the 2014 i2b2/UTHealth Challenge. This work was supported in part by “NIH P50 Neuropsychiatric Genome-Scale and RDoC Individualized Domains (N-GRID)” National Institutes of Health, NIH P50 MH106933, PI: Isaac Kohane, and by Philips Research NA, PI: Peter Szolovits. The content is solely the responsibility of the authors and does not necessarily represent the official views of the sponsors.
Appendix A
Figure A.1.
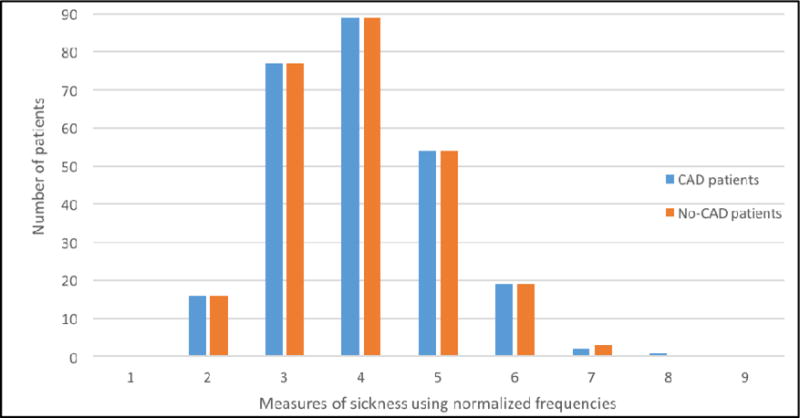
Measure of sickness in CAD and no-CAD patients.
Table A.1.
An analysis of word tokens and sentences in the corpus.
| Total | Average per document | ||
|---|---|---|---|
| # of tokens | # of sentences | # of tokens | # of sentences |
| 268,090 | 51,736 | 519 | 100 |
Table A.2.
A breakdown of the 45,695 total features in the final feature space by type.
| Feature type |
Positive CUIs |
Negated CUIs |
Positive CUI-histories |
Negated CUI-histories |
Positive CUI-TUIs |
Negated CUI-TUIs |
Unigrams |
Unigram- POS |
|---|---|---|---|---|---|---|---|---|
| # of features | 5,329 | 879 | 85 | 496 | 5,954 | 903 | 13,764 | 18,285 |
Table A.3.
Precision [P.], recall [R.], F1 score [F1] of full feature space.
| % of feature space | Total # of features | Naïve Bayes | SVM (linear kernel) | SVM (RBF kernel) | MaxEnt | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | ||
| 100.00 | 45,695 | 0.761 | 0.635 | 0.688 | 0.744 | 0.802 | 0.769 | 0.714 | 0.829 | 0.765 | 0.740 | 0.790 | 0.762 |
Note: Performance of shaded region is not significantly different from the top overall model (i.e., F1 of 77.4%).
Table A.4.
Precision [P.], recall [R.], F1 score [F1] results using PCA.
| POV | # of components | Naïve Bayes
|
SVM (linear kernel)
|
SVM (RBF kernel)
|
MaxEnt
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | ||
|
| |||||||||||||
| 0.99 | 426 | 0.556 | 0.907 | 0.689 | 0.739 | 0.798 | 0.765 | 0.709 | 0.825 | 0.760 | 0.737 | 0.790 | 0.760 |
| 0.95 | 333 | 0.541 | 0.957 | 0.691 | 0.743 | 0.774 | 0.753 | 0.712 | 0.802 | 0.752 | 0.731 | 0.790 | 0.757 |
| 0.90 | 259 | 0.532 | 0.969 | 0.687 | 0.709 | 0.717 | 0.710 | 0.714 | 0.783 | 0.745 | 0.731 | 0.782 | 0.753 |
| 0.85 | 205 | 0.528 | 0.965 | 0.682 | 0.713 | 0.721 | 0.714 | 0.686 | 0.786 | 0.731 | 0.728 | 0.779 | 0.750 |
| 0.80 | 163 | 0.526 | 0.965 | 0.681 | 0.701 | 0.732 | 0.714 | 0.698 | 0.791 | 0.741 | 0.711 | 0.740 | 0.723 |
| 0.75 | 130 | 0.528 | 0.965 | 0.682 | 0.700 | 0.740 | 0.717 | 0.672 | 0.771 | 0.717 | 0.712 | 0.744 | 0.725 |
| 0.70 | 102 | 0.532 | 0.961 | 0.685 | 0.686 | 0.721 | 0.700 | 0.665 | 0.771 | 0.714 | 0.681 | 0.736 | 0.706 |
| 0.65 | 80 | 0.536 | 0.950 | 0.685 | 0.699 | 0.725 | 0.710 | 0.670 | 0.764 | 0.713 | 0.703 | 0.736 | 0.717 |
| 0.60 | 62 | 0.546 | 0.930 | 0.688 | 0.692 | 0.702 | 0.693 | 0.678 | 0.752 | 0.712 | 0.687 | 0.713 | 0.698 |
Note: Performance of shaded region is not significantly different from the top overall model (i.e., F1 of 77.4%).
Table A.5.
Precision [P.], recall [R.], F1 score [F1] results using mutual information.
| % of feature space | Total # of features | Naïve Bayes
|
SVM (linear kernel)
|
SVM (RBF kernel)
|
MaxEnt
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | ||
|
| |||||||||||||
| 75.00 | 34,272 | 0.761 | 0.635 | 0.688 | 0.739 | 0.771 | 0.752 | 0.727 | 0.821 | 0.770 | 0.746 | 0.767 | 0.754 |
| 50.00 | 22,848 | 0.738 | 0.624 | 0.674 | 0.738 | 0.767 | 0.749 | 0.728 | 0.825 | 0.772 | 0.749 | 0.763 | 0.753 |
| 25.00 | 11,424 | 0.819 | 0.609 | 0.693 | 0.761 | 0.767 | 0.760 | 0.717 | 0.810 | 0.759 | 0.753 | 0.775 | 0.761 |
| 10.00 | 4,569 | 0.792 | 0.581 | 0.660 | 0.753 | 0.779 | 0.763 | 0.701 | 0.779 | 0.737 | 0.742 | 0.748 | 0.741 |
| 5.00 | 2,284 | 0.699 | 0.488 | 0.567 | 0.746 | 0.799 | 0.770 | 0.684 | 0.799 | 0.735 | 0.717 | 0.763 | 0.738 |
| 1.00 | 1142 | 0.676 | 0.542 | 0.595 | 0.705 | 0.775 | 0.738 | 0.657 | 0.810 | 0.723 | 0.685 | 0.760 | 0.720 |
| 0.50 | 456 | 0.688 | 0.728 | 0.705 | 0.685 | 0.782 | 0.729 | 0.637 | 0.830 | 0.720 | 0.671 | 0.786 | 0.723 |
| 0.10 | 228 | 0.628 | 0.821 | 0.710 | 0.703 | 0.763 | 0.731 | 0.618 | 0.841 | 0.711 | 0.650 | 0.771 | 0.705 |
| 0.05 | 45 | 0.561 | 0.856 | 0.677 | 0.767 | 0.767 | 0.703 | 0.596 | 0.899 | 0.715 | 0.611 | 0.806 | 0.694 |
Note: Performance of shaded region is not significantly different from the top overall model (i.e., F1 of 77.4%).
Table A.6.
Precision [P.], recall [R.], F1 score [F1] using UMLS ontology-guided feature extraction.
| % of feature space | Total # of features | Naïve Bayes
|
SVM (linear kernel)
|
SVM (RBF kernel)
|
MaxEnt
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | ||
|
| |||||||||||||
| 100.00 | 32,180 | 0.872 | 0.689 | 0.766 | 0.762 | 0.795 | 0.774 | 0.747 | 0.760 | 0.747 | 0.733 | 0.806 | 0.764 |
Note: Performance of shaded region is not significantly different from the top overall model (i.e., F1 of 77.4%).
Table A.7.
Precision [P.], recall [R.], F1 score [F1] results using UMLS ontology-guided feature extraction and PCA.
| POV | # of components | Naïve Bayes
|
SVM (linear kernel)
|
SVM (RBF kernel)
|
MaxEnt
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | ||
|
| |||||||||||||
| 0.99 | 405 | 0.533 | 0.950 | 0.683 | 0.748 | 0.779 | 0.758 | 0.718 | 0.775 | 0.741 | 0.726 | 0.810 | 0.763 |
| 0.95 | 289 | 0.539 | 0.938 | 0.684 | 0.719 | 0.767 | 0.738 | 0.717 | 0.744 | 0.725 | 0.719 | 0.790 | 0.749 |
| 0.90 | 210 | 0.538 | 0.942 | 0.684 | 0.710 | 0.751 | 0.725 | 0.703 | 0.736 | 0.713 | 0.704 | 0.775 | 0.734 |
| 0.85 | 158 | 0.535 | 0.934 | 0.680 | 0.695 | 0.729 | 0.708 | 0.688 | 0.728 | 0.698 | 0.690 | 0.760 | 0.722 |
| 0.80 | 121 | 0.537 | 0.934 | 0.681 | 0.679 | 0.740 | 0.706 | 0.675 | 0.732 | 0.695 | 0.686 | 0.768 | 0.722 |
| 0.75 | 93 | 0.537 | 0.926 | 0.680 | 0.682 | 0.756 | 0.715 | 0.641 | 0.736 | 0.681 | 0.674 | 0.764 | 0.715 |
| 0.70 | 71 | 0.539 | 0.915 | 0.678 | 0.683 | 0.764 | 0.720 | 0.651 | 0.725 | 0.681 | 0.671 | 0.752 | 0.708 |
| 0.65 | 54 | 0.544 | 0.896 | 0.676 | 0.676 | 0.744 | 0.706 | 0.637 | 0.741 | 0.682 | 0.664 | 0.744 | 0.701 |
| 0.60 | 41 | 0.556 | 0.888 | 0.683 | 0.669 | 0.764 | 0.711 | 0.645 | 0.772 | 0.700 | 0.661 | 0.748 | 0.701 |
Note: Performance of shaded region is not significantly different from the top overall model (i.e., F1 of 77.4%).
Table A.8.
Precision [P.], recall [R.], F1 score [F1] using UMLS ontology-guided feature extraction and mutual information.
| % of feature space | Total # of features | Naïve Bayes
|
SVM (linear kernel)
|
SVM (RBF kernel)
|
MaxEnt
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | P. | R. | F1 | ||
|
| |||||||||||||
| 75.00 | 24,136 | 0.879 | 0.693 | 0.771 | 0.752 | 0.791 | 0.768 | 0.747 | 0.744 | 0.738 | 0.720 | 0.786 | 0.749 |
| 50.00 | 16,054 | 0.858 | 0.693 | 0.763 | 0.753 | 0.791 | 0.768 | 0.746 | 0.740 | 0.736 | 0.721 | 0.790 | 0.751 |
| 25.00 | 8,027 | 0.868 | 0.605 | 0.710 | 0.746 | 0.787 | 0.763 | 0.749 | 0.748 | 0.742 | 0.723 | 0.790 | 0.752 |
| 10.00 | 3,210 | 0.769 | 0.531 | 0.620 | 0.738 | 0.771 | 0.751 | 0.729 | 0.763 | 0.738 | 0.708 | 0.775 | 0.737 |
| 5.00 | 1,605 | 0.713 | 0.515 | 0.590 | 0.747 | 0.798 | 0.769 | 0.715 | 0.795 | 0.747 | 0.693 | 0.787 | 0.736 |
| 1.00 | 321 | 0.670 | 0.612 | 0.635 | 0.703 | 0.783 | 0.740 | 0.688 | 0.794 | 0.735 | 0.665 | 0.767 | 0.712 |
| 0.50 | 160 | 0.671 | 0.748 | 0.704 | 0.691 | 0.806 | 0.743 | 0.665 | 0.821 | 0.731 | 0.660 | 0.810 | 0.726 |
| 0.10 | 32 | 0.613 | 0.833 | 0.705 | 0.685 | 0.771 | 0.725 | 0.646 | 0.791 | 0.707 | 0.643 | 0.810 | 0.715 |
| 0.05 | 16 | 0.575 | 0.849 | 0.685 | 0.650 | 0.767 | 0.702 | 0.614 | 0.845 | 0.707 | 0.614 | 0.810 | 0.698 |
Note: Performance of shaded region is not significantly different from the top overall model (i.e., F1 of 77.4%).
Table A.9.
Precision [P.], recall [R.], F1 score [F1] using ensemble classification through a weighted vote of classifiers for dimensionality reduction techniques under optimal settings.
| Dimensionality reduction technique | Total # of features | P. | R. | F1 |
|---|---|---|---|---|
|
| ||||
| Full feature space | 45,695 | 0.738 | 0.818 | 0.776 |
| PCA | 333 | 0.724 | 0.814 | 0.766 |
| Mutual Information | 2,284 | 0.741 | 0.798 | 0.769 |
| Ontology | 32,180 | 0.774 | 0.81 | 0.792 |
| Ontology + PCA | 289 | 0.723 | 0.798 | 0.759 |
| Ontology + MI | 1,605 | 0.749 | 0.798 | 0.773 |
Note: Performance of shaded region is not significantly different from the top overall model (i.e., F1 of 77.4%).
Table A.10.
Top sematntic features selected with Ontology + MI at 25% cut to feature space.
| Feature | CAD count | No-CAD count | % difference in corpus |
|---|---|---|---|
| Cerebrovascular accident | 20 | 64 | 17.05 |
| Simvastatin | 11 | 44 | 12.02 |
| Lisinopril | 55 | 86 | 12.02 |
| Metoprolol | 13 | 41 | 10.85 |
| Hypertension | 158 | 183 | 9.69 |
| Atenolol | 50 | 69 | 7.36 |
| Palpitation | 41 | 52 | 4.26 |
| Glyceryl trinitrate | 20 | 11 | 3.49 |
| Syncope | 25 | 17 | 3.10 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Some examples of smoking CUIs in UMLS are C0337664 (smoker), C0337666 (cigar smoker), C0337671 (former smoker), and C0337672 (non-smoker).
Conflict of Interest
This work was supported in part by “NIH P50 Neuropsychiatric Genome-Scale and RDoC Individualized Domains (N-GRID)” National Institutes of Health, NIH P50 MH106933, PI: Isaac Kohane, and by Philips Research NA, PI: Peter Szolovits. The content is solely the responsibility of the authors and does not necessarily represent the official views of the sponsors.
References
- 1.Coronary Artery Disease. MedlinePlus. 2015 https://www.nlm.nih.gov/medlineplus/coronaryarterydisease.html. Accessed: Accessed: 2015- 11- 18.
- 2.Gundlapalli AV, et al. Using natural language processing on the free text of clinical documents to screen for evidence of homelessness among US veterans. AMIA Annual Symposium Proceedings. 2013;2013:537. [PMC free article] [PubMed] [Google Scholar]
- 3.Stubbs A, Uzuner Ö. Annotating risk factors for heart disease in clinical narratives for diabetic patients. J Biomed Inform. 2015 doi: 10.1016/j.jbi.2015.05.009. 2015 May 21. pii: S1532-0464(15)00089-1. [Epub ahead of print]. http://www.ncbi.nlm.nih.gov/pubmed/26004790. [DOI] [PMC free article] [PubMed]
- 4.Chang N-W, Dai H-J, Jonnagaddala J, Chen C-W, Tsai RT-H, Hsu W-L. A context-aware approach for progression tracking of medical concepts in electronic medical records. Journal of Biomedical Informatics. 2015 Dec.58:S150–S157. doi: 10.1016/j.jbi.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalifa A, Meystre S. Adapting existing natural language processing resources for cardiovascular risk factors identification in clinical notes. Journal of Biomedical Informatics. 2015 Dec.58:S128–S132. doi: 10.1016/j.jbi.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack J, Nath C, Milward D, Raja K, Jonnalagadda SR. Agile text mining for the 2014 i2b2/UTHealth Cardiac risk factors challenge. Journal of Biomedical Informatics. 2015 Dec.58:S120–S127. doi: 10.1016/j.jbi.2015.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H, Garibaldi JM. A hybrid model for automatic identification of risk factors for heart disease. Journal of Biomedical Informatics. 2015 Dec.58:S171–S182. doi: 10.1016/j.jbi.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Li H, Tang B, Wang X, Liu X, Liu Z, Liu S, Wang W, Deng Q, Zhu S, Chen Y, Wang J. An automatic system to identify heart disease risk factors in clinical texts over time. Journal of Biomedical Informatics. 2015 Dec.58:S158–S163. doi: 10.1016/j.jbi.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Chen Y, Tang B, Wang X, Chen Q, Li H, Wang J, Deng Q, Zhu S. Automatic de-identification of electronic medical records using token-level and character-level conditional random fields. Journal of Biomedical Informatics. 2015 Dec.58:S47–S52. doi: 10.1016/j.jbi.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang H, Garibaldi JM. Automatic detection of protected health information from clinic narratives. Journal of Biomedical Informatics. 2015 Dec.58:S30–S38. doi: 10.1016/j.jbi.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grouin C, Moriceau V, Zweigenbaum P. Combining glass box and black box evaluations in the identification of heart disease risk factors and their temporal relations from clinical records. Journal of Biomedical Informatics. 2015 Dec.58:S133–S142. doi: 10.1016/j.jbi.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehghan A, Kovacevic A, Karystianis G, Keane JA, Nenadic G. Combining knowledge- and data-driven methods for de-identification of clinical narratives. Journal of Biomedical Informatics. 2015 Dec.58:S53–S59. doi: 10.1016/j.jbi.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shivade C, Malewadkar P, Fosler-Lussier E, Lai AM. Comparison of UMLS terminologies to identify risk of heart disease using clinical notes. Journal of Biomedical Informatics. 2015 Dec.58:S103–S110. doi: 10.1016/j.jbi.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonnagaddala J, Liaw ST, Ray P, Kumar M, Chang NW, Dai HJ. Coronary artery disease risk assessment from unstructured electronic health records using text mining. Journal of Biomedical Informatics. 2015 Dec.58:S203–S210. doi: 10.1016/j.jbi.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He B, Guan Y, Cheng J, Cen K, Hua W. CRFs based de-identification of medical records. Journal of Biomedical Informatics. 2015 Dec.58:S39–S46. doi: 10.1016/j.jbi.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen T, Cullen RM, Godwin M. Hidden Markov model using Dirichlet process for de-identification. Journal of Biomedical Informatics. 2015 Dec.58:S60–S66. doi: 10.1016/j.jbi.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urbain J. Mining heart disease risk factors in clinical text with named entity recognition and distributional semantic models. Journal of Biomedical Informatics. 2015 Dec.58:S143–S149. doi: 10.1016/j.jbi.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solomon JW, Nielsen RD. Predicting changes in systolic blood pressure using longitudinal patient records. Journal of Biomedical Informatics. 2015 Dec.58:S197–S202. doi: 10.1016/j.jbi.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torii M, Fan J, Yang W, Lee T, Wiley MT, Zisook DS, Huang Y. Risk factor detection for heart disease by applying text analytics in electronic medical records. Journal of Biomedical Informatics. 2015 Dec.58:S164–S170. doi: 10.1016/j.jbi.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shivade C, Hebert C, Lopetegui M, de Marneffe M-C, Fosler-Lussier E, Lai AM. Textual inference for eligibility criteria resolution in clinical trials. Journal of Biomedical Informatics. 2015 Dec.58:S211–S218. doi: 10.1016/j.jbi.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grouin C, Moriceau V, Rosset S, Zweigenbaum P. Risk factor identification from clinical records for diabetic patients. In: Stubbs Amber, Kotfila C, Xu H, Uzuner Ö., editors. Proceedings i2b2/UTHealth NLP Challenge. Washington, DC: 2014. i2b2. 5 pages. [Google Scholar]
- 22.Karystianis G, Dehghan A, Kovacevic A, Keane JA, Nenadic G. Using local lexicalized rules to identify heart disease risk factors in clinical notes. Journal of Biomedical Informatics. 2015 Dec.58:S183–S188. doi: 10.1016/j.jbi.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taslimitehrani V, Dong G, Pereira NL, Panahiazar M, Pathak J. Developing EHR-driven heart failure risk prediction models using CPXR (Log) with the probabilistic loss function. Journal of biomedical informatics. 2016;60:260–269. doi: 10.1016/j.jbi.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uzuner Ö, Stubbs A. Practical applications for natural language processing in clinical research: The 2014 i2b2/UTHealth shared tasks. Journal of Biomedical Informatics. 2015 Dec.58:S1–S5. doi: 10.1016/j.jbi.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coronary Heart Disease Risk Factors - NHLBI, NIH. Nhlbi.nih.gov. 2016 [Online]. Available: https://www.nhlbi.nih.gov/health/health-topics/topics/hd/atrisk. [Accessed: 2015- 11- 18]
- 26.López Pineda A, Ye Y, Visweswaran S, Cooper GF, Wagner MM, Tsui F(R) Comparison of machine learning classifiers for influenza detection from emergency department free-text reports. Journal of Biomedical Informatics. 2015 Dec.58:60–69. doi: 10.1016/j.jbi.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pham A-D, Névéol A, Lavergne T, Yasunaga D, Clément O, Meyer G, Morello R, Burgun A. Natural language processing of radiology reports for the detection of thromboembolic diseases and clinically relevant incidental findings. BMC bioinformatics. 2014;15(1):1. doi: 10.1186/1471-2105-15-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redd D, Frech TM, Murtaugh MA, Rhiannon J, Zeng QT. Informatics can identify systemic sclerosis (SSc) patients at risk for scleroderma renal crisis. Computers in Biology and Medicine. 2014 Oct.53:203–205. doi: 10.1016/j.compbiomed.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang BB, Mckay RIB, Abbass HA, Barlow M. A Comparative Study for Domain Ontology Guided Feature Extraction. Proceedings of the 26th Australasian Computer Science Conference - Volume 16; Darlinghurst, Australia, Australia. 2003. pp. 69–78. [Google Scholar]
- 30.Garla VN, Brandt C. Ontology-guided feature engineering for clinical text classification. Journal of Biomedical Informatics. 2012;45(5):992–998. doi: 10.1016/j.jbi.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar V, Stubbs A, Shaw S, Uzuner Ö. Creation of a new longitudinal corpus of clinical narratives. J Biomed Inform. 2015;58S:S6–S10. doi: 10.1016/j.jbi.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.cTAKES 3.0 - Apache cTAKES - Apache Software Foundation. Cwiki.apache.org. 2016 [Online]. Available: https://cwiki.apache.org/confluence/display/CTAKES.
- 33.sklearn.decomposition.PCA — scikit-learn 0.17.1 documentation. Scikit-learn.org. 2016 [Online]. Available: http://scikit-learn.org/stable/modules/generated/sklearn.decomposition.PCA.html. [Accessed: 22- Jun- 2016]
- 34.sklearn.metrics.normalized_mutual_info_score — scikit-learn 0.17.1 documentation. Scikit-learn.org. 2016 [Online]. Available: http://scikit-learn.org/stable/modules/generated/sklearn.metrics.normalized_mutual_info_score.html. [Accessed: 22- Jun- 2016]
- 35.Nancy Chinchor. The statistical significance of the MUC-4 results. Proceedings of the 4th conference on Message understanding. 1992:30–50. [Google Scholar]
- 36.Noreen EW. Computer-intensive methods for testing hypotheses: an introduction. New York: Wiley; 1989. [Google Scholar]
- 37.Stubbs A, Kotfila C, Uzuner Ö. Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/UTHealth shared task Track 1. Journal of biomedical informatics. 2015;58:S11–S19. doi: 10.1016/j.jbi.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.cTAKES 3.0 - Clinical Documents Pipeline - Apache cTAKES - Apache Software Foundation. Cwiki.apache.org. 2016 [Online]. Available: https://cwiki.apache.org/confluence/display/CTAKES/cTAKES+3.0+-+Clinical+Documents+Pipeline.
- 39.nltk.util — NLTK 3.0 documentation. Nltk.org. 2016 [Online]. Available: http://www.nltk.org/_modules/nltk/util.html. [Accessed: 09- Mar- 2016]
- 40.Metathesaurus. National Library of Medicine; US: 2009. [Google Scholar]
- 41.Term Frequency and Inverted Document Frequency. :2016. [Online]. Available: http://disi.unitn.it/~bernardi/Courses/DL/Slides_11_12/measures.pdf. [Accessed: 09- Mar- 2016]
- 42.Fan R-E, hang K-W, Hsieh C-J, Wang X-R, Lin C-J. LIBLINEAR: A library for large linear classification. The Journal of Machine Learning Research. 2008;9:1871–1874. [Google Scholar]
- 43.sklearn.grid_search.GridSearchCV — scikit-learn 0.17.1 documentation. Scikit-learn.org. 2016 [Online]. Available: http://scikit-learn.org/stable/modules/generated/sklearn.grid_search.GridSearchCV.html. [Accessed: 22- Jun- 2016.
- 44.Shlens J. A Tutorial on Principal Component Analysis. CoRR. 2014;abs/1404.1100 [Google Scholar]
- 45.Manning C, Raghavan P, Schütze H. Introduction to information retrieval. New York: Cambridge University Press; 2008. p. 252. [Google Scholar]
- 46.Searching the UMLS. Documentation.uts.nlm.nih.gov. :2016. [Online]. Available: https://documentation.uts.nlm.nih.gov/rest/search/index.html.
- 47.Treatment - Coronary artery disease - Mayo Clinic. Mayoclinic.org. :2016. [Online]. Available: http://www.mayoclinic.org/diseases-conditions/coronary-artery-disease/diagnosis-treatment/treatment/txc-20165340.
- 48.Uzuner Ö, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc. 2008;15(1):15–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savova GK, Ogren PV, Duffy PH, Buntrock JD, Chute CG. Mayo clinic NLP system for patient smoking status identification. Journal of the American Medical Informatics Association: JAMIA. 2008 Jan-Feb;15:25–8. doi: 10.1197/jamia.M2437. [DOI] [PMC free article] [PubMed] [Google Scholar]


