Abstract
Controlled releasing of regulations remains the most convenient method to deliver various drugs. In the present study, we precipitated gold nanoparticles with chrysophanol. The gold-chrysophanol into poly (DL-lactide-co-glycolide) nanoparticles was loaded and the biological activity of chrysophanol nanoparticles on human LNCap prostate cancer cells, was tested to acquire the sustained releasing property. The circular dichroism spectroscopy indicated that chrysophanol nanoparticles effectively resulted in conformational alterations in DNA and regulated different proteins associated with cell cycle arrest. The reactive oxygen species (ROS), apoptosis, cell cycle, DNA damage, Cyto-c and caspase-3 activity were analyzed, and the expression levels of different anti- and pro-apoptotic were studied using immunoblotting analysis. The cytotoxicity assay suggested that chrysophanol nanoparticles preferentially killed prostate cancer cells in comparison to the normal cells. Chrysophanol nanoparticles reduced histone deacetylases (HDACs) to suppress cell proliferation and induce apoptosis by arresting the cell cycle in sub-G phase. In addition, the cell cycle-related proteins, including p27, CHK1, cyclin D1, CDK1, p-AMP-activated protein kinase (AMPK) and p-protein kinase B (AKT), were regulated by chrysophanol nanoparticles to prevent human prostate cancer cell progression. Chrysophanol nanoparticles induced apoptosis in LNCap cells by promoting p53/ROS crosstalk to prevent proliferation. Pharmacokinetic study in mice indicated that chrysophanol nanoparticle injection showed high bioavailability compared to the free chrysophanol. Also, in vivo study revealed that chrysophanol nanoparticles obviously reduced tumor volume and weight. In conclusion, the data above suggested that chrysophanol nanoparticles might be effective to prevent human prostate cancer progression.
Keywords: chrysophanol nanoparticles, prostate cancer, apoptosis, ROS, bioavailability
Introduction
Currently, prostate cancer is the most commonly diagnosed cancer in male and is a major cause of death in men aged 40–70 years (1,2). The gland of prostate surrounds the base of the male bladder and is likely prone to tumorigenesis in aged men (3,4). The burden of human prostate cancer in the world is increasing annually (5). Thus, focusing on therapeutic strategies for prostate cancer is urgently necessary.
Naturally occurring compounds are considered as the most interesting agents to test for cancer prevention and therapy, due to their anticipated multimodal actions and limited toxicity, affecting the signaling pathways within the cells, including regulation of cell proliferation, activation of apoptosis and modulation of cell cycle arrest (6,7). Chrysophanol (1,8-dihydroxy-3-methylanthraquinone), extracted from plants of Rheum genus, is one of the anthraquinone compounds, which has been suggested to induce cell death in different types of cancer cells (8,9). The effects of chrysophanol on human prostate cancer cell death have not been studied. However, the naturally derived compounds have limitations of preservation, bioavailability and low water solubility. Thus, delivering the compound requires product formulations to maintain the active molecular form until consumption, as well as to preserve stability, bioactivity, and bio-availability, which is the central goal of developing a nanoparticle (NP)-based system. Nanoparticulate drug delivery system for drug intranasal administration needed less amounts of administrations to induce the required pharmacological reaction due to its ability to locate on the target region and supply controlled drug delivery for prolonged time periods (10,11). Accordingly, the concentrations of polyphenols, which appear to be effective in vitro, are usually an order of magnitude higher than the levels tested in vivo (12,13). Thus, delivering these natural compounds needs product formulations to keep the active form of the molecule until consumption, and to maintain stability, bioavailability, and bioactivity, an essential point to explore a nanoparticle-based system. Surface functionalization of gold nanoparticles (AuNPs) is important for biomedical applications, which target them to specific disease areas and selectively allow them to interact with biomolecules or cells. Surface conjugation is usually achieved by adsorption of the ligand to the surface of gold. Thus, they have been widely investigated for cancer because of their unique size and intrinsic optical properties, including localized surface plasmon resonance (14,15). Additionally, long-term circulating NPs are desirable in systemic applications, including passive targeting of tumors and inflammatory sites. Poly (ethylene glycol) (PEG)/poly (lactic-co-glycolic acid (PLGA)-modified NPs have a long-term circulating property, as they can evade macrophage-mediated uptake and removal from systemic circulation (16,17).
Inhibiting cancer cell cycle and proliferation rates relies on various parameters, including DNA structural alterations and suppressing the activities or expression of histone deacetylases (HDACs) (18). These anti-proliferation promoting activities can make drugs more specific for various cancers (19,20). As previously indicated, HDACs was highly expressed during the cellular oncogenesis (21). HDAC1 was the first identified mammalian HDAC and is considered the prototype of the HDAC family (22). Overexpression of HDAC1 is significantly associated with higher lymphatic metastases and decreased the survival rates in patients with gastric cancer (23). Recently studies showed that elevated levels of HDAC3 expression and activity caused epigenetic alterations associated with malignancies (24). HDAC6 is involved in protein trafficking and degradation, cell shape and migration. Deregulation of HDAC6 activity is associated with a variety of diseases including cancer leading to a growing interest for developing HDAC6 inhibitors (25,26). Increased HDAC6 expression and/or activity have been demonstrated to promote cell migration and tissue invasiveness. HDAC6 has also been shown to be required for oncogenic transformation and tumor formation. Upregulated HDAC6 has been observed in a number of different cancers and recently, specific HDAC6 inhibitors have been found to inhibit cell growth and prevent tumor formation in mouse models (27–29). Also, the use of HDACs inhibitors could suppress cancer cells both in vivo and in vitro through regulating gene expression, and protein levels to prevent tumor progression (30). We explored the effects of formulated chrysophanol nanoparticle on human prostate cancer cell lines in vitro and confirmed the possible molecular mechanisms involved in apoptosis induction in prostate cancer cells. We found that chrysophanol nanoparticle could reduce prostate cancer cell viability by the induction of apoptosis through ROS, which was associated with p53 expression. Chrysophanol nanoparticle also decreased the expression of HDACs, indicating its role in suppressing human prostate cancer cell proliferation. Also, in vivo, the mouse models revealed that chrysophanol nanoparticle showed high bioavailability compared to the free chrysophanol. The significantly reduced tumor volume and size was observed.
Materials and methods
The preparation of chrysophanol nanoparticle
High performance liquid chromatography-grade chrysophanol was purchased from SeeBio (Shanghai, China) in an anhydrous powder formation. AuNPs were synthesized by downregulating gold chloride (1 mM) with a freshly prepared chrysophanol solution in absolute alcohol. The pale-yellow solution changed to the deep red as chrysophanol nanoparticle (0, 10, 30, 50, 70, 90, 110 and 130 μM) were formed. PLGA (50 mg) was added into an AuNPs aqueous dispersion, which were added to an aqueous solution containing a stabilizer. The mixture was then stirred at 400 rpm, 4°C, until the organic solution had completely evaporated. The redundant stabilizer was diminished by washing and centrifugation at 25,000 × g, 4°C, for 30 min. Then, the pellet was re-suspended in Milli-Q water. Chrysophanol nanoparticles were stored at 4°C for the following study. Furthermore, the concentrations of chrysophanol nanoparticles used in our study were dependent on the doses of chrysophanol attached to nanoparticles. Thus, the mount of chrysophanol attached to nanomaterial was not higher than free chrysophanol used in each group. The mean size and the size distribution of the chrysophanol nanoparticle were determined by dynamic light scattering (DLS) with a Zetasizer Nano ZS90 by Malvern Instruments (UK). The surface morphology of the chrysophanol nanoparticle was determined by SEM (Jeol NeoScope JCM-5000 Benchtop, WA, USA).
Cells and culture
Human prostate cancer cell lines, DU145, LNCap and PC3, were purchased from American Type Tissue Collection (ATCC, USA). Human prostate normal cell line, RWPE-1, and human normal liver cell line, L02, were obtained from KeyGene Biotech (Nanjing, China). DU145, LNCap and PC3 cells were cultured in RPMI-1640 (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco). RWPE-1 and L02 were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco). All cells were incubated at 37°C and maintained at 5% CO2. The cells were cultivated in the absence or presence of various concentrations of chrysophanol nanoparticles for 24 h, which were harvested for the following research. All cancer cells were transfected with (100 nM) nonsense control and siRNA against p53 (#1:5′-GUA AGG AGA AUG GGU AUG GCG U; #2:5′-GUA UGG GUG AUG AGC AGG GAG AU) for 48 h using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer's protocol. The ROS scavenger NAC was purchased from Sigma-Aldrich (USA), which was treated in the culture medium at a final concentration of 1 mM for 1 h before chrysophanol nanoparticles administration. P53 inhibitor, pifithrin-α (PIF-α), was obtained from TargetMol (USA) and was administered to the culture medium at a final concentration of 30 μM in the absence or presence of chrysophanol nanoparticles (90 μM) for 24 h. Also, caspase-3 inhibitor, Z-VAD-FMK (Sigma-Aldrich), at dose of 10 μM, was administered to cells according to the manufacturer's instructions.
Survival rate analysis
Human prostate cancer cells (1×104 cells per well) were planted in triplicate on a 96-well plate and incubated overnight before various treatment for different time as indicated. After incubation, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye was added, and the mixture was then incubated for 4 h. The dye was solubilized with 100 μl of DMSO (KeyGen Biotech). The plates were read at 570 nm on an automated microtiter plate reader. A blank well that contained only media and drug was used as a control for all the experiments.
Flow cytometric analysis
The cells were seeded at 0.75×105 cells/ml. Cell cycle assays were carried out in LNCap cells. Chrysophanol nanoparticle (90 μM) was applied on LNCap cells for 24 h. Then, all cells were harvested, washed with PBS, fixed and subsequently incubated with RNaseA (KeyGen Biotech). Next, the cells were stained with 50 μg/ml propidium iodide (PI, Sigma-Aldrich) and the readings were obtained in flow cytometer (BD Biosciences, USA).
For apoptosis analysis, the cells were harvested after treatment with chrysophanol nanoparticle (90 μM) or PIF-α (30 μM) for 24 h and stained with the Annexin V/PI Cell Apoptosis Detection Kit (KeyGen Biotech) according to the manufacturer's instructions. The cells in early stages of apoptosis were Annexin V-positive and PI-negative, and the cells in the late stages of apoptosis were both Annexin V and PI-positive. The acquisition of results and analysis were carried out with a Becton-Dickinson FACSCalibur flow cytometer using Cell Quest software.
The number of apoptotic cells was calculated using the Fluorescein Active Caspase-3 Staining kit (Abcam, UK) and In Situ BrdU DNA Fragmentation assay kit (Abnova, USA) according to the manufacturer's instructions. The proportion of caspase-3 in active form and TUNEL-positive cells was analyzed by using a flow cytometer (BD Biosciences).
ROS generation was analyzed by flow cytometry. The cells treated under various conditions as described, and then were harvested and fixed with the ice-cold methanol (70%). Fixed cancer cells were incubated with H2DCFDA (Sigma-Aldrich) for 20 min in the dark according to the manufacturer's instructions. Fluorescence intensity was determined by the use of FL1H filter of the flow cytometer.
The cellular uptake and binding capability with DNA in cells
The cellular uptake of chrysophanol nanoparticle in LNCap cells was analyzed using fluorescence method. The cancer cells were washed with PBS twice. The cells after washing were re-suspended in medium and subsequently incubated with chrysophanol nanoparticle for different time intervals (0, 60, 90, 120, 180, and 480 min). Cells were then observed under a fluorescence microscope (Olympus, Japan), and the representative images were captured. For assessment of the interaction of chrysophanol nanoparticle with nuclear DNA, LNCap cells were treated with chrysophanol nanoparticle for different times as indicated. After incubation, the nuclear DNA of LNCap cells were extracted and purified using DNA and RNA Purification:Genomic DNA (Sigma-Aldrich). The collected DNA was applied to analyze the circular dichroism spectra using a spectroscope.
Hoechst analysis
The prostate cancer cells, after chrysophanol nanoparticle administration (0, 70, 90 and 110 μM) for 24 h on 12-well plates were stained with 10 μg/ml of Hoechst 33342 (Life Technologies Corp., USA) in 1 ml PBS for 30 min. Hoechst 33342 was applied for staining of nuclei of cells. After staining, samples were rinsed with PBS once and 1ml of PBS was added for observation using a fluorescent microscope.
Western blot analysis
The prostate cancer cells were harvested after various treatments and were homogenized into 10% (wt/vol) hypotonic buffer (pH 8.0, 1 mM EDTA, 5 μg/ml leupeptin, 25 mM Tris-HCl, 1 mM Pefabloc SC, 5 μg/ml soybean trypsin inhibitor, 50 μg/ml aprotinin, 4 mM benzamidine) to yield a homogenate. Also, then the final supernatants were obtained by centrifugation at 15,000 rpm for 15 min. Protein concentration was determined by BCA protein assay kit (Thermo Fisher Scientific, USA) with bovine serum albumin as a standard. The total protein extract was used for western blot analysis. Equal amounts (40 μg) of total protein were subjected to 10% SDS-PAGE followed by immunoblotting with the following primary polyclonal antibodies: rabbit anti-p53 (Abcam, USA), rabbit anti-Bax (1:1,000, Abcam), rabbit anti-caspase-3 (1:1,000, Abcam), rabbit anti-AMPK (1:1,000, Cell Signaling Technology), rabbit anti-p-AMPK (1:1,000, Cell Signaling Technology), rabbit anti-AKT (1:1,000, Cell Signaling Technology), rabbit anti-p-AKT (1:1,000, Cell Signaling Technology), rabbit anti-HDAC1 (1:1,000, Abcam), rabbit anti-HDAC3 (1:1,000, Abcam), rabbit anti-HDAC6 (1:1,000, Abcam), rabbit anti-Apaf-1 (1:1,000, Abcam), mouse anti-Bcl-2 (1:1,000, Cell Signaling Technology), rabbit anti-PARP (1:1,000, Cell Signaling Technology), rabbit anti-p27 (1:1,000, Cell Signaling Technology), mouse anti-Bcl-xl (1:1,000, Abcam), rabbit anti-Ac-p53 (K382) (1:1,000, Abcam), rabbit anti-Ac-p53 (K373) (1:1,000, Abcam), mouse anti-Ac-Histone H3 (AH3-120) (1:500, Santa Cruz Biotechnology, USA), rabbit anti-Cyto-c (1:1,000, Abcam) and anti-GAPDH (1:500, Abcam). Immunoreactive bands were visualized by ECL Immunoblot Detection system (Pierce Biotechnology, Inc., Rockford, IL, USA) and exposed to Kodak (Eastman Kodak Co., USA) X-ray film. Each protein expression level was defined as grey value (Version 1.4.2b, ImageJ, National Institutes of Health, USA) and standardized to housekeeping gene of GAPDH and expressed as a fold of control. All experiments were performed in triplicate and done three times independently.
Chrysophanol pharmacokinetics in mice
Eighty, 8-week-old C57BL/6J male mice (18–22 g), were purchased from Shanghai Slac Laboratory Animal Co. Ltd. (Shanghai, China). The mice were raised in air-conditioned pathogen-free rooms under controlled lighting (12 h light/day) and fed with water and standard laboratory food. The animal study was carried out according to the regulations of The Sixth Affliated Hospital of Sun Yat-Sen University (Guangzhou, China). The chrysophanol dose was 100 mg/kg for the free fisetin (n=35) and 50 mg/kg of its nanoemulsion (n=35). Mice were sacrificed at 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22 and 24 h. The blood was obtained by cardiac puncture, centrifuged at 10,000 × g for 10 min, and the harvested plasma was kept frozen at −20°C until HPLC analysis. Finally, liver, renal and lung tissue samples were isolated from mice, and then stored in 4% paraformaldehyde and embedded in paraffin. Paraffin sections (3-μm thick) were stained with hematoxylin-eosin (H&E) and observed by light microscopy.
Mouse therapeutic trial
Four to six-week-old, female, BALB/c athymic nude mice (nu/nu) were purchased from Shanghai Slac Laboratory Animal Co. Ltd. Mouse care and usage were performed in accordance to local ethics guidelines. The mice were raised in air-conditioned pathogen-free rooms under controlled lighting (12 h light/day) and fed with water and standard laboratory food. The animal study was carried out according to the regulations of The Sixth Affliated Hospital of Sun Yat-Sen University (Guangzhou, China). Single-tumor cell suspensions (LNCap, 2×105) were injected into the left flank of each mouse subcutaneously to obtain prostate cancer xenografts. The mice were divided into three groups (15 mice/group) 5 days after cell implantation. Mice in the experimental groups were intraperitoneally injected with chrysophanol nanoparticle at a dose of 25 and 50 mg/kg body weight per day. Mice in the control group received an equal volume of normal saline. The tumor volume was calculated every two days by two cross-sectional measurements, and the tumor size was measured as follows: tumor volume = width2 × length × 0.4. Mice were sacrificed after 28 days, and the tumors were weighed and then fixed in 10% formalin for the following experiments.
Immunofluorescent assays
All cells were seeded onto glass coverslips for the experiment. After various treatments with chrysophanol nanoparticle for 24 h, the cells were harvested and washed with PBS, fixed with 4% formaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 5 min, blocked with 5% goat serum for 1 h, incubated with primary antibody overnight and then with secondary antibody (Alexa Fluor 488 labeled anti-rabbit; Sigma-Aldrich). The antibody used in the immunofluorescence staining was Cyto-c (1:200; Abcam, UK). Localization of Cyto-c was visualized using a confocal microscopy.
Immunohistochemical analysis
Paraffin-embedded tumor sections were used for the blinded assessment of apoptosis levels. Mouse tumors were sectioned at 3 μM thickness, and terminal deoxynucleotidyl transferase (TdT) dUTP nick-end labeling (TUNEL) assay was carried out using light and electron microscope-based kits (R&D Systems, USA) for detecting DNA fragments. The tumor samples isolated from mice were collected, stored in 4% paraformaldehyde and embedded in paraffin. Paraffin sections (3-μm thick) were stained with hematoxylin-eosin (H&E) and observed by light microscopy.
Statistical analysis
Data are expressed as the means ± SEM. The treated cells, tissues and corresponding controls were compared using GraphPad PRI SM (version 6.0; GraphPad Software, USA) by a one-way ANOVA with Dunn's least significant difference tests or Student's t-tests. Differences between groups were considered significant at P<0.05.
Results
Gold-chrysophanol nanoparticle characterization and its cytotoxicity to prostate cancer cells
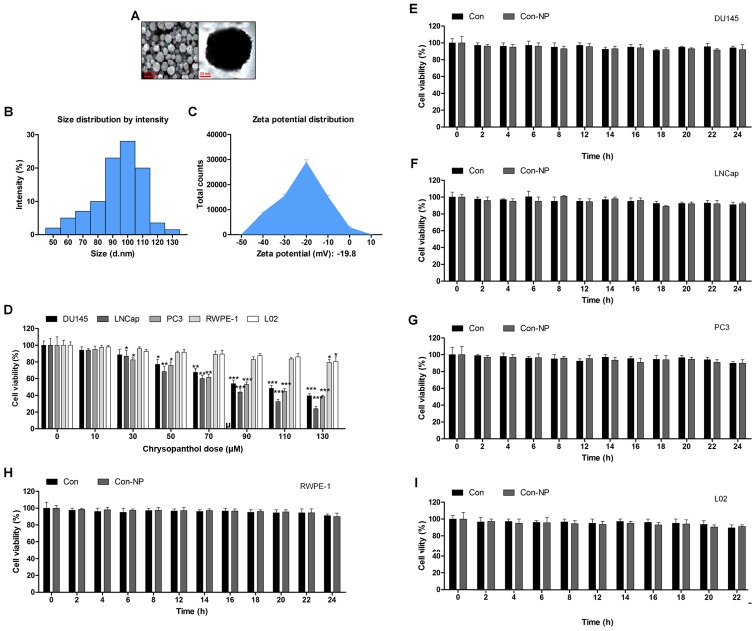
The surface morphology of chrysophanol nanoparticle was investigated under SEM. The images exhibited the spherically shaped chrysophanol nanoparticles with a smooth surface without cracks or pinholes (Fig. 1A). The DLS results indicated that mean chrysophanol nanoparticle diameter was 107.5 nm (Fig. 1B), and the zeta potential of chrysophanol nanoparticle was −18.8 mV (Fig. 1C). In this study, we attempted to explore if chrysophanol nanoparticle could be useful to prevent human prostate cancer development. Three prostate cancer cell lines were chosen to evaluate whether chrysophanol nanoparticle was effective and specific. Therefore, a dose-dependent (0–130 μM) study using human prostate cancer cell lines, DU145, LNCap, and PC3, were performed. DU145 cell viability was downregulated by chrysophanol nanoparticle from 100 to 48.5±1.8%, LNCap cells were reduced from 100 to 36.8±2.2%, and PC3 cells were decreased from 100 to 46.9±2.8% (Fig. 1D). As chrysophanol nanoparticle was more toxic to LNCap cells, all subsequent studies and experiments were conducted using LNCap cells. Prostate normal cells, RWPE-1, and human L02 normal liver cells were treated with chrysophanol nanoparticle to calculate the toxicity to normal cells. As shown in Fig. 1D, cell viability of the two cell types was decreased but showed a much lesser degree than that in the prostate cancer cell lines (Fig. 1D). As shown in Fig. 1E–I, DU145, LNCap, PC3, RWPE-1, and L02 cells were not given any treatment (Con) or only the nanoparticles (Con-NP) for different times (0, 2, 4, 6, 8, 12, 14, 16, 18, 20, 22 and 24 h). Then, the cell viability was evaluated using MTT analysis. The results indicated that there was no significant difference among various groups of cells treated under Con and Con-NP for different times, indicating insignificant difference in results between the control and the drug untreated groups (only with nanoparticles). Thus, the media as control was used for comparing the experimental data for the sake of clarity and brevity.
Figure 1.
Gold-chrysophanol nanoparticle characterization and its cytotoxicity to prostate cancer cells. (A) The scanning electron microscopy (SEM) images of chrysophanol nanoparticles. (B) Dynamic light scattering (DLS) image of cerium oxide nanoparticles. (C) Zeta potential of PLGA encapsulated chrysophanol nanoparticle. (D) Chrysophanol nanoparticle (0–130 μM) was added to human prostate cancer cell line cultures and human prostate normal epithelia cells, as well as normal liver cell culture for 24 h. The cell viability was calculated by MTT assay, and the graph exhibits a gradual downregulation in prostate cancer cell viability. (E) DU145, (F) LNCap, (G) PC3, (H) RWPE-1, and (I) L02 cells were not given any treatment (Con) or only the nanoparticles (Con-NP) for different times as indicated (0, 2, 4, 6, 8, 12, 14, 16, 18, 20, 22 and 24 h). Then, the cell viability was evaluated using MTT analysis. Data are shown as mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001 versus the control group in the absence of any treatment.
The release of chrysophanol in mice after intraperitoneal administration and its toxicity to mice
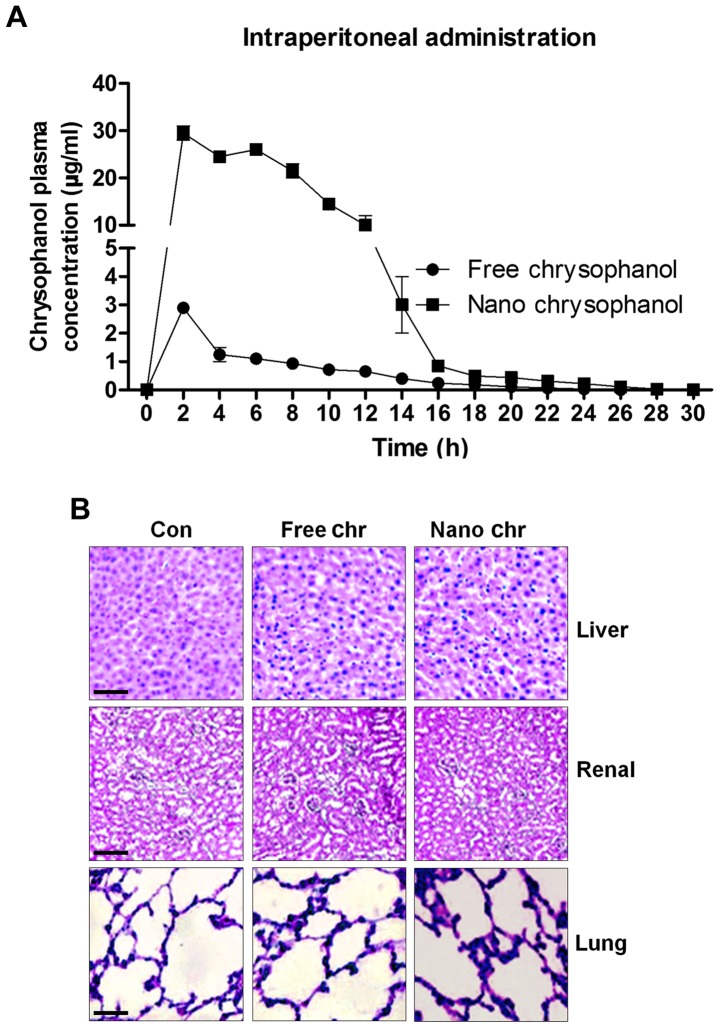
According to the results above, chrysophanol nanoparticle was effective to suppress the growth of human prostate cancer cells, especially LNCap. For the chrysophanol nanoemulsion, a comparison of its pharmacokinetics after intraperitoneal (i.p.) administration with the free chrysophanol injected by the same route is shown in Fig. 2A. Compared to the i.p. administration of free chrysophanol, the results indicated that injection of chrysophanol nanoparticle resulted in a significant upregulation of chrysophanol concentrations in plasma, even at the dose of 50 mg/kg half that of the free chrysophanol dose (100 mg/kg), indicating the effectiveness of chrysophanol nanoparticle in the organisms. Furthermore, H&E staining of liver, renal and lung were performed to investigate if chrysophanol nanoparticle showed any toxicity to animals. Compared to the Con groups, no significant histology was observed in different groups of liver, renal and lung tissue samples (Fig. 2B). The data above indicated that chrysophanol nanoparticle might be more effective than the free chrysophanol for human prostate cancer prevention with little toxicity to animals.
Figure 2.
The release of chrysophanol in mice after intraperitoneal administration and its toxicity to mice. (A) Chrysophanol pharmacokinetic in mice after intraperitoneal administration of free chrysophanol at 100 mg/kg, and after intraperitoneal administration of chrysophanol nanoemulsion at 50 mg/kg, the concentrations of chrysophanol in plasma of mice was evaluated. (B) Mice were administered with free chrysophanol and chrysophanol nanoemulsion at 100 and 50 mg/kg, respectively. Then, liver, renal and lung tissue samples were isolated for histology analysis using H&E staining. Data are shown as mean ± SEM.
The effects of chrysophanol nanoparticle on prostate cancer cell proliferation and cell arrest
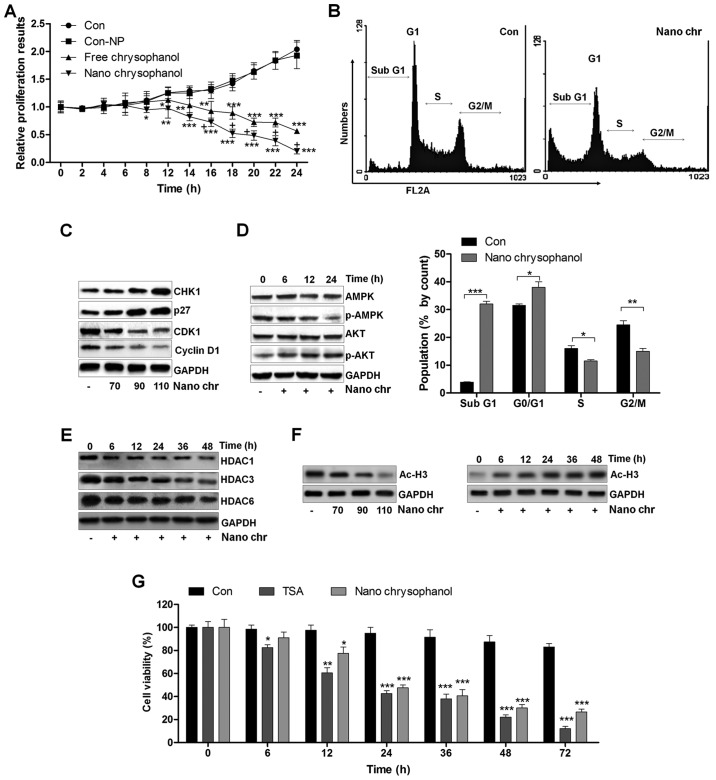
A proliferation assay was carried out to further determine the anti-proliferative action of chrysophanol nanoparticle and free chrysophanol. The results revealed that the growth rates of prostate cancer cells treated with chrysophanol nanoparticle decreased compared to that of the control, and similar results were observed in LNCap cells with free chrysophanol treatment. Also, consistently, no significant difference was observed between the Con and Con-NP group (Fig. 3A). Of note, chrysophanol nanoparticle exhibited higher anti-proliferative action, which was comparable to the free chrysophanol group. Chrysophanol nanoparticle increased the sub G-phase population and reduced the S-phase cell population, indicating induction of apoptosis in prostate cancer cells (Fig. 3B). Additionally, expression of CHK1 and p27, cell cycle-related proteins, increased, while CDK1 and cyclin D1 were decreased, indicating that these proteins were related to induction of cell cycle arrest in chrysophanol nanoparticle-treated cancer cells (Fig. 3C). We confirmed p-AMPK and p-AKT expression through western blotting. The results indicated that expression of the activated form of AMPK (p-AMPK) was upregulated in a time-dependent manner after chrysophanol nanoparticle treatment (Fig. 3D). In contrast, p-AKT was downregulated by chrysophanol nanoparticle administration. The result suggested that chrysophanol nanoparticle affected AMPK and AKT phosphorylation, reducing the survivability and proliferative potential of LNCap cells. Histone deacetylation activity has been reported to be involved in cell proliferation (14–16,18). Here, western blot analysis indicated that HDAC1, HDAC3 and HDAC6 were significantly reduced by chrysophanol nanoparticle treatment to cells in a time-dependent manner (Fig. 3E). We found that the global level acetylated Histone 3 (Ac-H3) was highly expressed with the increase of chrysophanol nanoparticle treatment (Fig. 3F). In addition, chrysophanol nanoparticle-induced Ac-H3 expression was in a time-dependent manner, which was contrary to the expression of HDACs in cancer cells treated under the same conditions. HDACs inhibitor of TSA was exposed to cells. MTT analysis indicated that the cell viability was reduced by TSA time-dependently, which was similar to chrysophanol nanoparticle administration, indicating that chrysophanol nanoparticle inhibited HDAC activity (Fig. 3F). The data above suggested that chrysophanol nanoparticle is a suppressor of transcription and proliferation.
Figure 3.
The effects of chrysophanol nanoparticles on prostate cancer cell proliferation and cell arrest. (A) LNCap cells were exposed to free chrysophanol, chrysophanol nanoparticle or only the nanoparticles in the absence of drug conjugation at 90 μM for 0, 2, 4, 6, 8, 12, 14, 16, 18, 20, 22 or 24 h. Then, the cell viability was evaluated using MTT analysis. *P<0.05, **P<0.01 and ***P<0.001 versus the control group in the absence of any treatments. (B) Upper, chrysophanol nanoparticle (90 μM) was added to LNCap cells for 24 h, and then cell cycle analysis was performed. Lower, the quantification of cells in different phases of cell cycle is exhibited. *P<0.05, **P<0.01 and ***P<0.001. LNCap cells were treated with 90 μM chrysophanol nanoparticle for 24 h. (C) Then, western blot analysis of CHK1, p27, CDK1 and cyclin D1 was carried out. (D) AMPK and AKT phosphorylation was calculated using western blot analysis. (E) LNCap cells were treated with 90 μM chrysophanol nanoparticle for different times, ranging from 0 to 48 h, followed by HDAC1, HDAC3 and HDAC6 measurement by western blot assays. (F) LNCap cells were treated as indicated. Then, western blot analysis was used to calculate the expression levels of Ac-H3. (G) LNCap cells were administered with HDACs inhibitor, TSA (5 μM), and chrysophanol nanoparticle (90 μM) for the described time, followed by MTT analysis. *P<0.05, **P<0.01 and ***P<0.001 versus the control group in the absence of any treatments. Data are shown as mean ± SEM.
The interaction of chrysophanol nanoparticle with DNA of prostate cancer cells
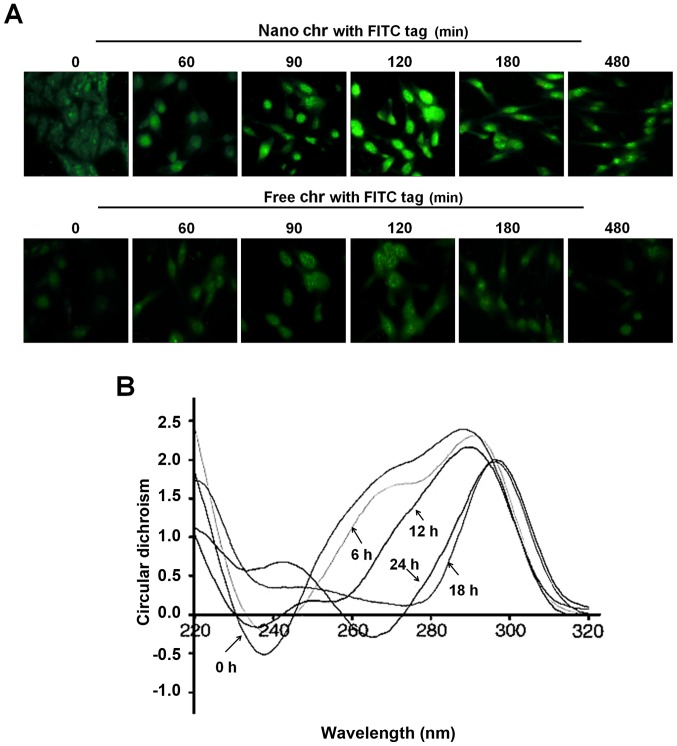
A drug entering into cells and its binding capacity with DNA are important ways to produce cytotoxicity. The immunofluorescent analysis suggested that chrysophanol nanoparticle reached a maximum level inside the cells after incubation for 120 min (Fig. 4A). However, in the cells treated with free chrysophanol exhibited weaker fluorescence compared to the group of cells with chrysophanol nanoparticles, indicating the more effective uptake of chrysophanol nanoparticles to cancer cells. In addition, the circular dichroism spectroscopic results indicated that chrysophanol nanoparticle injured the native B-confirmation of DNA in LNCap cells, with a gradually reduced negative signal and positive hypochromism signals with a peak shift. These alterations might to be the result of structural changes in the double helical DNA structure caused by chrysophanol nanoparticle (Fig. 4B).
Figure 4.
The interaction of chrysophanol nanoparticle with DNA of prostate cancer cells. (A) LNCap cells were treated with 90 μM chrysophanol nanoparticle (upper) or free chrysophanol (lower) for 0–480 min. The cells were visualized and analyzed at the described time intervals under a fluorescence microscope. Greenish cell fluorescence refer to the presence of chrysophanol nanoparticle inside the cancer cell. (B) DNA from LNCap cells was extracted at the indicated time intervals. The interaction between chrysophanol nanoparticle and DNA was investigated by circular dichroism (CD) spectroscopy. Data are shown as mean ± SEM.
Chrysophanol nanoparticle increases ROS generation, p53 expression and apoptosis
To reveal the molecular mechanism behind the cell death, ROS activity in chrysophanol nanoparticle-treated cells was evaluated using flow cytometry. The results indicated that ROS levels were produced in LNCap cells after chrysophanol nanoparticle treatment (Fig. 5A). P53 knockdown reduced ROS generation (Fig. 5B). This result confirmed that ROS production following chrysophanol nanoparticle treatment was an important event to regulate the cell death and production of ROS might be also controlled by p53 (Fig. 5B). p53 expression was determined using western blot analysis. As shown in Fig. 5C, p53 expression in LNCap cells was increased dose-dependently. P53 expression increased early in chrysophanol nanoparticle treatment and then returned to the lower level. In addition, we found that Ac-p53 both at K382 and K373 sites was highly induced by chrysophanol nanoparticles in a dose-dependent manner. Also, administration of chrysophanol nanoparticles at 110 μM, we found that both Ac-p53 (K382) and Ac-p53 (K373) were highly induced, especially when treated for 24 h. The results indicated that acetylation of p53 was also involved in chrysophanol nanoparticle-suppressed prostate cancer cells. According to previous studies, HDAC inhibitors are also reported to induce acetylation in non-histone proteins (31). For example, the HDAC1 inhibitor TSA leads to acetylation of p53 on K373 primarily under conditions in which cells are subjected to ionizing radiation, acting as a transcription factor regulating multiple genes important for the regulation of cell cycle arrest, senescence and apoptosis (32).
Figure 5.
Chrysophanol nanoparticle increases ROS generation and p53 expression. (A) LNCap cells were treated with 90 μM chrysophanol nanoparticle for 24 h. Then, flow cytometry analysis was performed to evaluate ROS generation. (B) Chrysophanol nanoparticle (90 μM) was added to LNCap cells, and ROS production was diminished when chrysophanol nanoparticle was added to p53-knockdown LNCap cells, but ROS were produced in LNCap cells expressing p53. (C) Left, 70, 90 and 110 μM chrysophanol nanoparticles were administered to LNCap cells for 24 h, which was followed by immunoblot analysis of p53 and Ac-p53. Right, p53 expression levels were calculated using western blot assay. P53 and Ac-p53 expression were upregulated at earlier treatment time, but downregulated further at late time intervals. Data are shown as mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001 versus the control group in the absence of any treatments.
LNCap cells were treated with the ROS scavenger NAC and p53 inhibitor PIF-α, as indicated in Fig. 6A to determine the relationship of p53 and ROS production. The data indicated that NAC treatment reduced p53 expression, which was enhanced by chrysophanol nanoparticle administration. Furthermore, the results showed that the NAC-treated cells combined with chrysophanol nanoparticle had a higher level of p53 expression compared to that of NAC-treated cells, while reduced p53 expression in comparison to the chrysophanol nanoparticle-treated cells. Also, in PIF-α-treated cancer cells, p53 was highly reduced, while its combination with chrysophanol nanoparticle upregulated p53 expression. Additionally, similar results were observed in p27 expression levels. Then, the cell viability was measured using MTT analysis. As shown in Fig. 6B and C, NAC and PIF-α treatment to cells showed upregulated cell viability, which were significantly reduced by treatment of chrysophanol nanoparticles. Also, NAC or PIF-α co-treated with chrysophanol nanoparticle increased the viability of LNCap cells. The data above indicated that chrysophanol nanoparticle-reduced cell proliferation was associated with ROS and p53 expression. Next, p53 expression was inhibited by PIF-α. Also, apoptosis of cancer cells was investigated using flow cytometry. The results indicated that PIF-α treatment decreased the number of apoptotic cells induced by chrysophanol nanoparticle (Fig. 6D). Thus, chrysophanol nanoparticle-regulated p53 activity was involved in apoptotic response in prostate cancer cells.
Figure 6.
The effects of chrysophanol nanoparticle on ROS, p53 and apoptosis. (A) Upper, LNCap cells were treated with 90 μM chrysophanol nanoparticle for 24 h with or without ROS scavenger of NAC (1 mM) pre-treatment for 1 h. Then, p53 and p27 protein levels were assessed using western blot analysis. Lower, LNCap cells were treated with 90 μM chrysophanol nanoparticle in the presence or absence of p53 inhibitor, PIF-α (30 μM), for 24 h. Then, all cells were harvested for p53 and p27 assessment through western blot analysis. Viability assay of LNCap cells using (B) NAC (1 mM) and (C) p53-inhibitor (PIF-α, 30 μM) with 90 μM chrysophanol nanoparticle was conducted via MTT analysis. (D) LNCap cells were exposed to 90 μM chrysophanol nanoparticle and 30 μM PIF-α in single or double treatments for 24 h. Then, flow cytometry analysis was carried out to evaluate apoptosis. Data are shown as mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001.
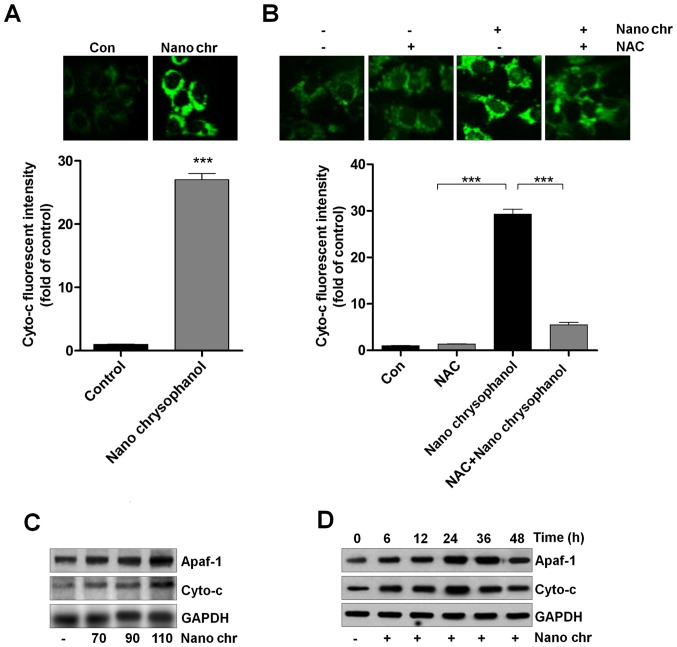
Chrysophanol nanoparticle promotes the release of Cyto-c into cytoplasm
Cyto-c expression has been reported to be linked with apoptosis induction in cancer cells, which is also related to ROS generation (33,34). Thus, here we attempted to explore its role in chrysophanol nanoparticle-induced apoptosis in LNCap cells. As shown in Fig. 7A, chrysophanol nanoparticle treatment significantly enhanced the fluorescent intensity, indicating the Cyto-c expression levels. Also, NAC treatment reduced chrysophanol nanoparticle-caused high expression of Cyto-c through immunofluorescent assays (Fig. 7B). Western blot analysis further confirmed that expression of Apaf-1 and Cyto-c in LNCap cells was upregulated by chrysophanol nanoparticle dose- and time-dependently (Fig. 7C and D). Together, the results above suggested that chrysophanol nanoparticle-reduced LNCap proliferation was related to intrinsic pathway through modulating Cyto-c expression.
Figure 7.
Chrysophanol nanoparticle promotes the release of Cyto-c into the cytoplasm. (A) LNCap cells were treated with 90 μM chrysophanol nanoparticle for 24 h, and then immunofluorescent analysis was used to evaluate Cyto-c expression levels. (B) LNCap cells were treated with 90 μM chrysophanol nanoparticle for 24 h with or without NAC (1 mM) pre-treatment for 1 h, followed by fluorescent intensity analysis of Cyto-c. (C) Chrysophanol nanoparticle (70, 90 and 110 μM) was administered to LNCap cells for 24 h. Then, Apaf-1 and Cyto-c expression levels were evaluated using western blot analysis. (D) LNCap cells were treated with 90 μM chrysophanol nanoparticle for 0, 6, 12, 24, 36 and 48 h, followed by Apaf-1 and Cyto-c measurement using western blot assays. Data are shown as mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001.
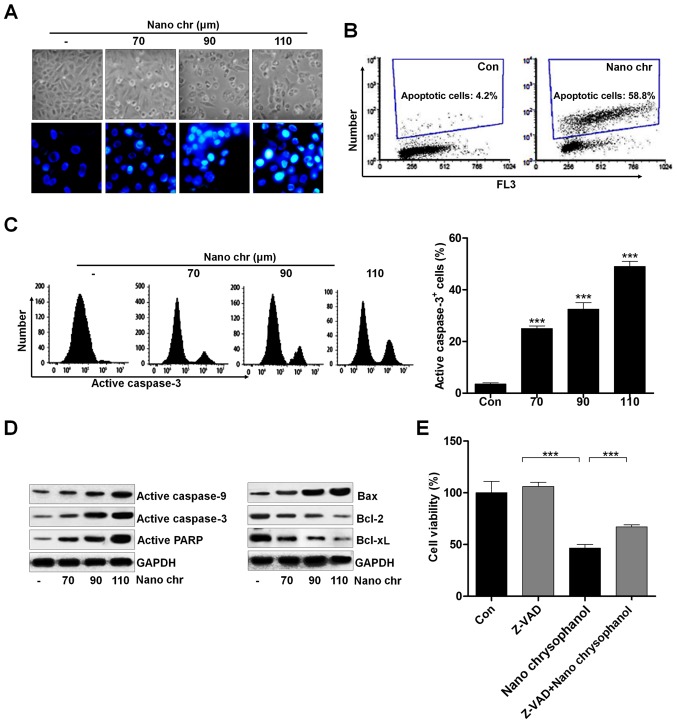
Chrysophanol nanoparticle induces apoptosis in human prostate cancer cells through activating caspase-3 signaling pathway
Activating apoptosis needs to inhibit anti-apoptotic proteins and enhance pro-apoptotic proteins, activating caspase-3 and PARP to induce apoptosis. We investigated the cellular morphological changes and DNA damage induced by NQ. NQ induced the formation of blebs and rounding off of cellular structures. The morphology and Hoechst 33342 staining of cancer cells indicated that chrysophanol nanoparticle dose-dependently induced cell death (Fig. 8A). Also, TUNEL analysis using flow cytometry revealed that apoptosis was induced by chrysophanol nanoparticle in LNCap cells (Fig. 8B). Caspase-3 activation is well reported to be essential for inducing apoptosis. Flow cytometry analysis indicated that active caspase-3 was dose-dependently induced by chrysophanol nanoparticle in prostate cancer cells (Fig. 8C). The immunoblotting was used here to calculate the role of different proteins associated with apoptosis. The overall results indicated the increased expression of caspase-9, and caspase-3, PARP, and Bax, and decreased expression of Bcl-2, and Bcl-xL when LNCap cells were incubated with chrysophanol nanoparticle for 24 h (Fig. 8D). Treatment with ZDVD-FMK (caspase-3 inhibitor) increased the cell viability, and chrysophanol nanoparticle-reduced cell survival rate was enhanced after caspase-3 inhibition (Fig. 8E). Together, these results above indicated that chrysophanol nanoparticle accelerates DNA damage and induces caspase-3-regulated apoptosis in LNCap cells.
Figure 8.
Chrysophanol nanoparticle induces apoptosis in human prostate cancer cells through activating caspase-3 signaling pathway. (A) LNCap cells were treated with various concentrations of chrysophanol nanoparticle for 24 h. Then, the morphology and Hoechst 33342 staining of cancer cells were performed. (B) The number of apoptotic cells treated with or without chrysophanol nanoparticle (90 μM) for 24 h were calculated using TUNEL assays. *P<0.05, **P<0.01 and ***P<0.001. (C) LNCap cells were treated with different concentrations of chrysophanol nanoparticles for 24 h, and then the active caspase-3 cells were analyzed using caspase-3 assay kit. (D) Active caspase-9, caspase-3 and PARP were assessed using western blot analysis. Pro- and anti-apoptotic signals of Bax, Bcl-2 and Bcl-xl were measured using immunoblotting analysis. (E) LNCap cells were exposed to 90 μM chrysophanol nanoparticle and caspase-3 inhibitor, Z-VAD (10 μM), for 24 h. Then, the cell viability was calculated using MTT analysis. Data are shown as mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001.
Chrysophanol nanoparticle reduces tumor growth in vivo
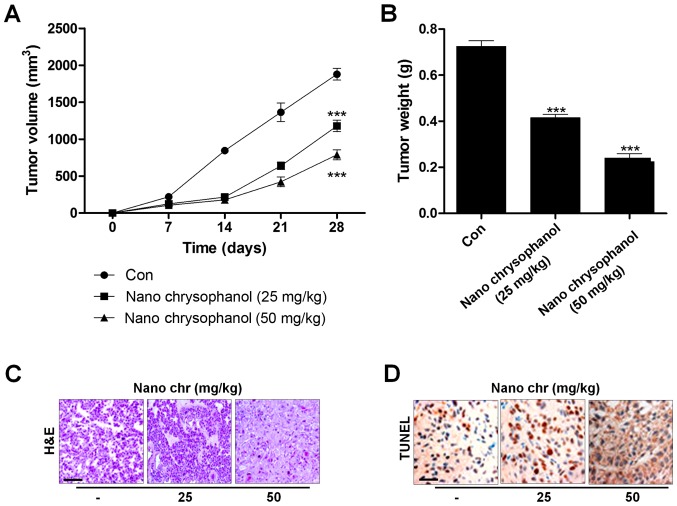
Next, xenograft tumor models were established in nude mice using LNCap cancer cells to further investigate the effects of chrysophanol nanoparticle in vivo. The tumorgenicity of the cells was calculated. LNCap cells at 2×105 were inoculated subcutaneously into nude mice. When tumors were obvious (tumor size 50 mm3), mice were randomly grouped to receive 25 and 50 mg/kg chrysophanol nanoparticle for 28 days. Then, all mice were sacrificed for tumor weight, and IHC assays. The tumor volumes were tested every 7 days. After 28 days, the tumor size and weight were significantly reduced by chrysophanol nanoparticle in mice (Fig. 9A and B). Subsequently, IHC analysis was performed to evaluate TUNEL levels in tumor tissue sections isolated from mice treated with different concentrations of chrysophanol nanoparticle. As shown in Fig. 9C, H&E staining indicated that the suppressed progression of tumor after chrysophanol nanoparticle administration. Furthermore, TUNEL-positive cells were found to be upregulated in tumor sections with chrysophanol nanoparticle treatments (Fig. 9D). Together, the data above indicated that chrysophanol nanoparticle inhibited tumor growth in vivo.
Figure 9.
Chrysophanol nanoparticle reduces tumor growth in vivo. LNCap cells (2×105) were inoculated subcutaneously into nude mice. When tumors were obvious (tumor size 50 mm3), mice were randomly grouped to receive 25 and 50 mg/kg chrysophanol nanoparticle for 28 days. Then, all mice were sacrificed for further research. The (A) volume and (B) weight of LNCap tumors were measured weekly and the tumor growth curves were plotted over time. Tumors were isolated from mice immediately after sacrifice. Tumor sections were analyzed using (C) H&E staining and (D) TUNEL assays. Data are shown as mean ± SEM (n=10). *P<0.05, **P<0.01 and ***P<0.001 versus the Con group.
Discussion
Prostate cancer has one of the highest incidence rates amongst all diagnosed cancers in males worldwide (35,36). Finding effective therapeutic strategies is urgently necessary to prevent or treat human prostate cancer progression. According to the role of chrysophanol in suppressing lung cancer, leukaemia and breast cancer, it was applied in our study to investigate if it could be a novel candidate for prostate cancer treatment (37–39). Also, the molecular mechanism revealing preventing prostate cancer by chrysophanol remains poorly understood.
Application of nanoparticles as carriers or delivery systems for chemotherapeutic drugs is attracting attention due to the specificity of nanoparticle to cancer cells, which improve drug efficiency and reduce systemic toxicity (40,41). AuNPs have some advantages, such as a bio-compatible core, making them an ideal initiating point for a nanocarrier system (42). Furthermore, AuNPs experience functionalization of multiple surfaces, highly rendering them multi-use for targeting. However, some studies have pointed out high toxicity of AuNPs (43,44). Hence, decreasing the undesirable toxicity and enhancing sustained releasing ability and bioavailability of drugs are essential considerations. In the present study, reduced AuNPs with chrysophanol and PLGA were encapsulated for the first time. PEG tagging or PLGA encapsulation could protect the compound inside from being attacked by immune cells (11). Following the results of our study, we found that nano-chrysophanol showed a significant enhancement of chrysophanol concentrations in serum, even at the half dose of the free chrysophanol, demonstrating the effectiveness of chrysophanol nanoparticle in organisms to prevent prostate cancer. In addition, chrysophanol nanoparticles exhibited more suppressive role in the cell proliferation compared to the free chrysophanol treatment. Thus, we supposed that chrysophanol nanoparticles have more advantages than its free form, which might be associated with the enhanced bioavailability, preservation, and water solubility. We attempted to calculate the underlying molecular mechanism of the tumor-killing role of the new drug in prostate cancer. We found that chrysophanol nanoparticles showed remarkable anticancer effects and toxicity towards human prostate cancer cells, and more effectively towards LNCap cells than other two prostate cancer cells. Furthermore, no significant difference was observed in human normal prostate cells and normal liver cells, indicating its low toxicity. We revealed that the i.p. administration of chrysophanol nanoparticles led to a significant improvement of bioavailability in comparison to the i.p. administration of free chrysophanol administered through the same route.
The therapy of targeting DNA was developed recently. DNA functions as a large receptor for a variety numbers of small molecules and thus becomes an important target for exploring anticancer agents (45,46). Circular dichroism spectroscopic data suggested that chrysophanol nanoparticles have the activity to interact with DNA. Therefore, chrysophanol nanoparticle cytotoxicity might be due to its inhibition of DNA synthetic process at least partly. The blocking capacity of chrysophanol nanoparticles made it effective to induce apoptosis in LNCap cells, and enhancing chrysophanol nanoparticles into nuclear DNA also caused nuclear damage. The circular dichroism spectroscopic data indicated that the interaction between chrysophanol nanoparticles and the cellular DNA in LNCap cells was more pronounced 120 min after treatment of chrysophanol nanoparticles. The interaction of chrysophanol nanoparticles with DNA promoted the ability of the drug to suppress the cell proliferation and to induce cell cycle arrest, which was accompanied with reduced cyclin D1 and CDK1, essential signals to promote cell proliferation through affecting G1 progression (47,48). In contrast, elevated p27 levels, regulating cell differentiation and cell cycle arrest, were found to be upregulated by chrysophanol nanoparticles (49).
Histone deacetylase inhibitors have been investigated as therapeutic strategy for various cancers (19,20). The exact molecular mechanisms by which the inhibitors work remained unclear. Histone deacetylase inhibitors induced p27 and p21 expression, associated with p53 activity to inhibit tumor progression via triggering cell differentiation, growth arrest, or apoptosis-related death in a variety of cultured cells, such as leukemia cells, neuroblastoma, and melanoma, and cells from prostate, ovary, liver, lung, and gastric cancers (50–52). Histone deacetylase inhibitors induce apoptosis, which could be assayed by DNA fragmentation, and the number of sub-G1 population was assessed using flow cytometry. Overexpression of HDACs, including HDAC1, HDAC3 and HDAC6, which are reported as the key histone deacetylation regulators, is associated with cancer development through silencing p53, p27 and p21 (53,54). In our study, we found that chrysophanol nanoparticles, similar to inhibitors of HDACs, could reduce HDAC1, HDAC3 and HDAC6 expression, a possible molecular mechanism by which chrysophanol nanoparticles performed their role in preventing human prostate cancer progression. The treatment of chrysophanol nanoparticles further resulted in the concomitant enrichment of acetyl histones such as Ac-H3.
Chrysophanol nanoparticle has a similar ability to trigger ROS generation. Chrysophanol has efficacy to induce apoptosis in p53-expressing cancer cells by promoting ROS. The molecular mechanism behind the enhancement in ROS is increased p53 activity (55). P53 expression and ROS generation are correlated, and p53 could modulate proliferative proteins, including cyclin D1 (56). Thus, elevated p53 activity and ROS production occurred in prostate cancer cells. ROS-dependent apoptosis involving p53 relies on damage to mitochondrial generally. The dramatic alterations in mitochondrial morphology during the early stages of apoptotic cell death, involve the network fragmentation and the remodeling of cristae. Activation-inactivation of p53 is also dependent on acetylation. P53 acetylation was found to be indispensable for its activation (57,58). In this study, we also found that expression levels of Ac-p53 were induced by chrysophanol nanoparticle, which suggested that p53 activation through its acetylation might be included in chrysophanol nanoparticle-prevention of the progression of prostate cancer. Translocation of Bax onto mitochondrial membrane produces pores, depolarizing the potential of mitochondrial membrane. Subsequently, cytochrome c (Cyto-c) moves into the cytoplasm and initiates the formation of apoptosomes along with adopter molecule Apaf-1, as well as other pro-caspase molecules, including caspase-9 and caspase-3 (59,60). Caspase-3 is regarded as the terminal mediator of cell apoptosis in caspase family. Caspase-3 activation has been suggested to activate PARP and induce apoptosis, which is also related to ROS production (61,62). In our study, we found that chrysophanol nanoparticles enhanced Cyto-c expression in cells and elevated caspase-9, caspase-3 and PARP activity. Accordingly, the anti-apoptotic signals, Bcl-2 and Bcl-xl, were reduced by chrysophanol nanoparticles in prostate cancer cells (63).
AMPK is a pleiotropic enzyme, playing a role in the regulation of numerous functions both peripherally and centrally (64). AMPK has now been considered as a potential therapeutic target for preventing cancer as a consequence of being a substrate (65). Furthermore, AKT signaling pathways are linked to the inhibition of proliferation, apoptosis, and the metastasis of cells, angiogenesis and various other processes (66,67). AKT is involved in the proliferation of cells through its activation of cyclin D1, and inactivates caspases (68,69). Our results indicated that chrysophanol nanoparticles upregulated AMPK activation, while downregulated AKT phosphorylation, contributing to the suppression of prostate cancer cells.
In conclusion, chrysophanol nanoparticles reduced the expression of HDACs, CDK1, cyclin D1 and p-AKT, which might be related to the cell cycle arrest. The upregulation of p27, CHK1 and p-AMPK may have led to the arrest of LNCap cell proliferation. Flow cytometry analysis indicated that chrysophanol nanoparticles arrested the LNCap cell cycle in sub-G phase. Additionally, treatment of chrysophanol nanoparticles resulted in ROS production, along with the release of Cyto-c and Apaf-1 expression, inducing apoptosis in LNCap cells through activating caspase-3. Hence, inhibition of proliferation and induction of ROS and apoptosis by chrysophanol nanoparticles may provide a potential therapeutic strategy to prevent human prostate cancer development.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (no. 81402111).
References
- 1.Øverbye A, Skotland T, Koehler CJ, Thiede B, Seierstad T, Berge V, Sandvig K, Llorente A. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget. 2015;6:30357–30376. doi: 10.18632/oncotarget.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanno T, Rabel A, Alleyne M, Lee YT, Dahut WL, Gulley JL, Miller JL. Hepcidin, anaemia, and prostate cancer. BJU Int. 2011;107:678–679. doi: 10.1111/j.1464-410X.2011.10108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu T, Wu LY, Kazak M, Berkman CE. Cell-Surface labeling and internalization by a fluorescent inhibitor of prostate-specific membrane antigen. Prostate. 2008;68:955–964. doi: 10.1002/pros.20753. [DOI] [PubMed] [Google Scholar]
- 4.Tagawa ST, Beltran H, Vallabhajosula S, Goldsmith SJ, Osborne J, Matulich D, Petrillo K, Parmar S, Nanus DM, Bander NH. Anti-prostate-specific membrane antigen-based radioimmunotherapy for prostate cancer. Cancer. 2010;116(Suppl):1075–1083. doi: 10.1002/cncr.24795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho SY, Gage KL, Mease RC, Senthamizhchelvan S, Holt DP, Jeffrey-Kwanisai A, Endres CJ, Dannals RF, Sgouros G, Lodge M, et al. Biodistribution, tumor detection, and radiation dosimetry of 18F-DCFBC, a low-molecular-weight inhibitor of prostate-specific membrane antigen, in patients with metastatic prostate cancer. J Nucl Med. 2012;53:1883–1891. doi: 10.2967/jnumed.112.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.López-Lázaro M. Distribution and biological activities of the flavonoid luteolin. Mini Rev Med Chem. 2009;9:31–59. doi: 10.2174/138955709787001712. [DOI] [PubMed] [Google Scholar]
- 7.Webb KM, DiRuggiero J. Role of Mn2+ and compatible solutes in the radiation resistance of thermophilic bacteria and archaea. Archaea. 2012;2012:845756. doi: 10.1155/2012/845756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu CC, Yang JS, Huang AC, Hsia TC, Chou ST, Kuo CL, Lu HF, Lee TH, Wood WG, Chung JG. Chrysophanol induces necrosis through the production of ROS and alteration of ATP levels in J5 human liver cancer cells. Mol Nutr Food Res. 2010;54:967–976. doi: 10.1002/mnfr.200900265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darzynkiewicz Z, Carter SP, Kapuscinski J, Watanabe KA. Effect of derivatives of chrysophanol, a new type of potential antitumor agents of anthraquinone family, on growth and cell cycle of L1210 leukemic cells. Cancer Lett. 1989;46:181–187. doi: 10.1016/0304-3835(89)90128-6. [DOI] [PubMed] [Google Scholar]
- 10.Zeng X, Tao W, Mei L, Huang L, Tan C, Feng SS. Cholic acid-functionalized nanoparticles of star-shaped PLGA-vitamin E TPGS copolymer for docetaxel delivery to cervical cancer. Biomaterials. 2013;34:6058–6067. doi: 10.1016/j.biomaterials.2013.04.052. [DOI] [PubMed] [Google Scholar]
- 11.Thamake SI, Raut SL, Gryczynski Z, Ranjan AP, Vishwanatha JK. Alendronate coated poly-lactic-co-glycolic acid (PLGA) nanoparticles for active targeting of metastatic breast cancer. Biomaterials. 2012;33:7164–7173. doi: 10.1016/j.biomaterials.2012.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Hrkach J, Von Hoff D, Mukkaram Ali M, Andrianova E, Auer J, Campbell T, De Witt D, Figa M, Figueiredo M, Horhota A, et al. Preclinical development and clinical translation of a PSMA-targeted docetaxel nanoparticle with a differentiated pharmacological profile. Sci Transl Med. 2012;4:128ra39. doi: 10.1126/scitranslmed.3003651. [DOI] [PubMed] [Google Scholar]
- 13.Ganju A, Yallapu MM, Khan S, Behrman SW, Chauhan SC, Jaggi M. Nanoways to overcome docetaxel resistance in prostate cancer. Drug Resist Updat. 2014;17:13–23. doi: 10.1016/j.drup.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X. Gold nanoparticles: Recent advances in the biomedical applications. Cell Biochem Biophys. 2015;72:771–775. doi: 10.1007/s12013-015-0529-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang CC, Wu SM, Li HW, Chang HT. Biomedical applications of DNA-conjugated gold nanoparticles. ChemBioChem. 2016;17:1052–1062. doi: 10.1002/cbic.201600014. [DOI] [PubMed] [Google Scholar]
- 16.Simon LC, Stout RW, Sabliov C. Bioavailability of orally delivered alpha-tocopherol by poly (lactic-co-glycolic) acid (PLGA) nanoparticles and chitosan covered PLGA nanoparticles in F344 rats. Nanobiomedicine. 2016;3:8. doi: 10.5772/63305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin TsT, Gao DY, Liu YC, Sung YC, Wan D, Liu JY, Chiang T, Wang L, Chen Y. Development and characterization of sorafenib-loaded PLGA nanoparticles for the systemic treatment of liver fibrosis. J Control Release. 2016;221:62–70. doi: 10.1016/j.jconrel.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/S0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 19.Horikoshi M. Histone acetylation: From code to web and router via intrinsically disordered regions. Curr Pharm Des. 2013;19:5019–5042. doi: 10.2174/1381612811319280002. [DOI] [PubMed] [Google Scholar]
- 20.Sachweh MC, Drummond CJ, Higgins M, Campbell J, Laín S. Incompatible effects of p53 and HDAC inhibition on p21 expression and cell cycle progression. Cell Death Dis. 2013;4:e533. doi: 10.1038/cddis.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T, Kuljaca S, Tee A, Marshall GM. Histone deacetylase inhibitors: Multifunctional anticancer agents. Cancer Treat Rev. 2006;32:157–165. doi: 10.1016/j.ctrv.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weichert W, Röske A, Gekeler V, Beckers T, Ebert MP, Pross M, Dietel M, Denkert C, Röcken C. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: A retrospective analysis. Lancet Oncol. 2008;9:139–148. doi: 10.1016/S1470-2045(08)70004-4. [DOI] [PubMed] [Google Scholar]
- 24.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 26.Zuo Q, Wu W, Li X, Zhao L, Chen W. HDAC6 and SIRT2 promote bladder cancer cell migration and invasion by targeting cortactin. Oncol Rep. 2012;27:819–824. doi: 10.3892/or.2011.1553. [DOI] [PubMed] [Google Scholar]
- 27.Park SY, Jun JA, Jeong KJ, Heo HJ, Sohn JS, Lee HY, Park CG, Kang J. Histone deacetylases 1, 6 and 8 are critical for invasion in breast cancer. Oncol Rep. 2011;25:1677–1681. doi: 10.3892/or.2011.1236. [DOI] [PubMed] [Google Scholar]
- 28.Aldana-Masangkay GI, Sakamoto KM. The role of HDAC6 in cancer. J Biomed Biotechnol. 2011;2011:875824. doi: 10.1155/2011/875824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henderson C, Mizzau M, Paroni G, Maestro R, Schneider C, Brancolini C. Role of caspases, Bid, and p53 in the apoptotic response triggered by histone deacetylase inhibitors trichostatin-A (TSA) and suberoylanilide hydroxamic acid (SAHA) J Biol Chem. 2003;278:12579–12589. doi: 10.1074/jbc.M213093200. [DOI] [PubMed] [Google Scholar]
- 31.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 32.Kloster MM, Naderi EH, Haaland I, Gjertsen BT, Blomhoff HK, Naderi S. cAMP signalling inhibits p53 acetylation and apoptosis via HDAC and SIRT deacetylases. Int J Oncol. 2013;42:1815–1821. doi: 10.3892/ijo.2013.1853. [DOI] [PubMed] [Google Scholar]
- 33.Jiang X, Wang X. Cytochrome c-mediated apoptosis. Annu Rev Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 34.Garrido C, Galluzzi L, Brunet M, Puig PE, Didelot C, Kroemer G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006;13:1423–1433. doi: 10.1038/sj.cdd.4401950. [DOI] [PubMed] [Google Scholar]
- 35.Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM, Magi-Galluzzi C, Mehra R, Sahu A, Siddiqui J, et al. RNA biomarkers associated with metastatic progression in prostate cancer: A multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15:1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Severi G, Morris HA, MacInnis RJ, English DR, Tilley W, Hopper JL, Boyle P, Giles GG. Circulating steroid hormones and the risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:86–91. doi: 10.1158/1055-9965.EPI-05-0633. [DOI] [PubMed] [Google Scholar]
- 37.Ni CH, Yu CS, Lu HF, Yang JS, Huang HY, Chen PY, Wu SH, Ip SW, Chiang SY, Lin JG, et al. Chrysophanol-induced cell death (necrosis) in human lung cancer A549 cells is mediated through increasing reactive oxygen species and decreasing the level of mitochondrial membrane potential. Environ Toxicol. 2014;29:740–749. doi: 10.1002/tox.21801. [DOI] [PubMed] [Google Scholar]
- 38.Hong JY, Chung HJ, Bae SY, Trung TN, Bae K, Lee SK. Induction of cell cycle arrest and apoptosis by physcion, an anthraquinone isolated from rhubarb (rhizomes of Rheum tanguticum), in MDA-MB-231 human breast cancer cells. J Cancer Prev. 2014;19:273–278. doi: 10.15430/JCP.2014.19.4.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozenver N, Saeed M, Guvenalp Z, et al. Chrysophanol-and nepodin-8-O-β-D-glucopyranoside from Rumex acetosella, the cytotoxicity towards drug sensitive and multi-drug resistant T leukaemia cancer cells. Planta Med. 2016;81:388. [Google Scholar]
- 40.Gradishar WJ, Tjulandin S, Davidson N, Shaw H, Desai N, Bhar P, Hawkins M, O'Shaughnessy J. Phase III trial of nanoparticle albumin-bound paclitaxel compared with polyethylated castor oil-based paclitaxel in women with breast cancer. J Clin Oncol. 2005;23:7794–7803. doi: 10.1200/JCO.2005.04.937. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Tan B, Wu Y. Mesoporous Co3O4 nanowire arrays for lithium ion batteries with high capacity and rate capability. Nano Lett. 2008;8:265–270. doi: 10.1021/nl0725906. [DOI] [PubMed] [Google Scholar]
- 42.Park SY, Chae SY, Park JO, Lee KJ, Park G. Gold-conjugated resveratrol nanoparticles attenuate the invasion and MMP-9 and COX-2 expression in breast cancer cells. Oncol Rep. 2016;35:3248–3256. doi: 10.3892/or.2016.4716. [DOI] [PubMed] [Google Scholar]
- 43.Saha K, Agasti SS, Kim C, Li X, Rotello VM. Gold nanoparticles in chemical and biological sensing. Chem Rev. 2012;112:2739–2779. doi: 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi J, Chan C, Pang Y, Ye W, Tian F, Lyu J, Zhang Y, Yang M. A fluorescence resonance energy transfer (FRET) biosensor based on graphene quantum dots (GQDs) and gold nanoparticles (AuNPs) for the detection of mecA gene sequence of Staphylococcus aureus. Biosens Bioelectron. 2015;67:595–600. doi: 10.1016/j.bios.2014.09.059. [DOI] [PubMed] [Google Scholar]
- 45.Zeng L, Wu GZ, Goh KJ, Lee YM, Ng CC, You AB, Wang J, Jia D, Hao A, Yu Q, et al. Saturated fatty acids modulate cell response to DNA damage: Implication for their role in tumorigenesis. PLoS One. 2008;3:e2329. doi: 10.1371/journal.pone.0002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fahrer J, Kaina B. O6-methylguanine-DNA methyltransferase in the defense against N-nitroso compounds and colorectal cancer. Carcinogenesis. 2013;34:2435–2442. doi: 10.1093/carcin/bgt275. [DOI] [PubMed] [Google Scholar]
- 47.Casagrande F, Darbon JM. Effects of structurally related flavonoids on cell cycle progression of human melanoma cells: Regulation of cyclin-dependent kinases CDK2 and CDK1. Biochem Pharmacol. 2001;61:1205–1215. doi: 10.1016/S0006-2952(01)00583-4. [DOI] [PubMed] [Google Scholar]
- 48.Hansel DE, Dhara S, Huang RC, Ashfaq R, Deasel M, Shimada Y, Bernstein HS, Harmon J, Brock M, Forastiere A, et al. CDC2/CDK1 expression in esophageal adenocarcinoma and precursor lesions serves as a diagnostic and cancer progression marker and potential novel drug target. Am J Surg Pathol. 2005;29:390–399. doi: 10.1097/00000478-200503000-00014. [DOI] [PubMed] [Google Scholar]
- 49.Lu Z, Hunter T. Ubiquitylation and proteasomal degradation of the p21(Cip1), p27(Kip1) and p57(Kip2) CDK inhibitors. Cell Cycle. 2010;9:2342–2352. doi: 10.4161/cc.9.12.11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pandey M, Kaur P, Shukla S, Abbas A, Fu P, Gupta S. Plant flavone apigenin inhibits HDAC and remodels chromatin to induce growth arrest and apoptosis in human prostate cancer cells: In vitro and in vivo study. Mol Carcinog. 2012;51:952–962. doi: 10.1002/mc.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schäfer C, Göder A, Beyer M, Kiweler N, Mahendrarajah N, Rauch A, Nikolova T, Stojanovic N, Wieczorek M, Reich TR, et al. Class I histone deacetylases regulate p53/NF-κB crosstalk in cancer cells. Cell Signal. 2017;29:218–225. doi: 10.1016/j.cellsig.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 52.Ververis K, Hiong A, Karagiannis TC, Licciardi PV. Histone deacetylase inhibitors (HDACIs): Multitargeted anticancer agents. Biologics. 2013;7:47–60. doi: 10.2147/BTT.S29965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varricchio L, Dell'Aversana C, Nebbioso A, Migliaccio G, Altucci L, Mai A, Grazzini G, Bieker JJ, Migliaccio AR. Identification of NuRSERY, a new functional HDAC complex composed by HDAC5, GATA1, EKLF and pERK present in human erythroid cells. Int J Biochem Cell Biol. 2014;50:112–122. doi: 10.1016/j.biocel.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan H, Li AJ, Ma SL, Cui LJ, Wu B, Yin L, Wu MC. Inhibition of autophagy significantly enhances combination therapy with sorafenib and HDAC inhibitors for human hepatoma cells. World J Gastroenterol. 2014;20:4953–4962. doi: 10.3748/wjg.v20.i17.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD, Clair DK. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res. 2005;65:3745–3750. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 56.Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol. 2004;24:6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Proietti S, Cucina A, Dobrowolny G, D'Anselmi F, Dinicola S, Masiello MG, Pasqualato A, Palombo A, Morini V, Reiter RJ, et al. Melatonin down-regulates MDM2 gene expression and enhances p53 acetylation in MCF-7 cells. J Pineal Res. 2014;57:120–129. doi: 10.1111/jpi.12150. [DOI] [PubMed] [Google Scholar]
- 58.Ono W, Hayashi Y, Yokoyama W, Kuroda T, Kishimoto H, Ito I, Kimura K, Akaogi K, Waku T, Yanagisawa J. The nucleolar protein Myb-binding protein 1A (MYBBP1A) enhances p53 tetramerization and acetylation in response to nucleolar disruption. J Biol Chem. 2014;289:4928–4940. doi: 10.1074/jbc.M113.474049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Estaquier J, Vallette F, Vayssiere JL, Mignotte B. The mitochondrial pathways of apoptosis. Adv Exp Med Biol. 2012;942:157–183. doi: 10.1007/978-94-007-2869-1_7. [DOI] [PubMed] [Google Scholar]
- 60.Pradelli LA, Bénéteau M, Ricci JE. Mitochondrial control of caspase-dependent and -independent cell death. Cell Mol Life Sci. 2010;67:1589–1597. doi: 10.1007/s00018-010-0285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi Y. caspase activation: Revisiting the induced proximity model. Cell. 2004;117:855–858. doi: 10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 62.Isabelle M, Moreel X, Gagné JP, Rouleau M, Ethier C, Gagné P, Hendzel MJ, Poirier GG. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci. 2010;8:22. doi: 10.1186/1477-5956-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: Implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 64.Li W, Saud SM, Young MR, Chen G, Hua B. Targeting AMPK for cancer prevention and treatment. Oncotarget. 2015;6:7365–7378. doi: 10.18632/oncotarget.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oakhill JS, Scott JW, Kemp BE. AMPK functions as an adenylate charge-regulated protein kinase. Trends Endocrinol Metab. 2012;23:125–132. doi: 10.1016/j.tem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 66.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 67.Benbrook DM, Masamha CP. The pro-survival function of Akt kinase can be overridden or altered to contribute to induction of apoptosis. Curr Cancer Drug Targets. 2011;11:586–599. doi: 10.2174/156800911795655994. [DOI] [PubMed] [Google Scholar]
- 68.Unger C, Popescu R, Giessrigl B, Rarova L, Herbacek I, Seelinger M, Diaz R, Wallnöfer B, Fritzer-Szekeres M, Szekeres T, et al. An apolar extract of Critonia morifolia inhibits c-Myc, cyclin D1, Cdc25A, Cdc25B, Cdc25C and Akt and induces apoptosis. Int J Oncol. 2012;40:2131–2139. doi: 10.3892/ijo.2012.1412. [DOI] [PubMed] [Google Scholar]
- 69.Polivka J, Jr, Janku F. Molecular targets for cancer therapy in the I3K/AKT/mTOR pathway. Pharmacol Ther. 2014;142:164–175. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]