Abstract
Though incidence of PI3K oncogenic mutation is prominent in breast cancer (20-30%), pharmacological targeting of this signaling pathway alone has failed to provide meaningful clinical benefit. To better understand and address this problem, we conducted genome-wide analysis to study the association of mutant PI3K with other gene amplification events. One of the most significant copy number gain events associated with PIK3CA mutation was the region within chromosome 17 containing HER2To investigate the oncogenic effect and cell signaling regulation of co-occurring PIK3CA-H1047R and or HER2 gene, we generated cell models ectopically expressing mutant PIK3CA, HER2 or both genetic alterations. We observed that cells with both genetic alterations demonstrate increased aggressiveness and invasive capabilities than cells with either genetic change alone. Furthermore, we found that the combination of the HER2 inhibitor (CP-724714) and pan PI3K inhibitor (LY294002) is more potent than either inhibitor alone in terms of inhibition of cell proliferation and colony formation. Significantly, four cell signaling pathways were found in common for cells with HER2, mutant PIK3CA and cells with both genetic alterations through an Affymetric microarray analysis. Moreover, the cells with both genetic alterations acquired more significant replication stress as shown by enriched signaling pathways of cell cycle checkpoint control and DNA damage response signaling. Our study suggests co-occurrence of oncogenic HER2 and mutant PIK3CA cooperatively drives breast cancer progression. The cells with both genetic alterations obtain additional features of replication stress which could open new opportunity for cancer diagnostics and treatment.
Keywords: PIK3CA, HER2, breast cancer, gene amplification
Introduction
The PI3K/PTEN/AKT signaling pathway has well-established roles in multiple cellular activities, including cell proliferation, survival, metabolism, cytoskeleton reorganization and membrane trafficking (1-3). The abnormal activation of this signaling pathway leads to various diseases such as diabetes, autoimmunity and cancer. About ten years ago, our group, along with several other research groups, simultaneously found that the somatic mutation frequency of the PIK3CA gene in breast cancer is 20-30% (4-7). Our research demonstrated that somatic mutation, rather than gain of PIK3CA copy number, is one of the most frequent genetic alterations contributing to human breast cancer progression (7). In another study, we comprehensively analyzed and compared the oncogenic properties of nine different PIK3CA somatic mutations, which localized in different domains of the PIK3CA gene and with different frequencies in human breast cancer (8). The results of our study are consistent with several other groups, using different research systems, and strongly indicate that different PI3KCA mutants exhibit different abilities in contributing to cell proliferation, EGF independent growth, cell morphogenesis, transformation, invasion and signaling (9-12). These findings collectively provide fundamental biological evidence to support the critical role of the PI3k/AkT signalling pathway in breast cancer progression. However, to date, there is insufficient clinical data to support that PI3K or AKT inhibitors can be powerful single agents for breast cancer patients (13,14).
HER2 (ErbB2), a member of the HER family of tyrosine kinase receptors (HER1-4), is a major driver of tumor growth in 20% of breast cancers. Due to the well-studied nature of the HER2 gene in breast cancer and the availability of the monoclonal targeting antibody trastuzumab, targeting HER2 has been the most successful targeted treatment for breast cancer patients (15,16). However, targeting HER2 alone was less effective for breast cancer patients with PIK3CA mutations in clinical studies (17,18). In line with these observations, several groups reported that HER2 amplification and mutation of PIK3CA genes could be co-occurring in certain breast cancer population (6,19-22). However, the cooperative effect of these two genetic alterations in comparison with either single genetic change on cell oncogenic properties has not been well investigated.
In this study, we performed a genome-wide analysis for amplification regions and corresponding genes that correlate to mutant PIK3CA in 51 human breast cancer cell lines. We also specifically examined the oncogenic properties driven by expressing both mutant PIK3CA and HER2 and compare the effects to cells with either genetic alteration alone. Additionally, we tested the drug treatment response in cells with ectopic expression of mutant PIK3CA and HER2 amplification. Finally, we investigated the downstream target genes and cell signalling pathways regulated by HER2, mutant PIK3CA and both of these genetic alterations.
Materials and methods
Bioinformatics analysis for amplification of regions that are correlated with mutant PIK3CA
A published database was used for bioinformatic analysis. This database contains gene expression and copy number information for 51 breast cancer cell lines (23) (http://caarraydb.nci.nih.gov/caarray/publicEx-perimentDetailAction.do?expId=1015897590151581 at http://cancer.lbl.gov/breastcancer/data.php). Among these 51 cell lines, 13 cell lines contain PIK3CA mutations. The other 38 are considered PIK3CA-wild type. We then used the strategy as follows to identify amplified gene that could synergize with mutant PIK3CA in breast cancer. a) Threshold aCGH and gene expression data: copy number variation (CNV) amplification based on a cut-off ≥0.2. Gene overexpression based on a cut-off >143.767 (3-fold of the median of all samples). b) CNV markers and genes with highly increased amplification/overexpression frequency based on the following criteria: i) frequency difference between cell line w/mutations and w/o ≥0.25 or ii) Fisher exact test P-value of the difference <0.05. c) Pairs of amplified CNV markers and overexpressed genes which are close to each other (distance ≤2 Mb). d) Pairs of amplified CNV markers and overexpressed genes which are close to each other (distance ≤2 Mb) and positively correlated.
Cell culture
MCF 10A and HCC1954 cells were obtained from the American Tissue culture collection. MCF10A cell lines expressing LacZ (negative control), PI3KCA-H1047R, HER2, and both PI3KCA-H1074R and HER2 genes were created in our laboratory at the Barbara Ann karmanos cancer Institute (KCI). Briefly, full-length PIK3CA-H1047R and HER2 were subcloned into a pENTR vector and recombinated into the pLenti-6/V5-DeST vector. The lentiviruses for the full-length genes were generated using the pLenti-virus-expression system (Invitrogen). The generated virus was used to infect targeted model cells. Stable cells were generated after being selected with blasticidin (10 μg/ml, Invivogen) and were grown at 37°C in an incubator with 5% CO2Most of the cell lines used were originally grown in standard SFIHE medium (serum-free medium with insulin, hydrocortisone, and epidermal growth factor) made by Ham's F12 media supplemented with 0.1% bovine serum albumin, 0.5 μg/ml fungizone, 5 μg/ml gentamycin, 5 mM ethanolamine, 10 mM HEPES, 5 μg/ml transferrin, 10 μM T3, 50 μM selenium, 1 μg/ml hydrocortisone, 10 ng/ml EGF, and 5 μg/ml insulin. MCF10A cells expressing HER2 were grown in SFHE medium (serum-free medium with hydrocortisone and EGF). For experimentation, each individual cell line with its own different overexpressed gene was first placed in SFIHE or SFHE medium for a few days, and then placed in either SFIHE medium, SFIH (lacking EGF or epidermal growth factor), SFHE (lacking insulin), or SFH (lacking both EGF and insulin) medium in order to observe cell growth and colony formation. For the invasion assay, cells were first plated in each well insert with SFIH medium, and then SFIHE with 5% FBS was placed outside in the well. Tests on inhibition were done in SFIHE medium.
Cell proliferation/MTT assay
McF10A/HER2, McF10A/PIK3CA-h1047r, McF10A/HER2/PIK3CA-H1047R and McF10A/LacZ were seeded in triplicate at a density of 2×103 cells per well in 96-well plates on day 0. Cells were grown in SFIHE (growth medium with insulin, hydrocortisone and EGF), SFIH (growth medium lacking EGF) or SFHE (growth medium lacking insulin) or SFH (lacking both EGF and insulin) medium. Cell proliferation was measured with an MTT assay kit (Fisher Scientific, Pittsburgh, PA, USA), trypsinized and counted with a particle counter (Beckman Coulter) with an absorbance value of 540 nm on days 1, 3, 5, and 7 to make the growth curve. Media was changed at each of these time points.
Colony formation
Each cell line was seeded in two wells of a 6-well plate with a density of 2×103 cells per well for 1-2 weeks in SFIHE, SFIH, SFHE, or SFH culture mediums. As soon as distinct clones were visible with a microscope and were almost starting to merge, all plates were stained with crystal violet on day 12 of growth for approximately 5 min and then exposed to water gently. The numbers of individual clones were then counted and recorded, and the plates were scanned for clear pictures of clones.
Migration and invasion assay
A cell migration and cell invasion assay was performed in BD control chamber and BioCoat Matrigel Invasion chambers (BD Bioscience, Corning, NY, USA) according to the manufacturer's instructions. Cells (5×104) were plated in the upper chamber in 500 μl of SFIH growth medium without FBS, and the lower chamber was filled with 750 μl of SFIHE growth medium with 5% FBS. After 48-h incubation, the cells that had invaded to the underside of the membrane were fixed and stained with the HEMA 3 kit (Fisher Scientific). The migration and invasive cells were then determined by counting the stained cells by microscopy of ×100 and images were taken.
Inhibition of cell proliferation or colony formation by inhibitors
LY294002 [pan PI3K inhibitor (24,25)], and CP-724714 [selective HER2 inhibitor (26,27)] were diluted to the concentrations of 3 and 10 μM and 0.1 and 1 μM in SFIHE medium, respectively, according to previous data. SFIHE medium with either concentration of the PI3K inhibitor, the HER2 inhibitor, or both inhibitors at both concentrations were added to only the MCF10A cell line expressing both the mutant PIK3CA-H1047R and the HER2 gene in a 96-well plate for cell growth, and in 6-well plates for colony formation with 2.5×104 cells/well. Medium with fresh inhibitors was replaced every other day. For the 96-well plate, cells were trypsinized and counted with a particle counter (Beckman coulter) with an absorbance value of 540 nm on the 9th day of growth. Clones in the 6-well plates were stained with crystal violet on the 12th day of experimentation, and clones were counted. The plates were also scanned for pictures of clones.
Affymetrix microarray
Gene expression levels for the four different cell lines were measured with Human Gene 2.1 ST Array Strip at the Genomics Core of University of Michigan. Microarray data reported herein have been deposited at the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99512) with the accession number GSE99512. Microarray analyses and pathway analysis were performed by the Biostatistics core of the KCI. The raw intensity probe-level values were background corrected, log2 transformed, quantile normalized, and summarized to probe set level expression values. The 'rma' function from R package 'oligo', based on robust multi-array average (RMA) method (28), was used for all above pre-processions. Control type of probe sets, such as 'Background probes', 'Poly-A controls' and 'Hybridization controls', were excluded from further analyses. Probe sets with log2 expression values less than 4 for any two cell lines compared were excluded from the analyses of differential expression (DE) probe sets. The k-mean clustering method was used for identifying DE probe sets based on each of three comparisons PIK3CA vs. MCF10A_ Control, HER2 vs. MCF10A_Control, and PIK3CA_HER2 vs. MCF10A_Control. The identified up-/down-regulated probe sets were subjected to ingenuity pathway analysis (IPA). IPA canonical pathway analysis identified the enriched pathways that were most significant to the data set. The significance of the association between the data set and the IPA canonical pathway was measured by the Fisher's exact test. The −Log (P-value) ≥3 was used as the cut-off for identifying the common or unique pathways across three comparisons.
Results
Bioinformatics analysis reveals the co-occurrence of mutant PIK3CA and HER2 amplification in breast cancer cells
Six distinct chromosomal regions were identified within chromosomes 3, 7, 12, 14, 16, and 17 as having increased frequency of amplification in PIK3CA-mutant versus PIK3CA-WT cell lines. The regions with the most amplification include chromosome 7, 14, 16, and 17 (P<0.05, Table I). In particular, the ERBB2/HER2 region was one of the most correlated regions. The percent difference of amplification of the HER2 region between the cell lines with and without the PIK3CA mutation was 37%, which indicated that there may be a synergistic relationship between the PIK3CA mutation and HER2 gene amplification. In addition to the chromosome 17 region, which contains HER2 and two other genes, multiple other chromosomal regions were also found to be correlated with PIK3CA mutation. Chromosome 7 region contains three genes. Chromosome 12 region contains ten genes. Chromosome 14 region contains seven genes and chromosome 16 contains 11 genes (Table II).
Table I.
Chromosome regions show different CGH frequency between cell lines with different status of PIK3CA.
| Chromosome | Difference in CGH frequency (%) | Fisher P-value |
|---|---|---|
| 3 | 33 | 0.07 |
| 7 | 39.40 | 0.04 |
| 12 | 30 | 0.09 |
| 14 | 36.60 | 0.04 |
| 16 | 46.10 | 0.002 |
| 17 | 38.40 | 0.04 |
Table II.
Differential amplified chromosome regions and genes.
| Clone | Chrom | HUGO | Description |
|---|---|---|---|
| CTD-2172D17 | 3 | EIF4G1 | Eukaryotic translation initiation factor 4 γ, 1 |
| CTD-2172D17 | 3 | ABCC5 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| CTD-2172D17 | 3 | EIF2B5 | Eukaryotic translation initiation factor 2B, subunit 5 (ε, 82 kD) |
| CTC-329F6 | 7 | FTSJ2 | FtsJ homolog 2 (E. coli) |
| GS1-165K6 | 7 | AHR | Aryl hydrocarbon receptor |
| GS1-165K6 | 7 | AGR2 | Anterior gradient 2 homolog (Xenepus laevis) |
| RP11-270J9 | 12 | U5-100K | prp28, U5 snRNP 100 kd protein |
| RP11-112M10 | 12 | GALNT6 | GalNAc-T6 |
| RP11-101H10 | 12 | TARBP2 | TAR (HIV) RNA binding protein 2 |
| RP11-101H10 | 12 | MGC11308 | Hypothetical protein MGC11308 |
| RP11-101H10 | 12 | AAAS | Achalasia, adrenocortical insufficiency, alacrimia |
| RP11-132H4 | 12 | HOXC10 | Homeo box C10 |
| VYS12P2692 | 12 | SAS | Sarcoma amplified sequence |
| RP11-234C19 | 12 | PXN | Paxillin |
| RP11-234C19 | 12 | MGC5139 | Hypothetical protein MGC5139 |
| RP11-234C19 | 12 | GCN1L1 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) |
| RP11-70F9 | 14 | THTP | Thiamine triphosphate |
| RP11-189N14 | 14 | PCK2 | Phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| RP11-189N14 | 14 | TM9SF1 | Transmembrane 9 superfamily member 1 |
| RP11-189N14 | 14 | FLJ23338 | Importin 4 |
| RP11-189N14 | 14 | LOC51016 | CGI-112 protein |
| RP11-189N14 | 14 | TINF2 | TERF1 (TRF1)-interacting nuclear factor 2 |
| RP11-111F22 | 14 | RNB6 | RNB6 |
| CTD-2271I17 | 16 | CES2 | Carboxylesterase 2 (intestine, liver) |
| CTD-2271I17 | 16 | ELMO3 | Engulfment and cell motility 3 (ced-12 homolog, C. elegans) |
| RP11-354N7 | 16 | CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) |
| RP11-354N7 | 16 | VPS4A | Vacuolar protein sorting factor 4A |
| RMC16P004 | 16 | LOC64146 | Peptide deformylase-like protein |
| RP11-253O10 | 16 | RAP1 | TRF2-interacting telomeric RAP1 protein |
| RP11-110L8 | 16 | MBTPS1 | Membrane-bound transcription factor protease, site 1 |
| RP11-162I18 | 16 | USP10 | Ubiquitin specific protease 10 |
| RP11-118F19 | 16 | LOC51659 | HSPC037 protein |
| RP11-122P17 | 16 | KIAA0182 | KIAA0182 protein |
| RMC16P026 | 16 | GALNS | Galactosamine (N-acetyl)-6-sulfate sulfatase |
| RMC17P077 | 17 | GRB7 | Growth factor receptor-bound protein 7 |
| RMC17P077 | 17 | ERBB2 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 |
| RMC17P077 | 17 | MGC9753 | Hypothetical gene MGC9753 |
Co-occuring HER2 and mutant PIK3CA-H1047R synergistically enhance cell proliferation, colony formation, and cell invasion in MCF10A cells
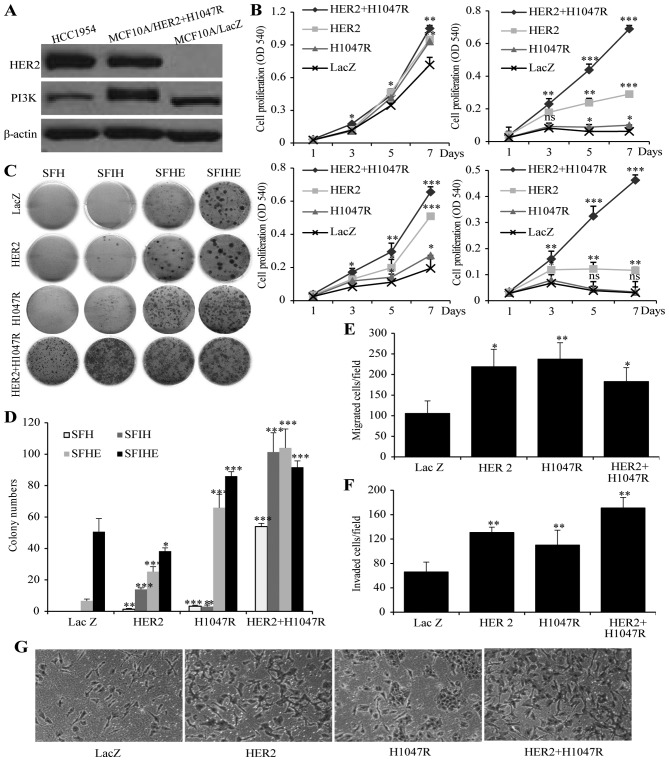
To specifically study the cooperative effect of mutant PIK3CA and HER2 genes in breast cancer cell growth, we chose to create MCF10A cell models expressing mutant PIK3CA-h1047r (MCF10A/PIK3CA-H1047R), HER2 (MCF10A/HER2) or both mutant PIK3CA-H1047R and HER2 genes (MCF10A/HER2/PIK3CA-H1047R). A MCF10A cell line expressing LacZ was used as a control (MCF10A/LacZ) (Fig. 1A). A cell proliferation assay was performed in SFIHE, SFIH, SFHE, and SFH media in order to address proliferation under different growth stimulating conditions. In SFIHE medium, all the cell models demonstrate similar proliferation rate (Fig. 1B). However, in media deprived of the growth factor EGF and insulin, the MCF10A/HER2/PIK3CA-H1047R cells exhibit the greatest growth advantage over MCF10A/PIK3CA-H1047R, MCF10A/HER2, and MCF10A/LacZ (negative control) (Fig. 1B). Epidermal growth factor (EGF) is an essential factor in the acceleration of cell growth in cancer cells (29-31). Insulin allows cells to use glucose for energy and maintains the glucose level in blood and is tightly associated with cancer cell growth and proliferation (32-34). This finding suggests a cooperative effect of HER2 and mutant PIK3CA that is insulin- and EGF-independent. A colony formation assay showed similar synergistic effect of the two genetic alterations on the promotion of colony formation (Fig. 1C and D).
Figure 1.
The effect of HER2 or/and PIK3CA-H1047R on cell proliferation, invasiveness and colony formation. (A) expression of PIK3CA and HER2 gene in MCF10A/HER2/PIK3CA-H1047 cells and HCC1954 cells. MCF10A/LacZ was used as negative control. (B) Cell proliferations of MCF10A/HER2/PIK3CA-H1047R, MCF10A/HER2, MCF10A/PIK3CA-H1047R and MCF10A/LacZ cells when cultured in SFIHE (top left), SFIH (top right), SFHE (bottom left), and SFH (bottom right) media. (c) Different capability in colony formation of MCF10A/HER2/PIK3CA-H1047R, MCF10A/HER2, MCF10A/PIK3CA-H1047R and MCF10A/LacZ cells when cultured in SFH, SFHE, SFIH and SFIHE media. (D) Summary of the colony formation assay under different cell culture conditions. Data are mean ± SD. (e) MCF10A/HER2/PIK3CA-H1047R, MCF10A/HER2 and MCF10A/PIK3CA-H1047R exhibited higher migration ability than MCF10A/Lac Z (negative control). (F) The effects of MCF10A/HER2/PIK3CA-H1047R, MCF10A/HER2, MCF10A/PIK3CA-H1047R and MCF10A/LacZ on cell invasion. Columns mean of three independent experiments. Bars mean SD. The MCF10A/LacZ was used as a control (100%). The invasion ability (%) was measured by the invading cell number of each cell line comparing to the invading cell number of MCF10A/LacZ cell line. Student's t-test was used for statistical analysis. ***P<0.001, **0.01>P>0.001; *0.05>P>0.01; ns, not significant. (g) representative images showing invading cells in Matrigel invasion chambers for four cell models. Original magnification, ×100.
We further investigated the effect of HER2, the mutant PIK3CA-H1047R, or both genetic alterations on the migration and invasion using a chamber assay. In the migration assay, all the oncogene expressing cells showed higher migration efficiency than control cell MCF10A/LacZ. MCF10A/HER2 and MCF10A/PIK3CA-H1047R showed a similar migration cell numbers, while MCF10A/HER2/PIK3CA-H1047R did not demonstrate higher migration capability than either oncogene alone (Fig. 1E). In the invasion assay, all the oncogene-expressing cells showed higher invasive efficiency than control cell MCF10A/LacZ. Specifically, MCF10A/HER2/PIK3CA-H1047R demonstrated a stronger invasion capability than MCF10A/HER2 and MCF10A/PIK3CA-H1047R cells (Fig. 1F and G).
Pharmacological inhibition of HER2 and PIK3CA-H1047R in MCF10A/HER2/PIK3CA-H1047R and HCC1954 cells
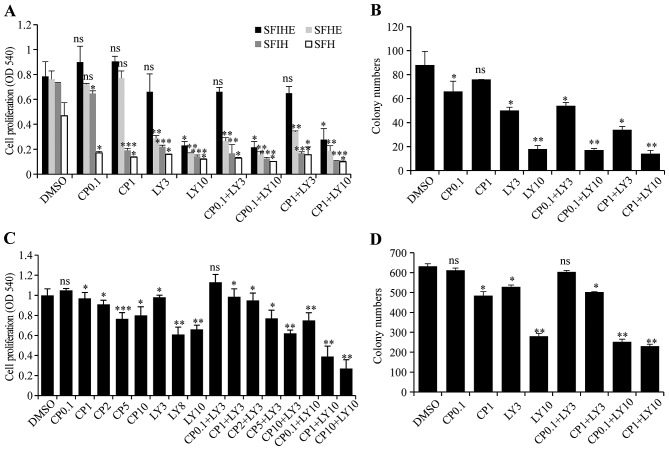
To test whether the cell model (MCF10A/HER2/PIK3CA-H1047R) with both genetic alterations can be synergistically inhibited by simultaneous targeting of both genes, LY294002, a pan-PI3K inhibitor, was used for targeting PIK3CA-H1047R and CP-724714, a selective HER2 inhibitor, was used for targeting active HER2. The results showed that the cells treated with CP-724714 had little to no change in cellular proliferation. Low dose LY294002 (3 μM) alone also had a minimal efficacy as an inhibitor of cell growth. However, cells exposed to a combination of both inhibitors with 10 μM LY294002 had a drastic decrease in cell proliferation. The combined inhibition effects were more significant in SFIH, SFHE, and SFH medium than in SFIHE medium (Fig. 2A and B).
Figure 2.
Inhibitory effects on cell proliferation and colony formation when the established MCF10A models and breast cancer cell were treated with PI3K and HER2 inhibitor individually and in combination. (A) cell proliferations were detected with treatment of LY294002 (LY) and/or CP724714 (CP) in four different media. Combination of both inhibitors played drastic role in decreasing cell viabilities of MCF10A/HER2/PIK3CA-H1047R cells. (B) The effect of LY294002 (LY) and/or CP724714 (CP) treatment on colony formations of MCF10A/HER2/PIK3CA-H1047R cell model. Evaluation of inhibitory effect of LY294002 (LY) or/and CP724714 (CP) on cell proliferation (c) and colony formation (D) of HCC1954 cells. For all the experiments in this figure, drug dose (μM) is shown next to the drug name. Data represent the mean ± SD of three independent experiments. Student t-test was used for statistical analysis. *0.05>P>0.01, **0.01>P>0.001, ***P<0.001; ns, not significant.
To validate the results obtained from the established MCF10A/HER2/PIK3CA-H1047R cell model, we repeated drug treatment using the HCC1954 cell line, which contains both endogenous PIK3CA mutation and HER2 amplification. The treatment with different doses of CP724714 (0.1, 1, 2, 5, and 10 μM) marginally changed cell growth. The treatment of HCC1954 cells with LY294002 only significantly inhibited cell proliferation at high doses (8 and 10 μM). The combination of these two drugs evinced a dose-dependent inhibition in cell proliferation. With a low dose of LY294002 (3 μM) treatment, increasing the dose of CP724714 from 0.1 to 10 μM resulted in a cell proliferation inhibition effect similar to the high dose of LY294002 (10 μM). However, when a high dose of LY294002 (10 μM) was used, further treatment with CP724714 at 0.1, 1, 10 μM obtained an even more significant reduction in cell growth (Fig. 2C). A similar result was obtained in colony formation assay when HCC1954 cells were treated with different combinations (Fig. 2D).
Common and distinct downstream genes and signaling pathways controlled by HER2 and/or mutant PIK3CA
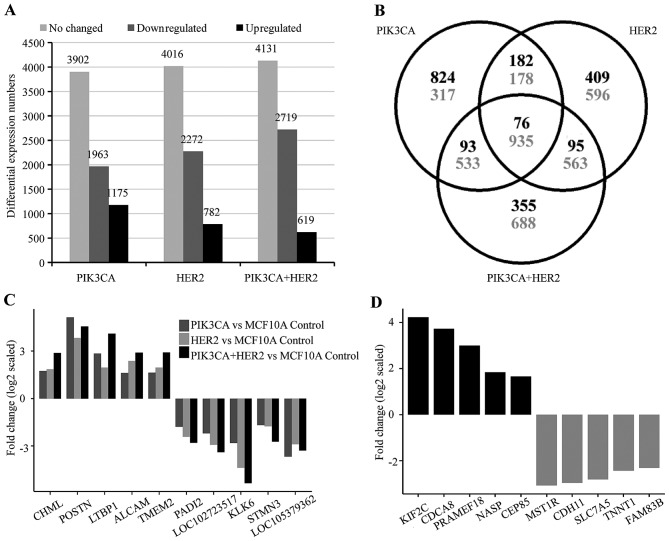
The study of the biological function of three different cell models showed that the MCF10A/HER2/PIK3CA-H1047R cells harbor unexpected characteristics in comparison to MCF10A/HER2 and MCF10A/PIK3CA-H1047R including EGF and insulin-independent growth, stronger invasion capability, and sensitive only to double inhibition. These results suggest that the co-occuring oncogenic HER2 and mutant PIK3CA could lead to activation of novel cell signaling pathways which drive unique oncogenic properties in cancer cells. To investigate the underlying mechanism of the functional differences between cells with mutant PIK3CA, HER2 or both mutant PIK3CA and HER2, we performed an Affymetrix microarray analysis. After normalization with MCF10A/LacZ control cells, MCF10A/PIK3CA-H1047R cells showed 1,175 upregulated, 1,963 downregulated and 3,902 no changed genes; MCF10A/HER2 cells showed 762 upregulated, 2,271 downregulated and 4,016 no changed genes. The MCF10A/mutant PIK3CA/-HER2 showed 619 upregulated, 2,719 downregulated and 4,131 no changed genes (Fig. 3A). Although there are 76 upregulated and 935 downregulated common genes shared in all three cell models, each cell model was shown to contain hundreds of unique genes (Fig. 3B–D).
Figure 3.
Downstream target genes regulated by HER2, mutant PIK3CA and both of HER2 and mutant PIK3CA. (A) The resulted number of probe sets that upregulated, downregulated, or no change in MCF10A/HER2, MCF10A/PIK3CA-H1047R and MCF10A/HER2/PIK3CA-H1047R models after compared to control cell model MCF10A/LacZ. (B) Venn diagram shows common and differential expressed probe sets among three cell models. Black: upregulated; gray: downregulated. (C) The top 5 upregulated and top 5 downregulated common downstream gene targets in three comparisons of MCF10A/HER2 vs. MCF10A/LacZ; MCF10A/PIK3CA-H1047R vs. MCF10A/LacZ, and MCF10A/HER2/PIK3CA-H1047R vs. MCF10A/LacZ control. (D) Specific deregulated genes were identified in MCF10A/HER2/PIK3CA-H1047R cells, but not in either MCF10A/HER2 or MCF10A/PIK3CA-H1047R cells, after compared to MCF10A/LacZ control cells. Top 10 up-/down-regulated genes are shown here. Probe sets with log2 expression values <4 for any two cell lines compared were excluded.
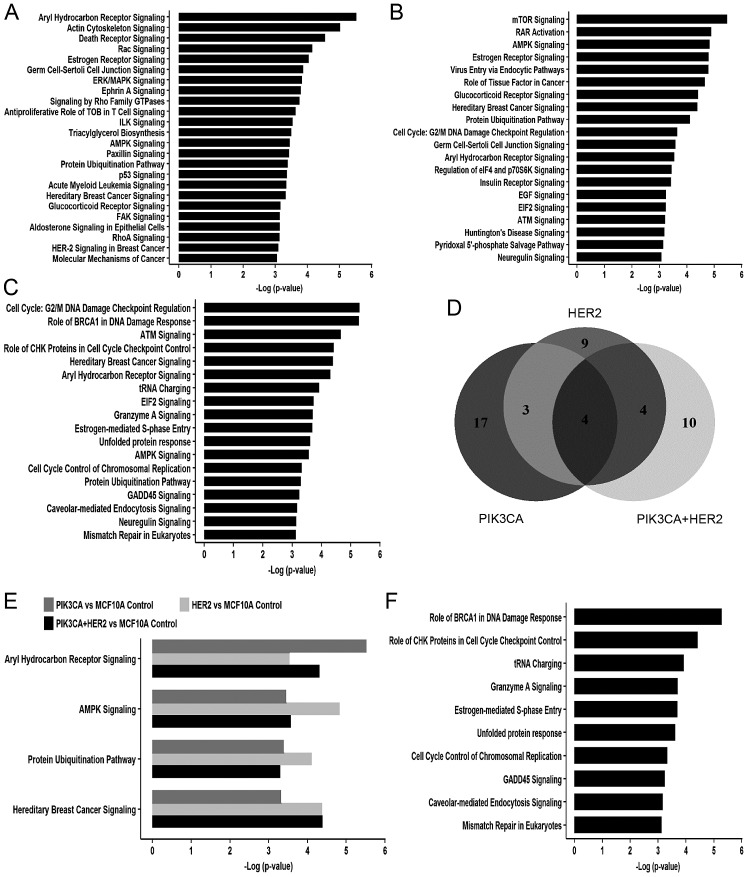
Further ingenuity pathway analysis showed that 24, 20 and 18 cell signaling pathways were enriched in MCF10A/PIK3CA-H1047R, MCF10A/HER2 and MCF10A/HER2/PIK3CA-H1047R, respectively, with a cut-off of -log (P-value) ≥3 (Fig. 4A–C). Under this condition, four common cell signaling were identified in all three cell models, including aryl hydrocarbon receptor (AhR) signaling, AMP-activated protein kinase (AMPK) signaling, protein ubiquitination pathways and hereditary breast cancer signaling (Fig. 4D and E, and Table III). Additionally, 25% (7 out of 24) signaling pathways in cells expressing PIK3CA-H1047R overlapped with 35% (7 out of 20) signaling pathways in HER2 overexpression cells (Fig. 4D). There is 55% (10 of 18) signaling pathways enriched only in cells with both PIK3CA-H1047R and HER2 and do not overlap with cells expressing either mutant PIK3CA or HER2 alone (Fig. 4F). Importantly, 50% (5 out of 10) of these cell signaling pathways are related to DNA replication stress, including role of BRCA1 in DNA damage response, role of CHK1 in cell cycle checkpoint control, estrogen-mediated S-phase entry, cell cycle control of chromosomal replication and mismatch repair in eukaryotes (Fig. 4F and Table IV).
Figure 4.
IPA canonical pathway analysis on targeted gene regulated by HER2, mutant PIK3CA and both of HER2 and mutant PIK3CA. (A–C) The signaling pathway controlled by mutant PIK3CA (A), HER2 (B) and both mutant PIK3CA and HER2 (C) with -log (P-value) ≥3. (D) Venn diagram shows common and differential cell signaling pathways regulated by mutant PIK3CA, HER2 and both mutant PIK3CA and HER2. (E) Four common signaling pathways with -Log (P-value) ≥3, among mutant PIK3CA, HER2, and both mutant PIK3CA and HER2 overexpression cells after compared to MCF10A control cell model. (F) Top ten signaling pathways with -Log (P-value) >3 in cells with both mutant PIK3CA and HER2 overexpression, but not in cells expressing either HER2 or mutant PIK3CA.
Table III.
Four common pathways in HER2, mutant PIK3CA and mutant PIK3CA+HER2 overexpression models.
| Ingenuity canonical pathways | Common genes |
|---|---|
| Aryl hydrocarbon receptor signaling | RELA, NFIX, MYC, CTSD, ALDH7A1, TP53, MED1, NQO1, BAX, CYP1B1, CDKN1A, ALDH3B1, NRIP1, HSPB1 |
| AMPK signaling | SMARCD2, MAPK13, ELAVL1, CRTC2, TSC2, PIK3R2, AKT1S1, CPT1A, GRB2, STK11, PPP2R1A, CDKN1A |
| Protein ubiquitination pathway | USP5, HSPA1A/HSPA1B, SACS, BAG1, USP36, USP19, PSMD3, SKP2, USP31, PSMD11, BAP1, HSPB1, USP21, PSMB3, HLA-A |
| Hereditary breast cancer signaling | SMARCD2, POLR2C, PIK3R2, TP53, GRB2, POLR2E, CDKN1A, SMARCC1, HLTF, RFC3 |
Table IV.
Top 10 signaling pathways enriched only in mutant PIK3CA and HER2 co-occuring model.
| Ingenuity canonical pathways | Genes |
|---|---|
| Role of BRCA1 in DNA damage response | ARID1A, FANCF, SMARGD2, RBL1, CHEK1, RB1, FANCD2, SMARCB1, RFC2, BRCA1, BRIP1, BLM, TP53, E2F4, PLK1, SMARCD1, RFC5, FANCL, NBN, MSH2, RFG4, CDKN1A, BRCA2, SMARCC1, HLTF, FANCA, RFC3 |
| Role of CHK proteins in cell cycle checkpoint control | TP53, GDG25G, E2F4, PLK1, RFG5, GDK1, GHEK1, NBN, PPP2R1A, PGNA, RFG4, GDKN1A, HUS1, ATMIN, RFG2, GLSPN, TLK2, BRGA1, GDK2, RFG3 |
| tRNA charging | GARS, MARS2, LARS2, MARS, QARS, FARSA, WARS, YARS, FARS2, VARS2, AARS, VARS, SARS, SARS2, IARS |
| Granzyme A signaling | HIST1H1B, SET, HIST1H1G, NME1, CREBBP, HMGB2, H1F0, APEX1, EP300 |
| Estrogen-mediated S-phase entry | MYG, RB1, GGNA2, E2F4, GGNE2, GDKN1A, RBL1, GGND1, GDK1, GDK2, SKP2 |
| Unfolded protein response | SCAP, MAP2K7, ERN1, INSIG1, HSPH1, HSPA1A/HSPA1B, HSPA9, GEBPD, GANX, OS9, MAP3K5, GEBPB, GEBPG, MBTPS2, SEL1L, MBTPS1, PPP1R15A, AMFR |
| Cell cycle control of chromosomal replication | MGM5, MGM3, MGM6, PGNA, PRIM1, POLA1, GDG6, GDG7, ORG6, TOP2A, MGM4, DBF4, GDK2, ORG1 |
| GADD45 signaling | TP53, PGNA, CCNE2, CDKN1A, BRGA1, CCND1, GDK1, GDK2, CCNB1 |
| Caveolar-mediated endocytosis signaling | B2M, FLNB, HLA-A, ITGA2, GOPE, ABL1, ITGA6, ITGA10, ITGB8, GD55, ITGA3, FLOT2, AGTA2, HLA-G, ITGB4, ITGB6, AGTG1, PTRF, DNM2, ITGB5, EGFR |
| Mismatch repair in eukaryotes | PGNA, MSH2, RFG4, RFG2, FEN1, RFG5, EXO1, RFG3 |
Discussion
Our current study represents the first large scale genome-wide screening of gene amplifications that correlate with the presence of activating PIK3CA mutations in a panel of 51 human breast cancer cell lines. Our results indicate that in the human genome there are multiple other amplification regions besides the HER2 region localized at different chromosomes that correlate with the PIK3CA mutation in human breast cancer. Further study of these novel regions could identify more novel therapeutic targets in the future for breast cancer treatment.
Using the cell models established, we showed that the co-occurrence of PIK3CA mutation and HER2 gene amplification greatly enhances proliferation. In the MTT assay, the MCF10A/HER2/PIK3CA-H1047R cell model exhibited highest proliferation in all four media, and a synergistic effect was evident in SFH (medium lacking insulin and EGF) and in SFIH (medium lacking EGF). In the colony formation assay, the MCF10A/HER2/PIK3CA-H1047R cell model had the most colonies than any other cell model in all media. We also determined that the cooperative action of HER2 and PIK3CA-H1047R genes is EGF and insulin-signaling pathway independently, because even in conditions lacking insulin and EGF, the growth of cells with both genetic alterations is similar to that in regular conditions (SFIHE medium) suggesting a strong self-sufficient capability (Growth factor independent proliferation). Consistent with previous studies (35,36), the treatment with two inhibitors is more potent in inhibiting cell proliferation and colony formation than using either inhibitor alone in cells with both HER2 and PIK3CA mutation.
We demonstrated that AhR signaling, AMPK signaling, protein ubiquitination pathway and hereditary breast cancer signaling are four common signaling pathways in cells expressing HER2, mutant PIK3CA and HER2 plus mutant PIK3CA. The AhR is a ligand-activated transcription factor that regulates a battery of genes in response to exposure to a broad class of environmental poly aromatic hydrocarbons (PAH). AhR is historically characterized for its role in mediating the toxicity and adaptive responses to these chemicals. However, mounting evidence has established a role for it in ligand-independent physiological processes and pathological conditions, including cancer. The AhR is overexpressed and constitutively activated in advanced breast cancer cases and was shown to drive the progression of breast cancer (37–39). The AMPK is a central regulator of cellular metabolism and energy homeostasis in mammalian tissues. The importance of such regulation of fundamental process poses the AMPK signaling pathway in one of the most attractive therapeutic targets in diabetes and cancer (40–42). Ubiquitination is a protein posttranslational modification reservedly regulated by a series of ubiquitination-associated enzymes. The cellular functions of ubiquitination span a wide spectrum that includes cell death, DNA damage repair, autophagy, proteasomal degradation of proteins and metabolism (43). Thus, dysregulation ubiquitination has broad consequences that may lead to aberration of tumor-promoting pathways and tumor-suppressing pathways (44,45). Consistent with these published results, the finding of our study highlights that these pathways are basic and essential fundamental signaling pathways in HER2 and PIK3CA driven breast cancer.
More importantly, we identified 10 signaling pathways that were specifically enriched in the cells with both mutant PIK3CA and HER2 overexpression. Among these pathways, we found that 50% (5/10) are related to cell cycle control and DNA damage response, including role of BRCA1 in DNA damage response, role of CHK proteins in cell cycle checkpoint control, estrogen-mediated S phase entry, cell cycle control of chromosomal replication and mismatch repair in eukaryotes. Oncogene expression has been shown to drive cell proliferation by interfering with the regulatory pathways of cell cycle progression control, which could further lead to replication stress, a term that is characterized by DNA synthesis slow down and/or replication fork stalling and is the primary cause of genome instability (46-49). Our study for the first time showed that co-occuring oncogenic HER2 and mutant PIK3CA could induce replication stress in mammary epithelial cells. This result highlights the acquisition of unique characteristics arising from co-occurrence of two prominent oncogenes, which will lead to us to re-think the tumor progression mechanism and open up new possibilities for breast cancer diagnostics and treatment.
Acknowledgments
We would like to thank Ms. Elizabeth A. Katz of the Karmanos Cancer Institute's Marketing and Communications Department for editing our manuscript. This work was supported in part by the NIH RO1 grant CA172480-01A1 (G.W.); MT program pilot grant (25RU21) from the Karmanos Cancer Institute, and Grant Boost award from Wayne State university (G.W.); the national natural Science Foundation of china (grant no. 81572601, F.M.); and the natural Science Foundation of Jiangsu Province (grant no. BK20151095) (F.M.). The Biostatistics core and the Biorepository core of the karmanos cancer Institute are supported by grant number P30-CA022453-29.
Abbreviations
- PI3k
phosphatidylinositol 3-kinase
- ErbB2
v-erb-b2 erythroblastic leukemia viral oncogene homolog
- aCGH
array comparative genomic hybridization
- CNV
copy number variation
- EGF
epidermal growth factor
- SFIHE
serum-free medium with insulin, hydrocortisone, and epidermal growth factor
- SFHE
serum-free medium with hydrocortisone and EGF
- SFIH
serum-free medium with insulin and hydrocortisone
- SFH
serum-free medium with hydrocortisone
- AhR
aryl hydrocarbon receptor
- IPA
ingenuity pathway analysis
- PAH
poly aromatic hydrocarbons
- AMPK
AMP-activated protein kinase
References
- 1.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 2.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 3.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase AkT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 4.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair Bg, Lin C, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 5.Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB, Phillips WA. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 6.Saal Lh, Holm K, Maurer M, Memeo L, Su T, Wang X, YU JS, Malmström PO, Mansukhani M, Enoksson J, et al. IK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 7.Wu G, Xing M, Mambo E, Huang X, Liu J, Guo Z, Chatterjee A, Goldenberg D, Gollin SM, Sukumar S, et al. Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res. 2005;7:R609–R616. doi: 10.1186/bcr1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Liu G, Dziubinski M, Yang Z, Ethier SP, Wu G. Comprehensive analysis of oncogenic effects of PIK3CA mutations in human mammary epithelial cells. Breast Cancer Res Treat. 2008;112:217–227. doi: 10.1007/s10549-007-9847-6. [DOI] [PubMed] [Google Scholar]
- 9.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, Cantley LC, Brugge JS. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 10.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuels Y, Diaz LA Jr, Schmidt-kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman JA. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 14.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Laurentiis M, Cancello G, Zinno L, Montagna E, Malorni L, Esposito A, Pennacchio R, Silvestro L, Giuliano M, Giordano A, et al. Targeting HER2 as a therapeutic strategy for breast cancer: A paradigmatic shift of drug development in oncology. Ann Oncol. 2005;16(Suppl 4):iv7–iv13. doi: 10.1093/annonc/mdi901. [DOI] [PubMed] [Google Scholar]
- 16.Li SG, Li L. Targeted therapy in HER2-positive breast cancer. Biomed Rep. 2013;1:499–505. doi: 10.3892/br.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gajria D, Chandarlapaty S. HER2-amplified breast cancer: Mechanisms of trastuzumab resistance and novel targeted therapies. Expert Rev Anticancer Ther. 2011;11:263–275. doi: 10.1586/era.10.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rexer BN, Arteaga CL. Optimal targeting of HER2-PI3K signaling in breast cancer: Mechanistic insights and clinical implications. Cancer Res. 2013;73:3817–3820. doi: 10.1158/0008-5472.CAN-13-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castaneda CA, Lopez-Ilasaca M, Pinto JA, Chirinos-Arias M, Doimi F, Neciosup SP, Rojas KI, Vidaurre T, Balko JM, Arteaga CL, et al. PIK3CA mutations in Peruvian patients with HER2-amplified and triple negative non-metastatic breast cancers. Hematol Oncol Stem Cell Ther. 2014;7:142–148. doi: 10.1016/j.hemonc.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Chakrabarty A, Rexer BN, Wang SE, Cook RS, Engelman JA, Arteaga CL. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene. 2010;29:5193–5203. doi: 10.1038/onc.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanker AB, Pfefferle AD, Balko JM, Kuba MG, Young CD, Sánchez V, Sutton CR, Cheng H, Perou CM, Zhao JJ, et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc Natl Acad Sci USA. 2013;110:14372–14377. doi: 10.1073/pnas.1303204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kataoka Y, Mukohara T, Shimada H, Saijo N, Hirai M, Minami H. Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines. Ann Oncol. 2010;21:255–262. doi: 10.1093/annonc/mdp304. [DOI] [PubMed] [Google Scholar]
- 23.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Searl TJ, Silinsky EM. LY 294002 inhibits adenosine receptor activation by a mechanism independent of effects on PI-3 kinase or casein kinase II. Purinergic Signal. 2005;1:389–394. doi: 10.1007/s11302-005-0778-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 26.Feng B, Xu JJ, Bi YA, Mireles R, Davidson R, Duignan DB, Campbell S, Kostrubsky VE, Dunn MC, Smith AR, et al. Role of hepatic transporters in the disposition and hepatotoxicity of a HER2 tyrosine kinase inhibitor CP-724,714. Toxicol Sci. 2009;108:492–500. doi: 10.1093/toxsci/kfp033. [DOI] [PubMed] [Google Scholar]
- 27.Jani JP, Finn RS, Campbell M, Coleman KG, Connell RD, Currier N, Emerson EO, Floyd E, Harriman S, Kath JC, et al. Discovery and pharmacologic characterization of CP-724,714, a selective ErbB2 tyrosine kinase inhibitor. Cancer Res. 2007;67:9887–9893. doi: 10.1158/0008-5472.CAN-06-3559. [DOI] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Han J, Lee SK, Koo M, Cho Dh, Bae Sy, Choi My, Kim JS, Kim JH, Choe JH, et al. Smad7 acts as a negative regulator of the epidermal growth factor (EGF) signaling pathway in breast cancer cells. Cancer Lett. 2012;314:147–154. doi: 10.1016/j.canlet.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 30.Chen JX, XU LL, Wang XC, Qin HY, Wang JL. Involvement of c-Src/STAT3 signal in EGF-induced proliferation of rat spermatogonial stem cells. Mol cell Biochem. 2011;358:67–73. doi: 10.1007/s11010-011-0922-2. [DOI] [PubMed] [Google Scholar]
- 31.Guo Y, Fu P, Zhu H, Reed E, Remick SC, Petros W, Mueller MD, YU JJ. correlations among ERCC1, XPB, UBE2I, EGF, TAL2 and ILF3 revealed by gene signatures of histological subtypes of patients with epithelial ovarian cancer. Oncol Rep. 2012;27:286–292. doi: 10.3892/or.2011.1483. [DOI] [PubMed] [Google Scholar]
- 32.Boyd DB. Insulin and cancer. Integr Cancer Ther. 2003;2:315–329. doi: 10.1177/1534735403259152. [DOI] [PubMed] [Google Scholar]
- 33.Müssig K, Häring HU. Insulin signal transduction in normal cells and its role in carcinogenesis. Exp Clin Endocrinol Diabetes. 2010;118:356–359. doi: 10.1055/s-0029-1243603. [DOI] [PubMed] [Google Scholar]
- 34.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: An update. Nat Rev Cancer. 2012;12:159–169. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 35.Rexer BN, Chanthaphaychith S, Dahlman K, Arteaga CL. Direct inhibition of PI3K in combination with dual HER2 inhibitors is required for optimal antitumor activity in HER2+ breast cancer cells. Breast Cancer Res. 2014;16:R9. doi: 10.1186/bcr3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez S, Cocco E, Black J, Bellone S, Bonazzoli E, Predolini F, Ferrari F, Schwab CL, English DP, Ratner E, et al. Dual HER2/IK3CA targeting overcomes single-agent acquired resistance in HER2-amplified uterine serous carcinoma cell lines in vitro and in vivo. Mol Cancer Ther. 2015;14:2519–2526. doi: 10.1158/1535-7163.MCT-15-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng S, Cao Z, Wang X. Role of aryl hydrocarbon receptor in cancer. Biochim Biophys Acta. 2013;1836:197–210. doi: 10.1016/j.bbcan.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Murray IA, Patterson AD, Perdew GH. Aryl hydrocarbon receptor ligands in cancer: Friend and foe. Nat Rev Cancer. 2014;14:801–814. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powell JB, Goode GD, Eltom SE. The aryl hydrocarbon receptor: A target for breast cancer therapy. J Cancer Ther. 2013;4:1177–1186. doi: 10.4236/jct.2013.47137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhajda FP. AMP-activated protein kinase and human cancer: Cancer metabolism revisited. Int J Obes. 2008;32(Suppl 4):S36–S41. doi: 10.1038/ijo.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faubert B, Vincent EE, Poffenberger MC, Jones RG. The AMP-activated protein kinase (AMPK) and cancer: Many faces of a metabolic regulator. Cancer Lett. 2015;356:165–170. doi: 10.1016/j.canlet.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 42.Jeon S-M, Hay N. The double-edged sword of AMPK signaling in cancer and its therapeutic implications. Arch Pharm Res. 2015;38:346–357. doi: 10.1007/s12272-015-0549-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20:1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 44.Ohta T, Fukuda M. Ubiquitin and breast cancer. Oncogene. 2004;23:2079–2088. doi: 10.1038/sj.onc.1207371. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Shaik S, Dai X, Wu Q, Zhou X, Wang Z, Wei W. Targeting the ubiquitin pathway for cancer treatment. Biochim Biophys Acta. 2015;1855:50–60. doi: 10.1016/j.bbcan.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hills SA, Diffley JF. DNA replication and oncogene-induced replicative stress. Curr Biol. 2014;24:R435–R444. doi: 10.1016/j.cub.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 47.Mazouzi A, Velimezi G, Loizou JI. DNA replication stress: causes, resolution and disease. Exp Cell Res. 2014;329:85–93. doi: 10.1016/j.yexcr.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 48.Gaillard H, García-Muse T, Aguilera A. replication stress and cancer. Nat Rev Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 49.Taylor EM, Lindsay HD. DNA replication stress and cancer: cause or cure? Future Oncol. 2016;12:221–237. doi: 10.2217/fon.15.292. [DOI] [PubMed] [Google Scholar]