Abstract
lncRNA-TUG1 (Taurine upregulated 1) is up regulated and highly correlated with poor prognosis and disease status in osteosarcoma. TUG1 knockdown inhibits osteosarcoma cell proliferation, migration and invasion, and promotes apoptosis. However, its mechanism of action has not been well addressed. Growing evidence documented that lncRNA works as competing endogenous (ce)RNAs to modulate the expression and biological functions of miRNA. As a putative combining target of TUG1, miR-144-3p has been associated with the progress of osteosarcoma. To verify whether TUG1 functions through regulating miR-144-3p, the expression levels of TUG1 and miR-144-3p in osteosarcoma tissues and cell lines were determined. TUG1 was upregulated in osteosarcoma tissues and cell lines, and negatively correlated with miR-144-3p. TUG1 knockdown induced miR-144-3p expression in MG63 and U2OS cell lines. Results from dual luciferase reporter assay, RNA-binding protein immuno precipitation (RIP) and applied biotin-avidin pull-down system confirmed TUG1 regulated miR-144-3p expression through direct binding. EZH2, a verified target of miR-144-3p was upregulated in osteosarcoma tissues and negatively correlated with miR-144-3p. EZH2 was negatively regulated by miR-144-3p and positively regulated by TUG1. Gain-and loss-of-function experiments were performed to analyze the role of TUG1, miR-144-3p and EZH2 in the migration and EMT of osteosarcoma cells. EZH2 overexpression partly abolished TUG1 knockdown or miR-144-3p overexpression induced inhibition of migration and EMT in osteosarcoma cells. In addition, TUG1 knockdown represses the activation of Wnt/β-catenin pathway, which was reversed by EZH2 over expression. The activator of Wnt/β-catenin pathway LiCl could partially block the TUG1-knockdown induced osteosarcoma cell migration and EMT inhibition. In conclusion, our results showed that TUG1 plays an important role in osteosarcoma development through miRNA-144-3p/EZH2/Wnt/β-catenin pathway.
Keywords: TUG1, miR-144-3p, EZH2, osteosarcoma, metastasis, epithelial-mesenchymal transition, Wnt/β-catenin
Introduction
Osteosarcoma is the most common primary malignant bone tumor with a similar incidence worldwide. It is highly aggressive and with early metastasis (1,2). Osteosarcoma occurs mainly among children and adolescents with low 5-year survival rate, high amputation rate and poor postoperative function recovery (3). Osteosarcoma is a major death-causing disease in adolescence due to its rapid progression and poor prognosis (4). The poor prognosis of osteosarcoma is partially due to the lack of a better molecular biomarker to detect it at early tumor stage. Therefore, it is necessary for us to further understand the underlying molecular mechanisms and find a useful biomarker of osteosarcoma to predict prognosis. Currently, there are no effective target drugs for treating osteosarcoma. Although chemotherapy largely improves the 5-year survival rate and life quality of osteosarcoma patients (5), intrinsic or acquired chemoresistance greatly impede further response to treatment (6). Similar to other tumors, osteosarcoma is a complicated disease with multi-genetic variations (7). Systemic studies on gene regulation network are essential for further understanding how osteosarcoma initiates and develops. It is urgent to develop effective targeted therapies to treat this disease. Hence, finding novel diagnostic biomarkers and therapeutic strategies to improve the prognosis of osteosarcoma is necessary.
Non-coding RNAs (ncRNAs) are divided based on their size into three groups: long non-coding RNAs (lncRNAs), which are longer than 200 base pairs; small ncRNAs, also knowns as microRNAs (miRNAs), which are usually 18–25 nucleotides in length; circular RNAs (circRNAs), which are small covalently closed circular loop structures with either 5′ to 3′ polarity or polyadenylation at the 3′ ends (8). The well-known and well-studied miRNAs are important post-transcriptional regulators involved in various biological processes (9,10). MicroRNAs regulate gene expression by inhibition of translation or destabilization of mRNA transcript through base pairing to the 3′-UTR regions of mRNAs (11). Although thousands of lncRNAs have been identified in the past decade, only a small number have been functionally characterized. However, emerging data demonstrate lncRNAs constitute a major part of human transcriptome and involve in various critical biological processes (12–15). Accumulating evidence has shown that both lncRNAs and microRNAs participate in the regulation of cell proliferation and apoptosis, and play key roles in tumori genesis and progression (16,17).
Taurine upregulated gene 1 (TUG1), a 7.1-kb lncRNA, was firstly identified as a transcript upregulated in response to taurine treatment, which affects mouse retinal development (18). TUG1 was found to be upregulated in various human tumors, such as osteosarcoma, breast cancer, urothelial carcinoma and esophageal squamous cell carcinoma (19–22). Accumulating evidence has verified that TUG1 expression was increased in osteosarcoma tissues and correlates with poor prognosis and disease status in osteosarcoma (21,23–26). Experiment in vitro indicated TUG1 inhibition strongly impaired proliferation of osteosarcoma cells and promoted apoptosis (21,27). Another ncRNA, miR-144-3p was shown to be downregulated in osteosarcoma (28). However, how and by which mechanism lncRNA TUG1 and miR-144-3p regulate osteosarcoma initiation and development is still poorly understood.
In this study, we aimed to elucidate the correlation between TUG1 and miR-144-3p expression in osteosarcoma and investigated their downstream targets. Our finding will provide new insights into the molecular function of TUG1 in osteosarcoma and find new biomarkers for osteosarcoma diagnosis and prognosis.
Materials and methods
Human osteosarcoma tissue collection
All the osteosarcoma and adjacent normal tissues were surgically resected from patients in the Sixth People's Hospital Affiliated to Shanghai Jiao Tong University with informed consent from 2010 to 2012. All specimens were confirmed by clinical, radiographic, and histological examination for osteosarcoma, and were not subjected to radiotherapy, chemotherapy, and blood transfusion therapy before the operation. All the clinical information was collected including sex, age, tumor size, tumor position, tumor stage, and initial metastasis. The present study was approved by the Research Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People's Hospital. All specimens were handled and made anonymous according to the ethical and legal standards.
Cell culture
Human normal osteoblast HFOB1.19 cells and osteosarcoma cell lines MG63, U2OS, HOS and Saos-2 were obtained from the Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). HFOB1.19 cells were cultured in F12 containing 10% fetal bovine serum (FBS). The HOS cell line was maintained in Eagle's minimum essential medium (MEM), and all other cell lines were cultured in RPMI-1640 medium containing 10% FBS. All the cell culture medium and FBS were purchased from Gibco, Invitrogen Corp. (Grand Island, NY, USA). The cell lines were incubated at 37°C in a 5% CO2 incubator.
Vector constructs
For shRNA-mediated TUG1 silencing, DNA oligonucleotides targeting TUG1 was inserted into a lentiviral pLKO.1 plasmid. DNA sequences were mainly obtained from Sigma MISSION shRNA library. For the overexpression of EZH2, the open reading frame of the EZH2 gene was amplified and cloned into a lentiviral pCDH vector.
Lentivirus production and transfection
pCDH-EZH2 or pLKO.1-shTUG1 was co-transfected with the packaging plasmid psPAX2 and the envelop plasmid pMD2.G into HEK-293FT cells using Calcium phosphate transfection. Virus particles were harvested 48 h after transfection. MG63 and U2OS cells were infected with lentivirus particles plus 4 g/ml Polybrene (Sigma).
RNA extraction and q-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). RNA (1 μg) was reversely transcribed into cDNA with M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). q-PCR was performed with SYBR Premix Ex Taq (Takara) on ABI 7500 fast real-time PCR system (Applied Biosystems). GAPDH mRNA was used as an endogenous control for mRNA.
Luciferase reporter assay
The luciferase assays were carried out using the Dual-luciferase Reporter Assay System (Promega). Briefly, cells were co-transfected with miR-144-3p mimics or miR-control and pMIR-reporter luciferase vector containing a specific sequence of wild-type or mutant TUG1 or EZH2 fragment, using Lipofectamine 2000 (Invitrogen). Cells were collected and lysed for luciferase detection 48 h after transfection. The relative luciferase activity was normalized against to the Renilla luciferase activity.
To test Wnt signaling, osteosarcoma cell lines were co-transfected with either the Wnt signaling reporter TOPFlash or the negative control FOPFlash plasmids (Upstate Biotechnology), together with Renilla plasmid. The cells were infected with TUG1 shRNA or EZH2 overexpression lentiviruses. The data are presented as normalized TOPFlash/FOPFlash values. After 48 h, the luciferase activities were measured by Dual-luciferase Reporter Assay System (Promega).
RNA-binding protein immunoprecipitation and RNA pull-down assay
RIP and RNA pull-down assay were performed as described by Lei et al (29).
Western blot analysis
The cells were suspended and lysed in RIPA buffer (Beyotime, Beijing, China) supplemented with protease inhibitor cocktail (Sigma). Protein extractions were separated by SDS-PAGE and transfected to a PVDF membrane (Millipore). The membrane was blocked with 5% (w/v) reagent-grade nonfat milk (Cell Signaling Technology) and incubated with primary antibodies at 4°C overnight followed by secondary antibody incubation. The protein bands were visualized using Clarity™ Western ECL substrate (Bio-Rad). The protein level was quantified using ImageJ software normalized with GAPDH.
Wound healing assays
Wound healing assays were performed to detect cell migration. MG63 and U2OS cells were seeded in 6-well plates and allowed to reach confluence. An artificial wound was made using a 200 μl pipette tip across the cell monolayer. Cells were rinsed with PBS and cultured in the medium. Wound closure was detected at 0, 24 and 48 h, and imaging performed under a microscope.
Transwell migration assays
Transwell migration assays were performed according to the manufacturer's protocol (BD Biosciences). Briefly, transfected cells in serum-free medium were added to the top chamber and incubated at 37°C in a humidified incubator containing 5% CO2. Cells that migrated into the lower chamber were stained with 10% crystal violet (Sigma) and quantitated by counting in five different areas under a light microscope.
Statistical analysis
All data are presented as the mean ± SD and derived from at least three independent experiments. Statistical analysis was performed by SPSS 18.0 software (SPSS, Chicago, IL, USA) and GraphPad Prism software (GraphPad Software, Inc., San Diego, CA, USA). For all comparisons, differences were considered significant at P<0.05.
Results
lnRNAs-TUG1 is upregulated in osteosarcoma and correlated with disease status
To identify the function of TUG1 in osteosarcoma, TUG1 expression in cancerous and adjacent normal tissues from 25 patients with osteosarcoma in different stages was detected using qPCR. The results showed that TUG1 levels were significantly higher in cancer tissues than normal tissues (P<0.0001) (Fig. 1A). In addition, higher TUG1 expression showed a strong correlation with osteosarcoma metastasis (Fig. 1B) and was associated with the late stage of osteosarcoma (Fig. 1C). TUG1 also showed a higher expression level in different osteosarcoma cell lines compared to the normal cells (Fig. 1D).
Figure 1.
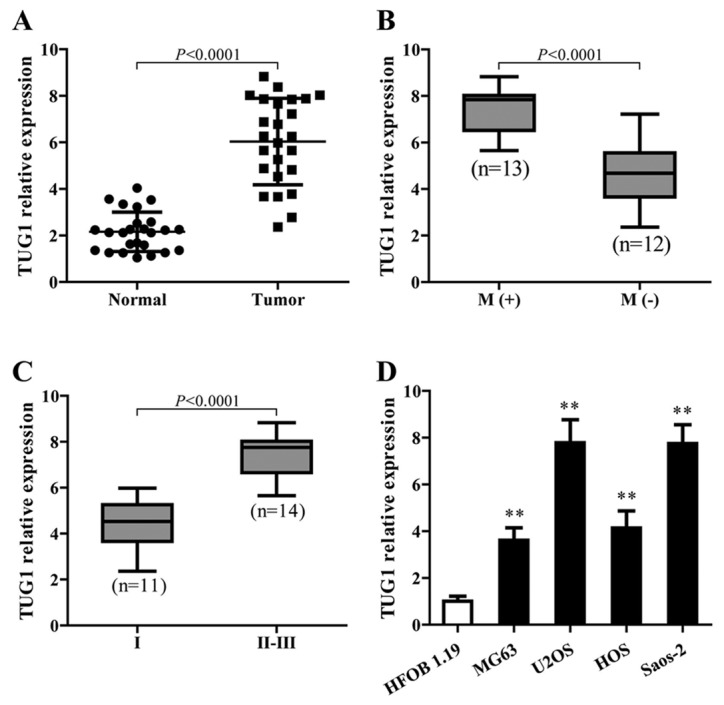
lncRNA-TUG1 is upregulated in osteosarcoma and correlated with disease status. (A) TUG1 expression levels were determined by RT-qPCR in osteosarcoma tissues and adjacent normal tissues (n=25, P<0.0001). (B) The relative TUG1 expressions were analyzed in metastatic and non-metastatic osteosarcoma cells (n=25, P<0.0001). (C) TUG1 expression in different stages of osteosarcoma (n=25, P<0.0001). (D) RT-qPCR was conducted to quantify the expression of TUG1 in osteosarcoma cell lines compared with the normal cells. GAPDH gene was used as the endogenous control for mRNA. Error bars indicate s.d. **P<0.01, Student's t-test.
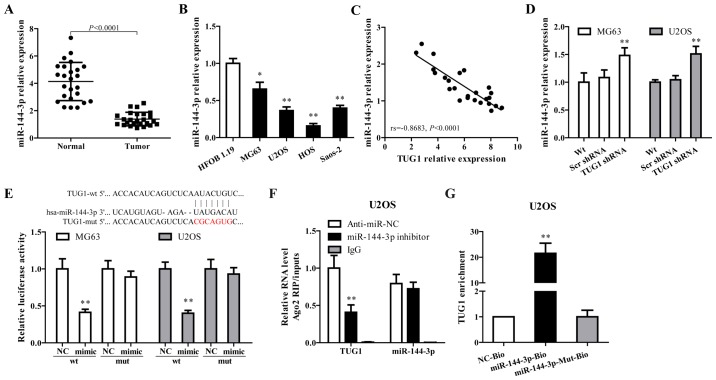
miR-144-3p is regulated by TUG1 in osteosarcoma cells
Growing evidence documented that TUG1 works competing with endogenous (ce)RNAs by serving as sponges that bind and sequester miRNAs (30,31). As a putative combining target of TUG1 (Fig. 2E, upper panel), miR-144-3p is downregulated in osteosarcoma and has been associated with the progress of this disease (32,33). To verify whether TUG1 functions through regulating miR-144-3p, miR-144-3p expression in osteosarcoma tissues and cell lines were determined. As shown in Fig. 2A–C, miR-144-3p was downregulated in tumor tissues and cell lines, and showed a negative correlation with TUG1 expression (rs= −0.8683, P<0.0001). TUG1 knockdown using TUG1 shRNA in MG63 and U2OS cells significantly increased miR-144-3p expression (Fig. 2D).
Figure 2.
miR-144-3p is regulated by TUG1 in osteosarcoma cells. (A) The expression of miR-144-3p was analyzed in osteosarcoma tissues compared with adjacent normal tissues by RT-qPCR (n=25, P<0.0001). (B) miR-144-3p expression levels were determined by RT-qPCR in osteosarcoma cell lines. (C) The correlation between TUG1 and miR-144-3p expression was determined in osteosarcoma tissues. (D) miR-144-3p was upregulated when TUG1 was knocked down. (E) Sequence alignment of miR-144-3p and TUG1 (in red). miR-144-3p reduced the activity of the luciferase reporter with the specific sequence of TUG1 but not with the mutant sequence. (F) Amount of TUG1 bound to Ago2 or IgG measured by RT-qPCR after RIP, IgG was used as a negative control and SNRNP70 was used as a positive control. (G) U2OS cells were transfected with biotinylated WT miR-144-3p (miR-144-3p-Bio) or biotinylated mutant miR-144-3p (miR-144-3p-Mut-Bio) or biotinylated NC (NC-Bio). Forty-eight hours after transfection, cells were collected for biotin-based pull-down assay. TUG1 expression levels were analyzed by RT-qPCR. Error bars indicate s.d. *P<0.05, **P<0.01, Student's t-test.
Dual luciferase reporter assay, RNA-binding protein immuno precipitation (RIP) and applied biotin-avidin pull-down system were performed to explore whether TUG1-mediated miR-144-3p expression through function as a ceRNA. As shown in Fig. 2E, co-transfection of pMIR-TUG1 wild-type and miR-144-3p mimics greatly reduced the luciferase activity compared with TUG1-wt+miR-144-3p NC group, whereas mutation of the miR-144-3p-binding site within TUG1 abrogated the inhibitory effect of miR-144-3p mimics on reporter gene expression. Results from RIP assay showed that TUG1 was detected in Ago2 immunoprecipitates from the control group, but was drastically reduced in Ago2 complexes purified from cells treated with miR-144-3p inhibitor (Fig. 2F), indicating that TUG1 is likely in the miR-144-3p RISC complex. Results from RNA pull-down showed that TUG1 was pulled down by miR-144-3p, while miR-144-3p-Mut with mutated binding site of TUG1 failed to pull-down TUG1 (Fig. 2G), indicating that the recognition of miR-144-3p to TUG1 is in a sequence-specific manner. Together, our findings confirmed miR-144-3p is regulated by TUG1 in osteosarcoma cells through direct binding.
EZH2, a target of miR-144-3p, is positively regulated by TUG1 in osteosarcoma
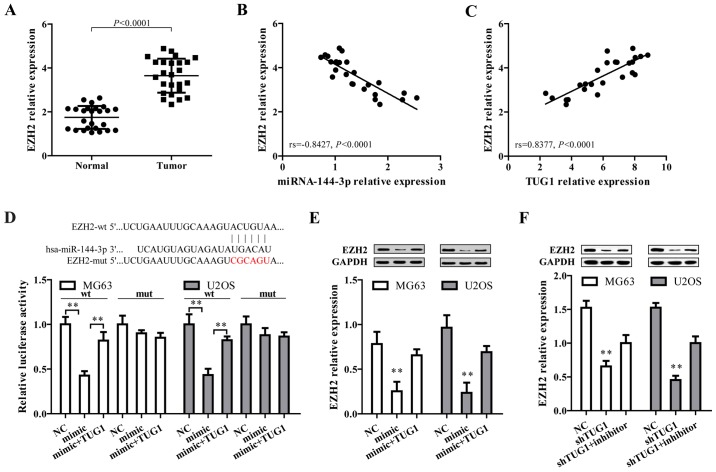
After the confirmation that TUG1 could regulate miR-144-3p in osteosarcoma cells, we proposed TUG1 may exert their function through regulating miR-144-3p target genes by functioning as a ceRNA. To address this, three bioinformatic databases (TargetScan, Miranda and PicTar) were employed to predict the potential target genes of miR-144-3p. Targets that were predicted in all databases and with PicTar score >2.40 are listed in Table I. Among these, EZH2 was selected as a target gene owing to its high expression and promoting effect on tumorigenesis and cancer progression in osteosarcoma (34–37). By comparing tumor and normal tissues, we found EZH2 expression level was higher in osteosarcoma tissues (Fig. 3A), and showed a negative correlation with miR-144-3p level (Fig. 3B) but positively associated with TUG1 expression (Fig. 3C). Additionally, bioinformatics analysis revealed that in the 3′UTR region of EZH2 contains a binding site for miR-144-3p (Fig. 3D, upper panel). Results from dual luciferase reporter assay showed that co-transfection of mature miR-144-3p and EZH2-wt significantly limited the luciferase activity in both MG63 and U2OS cells, which was abated by transfection of TUG1 overexpression (Fig. 2E, lower panel). Furthermore, miR-144-3p mimics markedly reduced EZH2 protein expression, which was reversed by TUG1 overexpression (Fig. 3E). Moreover, shRNA-mediated TUG1 knockdown significantly represses EZH2 expression, which could be reversed by miR-144-3p inhibitor (Fig. 3F). These data collectively demonstrated EZH2 is a target of miR-144-3p and is positively regulated by TUG1.
Table I.
Putative target genes of miR-144-3p predicted by TargetScan, Miranda and PicTar with PicTar score >2.40.
| Putative target | PicTar score | Putative target | PicTar score | Putative target | PicTar score |
|---|---|---|---|---|---|
| ARID1A | 9.44 | SHANK2 | 4.39 | ACBD3 | 3.10 |
| GLTSCR1 | 8.06 | BACH2 | 4.33 | ATP2B1 | 3.05 |
| SORCS3 | 8.01 | TRIO | 4.27 | USP38 | 3.04 |
| RARB | 7.99 | ALS2 | 4.24 | ANKRD17 | 3.03 |
| FBN2 | 7.89 | TJP1 | 4.23 | ATP2B2 | 3.02 |
| MAP3K4 | 7.67 | RBM12 | 4.23 | ZFX | 3.00 |
| UBE2D1 | 7.10 | PDE4D | 4.18 | GSPT1 | 2.92 |
| MYCN | 7.03 | STAG1 | 4.10 | ATP2B1 | 2.92 |
| QKI | 7.01 | ANK2 | 4.03 | SEMA6A | 2.87 |
| NFE2L2 | 6.82 | IDH2 | 3.82 | KCNH7 | 2.86 |
| MARK1 | 6.55 | FBXO32 | 3.75 | LHX2 | 2.86 |
| ADAMTSL3 | 6.53 | PDE7B | 3.75 | CAV2 | 2.86 |
| ABCA1 | 6.32 | NGFRAP1 | 3.61 | TFAP4 | 2.84 |
| ELL2 | 6.25 | STARD8 | 3.60 | CDH2 | 2.83 |
| MSI1 | 6.06 | CDYL | 3.59 | PPFIA1 | 2.81 |
| ZFHX4 | 6.05 | STC1 | 3.57 | MYB | 2.80 |
| DCBLD2 | 5.98 | ST18 | 3.54 | SLITRK4 | 2.78 |
| SOCS5 | 5.95 | C6orf62 | 3.51 | GDF10 | 2.78 |
| RIN2 | 5.84 | GOLGA4 | 3.50 | PAX3 | 2.78 |
| RNF111 | 5.82 | WTAP | 3.49 | STAT6 | 2.69 |
| SLC12A2 | 5.39 | SUCLA2 | 3.48 | MITF | 2.68 |
| SS18 | 5.37 | CXorf23 | 3.47 | MYO1E | 2.66 |
| APPBP2 | 5.35 | ZNF638 | 3.42 | VKORC1L1 | 2.64 |
| AEBP2 | 5.33 | ICK | 3.41 | PABPN1 | 2.64 |
| MEIS2 | 5.25 | HSF2 | 3.41 | ZDHHC17 | 2.64 |
| DLG5 | 5.24 | AHCY | 3.40 | PTHLH | 2.62 |
| APP | 5.21 | USP47 | 3.39 | CCNK | 2.57 |
| MBNL1 | 5.12 | EZH2 | 3.38 | ITSN2 | 2.53 |
| HDGFRP3 | 5.09 | ARRDC3 | 3.37 | CPEB1 | 2.52 |
| CCNT2 | 5.06 | SEMA6D | 3.37 | PPP2R2A | 2.51 |
| ATP1B1 | 5.00 | PHF3 | 3.35 | NID2 | 2.51 |
| PURA | 4.90 | STRN3 | 3.32 | EMP1 | 2.47 |
| KIF2A | 4.58 | SENP7 | 3.31 | DUSP1 | 2.46 |
| PDE4A | 4.58 | HAT1 | 3.29 | BTBD3 | 2.43 |
| FLRT3 | 4.47 | CPEB2 | 3.27 | MBNL2 | 2.43 |
| UBE2D2 | 4.42 | ACSL4 | 3.23 | CDH5 | 2.43 |
| BRPF1 | 4.42 | VLDLR | 3.17 | ||
| ATP5G2 | 4.40 | ESRRG | 3.16 |
Figure 3.
EZH2, a target of miR-144-3p, is positively regulated by TUG1 in osteosarcoma. (A) The EZH2 expression level was analyzed in osteosarcoma tissues compared with normal tissues (n=25, P<0.0001). (B and C) The expression correlation between EZH2 and miR-144-3p or TUG1 was analyzed. EZH2 expression was negatively correlated with miR-144-3p (B), but positively correlated with TUG1 (C). (D) miR-144-3p reduced the activity of the luciferase reporter with EZH2 wild-type 3′-UTR but not with the mutant 3′-UTR. Sequence alignment of miR-144-3p and EZH2 was shown. (E) miR-144-3p over expression reduced EZH2 expression. TUG1 overexpression reversed miR-144-3p-induced EZH2 repression. (F) TUG1 knockdown decreased EZH2 expression, which was reversed by the miR-144-3p inhibitor. Error bars indicate s.d. **P<0.01, Student's t-test.
TUG1-miR-144-3p-EZH2 axis is critical for osteosarcoma cell migration
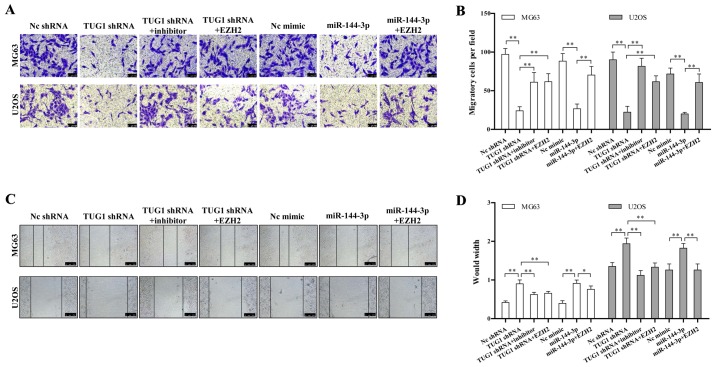
We determined whether the TUG1-miR-144-3p-EZH2 axis is involved in regulating osteosarcoma migration. We found TUG1 knockdown or miR-144-3p overexpression significantly repressed the migration of osteosarcoma cell line determined by both transwell and wound healing assay (Fig. 4A and C). The quantitative results are also shown in Fig. 4B and D. This migration inhibition could be rescued by miR-144-3p inhibitor or EZH2 overexpression (Fig. 4A–D). These data together provided a critical role of the TUG1-miR-144-3p-EZH2 axis in regulating osteosarcoma cell migration.
Figure 4.
TUG1-miR-144-3p-EZH2 axis is critical for osteosarcoma cell migration. (A) Osteosarcoma cell migration was determined by Transwell assay. TUG1 knockdown or miR-144-3p overexpression inhibited osteosarcoma cell migration, while EZH2 overexpression or miR-144-3p inhibitor reversed the above inhibition. (B) Quantification of cell migration is shown. (C) Wound healing assay was performed to analyze cell migration. TUG1 knockdown or miR-144-3p overexpression inhibited osteosarcoma cell migration, while EZH2 overexpression or miR-144-3p inhibitor reversed the above inhibition. (D) Quantification of cell migration is shown. Error bars indicate s.d. **P<0.01, Student's t-test.
TUG1-miR-144-3p-EZH2 axis is critical for osteosarcoma cell epithelial-mesenchymal transition (EMT)
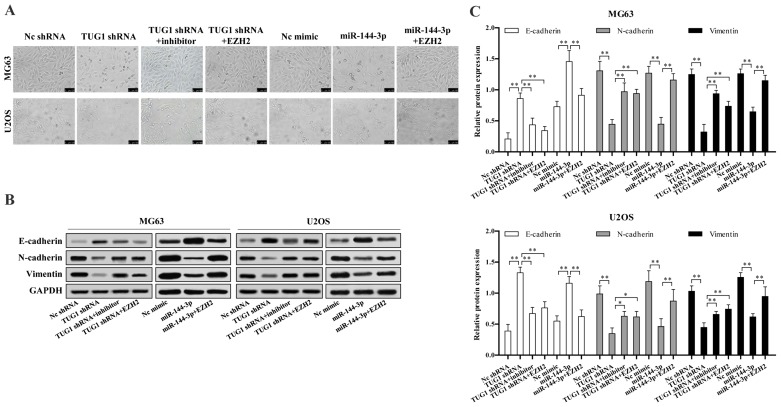
We next investigated whether TUG1-miR-144-3p-EZH2 axis is involved in osteosarcoma EMT. We found TUG1 knockdown or miR-144-3p overexpression significantly reduced osteosarcoma cell EMT, which could be reversed upon EZH2 overexpression or transfection of miR-144-3p inhibitor (Fig. 5A). TUG1 knockdown or miR-144-3p overexpression largely up regulated the protein level of epithelial marker E-cadherin, while decreased mesenchymal markers N-cadherin and vimentin expression, which were reversed upon EZH2 overexpression or miR-144-3p inhibition (Fig. 5B and C).
Figure 5.
TUG1-miR-144-3p-EZH2 axis is critical for osteosarcoma cell epithelial-mesenchymal transition (EMT). (A) TUG1 knockdown or miR-144-3p overexpression inhibited epithelial-mesenchymal transitions for osteosarcoma cells, while EZH2 overexpression or miR-144-3p inhibitor reversed the above inhibition. (B) TUG1 knockdown or miR-144-3p overexpression upregulated the protein level of epithelial marker E-cadherin, while decreased mesenchymal markers N-cadherin and vimentin expression, which was reversed by EZH2 overexpression or miR-144-3p inhibitor. (C) The protein quantification results are shown. Error bars indicate s.d. **P<0.01, Student's t-test.
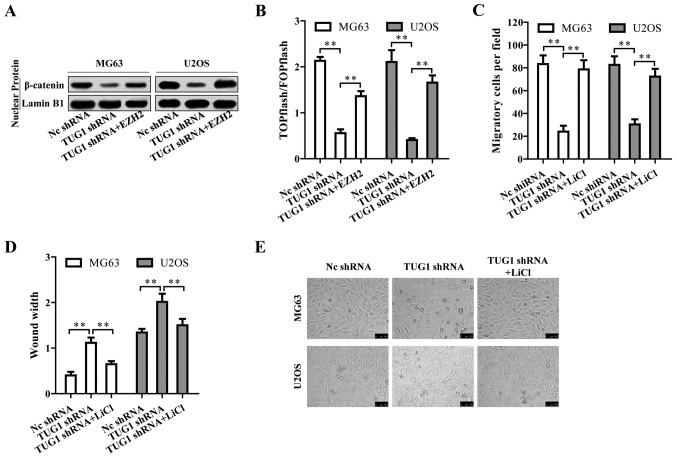
Wnt/β-catenin pathway is regulated by TUG1 through EZH2 and contributes to osteosarcoma tumorigenesis
We then investigated by which mechanism TUG1 regulates osteosarcoma progression. Surprisingly, TUG1 knockdown inactivated Wnt/β-catenin pathway indicated by the reduced nuclear β-catenin protein level (Fig. 6A). The TUG1-mediated inactivation of Wnt/β-catenin pathway could be reversed by EZH2 overexpression (Fig. 6A), suggesting that TUG1 promoted Wnt/β-catenin pathway activation through EZH2. This TUG1-EZH2 mediated Wnt/β-catenin pathway activation was also confirmed by TOPFlash luciferase reporter assay (Fig. 6B). We then treated the osteosarcoma cells with Wnt/β-catenin activator LiCl upon TUG1 knockdown. LiCl reversed the repressive cell migration and EMT caused by TUG1 knockdown (Fig. 6C–E). Based on the above results, TUG1 promotes osteosarcoma progression by Wnt/β-catenin pathway activation via upregulating EZH2.
Figure 6.
Wnt/β-catenin pathway is regulated by TUG1 through EZH2 and contributes to osteosarcoma tumorigenesis. (A) TUG1 knockdown reduced nuclear β-catenin level, which was restored by EZH2 overexpression. (B) TUG1 knockdown inhibited Wnt/β-catenin pathway activation measured by TOPFlash luciferase reporter assay. EZH2 overexpression re-activated Wnt/β-catenin pathway. (C-E) TUG1 knockdown reduced osteosarcoma cell migration and EMT, which was reversed by LiCl, an activator of Wnt/β-catenin pathway. Error bars indicate s.d. **P<0.01, Student's t-test.
Discussion
lncRNAs are a diverse set of RNA transcripts more than 200 nucleotides, but contain no substantial open reading frame, possessing no potential protein-coding capacity (38). lncRNAs are essential regulators at epigenetic, transcriptional, and post-transcriptional levels. Increasing studies have revealed that lncRNAs are expressed in a tissue-specific manner and play fundamental roles in the pathological processes related to tumorigenesis, angiogenesis, invasion, and metastasis (39). These properties make them valid candidates as diagnostic or prognostic cancer biomarkers. Osteosarcoma is the most common primary malignant bone tumor in adolescents (40). The high degree of malignancy makes the 5-year survival rate to decrease to approximately 20% for patients with metastasis (41,42). The majority of patients eventually died from complications related to pulmonary metastases. Previous studies have demonstrated that lncRNAs are key regulators in the initiation, development, and progression of osteosarcoma (43–45). However, the molecular mechanisms are still poorly understood. Moreover, powerful biomarkers to diagnose and predict osteosarcoma are strongly needed. The data presented in our study give a better understanding of how lncRNA-TUG1 regulates osteosarcoma metastasis and provide some potential biomarkers.
In this study, we revealed that lncRNA-TUG1 was up regulated in both osteosarcoma tissues and cell lines. The higher lncRNA-TUG1 expression was significantly correlated with metastasis and clinical stage. However, due to the difficulty of sample collection, the sample size of this study is relatively small, which is a limitation of our study. Fortunately, some other large sample research also verified that TUG1 expression was increased in osteosarcoma tissues and correlates with poor prognosis and disease status in osteosarcoma (21,23–26). Taken together, TUG1 may be a potential biomarker for osteosarcoma diagnosis, prognosis and stage classification.
Several studies have shown that lncRNA can influence post-transcriptional regulation by interfering with microRNAs through molecular sponge effect so as to silence miRNA expression and biological functions (46). These lncRNAs contain miRNA responsive elements (MRE), function as miRNA sponges to downregulated miRNAs, thus reducing the miRNA-induced repression of their target mRNAs (47). TUG1 has been shown to contribute to human osteosarcoma tumorigenesis by sponging miR-9-5p and regulating POU2F1 expression (10). It also functions as a miR-26a sponge in human glioma cells (48). TUG1 also targets miR-144/145 in bladder cancer and gastric cancer (48,49). To further clarify the underlying mechanism, we found that lncRNA-TUG1 could directly bind to the miR-144-3p and downregulate miR-144-3p expression in osteosarcoma cells. miR-144 was previously shown to be a tumor suppressor in osteosarcoma cells. Our results confirmed miR-144-3p was downregulated in osteosarcoma tissues. Moreover, the expression of miR-144-3p and TUG1 was negatively correlated. Specific shRNA-mediated TUG1 knockdown resulted in the upregulation of miR-144-3p.
By systemic analysis, we found miR-144-3p functioned by directly targeting EZH2, which was upregulated in osteosarcoma cells and showed a positive correlation with TUG1 expression. miR-144-3p mimics increased EZH2 expression at both mRNA and protein level. Furthermore, EZH2 overexpression enhanced migration and EMT of osteosarcoma even upon TUG1 knockdown, which confirmed TUG1 promoted tumor progression through EZH2. Consistent with our results, EZH2 was shown to promote osteosarcoma cell metastasis in several studies (37,38,50).
EZH2 is the catalytic subunits of polycomb repressive complex 2 (PRC2), acting as an H3K27me3 methyltransferase, leading to epigenetic silencing of target genes, mainly tumor suppressor genes (51,52). EZH2 has been proven to promote cell metastasis and malignancy (53–56). However, the downstream events involving EZH2 and osteosarcoma progression remains unclear. As a methyltransferase, EZH2 mainly functions in the nucleus, however, recent research revealed that EZH2 has been detected in the cytoplasm and implicated in the migration of cancer cells (54,57,58). In our study, we found EZH2 enhanced osteosarcoma migration by activation of Wnt/β-catenin pathway. It seems that EZH2 has other functions rather than as a methyltransferase functions in the nucleus. We proposed it would be of great interest to further check whether EZH2 affects tumor suppressor gene or EMT gene expression through the change of epigenetic modification in osteosarcoma cells.
In summary, we demonstrated that TUG1 and EZH2 were increased in osteosarcoma while miR-144-3p was down regulated. Our study further identified TUG1-miR-144-3p-EZH2 axis is critical for osteosarcoma cell migration and EMT by activating Wnt/β-catenin pathway. Hence, we elucidated the underlying mechanism and confirmed the role of TUG1 in osteosarcoma progression. Further studies need to be carried out to verify whether there are more TUG1 target genes and to determine how EZH2 affects osteosarcoma. Our study indicates TUG1 and EZH2 may be potential biomarkers for osteosarcoma diagnosis that need to be further developed.
References
- 1.Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer. 2009;125:229–234. doi: 10.1002/ijc.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valery PC, Laversanne M, Bray F. Bone cancer incidence by morphological subtype: A global assessment. Cancer Causes Control. 2015;26:1127–1139. doi: 10.1007/s10552-015-0607-3. [DOI] [PubMed] [Google Scholar]
- 3.Qureshi A, Ahmad Z, Azam M, Idrees R. Epidemiological data for common bone sarcomas. Asian Pac J Cancer Prev. 2010;11:393–395. [PubMed] [Google Scholar]
- 4.Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M, Winkelmann W, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: An analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20:776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 5.Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47:283–292. doi: 10.1016/j.ocl.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 6.Ferrari S, Serra M. An update on chemotherapy for osteosarcoma. Expert Opin Pharmacother. 2015;16:2727–2736. doi: 10.1517/14656566.2015.1102226. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama T, Dass CR, Choong PF. Novel therapeutic strategy for osteosarcoma targeting osteoclast differentiation, bone-resorbing activity, and apoptosis pathway. Mol Cancer Ther. 2008;7:3461–3469. doi: 10.1158/1535-7163.MCT-08-0530. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Fan S, Song E. Noncoding RNAs: New players in cancers. Adv Exp Med Biol. 2016;927:1–47. doi: 10.1007/978-981-10-1498-7_1. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Xie CH, Cao YM, Huang Y, Shi QW, Guo JH, Fan ZW, Li JG, Chen BW, Wu BY. Long non-coding RNA TUG1 contributes to tumorigenesis of human osteosarcoma by sponging miR-9-5p and regulating POU2F 1 expression. Tumour Biol. 2016;37:15031–15041. doi: 10.1007/s13277-016-5391-5. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:15–24. doi: 10.1016/j.addr.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859:128–138. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 18.Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15:501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 19.Han Y, Liu Y, Gui Y, Cai Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107:555–559. doi: 10.1002/jso.23264. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, Wang J, Qiu M, Xu L, Li M, Jiang F, Yin R, Xu L. Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumour Biol. 2015;36:1643–1651. doi: 10.1007/s13277-014-2763-6. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Geng PL, Yin P, Wang XL, Jia JP, Yao J. Downregulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev. 2013;14:2311–2315. doi: 10.7314/APJCP.2013.14.4.2311. [DOI] [PubMed] [Google Scholar]
- 22.Zhao XB, Ren GS. LncRNA taurine-upregulated gene 1 promotes cell proliferation by inhibiting microRNA-9 in MCF-7 cells. J Breast Cancer. 2016;19:349–357. doi: 10.4048/jbc.2016.19.4.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Yang T, Zhang Z, Lu M, Zhao W, Zeng X, Zhang W. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108:859–867. doi: 10.1111/cas.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Yu Y, Fan S, Luo L. Knockdown of long noncoding RNA TUG1 inhibits the proliferation and cellular invasion of osteosarcoma cells by sponging miR-153. Oncol Res. Apr;12:2017. doi: 10.3727/096504017X14908298412505. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Chen Q. Role of taurine upregulated gene 1 as a predictor of poor outcome in osteosarcoma. J Cancer Res Ther. doi: 10.4103/0973-1482.172585. (In press). http://www.cancerjournal.net/preprintarticle.asp?id=172585;type=0. [DOI] [PubMed]
- 26.Ma B, Li M, Zhang L, Huang M, Lei JB, Fu GH, Liu CX, Lai QW, Chen QQ, Wang YL. Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumour Biol. 2016;37:4445–4455. doi: 10.1007/s13277-015-4301-6. [DOI] [PubMed] [Google Scholar]
- 27.Feng YB, Liu XP, Li XL, Cao GL, Zhang P, Tian FM. LncRNA TUG1 is upregulated and promotes cell proliferation in osteosarcoma. Open Med (Wars) 2016;11:163–167. doi: 10.1515/med-2016-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Namløs HM, Meza-Zepeda LA, Barøy T, Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H, Cleton-Jansen AM, Myklebost O. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7:e48086. doi: 10.1371/journal.pone.0048086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei H, Gao Y, Xu X. LncRNA TUG1 influences papillary thyroid cancer cell proliferation, migration and EMT formation through targeting miR-145. Acta Biochim Biophys Sin (Shanghai) 2017;22:1–10. doi: 10.1093/abbs/gmx047. [DOI] [PubMed] [Google Scholar]
- 30.Liu L, Chen X, Zhang Y, Hu Y, Shen X, Zhu W. Long non-coding RNA TUG1 promotes endometrial cancer development via inhibiting miR-299 and miR-34a-5p. Oncotarget. 2017;8:31386–31394. doi: 10.18632/oncotarget.15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, An G, Zhang M, Ma Q. Long non-coding RNA TUG1 acts as a miR-26a sponge in human glioma cells. Biochem Biophys Res Commun. 2016;477:743–748. doi: 10.1016/j.bbrc.2016.06.129. [DOI] [PubMed] [Google Scholar]
- 32.Zhao M, Huang J, Gui K, Xiong M, Cai G, Xu J, Wang K, Liu D, Zhang X, Yin W. The downregulation of miR-144 is associated with the growth and invasion of osteosarcoma cells through the regulation of TAGLN expression. Int J Mol Med. 2014;34:1565–1572. doi: 10.3892/ijmm.2014.1963. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, Zhou X, Wei M. MicroRNA-144 suppresses osteosarcoma growth and metastasis by targeting ROCK1 and ROCK2. Oncotarget. 2015;6:10297–10308. doi: 10.18632/oncotarget.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang K, Zhang Y, Ren K, Zhao G, Yan K, Ma B. MicroRNA-101 inhibits the metastasis of osteosarcoma cells by downregulation of EZH2 expression. Oncol Rep. 2014;32:2143–2149. doi: 10.3892/or.2014.3459. [DOI] [PubMed] [Google Scholar]
- 35.Zhu Z, Tang J, Wang J, Duan G, Zhou L, Zhou X. MiR-138 acts as a tumor suppressor by targeting EZH2 and enhances cisplatin-induced apoptosis in osteosarcoma cells. PLoS One. 2016;11:e0150026. doi: 10.1371/journal.pone.0150026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun R, Shen J, Gao Y, Zhou Y, Yu Z, Hornicek F, Kan Q, Duan Z. Overexpression of EZH2 is associated with the poor prognosis in osteosarcoma and function analysis indicates a therapeutic potential. Oncotarget. 2016;7:38333–38346. doi: 10.18632/oncotarget.9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv Y-F, Yan G-N, Meng G, Zhang X, Guo Q-N. Enhancer of zeste homolog 2 silencing inhibits tumor growth and lung metastasis in osteosarcoma. Sci Rep. 2015;5:12999. doi: 10.1038/srep12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 40.Nagarajan R, Kamruzzaman A, Ness KK, Marchese VG, Sklar C, Mertens A, Yasui Y, Robison LL, Marina N. Twenty years of follow-up of survivors of childhood osteosarcoma: A report from the Childhood Cancer Survivor Study. Cancer. 2011;117:625–634. doi: 10.1002/cncr.25446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bielack S, Carrle D, Casali PG, ESMO Guidelines Working Group Osteosarcoma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2009;20(Suppl 4):137–139. doi: 10.1093/annonc/mdp154. [DOI] [PubMed] [Google Scholar]
- 42.Eccles SA, Welch DR. Metastasis: Recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan LH, Wang W, Yeung W, Deng Y, Yuan P, Mak KK. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene. 2014;33:4857–4866. doi: 10.1038/onc.2013.433. [DOI] [PubMed] [Google Scholar]
- 44.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Yao J, Meng H, Yu Z, Wang Z, Yuan X, Chen H, Wang A. A novel long non-coding RNA, hypoxia-inducible factor-2α promoter upstream transcript, functions as an inhibitor of osteosarcoma stem cells in vitro. Mol Med Rep. 2015;11:2534–2540. doi: 10.3892/mmr.2014.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lv J, Fan HX, Zhao XP, Lv P, Fan JY, Zhang Y, Liu M, Tang H. Long non-coding RNA Unigene56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140-5p in hepatocellular carcinoma cells. Cancer Lett. 2016;382:166–175. doi: 10.1016/j.canlet.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 47.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji TT, Huang X, Jin J, Pan SH, Zhuge XJ. Inhibition of long non-coding RNA TUG1 on gastric cancer cell transference and invasion through regulating and controlling the expression of miR-144/c-Met axis. Asian Pac J Trop Med. 2016;9:508–512. doi: 10.1016/j.apjtm.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 49.Tan J, Qiu K, Li M, Liang Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett. 2015;589:3175–3181. doi: 10.1016/j.febslet.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 50.Chochi Y, Kawauchi S, Nakao M, Furuya T, Hashimoto K, Oga A, Oka M, Sasaki K. A copy number gain of the 6p arm is linked with advanced hepatocellular carcinoma: An array-based comparative genomic hybridization study. J Pathol. 2009;217:677–684. doi: 10.1002/path.2491. [DOI] [PubMed] [Google Scholar]
- 51.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 52.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 53.Croonquist PA, Van Ness B. The polycomb group protein enhancer of zeste homolog 2 (EZH 2) is an oncogene that influences myeloma cell growth and the mutant ras phenotype. Oncogene. 2005;24:6269–6280. doi: 10.1038/sj.onc.1208771. [DOI] [PubMed] [Google Scholar]
- 54.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richter GH, Plehm S, Fasan A, Rössler S, Unland R, Bennani-Baiti IM, Hotfilder M, Löwel D, von Luettichau I, Mossbrugger I, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuroectodermal differentiation. Proc Natl Acad Sci USA. 2009;106:5324–5329. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 57.Nolz JC, Gomez TS, Billadeau DD. The Ezh2 methyltransferase complex: Actin up in the cytosol. Trends Cell Biol. 2005;15:514–517. doi: 10.1016/j.tcb.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–289. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]