Abstract
Candida krusei is a diploid, heterozygous yeast that is an opportunistic fungal pathogen in immunocompromised patients. This species also is utilized for fermenting cocoa beans during chocolate production. One major concern in the clinical setting is the innate resistance of this species to the most commonly used antifungal drug fluconazole. Here, we report a high-quality genome sequence and assembly for the first clinical isolate of C. krusei, strain 81-B-5, into 11 scaffolds generated with PacBio sequencing technology. Gene annotation and comparative analysis revealed a unique profile of transporters that could play a role in drug resistance or adaptation to different environments. In addition, we show that, while 82% of the genome is highly heterozygous, a 2.0 Mb region of the largest scaffold has undergone loss of heterozygosity. This genome will serve as a reference for further genetic studies of this pathogen.
Keywords: Candida krusei, 81-B-5, heterozygosity, LOH, mating type locus, transporters, Genome Report
Candida krusei is a diploid, heterozygous yeast with an estimated chromosome number of six (Whelan and Kwon-Chung 1988; Samaranayake and Samaranayake 1994; Essayag et al. 1996; Jacobsen et al. 2007). C. krusei is an opportunistic fungal pathogen in immunocompromised patients, and, unlike other major pathogenic Candida species (e.g., C. albicans), does not belong to the CUG clade (CTG is translated as serine rather than leucine) (Mühlhausen and Kollmar 2014). Pichia kudriavzevii (synomyn Issatschenkia orientalis) is the teleomorphic (sexual) state of C. krusei (Kurtzman et al. 1980); it is one of the main fermenters of cocoa beans important for the development of chocolate aroma (Jespersen et al. 2005; Nielsen et al. 2005; Pedersen et al. 2012), and is a potential producer of bioethanol and phytase (Chan et al. 2012).
In recent years, human fungal infections caused by C. krusei have increased in the clinic, due mainly to its innate resistance to the azole class of antifungal drugs, specifically to fluconazole (FLU) (Orozco et al. 1998; Guinea et al. 2006; Desnos-Ollivier et al. 2008; Lamping et al. 2009; Ricardo et al. 2014). FLU is a first line antifungal, also used as a prophylactic treatment in the intensive care unit, and breakthrough Candidemia is increasingly caused by non-albicans species including C. krusei (Lischewski et al. 1995; Chaudhary et al. 2015; Cuervo et al. 2016). Moreover, there are incidences of resistance to the echinocandin class of antifungals, which are the drug of choice to fight C. krusei infections (Forastiero et al. 2015). Therefore, identifying the exact mechanisms that underlie drug resistance, and in particular azole resistance, is of utmost importance.
The mechanisms causing C. krusei to be innately resistant to fluconazole are not well understood. Studies have shown that C. krusei Erg11p, the drug target, is significantly less susceptible to FLU inhibition than most other fungal Erg11p proteins (Orozco et al. 1998; Fukuoka et al. 2003), and that efflux pumps such as Abc1p are at least partially responsible for the innate FLU resistance of C. krusei (Lamping et al. 2009). Other studies have shown that overexpression of both ERG11 and ABC2 genes might be responsible for resistance to other azole drugs (He et al. 2015).
One approach to examine the basis of drug resistance of C. krusei is to mine the genome sequence for genes with potential roles in resistance such as novel drug pumps or transporters. To date, genome sequences have been generated for five environmental strains of C. krusei (P. kudriavzevii); the only available high quality assembly is of strain 129 isolated from fermented masau fruits (Van Rijswijck et al. 2017). A genome sequence for clinical isolates is still lacking. Here we report a high-quality genome sequence and assembly for clinical isolate C. krusei 81-B-5 (Scherer and Stevens 1987; Beckerman et al. 2001) into 11 scaffolds generated with PacBio sequencing technology. Gene annotation and comparative analysis revealed a unique profile of transporters that could play a role in drug resistance or adaptation to different environments. In addition, we show that while 82% of the genome is highly heterozygous, a 2.0 Mb region of the largest scaffold has undergone loss of heterozygosity.
Materials and Methods
Sequencing methods and preparation
High molecular weight genomic DNA was isolated from C. krusei strain 81-B-5 (Scherer and Stevens 1987; Beckerman et al. 2001) using a QIAGEN Genomic-tip 500/G kit (catalog # 10262). DNA was adapted using the SMRTbell Template Prep Kit, and sequenced using PacBio Technology (P6-C4 chemistry). A total of three SMRTcells were run, generating a total of 266,621 subreads with mean read length 5758, with a total of 1,535,304,314 bases (∼140× coverage). DNA was also adapted for Illumina sequencing, and a total of 16,953,446 paired 101b reads was generated on a HiSeq 2500.
Assembly and annotation
An initial assembly was generated using HGAP (Chin et al. 2013) version 3 with smrtanalysis-2.3.0; HGAP was run with an estimated genome size of 14 Mb. As the genome was highly heterozygous, we also evaluated Falcon and Falcon-unzip (Chin et al. 2016) assemblies after Quiver polishing (using smrtanalysis-2.3.0). Falcon assembly settings were as follows: length_cutoff = 10,000; length_cutoff_pr = 500; pa_HPCdaligner_option = –v –dal4 –t16 –e.70 –11000 –s1000 –M32; ovlp_HPCdaligner_option = –v –dal4 –t32 –h60 –e.96 –1500 –s1000 –M32; pa_DBsplit_option = –x500 –s1000; ovlp_DBsplit_option = –x500 –s1000; falcon_sense_option = –output_multi–min_idt 0.70–min_cov 2–max_n_read 15–n_core 6; overlap_filtering_setting = –max_diff 72–max_cov 100–min_cov 2–bestn 12–n_core 24. Falcon-unzip was run with default settings other than specifying settings for the Sun Grid Engine (SGE) compute environment. Quiver (Chin et al. 2013) was then run on both assemblies to improve the consensus accuracy; initial evaluation of assemblies prior to Quiver polishing revealed a high rate of base errors. In both the HGAP and Falcon assemblies, contigs representing the alternative haplotype were identified based on high identity alignments to larger contigs in the assembly and roughly half the sequence depth in these regions; these alternative contigs were removed from both assemblies. Mitochondrial contigs were identified in all assemblies and set aside; the largest mitochondrial contig of 51.3 kb was assembled by HGAP assembly and smaller mitochondrial sequences were also identified in the Falcon or Falcon-unzip assemblies.
All assemblies were annotated to evaluate gene set completeness. An initial gene set was predicted using BRAKER (Hoff et al. 2016) to execute Genemark-ET with the parameter –fungus; tRNAs were predicted using tRNAscan (Lowe and Eddy 1997) and rRNAs predicted using RNAmmer (Lagesen et al. 2007). Genes containing PFAM domains found in repetitive elements or overlapping tRNA/rRNA features were removed. Genes were named and numbered sequentially.
SNP calling
Illumina reads were aligned to the HGAP C. krusei genome assembly using the Burrows-Wheeler Aligner (BWA) 0.7.12 mem algorithm (Li 2013) with default parameters. Across the total of 16,306,945 aligned reads, the average depth was 140.0×. BAM files were sorted and indexed using Samtools (Li et al. 2009) version 1.2. Picard version 1.72 was used to identify duplicate reads and assign correct read groups to BAM files. BAM files were locally realigned around INDELs using GATK (McKenna et al. 2010) version 3.4-46 RealignerTargetCreator and IndelRealigner. SNPs and INDELs were called from all alignments using GATK version 3.4-46 HaplotypeCaller in GVCF mode with ploidy = 2, and genotypeGVCFs was used to predict variants in each isolate. Sites were filtered using variantFiltration with QD < 2.0, FS > 60.0, MQ < 40.0, and ReadPosRankSum < −8.0. Individual genotypes were then filtered if the minimum genotype quality <50, percent alternate allele <0.8, or depth <10.
Repeat analysis
De novo repetitive elements were identified with RepeatModeler version open-1.0.7 (www.repeatmasker.org/RepeatModeler/); this identified only one unclassified element of length 1.3 kb and further analysis revealed that this region contains an Arg-tRNA. To identify copies of previously identified elements, RepeatMasker version 4.0.5 (www.repeatmasker.org) was used to identify copies of the RepBase22.04 fungal elements. C. albicans major repeat sequences were retrieved from the Candida Genome Database assembly version 22 (Skrzypek et al. 2017). Sequences were compared to the C. krusei assembly using BLAST; no similarity was found at 1e−5, requiring an alignment length of ≥100 bases.
Comparative genomic analysis
Gene sets of C. krusei, C. lusitaniae (Butler et al. 2009), C. albicans (Jones et al. 2004; Van Het Hoog et al. 2007), P. pastoris (Love et al. 2016), C. glabrata (Dujon et al. 2004), and S. cerevisiae (Cherry, et al. 2012) were compared using BLASTP (e < 1e−10) and orthologs identified from the BLASTP hits using Orthomcl (Li et al. 2003). For the CDR/MDR gene family, protein sequences were aligned using MUSCLE (Edgar 2004) and alignments trimmed using TrimAl (Capella-Gutiérrez et al. 2009) with setting –gappyout. The best amino acid replacement model was selected using ProtTest version 3.4.2 (Darriba et al. 2011). A phylogeny was inferred using RAxML version 8.2.4 (Stamatakis 2014) with model GAMMALG and 1000 bootstrap replicates.
Karyotype analysis
Chromosome plugs were prepared using the CHEF Genomic DNA plug kit (Bio-Rad) with the following modifications: single colonies were transferred to 5 ml YPD broth (1% yeast extract, 2% bacto peptone, and 2% glucose), and incubated at 30° for 18 hr in a roller incubator. The lyticase incubation step was done for 24 hr, and the CHEF plugs were incubated with Proteinase K for 48 hr. For the final washing steps, plugs were transferred to 5 ml tubes containing 3 ml of wash buffer. Chromosomes were separated in a 0.8% agarose gel (certified Megabase agarose (Bio-Rad), in 0.5× TBE buffer) with a DRII pulsed-field gel electrophoresis (PFGE) apparatus (Bio-Rad) using the following run parameters: Block1; 300 sec initial and final switch, 3.9 V/cm, at a 120° angle for 24 hr at 10°, Block 2; 1000 sec initial and final switch at 2.7 V/cm at a 120° angle for 48 hr at 10°. The gel was stained with ethidium bromide (0.5 µl/ml) for 15 min, destained in distilled water for 15 min and photographed. S. cerevisiae and Hansenula wingei chromosome size markers (Bio-Rad) were used for size estimates.
Phenotypic analyses
Standard growth and media conditions have been previously described (Chauhan and Kruppa 2009). An Etest was used to determine the MIC for fluconazole (Pfaller et al. 2003). Briefly, overnight cultures were grown in YPD, washed and diluted to a final A600 of 0.1, then 500 μl were spread onto RPMI1640 agar plates (buffered with MOPS). After a 30 min preincubation, an Etest strip was applied and plates were incubated at 30° for 48 hr. The susceptibility endpoint reported was read at the first growth inhibition ellipse.
To confirm the nonfilamentous phenotype of C. krusei, 3 ml of YPD overnight cultures were washed, cells were counted, and 103 cells were transferred to wells of a 12-well Petri plate containing 1 ml RPMI1640 with 10% fetal bovine serum. Plates were incubated at 37° and microscopic images were taken at 2, 4, and 8 hr. C. albicans (SC5314) and S. cerevisiae (S288c) were used for positive (filamenting) and negative (nonfilamenting) controls, respectively.
Data availability
All genome sequence data (reads, assembly, and annotation) is available in GenBank under BioProject PRJNA381554. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession NHMM00000000. The version described in this paper is version NHMM01000000.
Results and Discussion
Strain sequenced and phenotypic characterization
The sequenced isolate C. krusei 81-B-5 (number 653 in Scherer strain collection) was collected from a clinical source prior to 1987 (Scherer and Stevens 1987). To confirm that strain 81-B-5 is resistant to FLU, this isolate was grown in the presence of FLU and an Etest was done confirming the drug resistant phenotype with a minimum inhibitory concentration (MIC) of 32 µg/ml (Supplemental Material, Figure S1), which is considered highly resistant (Pfaller et al. 2003; Espinel-Ingroff et al. 2014). To verify the nonfilamentous phenotype of C. krusei, cells were exposed to serum, a potent inducer of filamentation and microscopically observed over time. Our results confirm that C. krusei does not filament as compared to C. albicans (Figure S2).
Genome sequencing and assembly
We sequenced the genome of C. krusei using PacBio technology to generate long reads. Early attempts to assemble the genome using Illumina or 454 data had resulted in highly fragmented assemblies (Chan et al. 2012; JQFK00000000 and BBOI00000000), and we reasoned that the heterozygosity detected in multilocus sequence typing (MLST) analyses (Jacobsen et al. 2007) could likely complicate short read assembly. In assembling the genome, we compared assemblies generated using three methods, hierarchical genome assembly process (HGAP), Falcon, and Falcon-unzip, and evaluated metrics for the haploid consensus produced by HGAP and Falcon to the diploid assembly produced by Falcon-unzip. In addition to evaluating assembly metrics, we predicted gene calls on each assembly and evaluated gene set completeness as an additional metric.
While overall assembly statistics were similar, both assembly and gene metrics were superior on the HGAP version (Table S1). The HGAP assembly contained only 11 scaffolds, whereas nearly twice this number was generated by Falcon or in the Falcon-unzip primary contigs. The total assembly size in these assemblies was very similar, with 63 kb more sequence in the Falcon-unzip assembly compared to the HGAP assembly. As our prior experience in assembling diploid Candida genomes revealed that consensus errors can result in gene truncations where haplotypes are merged in a haploid assembly (Butler et al. 2009), we compared gene metrics across the three assemblies. Gene sets were compared to C. albicans to evaluate completeness. By this measure of gene content, the gene set on the HGAP assembly appears to be of higher quality, with 135 more C. albicans orthologs compared to the Falcon assembly, and 303 more than the Falcon-unzip. Gene length was also compared and not found to be very different; genes in the Falcon-unzip assembly were 16 bases larger on average than those in the HGAP. We also evaluated gene content on the second haplotype assembled by Falcon-unzip; these scaffolds totaled 2.1 Mb less than the other assemblies, and correspondingly fewer genes were predicted (Table S1). The completeness of the HGAP gene set was also evaluated by comparing to the BUSCO set of 1438 fungal orthologs (Simão et al. 2015). A total of 1278 appear complete in the C. krusei gene set. By comparison, this count is similar to the 1296 complete orthologs in C. lusitaniae, but fewer than the 1412 orthologs identified in the C. albicans genome, which has been extensively annotated (Braun et al. 2005; Butler et al. 2009; Bruno et al. 2010; Skrzypek et al. 2017). Based on considering both the assembly and gene metrics, we selected the HGAP assembly to represent the genome (Table 1). Compared to a previously reported draft genome (Chan et al. 2012), our assembly is more contiguous (11 contigs compared to 626 contigs for the PA12 assembly); the total size and gene number are comparable, with our assembly including 0.5 Mb more of sequence and a slightly higher gene count. A recently reported genome of isolate 129 using a hybrid of PacBio and Illumina in the assembly was also more fragmented (260 contigs) (Van Rijswijck et al. 2017); this assembly was larger in terms of total size (0.77 Mb), suggesting that some regions may be represented by both haplotypes in this assembly.
Table 1. C. krusei genome statistics.
| Scaffolds | 11 |
|---|---|
| Contigs | 11 |
| Total bases | 10,910,993 |
| Contig N50 length | 1.36 Mb |
| Contig N90 length | 543 kb |
| SNP rate | 1 SNP/ 340 bases |
| GC content | 38.42% |
| Repeat content | 2.15% |
| Protein coding genes | 4,949 |
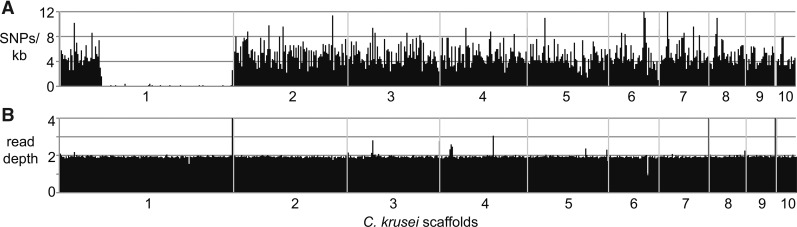
This C. krusei genome shows a high rate of heterozygous SNP variants, and one large region of loss of heterozygosity on scaffold 1. Using Illumina sequence, a total of 32,131 heterozygous SNPs was identified, for an average rate of 1 SNP every 340 positions, which is higher than rate reported in many C. albicans isolates (Butler et al. 2009, Hirakawa et al. 2015). While SNPs were distributed across the genome assembly, a 2.0 Mb region of scaffold 1 has undergone loss of heterozygosity; the first 0.6 Mb of scaffold 1 has a typical frequency of SNP variants; however, very few variants were detected across the remainder of the scaffold (Figure 1A). This homozygous region is not represented in the alternate haplotype contigs assembled by Falcon-unzip, and this difference explains the smaller assembly size of the Falcon-unzip alternate haplotype (Table S1). All of scaffold 1 is present at diploid levels, and we detect no large regions of aneuploidy in this isolate (Figure 1B).
Figure 1.
Genome-wide heterozygosity and genome coverage. (A) Heterozygous SNP positions are plotted across the assembly scaffolds in windows of 5 kb. (B) Normalized read depth is plotted across the assembly scaffolds in windows of 5 kb. Scaffold 11, consisting of around six ribosomal DNA repeats, is not depicted.
The C. krusei genome contains very few repetitive sequences. A search for conserved repetitive elements classified only 0.40% of the assembly as interspersed repeats, with an additional 1.89% of sequence representing simple repeats. There are no regions with significant similarity (BLAST, 1e−5) to the C. albicans major repeat sequences (Materials and Methods). The average GC content is 38.4%, which is intermediate compared to related species such as C. albicans (33.5%) or C. lusitaniae (44.5%) (Jones et al. 2004; Van Het Hoog et al. 2007; Butler et al. 2009).
Chromosome structure
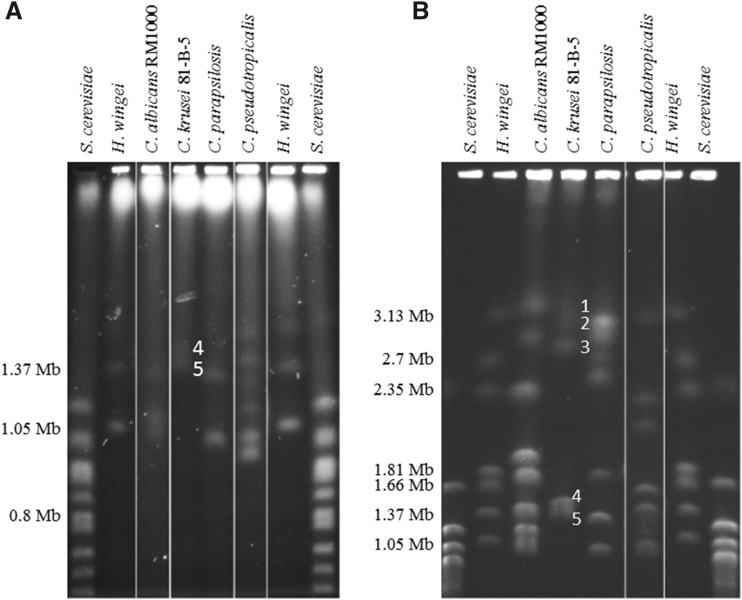
PFGE was used previously to estimate the number of chromosomes for clinical and environmental isolates of C. krusei (Iwaguchi et al. 1990; Doi et al. 1992; Dassanayake et al. 2000; Jespersen et al. 2005). Based on the chromosomal patterns, it was estimated that C. krusei has a total of four to six chromosomes: around two to four large chromosomes (∼2.8–3.5 Mb), and two small chromosomes (∼1.4 Mb). PFGE for C. krusei strain 81-B-5 showed around five chromosomal bands, which were numbered based on size with one being the largest chromosome (Chr1) (Figure 2). Chromosome sizes were estimated based on the H. wingei and S. cerevisiae chromosome standards, and three non-krusei Candida species with known chromosome sizes (Doi et al. 1992; Butler et al. 2009): Chr1 (3.1 Mb), Chr2 (2.9 Mb), Chr3 (2.7 Mb), Chr4 (1.4 Mb), and Chr5 (1.3 Mb) (Figure 2). Based on these sizes the estimated genome size is 11.4 Mb, which is in good agreement with the size of the genome assembly. CHEF Southern analysis will be required to assign each scaffold to its appropriate chromosome, and additional work would be needed to establish the order and orientation of scaffolds along each chromosome.
Figure 2.
Karyotype analysis of C. krusei strain 81-B-5 reveals five chromosomal bands. (A) Short run to separate chromosomes smaller than 2 Mb. (B) Long run to separate all chromosomes. The chromosomes for C. krusei are labeled one through five. Several other Candida species were run as references; S. cerevisiae and H. wingei standards (Bio-Rad) were used for chromosome size estimation of C. krusei chromosomes.
By searching for tandem repeats at scaffold ends, we identified a candidate telomeric repeat (ATTGTAACACACCTCGCTCCTAGTTCAT). This repeat is found at five scaffold ends, including the start of scaffold 1, the end of scaffold 3, both ends of scaffold 4, and the start of scaffold 10. This suggests that scaffold 4 is a complete chromosome, and that four other scaffolds extend to the telomeres. rDNA repeats are detected at the end of scaffold 1, across scaffold 11, and the end of scaffold 9, suggesting that these scaffolds may be joined in a single chromosome to form a continuous rDNA array.
Comparative genomics
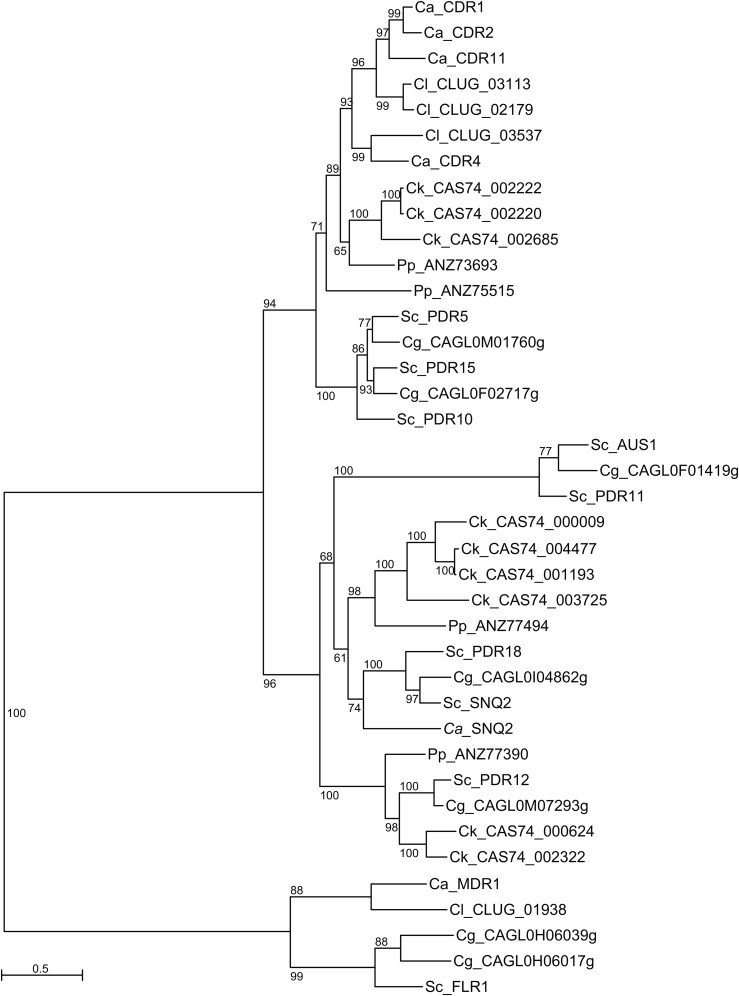
To provide a preliminary view of the genes involved in pathogenesis and drug resistance, we identified orthologs of C. albicans genes in the C. krusei genome. Overall, gene families involved in pathogenesis in C. albicans are present in fewer copies in C. krusei. We identified fewer copies of the secreted aspartyl proteases, oligopeptide transporters, and phospholipase B genes (Table S2). In addition, we found no copies of genes similar to the secreted lipase or ALS cell surface families of proteins from C. albicans. This result is consistent with prior comparison to a wider set of pathogenic Candida more closely related to C. albicans, which observed expansion of several of these families in the more commonly pathogenic species (Butler et al. 2009). We also identified orthologs of genes noted to be involved in drug resistance in C. albicans, via point mutations, increased transcription, or copy number variation. C. krusei contains a single copy of the ERG11 azole target and of each of the TAC1 and UPC2 transcription factors. Several of the sites often subject to drug resistant mutations in C. albicans are conserved in C. krusei (i.e., Y132, K143, and F126), suggesting no intrinsic azole resistance due to mutation of these sites in C. krusei. While we did not identify a copy of the MDR1 drug transporter, we identified nine candidate transporters related to CDR1, CDR2, and related genes (Figure 3). These include three C. krusei genes related to CDR1/CDR2/CDR11/CDR4, four genes related to SNQ2/PDR18, and two genes related to PDR12. This may suggest a very different capability for drug efflux.
Figure 3.
Phylogeny of Cdr and Mdr proteins in C. krusei and related species. Cdr and Mdr proteins identified across six species were aligned and used to infer a phylogeny using RAxML (Materials and Methods). Prefix for each protein corresponds to the species as follows: Ca, C. albicans; Cl, C. lusitaniae; Ck, C. krusei; Pp, P. pastoris; Cg, C. glabrata; Sc, S. cerevisiae.
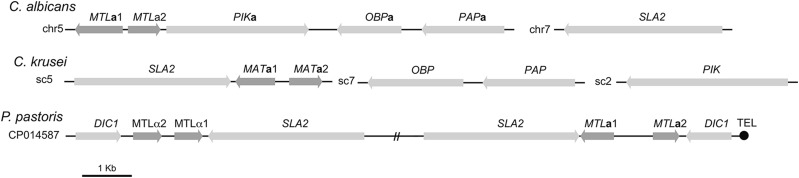
While previous genomic studies have revealed the highly variable content of the mating type locus in pathogenic Candida species (Butler et al. 2009), the mating type locus in C. krusei appears complete, and is more similar to that of Saccharomycetaceae yeasts than the CTG clade Candida. The mating type locus in C. krusei is found on scaffold 5, and includes the MTLa1 gene and MTLa2 located adjacent to SLA2 (Figure 4), similar to the configuration in many Saccharomycetaceae yeasts (Gordon et al. 2011). The mating type locus is close to the start of scaffold 7, separated from the end by four genes. Three other genes typically found at the mating locus of CTG clade Candida species (Butler et al. 2009) are located on adjacent scaffolds; PAP1 and OBPA are adjacent on scaffold 7 and PIKA is on scaffold 2. While the related species Pichia pastoris and Hansenula polymorpha contain two MAT loci (Hanson et al. 2014), only one copy of MTLa1, MTLa2, and SLA2 were found in the C. krusei assembly. This locus is potentially subtelomeric, as the start of the SLA2 gene is 7.4 kb from the start of scaffold 5. The MTL region is heterozygous (Figure 1), as observed in some MTLa/a and MTLα/α C. albicans isolates (Hirakawa et al. 2015). Both of the other assembled genomes of C. krusei also contain the MTLa idiomorph, based on blastp to the available gene set for the 129 assembly or tblastn to the available assembly for M12. This information could guide a search for isolates of the opposite mating type, to begin to study whether C. krusei is capable of sexual reproduction.
Figure 4.
Mating type locus of C. krusei. Genes adjacent to the mating type locus of C. krusei differ from the CTG clade Candida and other related species; there is a single copy of MATa1 and MATa2 found in the assembly, adjacent to the SLA2 gene, whereas the OBP, PIK, and PAP genes are found on other scaffolds in the assembly.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.043547/-/DC1.
Acknowledgments
We thank the Broad Technology Laboratories and Broad Genomics Platform for generating the genome sequence for Candida krusei. This project has been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Department of Health and Human Services, under Grant Number U19AI110818 to the Broad Institute and by NIAID grant R15 AI090633 to A.F.
Footnotes
Communicating editor: J. C. Fay
Literature Cited
- Beckerman J., Chibana H., Turner J., Magee P. T., 2001. Single-copy IMH3 allele is sufficient to confer resistance to mycophenolic acid in Candida albicans and to mediate transformation of clinical Candida species. Infect. Immun. 69: 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B. R., Van Het Hoog M., d’Enfert C., Martchenko M., Dungan J., et al. , 2005. A human-curated annotation of the Candida albicans genome. PLoS Genet. 1: 36–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno V. M., Wang Z., Marjani S. L., Euskirchen G. M., Martin J., et al. , 2010. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 20: 1451–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G., Rasmussen M. D., Lin M. F., Santos M. S., Sakthikumar S., et al. , 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G. F., Gan H. M., Ling H. L., Rashid N. A., 2012. Genome sequence of Pichia kudriavzevii M12, a potential producer of bioethanol and phytase. Eukaryot. Cell 11: 1300–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary U., Goel S., Mittal S., 2015. Changing trends of candidemia and antifungal susceptibility patterns in a tertiary health care centre. Infect. Disord. Drug Targets 15: 171–176. [DOI] [PubMed] [Google Scholar]
- Chauhan N., Kruppa M. D., 2009. Standard growth media and common techniques for use with Candida albicans, pp. 197–201 in Candida albicans: Methods and Protocols, edited by Cihlar R. L., Calderone R. A. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Cherry, J. M., E. L. Hong, C. Amundsen, R. Balakrishnan, G. Binkley, et al., 2012 Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40: D700–705. [DOI] [PMC free article] [PubMed]
- Chin C.-S., Alexander D. H., Marks P., Klammer A. A., Drake J., et al. , 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10: 563–569. [DOI] [PubMed] [Google Scholar]
- Chin C.-S., Peluso P., Sedlazeck F. J., Nattestad M., Concepcion G. T., et al. , 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13: 1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo G., Garcia-Vidal C., Nucci M., Puchades F., Fernández-Ruiz M., et al. , 2016. Breakthrough candidaemia in the era of broad-spectrum antifungal therapies. Clin. Microbiol. Infect. 22: 181–188. [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R., Posada D., 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake R. S., Samaranayake Y. H., Samaranayake L. P., 2000. Genomic diversity of oral Candida krusei isolates as revealed by DNA fingerprinting and electrophoretic karyotyping. APMIS 108: 697–704. [DOI] [PubMed] [Google Scholar]
- Desnos-Ollivier M., Bretagne S., Raoux D., Hoinard D., Dromer F., et al. , 2008. Mutations in the fks1 gene in Candida albicans, C. tropicalis and C. krusei correlate with elevated caspofungin MICs uncovered in AM3 medium using the method of the European Commitee on Antibiotic Susceptibility Testing. Antimicrob Agents Chemother 52: 3092–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M., Homma M., Chindamporn A., Tanaka K., 1992. Estimation of chromosome number and size by pulsed-field gel electrophoresis (PFGE) in medically important Candida species. J. Gen. Microbiol. 138: 2243–2251. [DOI] [PubMed] [Google Scholar]
- Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., et al. , 2004. Genome evolution in yeasts. Nature 430: 35–44. [DOI] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinel-Ingroff A., Pfaller M. A., Bustamante B., Canton E., Fothergill A., et al. , 2014. Multilaboratory study of epidemiological cutoff values for detection of resistance in eight Candida species to fluconazole, posaconazole, and voriconazole. Antimicrob. Agents Chemother. 58: 2006–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essayag S. M., Baily G. G., Denning D. W., Burnie J. P., 1996. Karyotyping of fluconazole-resistant yeasts with phenotype reported as Candida krusei or Candida inconspicua. Int. J. Syst. Bacteriol. 46: 35–40. [DOI] [PubMed] [Google Scholar]
- Forastiero A., Garcia-Gil V., Rivero-Menendez O., Garcia-Rubio R., Monteiro M. C., et al. , 2015. Rapid development of Candida krusei echinocandin resistance during caspofungin therapy. Antimicrob. Agents Chemother. 59: 6975–6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka T., Johnston D. A., Winslow C. A., De Groot M. J., Burt C., et al. , 2003. Genetic basis for differential activities of fluconazole and voriconazole against Candida krusei. Antimicrob. Agents Chemother. 47: 1213–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J. L., Armisén D., Proux-Wéra E., Óhéigeartaigh S. S., Byrne K. P., et al. , 2011. Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc. Natl. Acad. Sci. USA 108: 20024–20029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinea J., Sánchez-Somolinos M., Cuevas O., Peláez T., Bouza E., 2006. Fluconazole resistance mechanisms in Candida krusei: the contribution of efflux-pumps. Med. Mycol. 44: 575–578. [DOI] [PubMed] [Google Scholar]
- Hanson S. J., Byrne K. P., Wolfe K. H., 2014. Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system. Proc. Natl. Acad. Sci. USA 111: E4851–E4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Zhao M., Chen J., Wu R., Zhang J., et al. , 2015. Overexpression of both ERG11 and ABC2 genes might be responsible for itraconazole resistance in clinical isolates of Candida krusei. PLoS One 10: e0136185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa M. P., Martinez D. A., Sakthikumar S., Anderson M. Z., Berlin A., et al. , 2015. Genetic and phentoypic intra-species variation in Candida albicans. Genome Res. 25: 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff K. J., Lange S., Lomsadze A., Borodovsky M., Stanke M., 2016. BRAKER1: unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32: 767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaguchi S., Homma M., Tanaka K., 1990. Variation in the electrophoretic karyotype analysed by the assignment of DNA probes in Candida albicans. J. Gen. Microbiol. 136: 2433–2442. [DOI] [PubMed] [Google Scholar]
- Jacobsen M. D., Gow N. R., Maiden M. C. J., Shaw D. J., Odds F. C., 2007. Strain typing and determination of population structure of Candida krusei by multilocus sequence typing. J. Clin. Microbiol. 45: 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen L., Nielsen D. S., Hønholt S., Jakobsen M., 2005. Occurrence and diversity of yeasts involved in fermentation of West African cocoa beans. FEMS Yeast Res. 5: 441–453. [DOI] [PubMed] [Google Scholar]
- Jones T., Federspiel N. A., Chibana H., Dungan J., Kalman S., et al. , 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101: 7329–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C. P., Smiley M. J., Johnson C. J., 1980. Emendation of the genus Issatchenkia Kudriavzev and comparison of species by deoxyribonucleic acid reassociation, mating reaction, and ascospore ultrastructure. Int. J. Syst. Evol. Microbiol. 30: 503–513. [Google Scholar]
- Lagesen K., Hallin P., Rødland E. A., Stærfeldt H.-H., Rognes T., et al. , 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35: 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamping E., Ranchod A., Nakamura K., Tyndall J. D. A., Niimi K., et al. , 2009. Abc1p is a multidrug efflux transporter that tips the balance in favor of innate azole resistance in Candida krusei. Antimicrob. Agents Chemother. 53: 354–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., 2013 Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. Available at: https://arxiv.org/abs/1303.3997.
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J., Roos D. S., 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischewski A., Ruhnke M., Tennagen I., Schönian G., Morschhäuser J., et al. , 1995. Molecular epidemiology of Candida isolates from AIDS patients showing different fluconazole resistance profiles. J. Clin. Microbiol. 33: 769–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love K. R., Shah K. A., Whittaker C. A., Wu J., Bartlett M. C., et al. , 2016. Comparative genomics and transcriptomics of Pichia pastoris. BMC Genomics 17: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R., 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhausen S., Kollmar M., 2014. Molecular phylogeny of sequenced Saccharomycetes reveals polyphyly of the alternative yeast codon usage. Genome Biol. Evol. 6: 3222–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen D. S., Hønholt S., Tano-Debrah K., Jespersen L., 2005. Yeast populations associated with Ghanaian cocoa fermentations analysed using denaturing gradient gel electrophoresis (DGGE). Yeast 22: 271–284. [DOI] [PubMed] [Google Scholar]
- Orozco A. S., Higginbotham L. M., Hitchcock C. A., Parkinson T., Falconer D., et al. , 1998. Mechanism of fluconazole resistance in Candida krusei. Antimicrob. Agents Chemother. 42: 2645–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L. L., Owusu-Kwarteng J., Thorsen L., Jespersen L., 2012. Biodiversity and probiotic potential of yeasts isolated from Fura, a West African spontaneously fermented cereal. Int. J. Food Microbiol. 159: 144–151. [DOI] [PubMed] [Google Scholar]
- Pfaller M. A., Diekema D. J., Messer S. A., Boyken L., Hollis R. J., 2003. Activities of fluconazole and voriconazole against 1,586 recent clinical isolates of Candida species determined by broth microdilution, disk diffusion, and Etest methods: report from the ARTEMIS global antifungal susceptibility program, 2001. J. Clin. Microbiol. 41: 1440–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo E., Miranda I. M., Faria-Ramos I., Silva R. M., Rodrigues A. G., et al. , 2014. In vivo and in vitro acquisition of resistance to voriconazole by Candida krusei. Antimicrob. Agents Chemother. 58: 4604–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaranayake Y. H., Samaranayake L. P., 1994. Candida krusei: biology, epidemiology, pathogenicity and clinical manifestations of an emerging pathogen. J. Med. Microbiol. 41: 295–310. [DOI] [PubMed] [Google Scholar]
- Scherer S., Stevens D. A., 1987. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J. Clin. Microbiol. 25: 675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Skrzypek M. S., Binkley J., Binkley G., Miyasato S. R., Simison M., et al. , 2017. The Candida Genome Fatabase (CGD): incorporation of assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 45: D592–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Het Hoog M., Rast T. J., Martchenko M., Grindle S., Dignard D., et al. , 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 8: R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rijswijck I. M. H., Derks M. F. L., Abee T., De Ridder D., Smid E. J., 2017. Genome sequences of Cyberlindnera fabianii 65, Pichia kudriavzevii 129, and Saccharomyces cerevisiae 131 isolated from fermented masau fruits in Zimbabwe. Genome Announc. 5: e00064–e00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan W. L., Kwon-Chung K. J., 1988. Auxotrophic heterozygosities and the ploidy of Candida parapsilosis and Candida krusei. J. Med. Vet. Mycol. 26: 163–171. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genome sequence data (reads, assembly, and annotation) is available in GenBank under BioProject PRJNA381554. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession NHMM00000000. The version described in this paper is version NHMM01000000.