Abstract
Small mitochondrial genomes can behave as selfish elements by displacing wild-type genomes regardless of their detriment to the host organism. In the budding yeast Saccharomyces cerevisiae, small hypersuppressive mtDNA transiently coexist with wild-type in a state of heteroplasmy, wherein the replicative advantage of the small mtDNA outcompetes wild-type and produces offspring without respiratory capacity in >95% of colonies. The cytosolic enzyme ribonucleotide reductase (RNR) catalyzes the rate-limiting step in dNTP synthesis and its inhibition has been correlated with increased petite colony formation, reflecting loss of respiratory function. Here, we used heteroplasmic diploids containing wild-type (rho+) and suppressive (rho−) or hypersuppressive (HS rho−) mitochondrial genomes to explore the effects of RNR activity on mtDNA heteroplasmy in offspring. We found that the proportion of rho+ offspring was significantly increased by RNR overexpression or deletion of its inhibitor, SML1, while reducing RNR activity via SML1 overexpression produced the opposite effects. In addition, using Ex Taq and KOD Dash polymerases, we observed a replicative advantage for small over large template DNA in vitro, but only at low dNTP concentrations. These results suggest that dNTP insufficiency contributes to the replicative advantage of small mtDNA over wild-type and cytosolic dNTP synthesis by RNR is an important regulator of heteroplasmy involving small mtDNA molecules in yeast.
Keywords: DNA replication, heteroplasmy, mitochondrial mutations, ribonucleotide reductase, suppressive mtDNA
Eukaryotic cells generally contain multiple copies of mitochondrial DNA (mtDNA), which encodes tRNAs, ribosomal RNAs, and electron transport chain subunits essential for mitochondrial respiratory function. These mitochondrial genomes may exist as a mixture of wild-type and mutant copies within a cell, known as heteroplasmy. Within a heteroplasmic cell, unbalanced replication of wild-type and mutant mtDNA alleles may lead to changes in heteroplasmy level, causing deleterious effects on the health of the host organism (Stewart and Chinnery 2015). Deletion mutations, which can shorten the unit length of an mtDNA molecule by several kilobases, can allow a small genome to be copied more quickly and frequently relative to a full-length molecule and thus may possess a replicative advantage. The accumulation of deleted mtDNAs has been observed in tissue-derived and cultured human cells, mice, nematodes, and yeast (Blanc and Dujon 1980; Cortopassi et al. 1992; Melov et al. 1995; Diaz et al. 2002; Fukui and Moraes 2009). Saccharomyces cerevisiae has served as a potent example for the rapid expansion of small mtDNA. Fragments of mtDNA containing an active origin of replication (ori) sequence were discovered to be hypersuppressive (HS), as they rapidly replicate during heteroplasmy with wild-type mtDNA and become the major mtDNA allele within a few generations, which causes loss of respiratory function in >95% of colonies (Blanc and Dujon 1980).
The complex nature of mtDNA metabolism in yeast is highlighted by the fact that proteins indispensable for rho+ mtDNA replication are not always required for the propagation of rho− mtDNA molecules. The mitochondrial RNA polymerase Rpo41 catalyzes mtDNA transcription, initiates RNA-primed mtDNA replication at ori sequences, and is required for the stable maintenance of rho+ mtDNA (Greenleaf et al. 1986). On the other hand, replication of rho− mtDNA molecules can occur independently of RNA priming regardless of ori content (Fangman et al. 1990). Since one defining characteristic of HS rho− mtDNA is the possession of an active ori, a requirement for Rpo41 for the hypersuppressive phenotype would seem obvious, yet crosses between a ∆rpo41 rho− strain lacking an ori sequence with a ∆rpo41 HS rho− strain were discovered to result in strongly biased inheritance in favor of the HS rho− allele (Lorimer et al. 1995). The question of how the biased inheritance of HS rho− mtDNA is maintained in the absence of RNA priming from ori sequences remained. Similarly, the ssDNA binding protein and yeast mitochondrial nucleoid component, Mgm101 (Meeusen et al. 1999), is essential for maintenance of rho+ genomes containing ori and rho− genomes lacking ori. Interestingly, maintenance of HS rho− mtDNA was found to occur even in ∆mgm101 ∆rpo41 double-mutant cells (Zuo et al. 2002), strongly suggesting the existence of an alternative mtDNA replication pathway.
The mtDNA recombinase Mhr1 catalyzes homologous pairing of nascent ssDNA ends with circular DNA to form a recombination intermediate in which the 3′-ssDNA tail initiates a rolling-circle mode of replication that produces concatemers, linear mtDNA molecules of multiple-unit length (Ling et al. 1995; Ling and Shibata 2002, 2004). Ntg1 is a mitochondrial endonuclease that induces DNA double-stranded breaks (DSBs) at ori5 in response to oxidative stress and, together with Mhr1, contributes to HS rho− mtDNA replication through the initiation of rolling-circle mtDNA replication (Ling et al. 2007; Hori et al. 2009). Additionally, Din7 is a mitochondrial 5′ to 3′ exodeoxyribonuclease (Fikus et al. 2000) that generates 3′ single-stranded DNA tails and was also shown to promote recombination and replication at ori5 (Ling et al. 2013). The Mhr1-catalyzed recombination-dependent rolling-circle replication (RDR) pathway utilizes DSBs, instead of RNA priming at ori sequences. Evidence now suggests that the DSB-mediated form of mtDNA replication may be the predominant form of rho+ mtDNA maintenance in budding yeast cells. Blocking mtDNA DSBs by binding of mitochondrial-targeted MmKu, which prevents access by eukaryotic repair factors, triggered rho0 formation (Prasai et al. 2017). Because DSBs frequently occur at ori5, the RDR pathway could have a role in the replicative advantage of HS rho− mtDNA.
The conserved Mec1/Rad53 nuclear checkpoint pathway was the first signaling pathway identified to control mtDNA copy number (Taylor et al. 2005). Checkpoint activation slows the cell cycle during S phase (Paulovich and Hartwell 1995) and increases cytosolic dNTP synthesis by ribonucleotide reductase (RNR) complex. RNR catalyzes the rate-limiting step of cellular dNTP synthesis through the conversion of ribonucleoside 5′-diphosphates to deoxyribonucleoside 5′-diphosphates and in yeast mainly consists of a large Rnr1-Rnr1 homodimer containing the allosteric feedback and catalytic sites, and a small Rnr2-Rnr4 heterodimer housing the diferric-tyrosyl radical cofactor required for the reduction reaction (Zhang et al. 2006). Control of RNR activity in S. cerevisiae occurs at four levels: regulation by the transcriptional repressor Crt1 (Huang et al. 1998), prevention of Rnr1p homodimerization by binding of the inhibitor Sml1 (Chabes et al. 1999), sequestration of the Rnr2-Rnr4 heterodimer in the nucleus (Zhang et al. 2006), and allosteric inhibition on the Rnr1 subunit (Chabes et al. 2003).
How the replicative advantage of short mtDNA over wild-type is affected by alterations in the RNR pathway remains unexplored. In this study, we have collected evidence to demonstrate a negative correlation between dNTP synthesis by RNR and the replicative advantage for small moderately suppressive or hypersuppressive mtDNA molecules during heteroplasmy with wild-type mtDNA.
Materials and Methods
Yeast transformation
Yeast transformation was performed using the lithium-cesium acetate method (Ito et al. 1983) using a High Efficiency Yeast Transformation Kit (MoBiTec GmbH). Cloning and overexpression of RNR1 and SML1 was carried out with the plasmid pVT100U (Westermann and Neupert 2000) containing the 397-bp constitutive ADH promoter. Selection for cells harboring the desired plasmids was carried out on synthetic dropout minus uracil (SD-U) plates.
Yeast crossing experiments
Parental haploid strains were cultivated separately in rich media at 30° overnight to mid log-phase, using YPGlycerol (yeast extract, peptone, 50mM KH2PO4, 3% glycerol v/v, pH 6.4) for rho+ or YPD (yeast extract, peptone, dextrose) medium for rho− cells. Cell concentrations were counted by hemocytometer and 107 cells from each parental strain were added to 1 ml of YPD medium and crossed for 6 hr at 30°. Mated cells were diluted and spread on synthetic defined minimal medium plus leucine (SD+L) or leucine and uracil (SD+LU) agar plates to select for diploid cells (Supplemental Material, Figure S1). Diploid selection plates were incubated at 30° for 2 d and photographed with a LAS-4000 imaging system (GE Healthcare). The diploid selection plates were then replica-plated onto YPGlycerol plates, which were incubated for another 2 d at 30° and then photographed. Images of the SD master plate and its respective YPGlycerol plate were overlaid in Adobe Photoshop Elements and colonies were counted to determine the percentage of rho+ colony-forming units (CFUs) formed.
Quantification of mtDNA levels in heteroplasmic cells
Diploid colonies obtained during crossing experiments were pooled by elution from diploid selection plates with 1× PBS. Samples were pelleted and stored at −80°. Whole yeast DNA including mtDNA was prepared and DNA concentration was measured with a NanoVue spectrophotometer (GE Healthcare). Ninety-five nanograms of whole yeast DNA was used as the standard template concentration for PCR. Primers used for specific detection of nuclear, rho+, or HS rho− mtDNA, respectively, were: NUC1-Fwd, 5′-GATACTCTTGTCCGGTTTAGTCG-3′; NUC1-Rev, 5′-ATCTTTCGACTGTTTGATCGCC-3′; COX3-Fwd, 5′-ATGCCTTCACCATGACCTATTG-3′; COX3-Rev, 5′-CCAACATGATGTCCAGCTGTTA-3′; HSC1-Fwd, 5′-GAAGATATCCGGGTCCCAATAATAA-3′; HSC1-Rev, 5′-AATATAATAGTCCCCACTCCGCG-3′. Gels were photographed with a FAS-IV gel imaging system (Nippon Genetics) and band intensities were measured with ImageJ software. MtDNA level was calculated relative to the nuclear DNA signal by the 2−∆CT method (Livak and Schmittgen 2001).
Western blotting
Parental rho+ and HS rho− haploid yeast strains expressing the plasmids pVT100U-Empty, pVT100U-RNR1-FLAG, or pVT100U-SML1-FLAG were grown on SD-U selection plates. Diploid cells were obtained under conditions identical to the crossing experiments and selectively grown to mid log-phase by transferring 20 µl of mated cells to SD+L liquid medium and cultivating overnight at 30°. Protein extraction was performed by the LiAc/NaOH method on ice (Zhang et al. 2011). Ten microliters of protein extracts were run on 10 or 15% PAGE gels and semidry transferred to Immobilon-P transfer membranes (Millipore). Primary antibodies used for protein detection were anti-FLAG M2 (Sigma-Aldrich) and yeast anti-Phosphoglycerate Kinase (Invitrogen).
PCR assay for template amplification rates under increasing dNTP concentrations
Templates used in the PCR assay were generated by inserting tdTomato (Clontech) between the KpnI and XbaI cutting sites, or RNR1 between the KpnI and XhoI cutting sites of the plasmid pUC119. Template DNA concentrations were optimized to give signals of nearly equivalent apparent strength under ethidium bromide (EtBr) staining. All PCR reactions were performed with ScaI-linearized pUC119 templates for 12 cycles. The set of primers flanking the multi-cloning site of plasmid pUC119 used for all PCR reactions was: pUC119-MCS-Fwd, 5′-TTGTGTGGAATTGTGAGCGG-3′; pUC119-MCS-Rev, 5′-TGCAAGGCGATTAAGTTGGG-3′. KOD Dash (TOYOBO) or Ex Taq (Takara Bio.) polymerases were used for PCR. Gels were photographed with a FAS-IV gel imaging system (Nippon Genetics) and band intensities were measured with ImageJ software. All band intensities were normalized against the control band with the weakest EtBr signal (left side of gel; amplified with 200 µM dNTPs). Relative DNA levels (EtBr signal %) at each dNTP concentration were calculated as Anorm/Anorm + Bnorm.
Microscopy
Parental haploid cells were cultivated in SD-U liquid medium overnight then transferred to rich media and grown to mid log-phase at 30° on a rotary shaker at 120 rpm for 2–6 hr, using YPGlycerol (pH 6.9) for rho+ or YPD medium for rho− cells. Cells were treated with 1 µg/ml DAPI and incubated at 30° for 15 min. DAPI-stained cells were then mixed at a 1:1 ratio with 1% low-melting agarose (Lonza), mounted on glass slides and observed with a Deltavision microscope system (Applied Precision) equipped with an IX71 microscope (Olympus). Mitochondria were tagged with GFP for parental rho− cells using the plasmid pVT100U-mtGFP (Westermann and Neupert 2000), or pVT100U-mtTomato, which was constructed by cloning tdTomato (Clontech) into pVT100U, for parental rho+ cells.
Data availability
Yeast strains used in this study are listed in Table S1 and are available upon request.
Results
Sml1 is required for the hypersuppressive phenotype
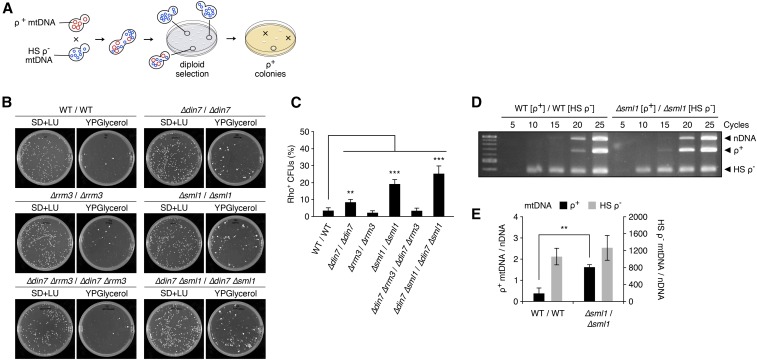
To investigate the possible effects of checkpoint signaling on heteroplasmy with HS rho− mtDNA and the involvement of the Mhr1 pathway, we crossed parental strains with the mutations: ∆din7, ∆rrm3, ∆sml1, ∆din7 ∆rrm3, and ∆din7 ∆sml1. Heteroplasmic diploids containing the 85.7-kbp wild-type (rho+) and 1.1-kbp hypersuppressive (HS rho−) mitochondrial genomes were produced after mating (Figure 1A) as previously described (Ling et al. 2007). The ∆sml1 diploids displayed a marked increase in the proportion of heteroplasmic rho+ CFUs, from 3.5 ± 1.7% in wild-type cells to 19.2 ± 2.6% and 25.3 ± 4.5% in ∆sml1 and ∆din7 ∆sml1 mutants, respectively (Figure 1, B and C). ∆din7 cells showed a small increase to 8.5 ± 1.6%, while ∆rrm3 cells gave a slight decrease in rho+ CFU formation to 2.3 ± 1.1%, suggesting that neither lack of mitochondrial 5′ to 3′ exonuclease activity nor checkpoint activation induced by nuclear replication-fork stalling, respectively, strongly affected the replicative advantage of HS rho− mtDNA.
Figure 1.
SML1 deletion increases the proportion of rho+ colonies and the relative amount of rho+ mtDNA during heteroplasmy with hyper-suppressive mtDNA. (A) Scheme of hypersuppressive crossing experiments. Diploid colonies selectively grown on synthetic media contain different amounts of parental rho+ and HS rho− mtDNA. Following replica plating, diploid colonies with sufficient rho+ mtDNA content are able to grow on YPGlycerol media. (B) Representative images of master and replica plates from genetic crossing experiments. (C) Quantified results of crossing experiments for the genotypes: WT/WT (n = 7); ∆din7/∆din7 (n = 3); ∆rrm3/∆rrm3 (n = 3); ∆sml1/∆sml1 (n = 6); ∆din7∆rrm3/∆din7∆rrm3 (n = 3); ∆din7∆sml1/∆din7∆sml1 (n = 3). (D) PCR amplified nuclear and mitochondrial DNA from WT/WT and ∆sml1/∆sml1 heteroplasmic cells collected from master plates 2 d after crossing. (E) Rho+ and HS rho− mtDNA levels were calculated relative to nDNA signals. Quantified mtDNA levels are from three independent PCR experiments. Error bars indicate SD. ** P <0.005, *** P <0.0005.
To rule out effects from altered mitochondrial morphology or mtDNA nucleoid size in ∆sml1 cells, which could potentially affect mtDNA transmission (Lockshon et al. 1995; Westermann 2010), we tagged mitochondria in rho+ parental cells with tdTomato and in HS rho− cells with GFP, and stained mtDNA nucleoids with DAPI. We did not observe any apparent differences in mitochondrial morphology or nucleoid size among wild-type and ∆sml1 cells of the same parental background (Figure S2), indicating that the phenotype of ∆sml1 cells is not likely due to irregular transmission of mitochondria or nucleoids.
We detected the proportional amounts of rho+ and HS rho− mtDNA in heteroplasmic cells using PCR primers specific for a nuclear gene, rho+ mtDNA and HS rho− mtDNA (Figure S3). Analysis of total genomic DNA including mtDNA, obtained by washing all colonies from diploid selection plates, revealed that relative to nuclear DNA (nDNA), levels of rho+ mtDNA were 0.38 ± 0.26-fold in wild-type cells compared with 1.64 ± 0.12-fold in ∆sml1 cells (Figure 1, D and E), indicating that the ∆sml1 mutation significantly increased the proportional level of rho+ mtDNA in heteroplasmic cells containing HS rho− mtDNA. On the other hand, we observed no significant difference in HS rho− mtDNA levels among the wild-type and ∆sml1 backgrounds, indicating that replication of wild-type mtDNA is increased to a greater extent than HS rho− mtDNA in ∆sml1 cells.
To address the possibility that the increased rho+ phenotype was unique to crosses involving our HS rho− strain (YKN1423-C1), we conducted additional crossing experiments with two additional rho− strains: another HS rho− strain (YKN1423-A1) and a moderately suppressive rho− strain (YKN1423-A2). In A1 wild-type crosses, 4.4 ± 0.9% of CFUs were rho+ while A1 ∆sml1 crosses gave rise to 75.8 ± 7.8% rho+ CFUs. A2 wild-type crosses yielded 66.4 ± 4.0% rho+ CFUs while A2 ∆sml1 crosses yielded 79.6 ± 8.1% rho+ CFUs (Figure S4, A and B). Attempts to find unique restriction sites in these rho− genomes as a necessary step for sequencing and designing specific PCR primers were unsuccessful. However, we did observe significant increases in rho+ mtDNA content in diploids from both crosses (Figure S4, C and D). Taken together, the ∆sml1 mutation increased wild-type mtDNA levels in heteroplasmic cells containing either moderately suppressive or hypersuppressive mtDNA.
RNR1 overexpression enhances rho+ mtDNA replication in hypersuppressive crosses
RNR1 overexpression is sufficient to rescue the temperature-sensitive mtDNA loss phenotype of mitochondrial DNA polymerase mip1-1 mutants, demonstrating a close relationship between RNR activity and mtDNA maintenance (Lecrenier and Foury 1995). Both RNR1 overexpression and the ∆sml1 mutation increase cellular dNTP concentration (Zhao et al. 1998; Chabes and Stillman 2007) and mtDNA copy number (Taylor et al. 2005; Lebedeva and Shadel 2007). Furthermore, the ∆sml1 mutation was shown to reduce rates of spontaneous petite formation (Zhao et al. 1998). Sml1 inhibits RNR by binding and preventing Rnr1 homodimerization (Chabes et al. 1999; Chabes and Stillman 2007), therefore we hypothesized that increased dNTP synthesis by RNR was responsible for the observed increases in rho+ CFU formation.
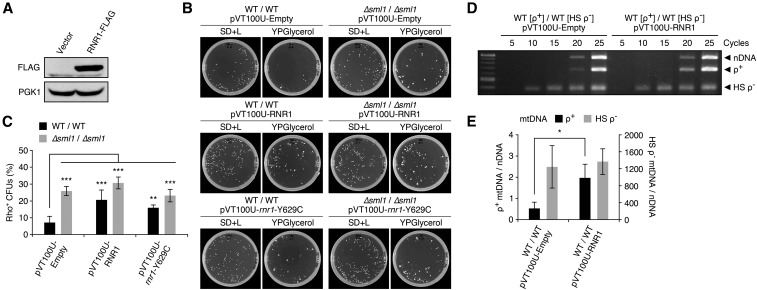
To confirm the role of elevated dNTP synthesis, we overexpressed RNR1 via plasmid and confirmed by immunoblot analysis (Figure 2A). In agreement with the behavior of ∆sml1 crosses, we observed that 20.6 ± 5.9% of heteroplasmic diploid CFUs overexpressing RNR1 were rho+, compared with 7.2 ± 3.7% of diploid CFUs expressing the empty vector (Figure 2, B and C). In the strain containing the empty vector, levels of rho+ mtDNA were 0.53 ± 0.29-fold relative to NUC1, compared with 1.97 ± 0.64-fold in the RNR1-overexpressing strain (Figure 2, D and E). There was no significant change in levels of HS rho− mtDNA among cells expressing the empty or RNR plasmids. In addition, we expressed a mutant isoform, rnr1-Y629C, and observed that 15.9 ± 1.7% and 23.2 ± 3.7% of wild-type and ∆sml1 diploid CFUs, respectively, were rho+ in rnr1-Y629C expressing cells. The lower rho+ CFU formation rate suggests a lower catalytic activity of the rnr1-Y629C mutant gene product and supports the notion that elevated RNR activity contributes to the replication of rho+ genomes in the presence of HS rho− mtDNA, consistent with the observation of cells lacking Sml1.
Figure 2.
RNR1 overexpression increases rho+ mtDNA levels and respiratory function in heteroplasmic cells. (A) Immunoblot of RNR1-FLAG overexpression in heteroplasmic diploid cells following crossing. Anti-PGK is shown as a loading control. (B) Representative images of master and replica plates from genetic crossing experiments of strains expressing an empty vector, overexpressing RNR1, or the mutant rnr1-Y629C isoform. (C) Quantified crossing results were obtained from multiple crossing experiments yielding the genotypes: WT/WT/pVT100U-Empty (n = 14); ∆sml1/∆sml1/pVT100U-Empty (n = 3); WT/WT/pVT100U-RNR1 (n = 12); ∆sml1/∆sml1/pVT100U-RNR1 (n = 3); WT/WT/pVT100U-rnr1-Y629C (n = 3); ∆sml1/∆sml1/pVT100U-rnr1-Y629C (n = 3). (D) PCR amplified nuclear and mitochondrial DNA from WT/WT heteroplasmic cells expressing pVT100U-Empty or pVT100U-RNR1, collected 2 d after crossing. (E) Rho+ and HS rho− mtDNA levels were calculated relative to nDNA signals. Quantified mtDNA levels are from four independent PCR experiments. Error bars indicate SD. * P <0.05, ** P <0.005, *** P <0.0005.
Overproducing Sml1 in ∆sml1 cells restores the hypersuppressive phenotype
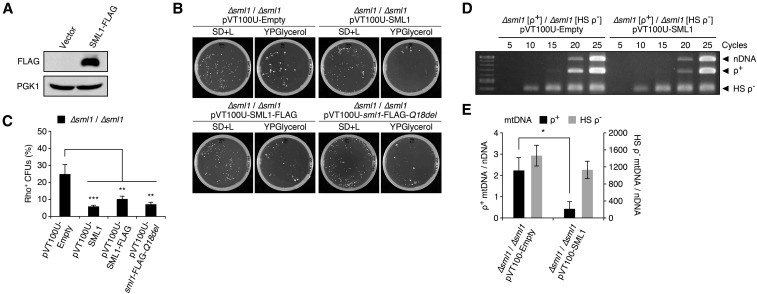
Sml1 inhibits RNR outside of S phase when demand for dNTP synthesis is low and is removed during S phase or in response to DNA damage (Zhao et al. 2001; Chabes et al. 2003). SML1 overexpression increases the frequency of spontaneous petite colony formation compared to wild-type cells, indicating that mitochondrial genome maintenance is impaired by the inhibition of cytosolic dNTP synthesis by RNR (Zhao et al. 1998). To further demonstrate the relationship between the RNR pathway and selfish mtDNA dynamics, we examined whether artificially lowering RNR activity by increasing its inhibition can restore the replicative advantage of small hypersuppressive mtDNA. We confirmed SML1 overexpression by immunoblot (Figure 3A) and found a significant decrease in the proportion of rho+ CFUs from 24.9 ± 5.6% in ∆sml1 cells containing the empty vector to 5.9 ± 0.7%, 10.3 ± 1.6%, and 7.2 ± 1.1% in ∆sml1 cells overexpressing SML1, SML1-FLAG, and sml1-FLAG-Q18del, respectively (Figure 3, B and C). Since the Sml1 protein consists of only 111 amino acids, the relative size of the FLAG tag may have lowered its binding and inhibitory effect on Rnr1p, while the Q18 deletion appears to have slightly improved inhibitory function. Consistent with the drop in rho+ CFU formation rate, rho+ mtDNA level declined approximately fivefold, from 2.23 ± 0.61-fold relative to NUC1 in ∆sml1 cells expressing the empty plasmid to 0.43 ± 0.34-fold upon SML1 expression (Figure 3, D and E). On the other hand, we observed a small but not statistically significant decrease in the level of HS rho− mtDNA level upon SML1 expression. Together, these results indicate that artificially lowering RNR activity enhances the replicative advantage of hypersuppressive over wild-type mtDNA.
Figure 3.
SML1 overexpression suppresses rho+ mtDNA levels and respiratory function in heteroplasmic cells. (A) Immunoblot of SML1-FLAG overexpression in heteroplasmic diploid cells following crossing. Anti-PGK is shown as a loading control. (B) Representative images of master and replica plates from genetic crossing experiments of strains expressing an empty vector, or overexpressing SML1, SML1-FLAG, or the mutant sml1-FLAG-Q18del isoform. (C) Quantified crossing results were obtained from multiple crossing experiments yielding the genotypes: ∆sml1/∆sml1/pVT100U-Empty (n = 8); ∆sml1/∆sml1/pVT100U-SML1 (n = 8); ∆sml1/∆sml1/pVT100U-SML1-FLAG (n = 3); ∆sml1/∆sml1/pVT100U-sml1-FLAG-Q18del (n = 3). (D) PCR amplified nuclear and mitochondrial DNA from ∆sml1/∆sml1 heteroplasmic cells expressing pVT100U-Empty or pVT100U-SML1, collected 2 d after crossing. (E) Rho+ and HS rho− mtDNA levels were calculated relative to nDNA signals. Quantified mtDNA levels are from three independent PCR experiments. Error bars indicate SD. * P <0.05, ** P <0.005, *** P <0.0005.
Low dNTP concentration enhances the replicative advantage of small template DNA over large in vitro
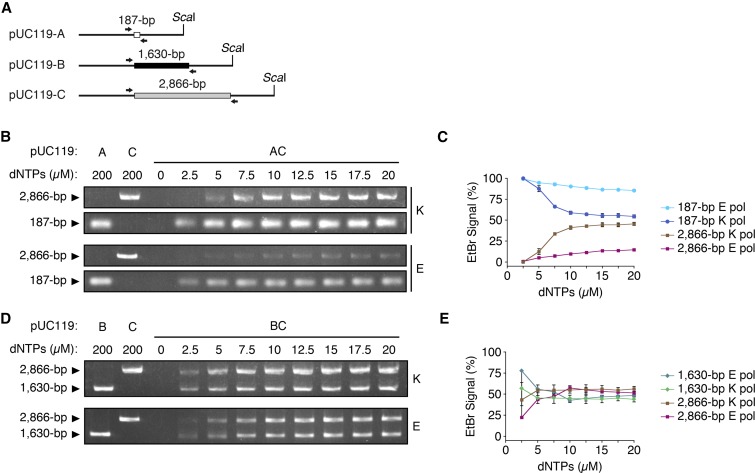
Overexpression of RNR1 or the ∆sml1 mutation are known to positively regulate dNTP concentration and mtDNA copy number in yeast, and our experimental results suggest that relatively low dNTP concentration may contribute to the replicative advantage of small mtDNA. To further illustrate the effect of low dNTP concentration, we tested competitive amplification by PCR using Ex Taq or KOD Dash polymerases and templates of different size (Figure 4A). We examined a dNTP concentration range of 0 to 20 µM, as these levels reflect the physiological dNTP concentrations within mammalian mitochondria (Song et al. 2005).
Figure 4.
Competitive amplification of DNA templates of disparate lengths under increasing dNTP concentrations in vitro. (A) Schematic of ScaI-linearized pUC119 DNA templates. (B) DNA amplification following 12 PCR cycles with templates A and C in isolation at a dNTP concentration of 200 µM (left), or a mixture of pUC119-A and pUC119-C at dNTP concentrations from 0 to 20 µM. K, KOD Dash polymerase; E, Ex Taq polymerase. (C) Percentage of the total signal representing the relative amount of the 187- or 2866-bp template amplified at the indicated dNTP concentrations. (D) DNA amplification following 12 PCR cycles with templates B and C in isolation at a dNTP concentration of 200 µM (left), or a mixture of pUC119-B and pUC119-C at dNTP concentrations from 0 to 20 µM. K, KOD Dash polymerase; E, Ex Taq polymerase. (E) Percentage of the total signal representing the relative amount of the 2866- or 1630-bp template amplified at the indicated dNTP concentrations. Results in C and E represent three independent experiments using KOD Dash polymerase and two independent experiments using Ex Taq polymerase. Error bars indicate SD.
Consistent with our observations of suppressive mtDNA in yeast crossing experiments, the small template was amplified much more readily compared to the large at dNTP concentrations of <10 µM (Figure 4, B and C and Figure S5, A and B). The strength of this effect varied between the two polymerases tested; however, the replicative advantage of the smallest template decreased with increasing dNTP concentration and signals from either large PCR product (2866 or 1630-bp) did not exceed those of the small (187-bp) PCR product at any dNTP concentration. On the other hand, during PCR amplification of two templates of closer size, the small template only displayed a replicative advantage at 2.5 µM with one of the two polymerases under our experimental conditions (Figure 4, D and E). Additionally, PCR reactions using a mixture of all three templates showed that the smallest template was amplified almost exclusively at dNTP concentrations of <7.5 µM (Figure S5, C and D). These data show that dNTP concentration and the relative sizes of templates are important factors in replicative advantage during PCR.
Discussion
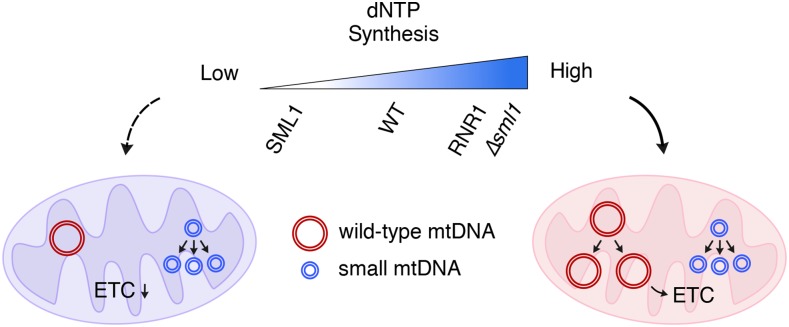
Disruption of dNTP balance or availability within mitochondria has been linked to mtDNA depletion and disease (Gonzalez-Vioque et al. 2011; Dalla Rosa et al. 2016) and promotes mtDNA deletion mutagenesis in cultured cells (Song et al. 2005). In this study, we demonstrated that the replicative advantage of moderately suppressive or hypersuppressive mtDNA molecules is partially due to insufficient dNTP synthesis by RNR. Competition between small and full-length mtDNA in heteroplasmic cells is naturally weighted against a larger allele; however, reducing RNR activity appears to enhance the replicative advantage of small mtDNA. Indeed, competitive amplification of a mixture of small and large templates via PCR showed that the replicative advantage of small DNA is affected by relative template size and dNTP concentration in vitro. Though replication in vivo by mtDNA polymerase γ occurs under physiological conditions and in conjunction with the mitochondrial replisome, both mtDNA polymerase γ and Taq polymerases are derived from the A family of DNA polymerases (Steitz 1999), while KOD enzymes belong to family B (Elshawadfy et al. 2014), which possess a catalytic “palm” domain homologous to family A polymerases. Both PCR enzymes showed a general trend of increasing replicative advantage for smaller templates as dNTP concentrations decrease. Taken together, these results support a model wherein dNTP synthesis by RNR influences the extent of the replicative advantage of small mtDNA in yeast, and therefore affects mtDNA heteroplasmy level and respiratory function (Figure 5).
Figure 5.
Model for the role of cytosolic dNTP synthesis in regulating rho+ mtDNA replication during heteroplasmy with small mtDNA in yeast. In heteroplasmic cells containing a mixture of small and full-length mtDNA, the wild-type or SML1-overexpressing backgrounds have dNTP synthesis levels only sufficient for small mtDNA replication, causing most cells to lose respiratory function. Increased dNTP synthesis by RNR1 overexpression or SML1 deletion allows more full-length mtDNA replication, resulting in improved respiratory growth via electron transport chain function.
Nuclear DNA damage in yeast activates the Mec1/Rad53 nuclear checkpoint pathway, halts the cell cycle (Paulovich and Hartwell 1995), and increases dNTP production through the removal of Sml1 (Zhao et al. 2001; Chabes et al. 2003) and increased transcription of the RNR genes (Huang et al. 1998). Importantly, checkpoint activation was also shown to increase mtDNA copy number as much as twofold in ∆rrm3 and ∆sml1 deletion mutants (Taylor et al. 2005). However, as shown in Figure 1, the ∆rrm3 mutation did not increase the proportion of rho+ CFUs formed during heteroplasmy with HS rho− mtDNA, suggesting that increased mtDNA point mutagenesis in the ∆rrm3 background (O’Rourke et al. 2005) may strongly affect heteroplasmic cells.
We previously showed that the Mhr1 pathway regulates DSB-induced RDR in response to oxidative stress. The ori5 region is particularly sensitive to oxidative modification, and following exposure to hydrogen peroxide, Ntg1 was shown to increase DSB formation in this locus (Ling et al. 2007; Hori et al. 2009). Supporting this notion, the DSB-binding protein MmKu binds preferentially to the ori5 region, though ∆ntg1 cells showed only a slight decrease in MmKu binding, indicating that additional factors likely contribute to DSB formation at ori5 (Prasai et al. 2017). DSBs are substrates of Din7, which catalyzes 5′ end resection to yield 3′-ssDNA tails, which can then be used for homologous pairing by Mhr1 to initiate RDR (Ling et al. 2013). Compared to 85.7-kbp wild-type mtDNA, the 1.1-kbp HS [ori5] rho− mtDNA molecule has a much higher density of ori5 sequences. Therefore, reducing Mhr1 pathway activity through the ∆din7 mutation could be expected to disproportionately inhibit replication of HS [ori5] rho− mtDNA compared to wild-type. In our experiments, crosses of din7 null mutants did show a small but significant (P = 0.002) increase in the rho+ CFU formation rate compared to wild-type crosses. However, due to the presence of a functional SML1 gene, any benefit for wild-type mtDNA synthesis in the ∆din7 background was likely reduced due to the suppression of dNTP synthesis. Indeed, the ∆din7 ∆sml1 background showed an additive effect compared with ∆din7 (P = 0.003) or ∆sml1 (P = 0.032) single-mutants, indicating that dNTP availability plays an important regulatory role in the replicative advantage of HS rho− mtDNA.
Ribonucleotide reductase is considered an attractive target for inhibiting cell proliferation in cancer therapy and other disease. Here, our study in yeast suggests that inhibiting dNTP synthesis may produce the undesirable effect of increasing the replicative advantage of small mtDNAs, which have been associated with aging and several aging-related diseases in humans (see review by Kauppila et al. 2017). Precisely how RNR may contribute to human aging and disease in this context remains for future study.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.043851/-/DC1.
Acknowledgments
The authors thank Dr. Tilman Schneider-Poetsch who works in the Chemical Genetics Laboratory at RIKEN for helpful technical advice. This work was supported in part by a Grant-in-Aid for Scientific Research (C) (No. 23510237) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to F.L.; an Incentive Research Grant from RIKEN to F.L.; a grant from the RIKEN Strategic Research Program; and a grant from the Japan Agency for Medical Research and Development-Core Research for Evolutional Science and Technology to F.L.
Footnotes
Communicating editor: R. A. Sclafani
Literature Cited
- Blanc H., Dujon B., 1980. Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc. Natl. Acad. Sci. USA 77: 3942–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A., Stillman B., 2007. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 104: 1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A., Domkin V., Thelander L., 1999. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274: 36679–36683. [DOI] [PubMed] [Google Scholar]
- Chabes A., Georgieva B., Domkin V., Zhao X., Rothstein R., et al. , 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391–401. [DOI] [PubMed] [Google Scholar]
- Cortopassi G. A., Shibata D., Soong N. W., Arnheim N., 1992. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc. Natl. Acad. Sci. USA 89: 7370–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Rosa I., Camara Y., Durigon R., Moss C. F., Vidoni S., et al. , 2016. MPV17 loss causes deoxynucleotide insufficiency and slow DNA replication in mitochondria. PLoS Genet. 12: e1005779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F., Bayona-Bafaluy M. P., Rana M., Mora M., Hao H., et al. , 2002. Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control. Nucleic Acids Res. 30: 4626–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshawadfy A. M., Keith B. J., Ee Ooi H., Kinsman T., Heslop P., et al. , 2014. DNA polymerase hybrids derived from the family-B enzymes of Pyrococcus furiosus and Thermococcus kodakarensis: improving performance in the polymerase chain reaction. Front. Microbiol. 5: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W. L., Henly J. W., Brewer B. J., 1990. RPO41-independent maintenance of [rho-] mitochondrial DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fikus M. U., Mieczkowski P. A., Koprowski P., Rytka J., Sledziewska-Gojska E., et al. , 2000. The product of the DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, specifically functions in mitochondria. Genetics 154: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H., Moraes C. T., 2009. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum. Mol. Genet. 18: 1028–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Vioque E., Torres-Torronteras J., Andreu A. L., Marti R., 2011. Limited dCTP availability accounts for mitochondrial DNA depletion in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). PLoS Genet. 7: e1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf A. L., Kelly J. L., Lehman I. R., 1986. Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc. Natl. Acad. Sci. USA 83: 3391–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A., Yoshida M., Shibata T., Ling F., 2009. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication. Nucleic Acids Res. 37: 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Zhou Z., Elledge S. J., 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A., 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppila T. E., Kauppila J. H., Larsson N. G., 2017. Mammalian mitochondria and aging: an update. Cell Metab. 25: 57–71. [DOI] [PubMed] [Google Scholar]
- Lebedeva M. A., Shadel G. S., 2007. Cell cycle- and ribonucleotide reductase-driven changes in mtDNA copy number influence mtDNA inheritance without compromising mitochondrial gene expression. Cell Cycle 6: 2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecrenier N., Foury F., 1995. Overexpression of the RNR1 gene rescues Saccharomyces cerevisiae mutants in the mitochondrial DNA polymerase-encoding MIP1 gene. Mol. Gen. Genet. 249: 1–7. [DOI] [PubMed] [Google Scholar]
- Ling F., Shibata T., 2002. Recombination-dependent mtDNA partitioning: in vivo role of Mhr1p to promote pairing of homologous DNA. EMBO J. 21: 4730–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F., Shibata T., 2004. Mhr1p-dependent concatemeric mitochondrial DNA formation for generating yeast mitochondrial homoplasmic cells. Mol. Biol. Cell 15: 310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F., Makishima F., Morishima N., Shibata T., 1995. A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J. 14: 4090–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F., Hori A., Shibata T., 2007. DNA recombination-initiation plays a role in the extremely biased inheritance of yeast [rho-] mitochondrial DNA that contains the replication origin ori5. Mol. Cell. Biol. 27: 1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F., Hori A., Yoshitani A., Niu R., Yoshida M., et al. , 2013. Din7 and Mhr1 expression levels regulate double-strand-break-induced replication and recombination of mtDNA at ori5 in yeast. Nucleic Acids Res. 41: 5799–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lockshon D., Zweifel S. G., Freeman-Cook L. L., Lorimer H. E., Brewer B. J., et al. , 1995. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell 81: 947–955. [DOI] [PubMed] [Google Scholar]
- Lorimer H. E., Brewer B. J., Fangman W. L., 1995. A test of the transcription model for biased inheritance of yeast mitochondrial DNA. Mol. Cell. Biol. 15: 4803–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., Tieu Q., Wong E., Weiss E., Schieltz D., et al. , 1999. Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA. J. Cell Biol. 145: 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melov S., Lithgow G. J., Fischer D. R., Tedesco P. M., Johnson T. E., 1995. Increased frequency of deletions in the mitochondrial genome with age of Caenorhabditis elegans. Nucleic Acids Res. 23: 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke T. W., Doudican N. A., Zhang H., Eaton J. S., Doetsch P. W., et al. , 2005. Differential involvement of the related DNA helicases Pif1p and Rrm3p in mtDNA point mutagenesis and stability. Gene 354: 86–92. [DOI] [PubMed] [Google Scholar]
- Paulovich A. G., Hartwell L. H., 1995. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82: 841–847. [DOI] [PubMed] [Google Scholar]
- Prasai K., Robinson L. C., Scott R. S., Tatchell K., Harrison L., 2017. Evidence for double-strand break mediated mitochondrial DNA replication in Saccharomyces cerevisiae. Nucleic Acids Res. DOI: 10.1093/nar/gkx443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Pursell Z. F., Copeland W. C., Longley M. J., Kunkel T. A., et al. , 2005. DNA precursor asymmetries in mammalian tissue mitochondria and possible contribution to mutagenesis through reduced replication fidelity. Proc. Natl. Acad. Sci. USA 102: 4990–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., 1999. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 274: 17395–17398. [DOI] [PubMed] [Google Scholar]
- Stewart J. B., Chinnery P. F., 2015. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet. 16: 530–542. [DOI] [PubMed] [Google Scholar]
- Taylor S. D., Zhang H., Eaton J. S., Rodeheffer M. S., Lebedeva M. A., et al. , 2005. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol. Biol. Cell 16: 3010–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B., 2010. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11: 872–884. [DOI] [PubMed] [Google Scholar]
- Westermann B., Neupert W., 2000. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16: 1421–1427. [DOI] [PubMed] [Google Scholar]
- Zhang T., Lei J., Yang H., Xu K., Wang R., et al. , 2011. An improved method for whole protein extraction from yeast Saccharomyces cerevisiae. Yeast 28: 795–798. [DOI] [PubMed] [Google Scholar]
- Zhang Z., An X., Yang K., Perlstein D. L., Hicks L., et al. , 2006. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc. Natl. Acad. Sci. USA 103: 1422–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Muller E. G., Rothstein R., 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2: 329–340. [DOI] [PubMed] [Google Scholar]
- Zhao X., Chabes A., Domkin V., Thelander L., Rothstein R., 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20: 3544–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X. M., Clark-Walker G. D., Chen X. J., 2002. The mitochondrial nucleoid protein, Mgm101p, of Saccharomyces cerevisiae is involved in the maintenance of rho(+) and ori/rep-devoid petite genomes but is not required for hypersuppressive rho(-) mtDNA. Genetics 160: 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Yeast strains used in this study are listed in Table S1 and are available upon request.