Abstract
Double-strand breaks (DSBs) are lethal DNA lesions, which are repaired by homologous recombination in Escherichia coli. To study DSB processing in vivo, we induced DSBs into the E. coli chromosome by γ-irradiation and measured chromosomal degradation. We show that the DNA degradation is regulated by RecA protein concentration and its rate of association with single-stranded DNA (ssDNA). RecA decreased DNA degradation in wild-type, recB, and recD strains, indicating that it is a general phenomenon in E. coli. On the other hand, DNA degradation was greatly reduced and unaffected by RecA in the recB1080 mutant (which produces long overhangs) and in a strain devoid of four exonucleases that degrade a 3′ tail (ssExos). 3′–5′ ssExos deficiency is epistatic to RecA deficiency concerning DNA degradation, suggesting that bound RecA is shielding the 3′ tail from degradation by 3′–5′ ssExos. Since 3′ tail preservation is common to all these situations, we infer that RecA polymerization constitutes a subset of mechanisms for preserving the integrity of 3′ tails emanating from DSBs, along with 3′ tail’s massive length, or prevention of their degradation by inactivation of 3′–5′ ssExos. Thus, we conclude that 3′ overhangs are crucial in controlling the extent of DSB processing in E. coli. This study suggests a regulatory mechanism for DSB processing in E. coli, wherein 3′ tails impose a negative feedback loop on DSB processing reactions, specifically on helicase reloading onto dsDNA ends.
Keywords: DNA degradation, exonuclease activity, RecA protein, single-strand-specific exonucleases, recB1080 mutant
A DSB is an adverse DNA lesion that has to be repaired in order for a cell to survive. DSBs are repaired in all living organisms by either mutagenic nonhomologous end joining or by much more universally distributed and precise homologous recombination (HR). During HR, a single 3′-terminated strand is produced from each of two double-strand DNA (dsDNA) ends of a DSB by a process called DNA end resection, wherein a combination of helicase and nuclease activities result in degradation of complementary 5′-terminated strands (Symington 2014). The 3′-end overhangs emanating from a DSB are bound by a recombinase protein, thus creating the central recombination intermediate, the nucleoprotein filament. A recombinase nucleoprotein filament searches for an intact homologous sequence and invades it, hence restoring continuity of genomic information.
Since evolutionarily conserved recombinase proteins [RecA, RadA, and Rad51 (Dmc1) from bacteria, archaea, and eukaryotes, respectively] have a lower affinity of binding to ssDNA than their cognate ssDNA-binding proteins SSB/RPA, a recombination-mediator class of proteins (RecBCD and RecFOR proteins in bacteria and BRCA2, PALB2, and Rad52 in eukaryotes) facilitates recombinase polymerization on ssDNA (Zelensky et al. 2014).
In addition to its role in HR, the RecA nucleoprotein filament in Escherichia coli serves as a coprotease to promote autocatalytic cleavage of the LexA repressor, leading to induction of an SOS response (Little 1991). RecA also activates a mutagenic DNA polymerase V during SOS induction (Shinagawa et al. 1988).
In bacteria, both helicase and nuclease activities for DNA end resection are provided by the functionally related RecBCD, AddAB, and AdnAB enzymes (Wigley 2013). In E. coli, the RecBCD enzyme binds to a flush dsDNA end and unwinds a duplex molecule with its fast and processive helicase activity (Dillingham and Kowalczykowski 2008). Both of the unwound strands are degraded by a single nuclease center of the enzyme (residing in its RecB subunit) (Yu et al. 1998) until the enzyme encounters a regulatory octanucleotide sequence designated χ. Interaction with χ changes RecBCD’s behavior so that it ceases degradation of the 3′-terminated strand, while continuing DNA unwinding and degradation of the 5′-terminated strand (Anderson and Kowalczykowski 1997a). Also, the χ-modified RecBCD starts facilitating RecA polymerization onto the post-χ 3′ strand, hence producing a RecA-nucleoprotein filament (Anderson and Kowalczykowski 1997b). In this way, χ switches RecBCD enzyme degradation activity into a repair activity.
DSB repair in E. coli is active even in the absence of RecBCD due to RecQ helicase unwinding of duplex DNA, RecJ exonuclease trimming of ssDNA tails (ssExo) from the 5′-end, and RecFOR proteins mediating RecA polymerization onto the unwound 3′ overhangs. This pathway is operative only when ssExos that degrade 3′-terminated overhangs [e.g., Exonuclease I (ExoI) and SbcCD] are inactive, which enables stabilization of the recombinogenic substrate (Persky and Lovett 2008).
There are some mutants that show only partial function of RecBCD, yet are DSB repair proficient as well. When RecBCD lacks its RecD subunit, the resulting RecBC enzyme shows reduced helicase rate and processivity compared to RecBCD, and is completely devoid of nuclease activity and χ interaction (Dillingham and Kowalczykowski 2008). In the recD mutant, RecBC enzyme unwinds duplex DNA and constitutively loads RecA protein onto the unwound 3′ tail (Churchill et al. 1999), while its 5′ complement is trimmed by RecJ and Exonuclease VII (ExoVII) ssExos (Đermić 2006; Đermić et al. 2006a).
The recB1080 mutation renders the RecBCD enzyme deficient in nuclease and RecA loading activity, whereas the enzyme’s binding to DNA as well as rate and processivity of its helicase activity is unaffected (Yu et al. 1998; Anderson et al. 1999). In vitro, the RecB1080CD enzyme unwinds the linear DNA duplex, producing full length, RecA-free ss tails in the presence of SSB (Yu et al. 1998; Anderson et al. 1999). In the recB1080 mutant, the 5′-ended tail is clipped by RecJ and ExoVII ssExos, while its 3′ complement is covered with RecA protein with the help of RecFOR proteins (Jockovich and Myers 2001; Ivančić-Baće et al. 2003; Ivanković et al. 2017). The recB1080 mutant is recombination proficient; however, the efficiency of HR depends on trimming of its excessively long 3′ tails, and is lower than in wild-type bacteria (Ivanković and Đermić 2012; Ivanković et al. 2017). HR is not regulated by χ in the recB1080 mutant (Jockovich and Myers 2001).
An E. coli mutant lacking RecA protein is HR-deficient and is therefore unable to repair DSBs, which is reflected in its extreme sensitivity to various genotoxic agents and reduced viability (∼60% of the wild-type) (Kuzminov 1999). This relatively substantial decrease in viability is due to the exonuclease (ExoV) activity of the RecBCD enzyme (Miranda and Kuzminov 2003), which is actually unregulated, or “reckless”, in a recA mutant (Willets and Clark 1969). In this mutant, RecBCD degrades DNA duplexes with free dsDNA ends (either damaged or linear molecules introduced exogenously) so heavily that a fraction of the recA mutant population is devoid of a chromosome due to its complete degradation (Capaldo and Barbour 1975; Skarstad and Boye 1993).
In this study, we have characterized processing of DSBs introduced synchronously into a radioactively-labeled E. coli chromosome by γ-irradiation. Ionizing radiation, such as γ-rays, induce DSBs in DNA by either directly transferring energy to it or indirectly, by creating reactive oxygen species in the cell’s cytoplasm that then damage DNA; during repair of these clustered lesions, DSBs are produced (Bresler et al. 1979; Wallace 1998). DSB processing was assessed by measuring degradation of the fragmented chromosome. We show that DNA degradation in γ-irradiated E. coli is inhibited by RecA protein concentration and its ssDNA binding affinity. In fact, we show that binding of RecA to ssDNA is sufficient to protect the DNA duplex from degradation. However, DNA degradation as well as its RecA dependence was diminished in cells with preserved 3′ overhangs. Our results suggest that 3′ overhangs exert their influence on DSB processing by inhibiting helicase reloading onto dsDNA ends.
Materials and Methods
Bacterial strains, media, growth conditions, phage plating, and microscopy
We used AB1157, a standard HR-, DNA repair-, and DNA degradation-proficient strain (Bachmann 1972). Its derivatives were constructed by P1 transduction, as described earlier (Miller 1992), and are listed in Table 1. Bacteria were grown at 37° in LB medium and on LB agar plates (Miller 1992), supplemented with antibiotics when required.
Table 1. Bacterial strains used in this study.
| Strain | Relevant Genotype | Reference or Construction |
|---|---|---|
| AB1157 | Wild-type, rec+, ExoV+ | Bachmann (1972) |
| DE586 | ΔrecA774::kan | P1.JW2669 × AB1157 to Kanr |
| DE127 | lexA3 (ind−) malB::Tn9 | Laboratory collection |
| DE583 | recA730 (E38K) srlD300::Tn10 sulA::Tn5 | Laboratory collection |
| DE584 | recA730 sulA::Tn5 lexA3 malB::Tn9 | P1.DE202 × DE583 to Cmr, UVs |
| DE656 | ΔrecB1910::dhfr | Laboratory collection |
| DE657 | recA730 srl::Tn10 sulA::Tn5 ΔrecB1910::dhfr | P1.DE656 × DE583 to Tmr |
| DE393 | recAo281 srlD300::Tn10 lexA3 malB::Tn9 | Laboratory collection |
| DE628 | recAo281 srlD300::Tn10 ΔmalB732::kan | P1.JW3996 × DE393 to Kanr, UVr |
| DE189 | ΔumuDC::cat | Laboratory collection |
| DE637 | ΔumuDC::cat ΔrecA774::kan | P1.DE586 × DE189 to Kanr, |
| DE390 | ΔrecD744::kan | P1.JW2787 × AB1157 to Kanr |
| DE595 | ΔrecD744::kan recA269::Tn10 | P1.DE177 × DE390 to Tcr |
| DE101 | recB268::Tn10 | Đermić et al. (2005) |
| DE589 | recB268::Tn10 ΔrecA774::kan | P1.DE586 × DE101 to Kanr |
| DE590 | recQ1803::Tn3 | P1.JJC405 × AB1157 to Apr |
| DE591 | recQ1803::Tn3 recB268::Tn10 | P1.DE101 × DE590 to Tcr |
| DE592 | recQ1803::Tn3 recB268::Tn10 ΔrecA774::kan | P1.DE586 × DE591 to Kanr |
| DE110 | ΔrecQ::kan | Ivanković and Đermić (2012) |
| DE623 | ΔrecQ::kan recA269::Tn10 | P1.DE177 × DE110 to Tcr |
| RIK174 | recBD1080A | Jockovich and Myers (2001) |
| DE596 | recB1080 ΔrecA774::kan | P1.DE586 × RIK174 to Kanr |
| DE303 | recJ2052::Tn10 kan ΔxseA18::amp | Đermić et al. (2006a) |
| DE457 | sbcC201 ΔexoX769::frt ΔsbcB780::frt | Laboratory collection |
| DE585 | sbcC201 ΔexoX769::frt ΔsbcB780::frt ΔxseA758::kan | P1.JW2493 × DE457 to Kanr |
| DE587 | sbcC201 ΔexoX769::frt ΔsbcB780::frt ΔxseA758::kan recA269::Tn10 | P1.DE177 × DE585 to Tcr |
| DE460 | sbcC201 ΔexoX769::frt ΔsbcB780::frt ΔxseA18::amp | P1.STL4537 × DE457 to Apr |
| DE630 | sbcC201 ΔexoX769::frt ΔsbcB780::frt ΔxseA18::amp ΔrecD744::kan recA269::Tn10 | P1.DE177 × DE624 to Tcr |
Kan, kanamycin; Cm, chloramphenicol; Tm, trimetoprim; Tc, tetracycline; Ap, ampicillin; UV, ultraviolet light.
Exponentially growing bacteria (OD600∼0.3) were mixed with serially diluted phage T4 2 stock and incubated for 15 min at 37°. Soft LB agar was added to each mixture, the mixtures were poured onto LB plates, and incubated for 24 hr at 37°. Determinations were repeated three times.
Bacterial cells (grown identically to those for DNA degradation experiments, except for lacking [3H]thymidine) and their chromosomes were visualized in three independent experiments by combined phase-contrast and fluorescent microscopy, as described earlier (Zahradka et al. 2009).
γ-irradiation
Bacteria were exposed to various doses of γ-rays from a 60Co source, with a dose rate of ∼2.2 Gy s−1. For a bacterial survival assay, bacterial cultures were grown to midexponential phase (OD600∼0.3), serially diluted in 67 mM phosphate buffer (pH 7.0), and aliquots spread onto LB plates. The plates were immediately irradiated at room temperature and then incubated at 37° for 24–48 hr.
Chromosomal DNA degradation
We used the procedure described earlier (Đermić et al. 2005, 2006b). The cells were grown overnight in LB medium supplemented with 0.07 MBq ml−1 [3H]thymidine (specific activity 962 GBq mmol−1; Amersham, UK) and 100 µg ml−1 deoxyadenosine. Unincorporated [3H]thymidine was washed out with phosphate buffer; the cultures were suspended in a double volume of fresh LB medium and divided into two counterparts. One served as an unirradiated control, while the other was irradiated with 400 Gy at 0°. After irradiation, the cultures were incubated at 37° and at intervals duplicate samples were spread onto Whatman filters pretreated with 0.3 M NaOH. The filters were allowed to dry at room temperature, and then were suspended for 30 min in 10% trichloroacetic acid and twice in 5% trichloroacetic acid at 4°. Trichloroacetic acid precipitates high molecular weight DNA, while the low molecular weight DNA is washed away. The filters were then washed in 1:1 solution of ether and ethanol at 4° for 30 min, and then in ether at room temperature. Acid-precipitable radioactivity of the filters represents the amount of the intact chromosomal DNA, and was measured by scintillation counting (Liquid Scintillation Analyzer, Tri-Carb 2810 TR, Perkin Elmer). On average, 1500–3000 cpm was measured in unirradiated samples.
The frequency of genomic DSBs inflicted by γ-irradiation ranges from 0.004 to 0.01 DSBs/Gy/Mbp in various organisms (Daly 2009). The dose of 400 Gy is therefore expected to produce at least eight DSBs per E. coli chromosome. This is in accord with a measured rate of DSB induction in the E. coli chromosome of 2.7 DSBs per 100 Gy (Ulmer et al. 1979) (which would amount to ∼10 DSBs at 400 Gy), and also with the prediction of ∼9 DSBs per cell at 400 Gy (Shee et al. 2013), based on direct detection of DSBs in vivo using a fluorescent DSB binding protein.
Preparation of cell-free extracts and western analysis
Bacterial cultures (grown identically as those for DNA degradation experiments, except for lacking [3H]thymidine) were split into two parts, one of which was irradiated with 400 Gy at 0°. After irradiation, both cultures were incubated at 37° for an additional 35 min. Cell-free extracts were prepared as previously described (Vujaklija and Maček 2012), with some modifications. Briefly, the cells were harvested from 20 ml of culture by centrifugation at 5000 × g and washed with 25 mM Tris-HCl pH 8.3, 1 mM NaCl, 1 mM EDTA, 1 mM DTT, and 0.5 mM PMSF. The cells were suspended in the same buffer and disrupted by sonication. Cell debris was removed by centrifugation at 12000 × g and the supernatant was used as a cell-free extract. Next, 15 µg of total protein were loaded into each lane and resolved by SDS-PAGE under reducing conditions in 10% gels. After electrophoresis, the separated proteins were transferred to an Amersham Hybond-P PVDF membrane (GE Healthcare). The membrane was stained with Amido Black (Sigma Aldrich, St. Louis) for protein visualization. This was a loading control and allowed determination of protein transfer efficiency across the blot. Western analysis performed as described (Vujaklija and Maček 2012) was used to estimate the relative concentrations of RecA protein in each sample. RecA was detected on western blots with the polyclonal anti-RecA antibody ab63797 (Abcam) diluted 1:6000. Antibody binding was visualized with peroxidase-coupled goat anti-rabbit antibody diluted 1:30,000 and Amersham ECLTM Western Blotting Reagent Pack (GE Healthcare). The specificity of the antibody was examined using an E. coli strain lacking the recA gene. Two biological replicates of each strain were analyzed by immunoblotting. ImageJ, a Java-based image processing program (Schneider et al. 2012), was applied to analyze western blot signal intensity.
Data availability
Strains are available upon request. All the data necessary for confirming the conclusions presented in this paper are represented in the paper.
Results
Synchronous DSBs were induced into E. coli DNA by γ-irradiation and their processing was monitored by measuring degradation of the fragmented chromosome, which was labeled with [3H]thymidine.
RecA inhibits DNA degradation in γ-irradiated E. coli
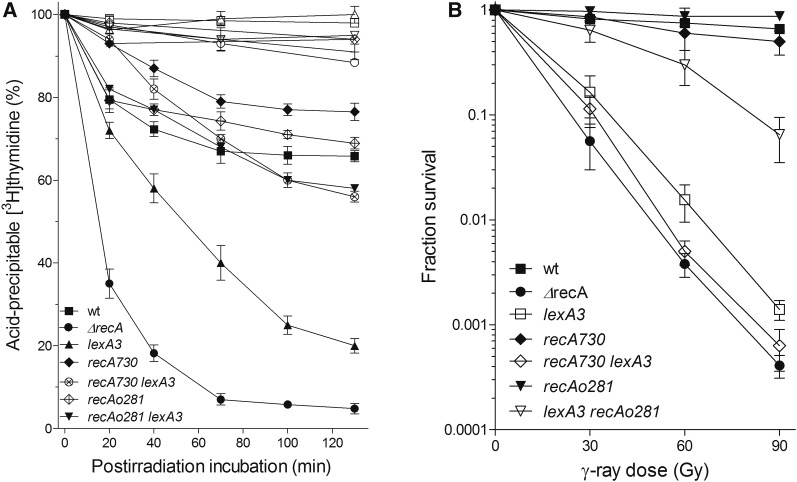
Figure 1A shows a time course of chromosomal DNA degradation in E. coli irradiated with 400 Gy of γ-rays. Irradiated wild-type strain AB1157 degraded ∼30% of its DNA during ∼60 min, after which degradation ceased and the remaining DNA was stable for another ∼60 min of postirradiation incubation at 37° (Figure 1A). This result is in accord with previous studies (Đermić et al. 2005, 2006b).
Figure 1.
(A) DNA degradation in γ-irradiated E. coli depends on RecA protein concentration and its rate of association with ssDNA. Bacterial cultures were divided into two counterparts; one served as a control (open symbols), while the other was irradiated with 400 Gy (closed symbols). The cultures contained [3H]thymidine-labeled chromosome; kinetics of their degradation were monitored during incubation at 37°. AB1157 (□,▪); ∆recA (○,●); lexA3 (▵,▴); recAo281 (+, ); lexA3 recAo281 (▿,▾); recA730 (◊,♦); and recA730 lexA3 (×,⊗). Each value is a mean of three independent experiments, with error bars representing SD. (B) Survival of γ-irradiated bacteria. Wild-type strain AB1157 (▪) and its recA (●), lexA3 (h), recAo281 (▾), lexA3 recAo281 (▿), recA730 (♦), and recA730 lexA3 (◊) derivatives. Fraction survival is given as a fraction of the unirradiated control. Each value is a mean of three independent experiments, with error bars representing SD. ssDNA, single-stranded DNA; wt, wild-type.
); lexA3 recAo281 (▿,▾); recA730 (◊,♦); and recA730 lexA3 (×,⊗). Each value is a mean of three independent experiments, with error bars representing SD. (B) Survival of γ-irradiated bacteria. Wild-type strain AB1157 (▪) and its recA (●), lexA3 (h), recAo281 (▾), lexA3 recAo281 (▿), recA730 (♦), and recA730 lexA3 (◊) derivatives. Fraction survival is given as a fraction of the unirradiated control. Each value is a mean of three independent experiments, with error bars representing SD. ssDNA, single-stranded DNA; wt, wild-type.
On the other hand, a recA null mutant (DE586) showed much stronger, continuous DNA degradation that resulted in degradation of ∼95% of its cellular DNA after ∼120 min postirradiation incubation (Figure 1A). Therefore, we confirmed a “reckless” DNA degradation in the recA mutant.
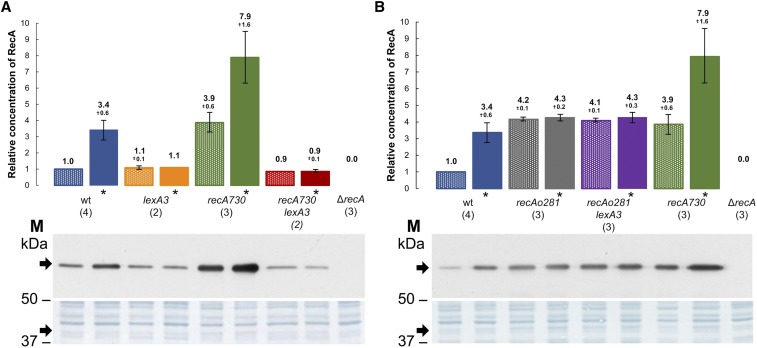
Since RecA protein regulates SOS induction and is itself regulated by SOS, we wanted to assess the effect of the SOS response on DNA degradation in γ-irradiated E. coli. For that, we made the SOS system in AB1157 uninducible by introducing a lexA3 allele, coding for a noncleavable SOS repressor. By western analysis, we estimated the relative concentrations of RecA protein in the wild-type cells that were or were not exposed to γ-rays. Our result revealed an increase of around threefold the basal RecA concentration in the irradiated wild-type strain (35 min after irradiation) compared to the unirradiated control (Figure 2). The lower level of SOS induction compared to the previous study (Sassanfar and Roberts 1990) is likely due to short postirradiation incubation time and different SOS-inducing agents used. As expected, no change of (basal) RecA concentration was observed in the lexA3 mutant upon irradiation (Figure 2A). As shown in Figure 1A, the lexA3 mutant also showed continuous DNA degradation during ∼120 min postirradiation incubation, but the amount of degradation was lower than in the recA mutant, with ∼20% of its DNA being spared. This result shows that the SOS system influences DNA degradation in γ-irradiated E. coli. In order to check which function of the SOS is important for suppression of DNA degradation, we made use of recAo281, an operator mutation that enables SOS-independent constitutive overexpression of RecA protein (Clark 1982). RecA concentration was ∼4 fold higher in a recAo281 mutant compared to the unirradiated wild-type strain, while being about the same in their irradiated counterparts (Figure 2B). DNA degradation was similar in the recAo281 mutant and wild-type strain (Figure 1A). A lexA3 recAo281 mutant is SOS-deficient, yet expresses similarly elevated (around fourfold) concentration of RecA protein (Figure 2B). After 2 hr of postirradiation incubation, it degraded ∼40% of its DNA, which is similar to degradation in the wild-type strain, whereas about threefold more of its DNA was preserved compared to DNA in the irradiated single lexA3 mutant (Figure 1A). This suggests that RecA concentration is important for suppressing DNA degradation, while other parts of the SOS response have a minor effect. Therefore, our results reveal an inverse relationship between RecA concentration and DNA degradation in irradiated E. coli, which led us to conclude that RecA protein suppresses DNA degradation by mass effect.
Figure 2.
Immunoblotting analysis of relative RecA protein concentrations in E. coli (A and B). Protein samples obtained from the cells that were irradiated with 400 Gy of γ-rays and subsequently incubated for 35 min at 37° are marked with *. PVDF membranes stained with Amido Black were used as a loading control (lower panels). SDS-PAGE gels and PVDF membranes are from one experiment, while graph depicts mean and SD (error bars) from multiple replicates (numbered in parentheses) from two independent experiments. PVDF, polyvinylidene fluoride; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; wt, wild-type.
Next, we wanted to determine how the RecA rate of association with ssDNA influences DNA degradation in irradiated cells. For that, we used a recA730 mutant, which produces the RecAE38K mutant protein that shows an increased rate of association with ssDNA and hence competes more efficiently with SSB protein for binding to ssDNA (Lavery and Kowalczykowski 1992). When introduced into AB1157, the recA730 mutation reduced DNA degradation in the resulting strain ∼1.5-fold (Figure 1A). This effect can be caused by an increased affinity of the RecAE38K for ssDNA, but also by an increased concentration of the enzyme in that mutant due to constitutive SOS induction. RecAE38K concentration in unirradiated and irradiated cells of the recA730 mutant was around four- and eightfold higher, respectively, than the RecA concentration in an unirradiated wild-type strain (Figure 2). We measured the effect of the increased affinity of RecAE38K for ssDNA by using a lexA3 recA730 mutant, which produces a basal level of RecA E38K that is equal to that of RecA in the lexA3 mutant (Figure 2A). The lexA3 recA730 mutant showed greatly reduced (about twofold) DNA degradation compared to that in the lexA3 mutant (Figure 1A), revealing that RecA dependency of DNA degradation is proportionate to RecA’s rate of association with ssDNA. In order to check whether the protective effect of RecAE38K protein on DNA degradation is indeed due to its high affinity to ssDNA, and not to its possible nonspecific binding to dsDNA, we determined the titer of T4 2 phage in recA730 and recA730 recB mutants. The T4 2 mutant phage genome is a linear dsDNA with free, blunt ends and hence susceptible to RecBCD binding and nucleolytic degradation. If RecAE38K would bind to dsDNA, hence blocking the access and activity of RecBCD on it, the titer of T4 2 phage should increase. In comparison to a recB mutant (which cannot degrade the T4 2 phage genome and therefore enables maximal phage yield), the T4 2 phage plating efficiencies in the recA730 and wild-type strains were 0.0019 ± 0.001 and 0.0017 ± 0.0007, respectively, which is significantly different (P = 0.001, two-tailed t-test) from the recB derivative of recA730 mutant that had 0.94 ± 0.19 of recB’s titer (n = 3), suggesting that RecBCD inhibits phage propagation in the recA730 mutant and that phage genome metabolism is otherwise not impaired in that mutant. Hence, we infer that RecAE38K protein does not interfere with degradation of purely dsDNA.
The difference in degradation between recA730 and recA730 lexA3 mutants (Figure 1A) is likely mostly due to different concentrations of the RecAE38K protein in these strains. The same, moderate amount of DNA degradation observed in the lexA3 recA730 and lexA3 recAo281 mutants (Figure 1A) indicates that, for DNA degradation inhibition, the higher RecA rate of association with ssDNA can compensate for lower RecA concentration, and that SOS induction is not required per se for protection of cellular DNA from degradation.
In summary, we have shown that RecA protein protects a fragmented chromosome from degradation in γ-irradiated wild-type E. coli. The protective activity depends on RecA concentration and its rate of association with ssDNA.
DNA degradation is distributed uniformly in wild-type cells
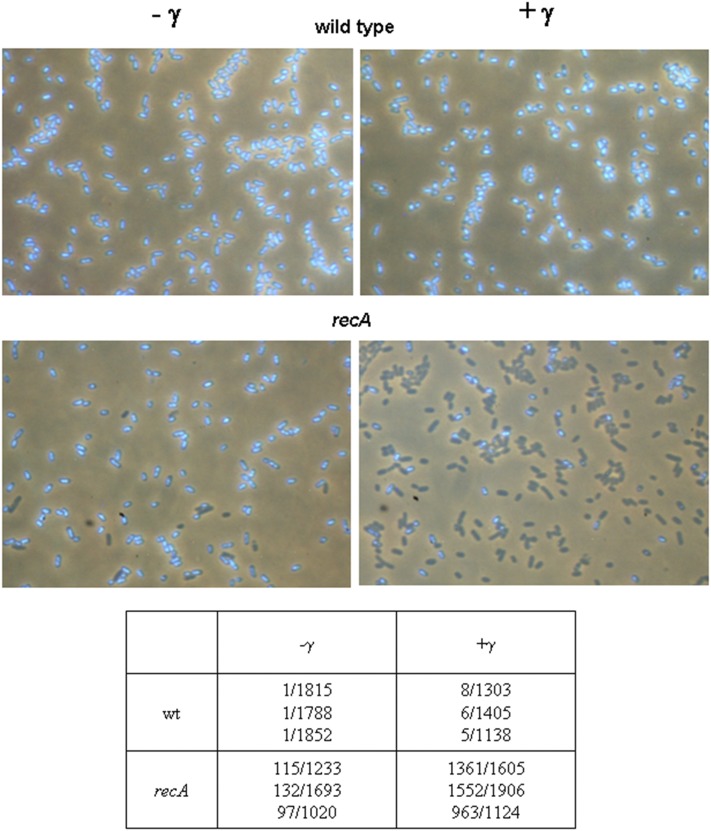
The DNA degradation experiments used in this study provide bulk population data that do not give us information on degradation distribution in a population. To assess the distribution of DNA degradation in a cell population, we visualized bacteria by microscopy and determined the fraction of cells lacking DAPI staining material in a population of wild-type (AB1157) and recA bacteria after 90 min postirradiation incubation. Unirradiated populations of wild-type and recA cells contained 0.055 ± 0.001% and 8.87 ± 0.93% anucleate cells, respectively (Figure 3). On the other hand, wild-type and recA populations γ-irradiated with 400 Gy had 0.49 ± 0.10% and 84.1 ± 2.4% of cells lacking a DAPI signal, respectively, after 90 min incubation (Figure 3). These data suggest that DNA degradation in wild-type cells is tightly regulated since it is very uniform, so that ∼35% of their degraded cellular DNA (Figure 1A) is reflected in <1% of chromosomeless cells. Conversely, only ∼16% of irradiated recA cells contained DNA, which is consistent with heavy, uncontrolled DNA degradation in them, sparing just ∼5% of their DNA (Figure 1A).
Figure 3.
DNA degradation is strictly controlled in wild-type, but not in recA cells. A fraction of anucleate cells was determined in wild-type (AB1157) and recA populations, which were either unirradiated (left side of the panel) or irradiated with 400 Gy of γ-rays and then incubated for 90 min at 37° (right side of the panel). Cells were fixed with osmium tetroxide, their DNA stained with DAPI, and visualized by fluorescence microscopy. An anucleated cell count in an analyzed population from three experiments is represented in a table. DAPI, 4’,6-diamidino-2-phenylindole; wt, wild-type.
Role of RecA protein in survival of γ-irradiated cells
Since degradation of a fragmented chromosome reflects DSB processing reactions, we wanted to relate it to a DSB repair process. Therefore, we determined survival of γ-irradiated bacteria as a measure of the efficiency of their DSB repair. As expected, the recA mutant showed an extreme sensitivity to γ-rays; at 90 Gy dose, its survival was more than three orders of magnitude lower than that of the wild-type strain AB1157 (Figure 1B). The lexA3 mutant had somewhat higher survival than the recA mutant (Figure 1B), which is not surprising considering that it retains basal RecA concentration. Survival of the lexA3 recAo281 mutant was considerably higher than that of the lexA3 mutant (also due to higher RecA concentration), whereas the recAo281 mutant had the same survival as the wild-type strain (Figure 1B).
While the recA730 mutant had essentially the same survival as the wild-type strain, its lexA3 derivative showed extreme sensitivity to γ-rays, similar to that of the recA and lexA3 mutants (Figure 1B). Since the recA730 lexA3 mutant showed highly reduced DNA degradation compared to the recA (and lexA3) mutant, whereas it displayed almost identical γ-survival (compare Figure 1, A and B), we conclude that reducing “reckless” degradation is insufficient to enable DNA repair.
A comparison of γ-survivals shows that the important factors for higher survival of γ-irradiated cells are: higher RecA protein concentration (hence the higher survival of recAo281 lexA3 than lexA3 and recA730 lexA3 mutants) and SOS induction (causing increased survival of recAo281 compared to the recAo281 lexA3 mutant).
RecA inhibition of DNA degradation is independent of DNA polymerase V
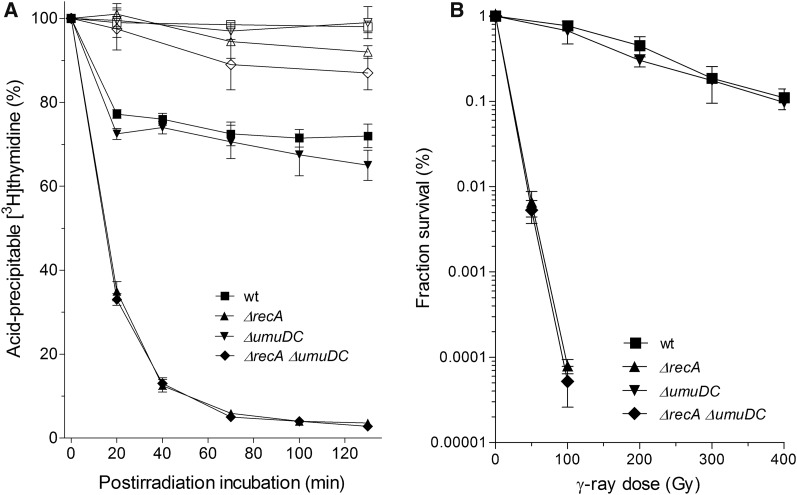
In E. coli, RecA is required for three processes, namely for HR, SOS induction, and activation of mutagenic DNA polymerase V (PolV) (see Introduction). Since we have shown that RecA’s function in inhibition of DNA degradation does not necessarily correspond to its role in DSB repair and SOS induction, we wanted to determine its relationship with PolV activation. For that goal, we determined DNA degradation and γ-survival of a PolV-deficient umuDC mutant and its recA derivative. As shown in Figure 4, the PolV-deficient mutant had mildly increased DNA degradation compared to the wild-type strain, while its γ-survival was the same as that of the wild-type strain. Furthermore, no difference was observed in DNA degradation and survival of umuDC recA and recA mutants (Figure 4). These results indicate that the RecA protective role during DNA degradation does not depend on PolV.
Figure 4.
DNA degradation is unrelated to DNA polymerase V. Kinetics of [3H]thymidine-labeled DNA degradation (A), and survival (B) of γ-irradiated wild-type strain AB1157 (▪) and its umuDC (▾), recA (▴), and umuDC recA (♦) derivatives. For DNA degradation assay, bacterial cultures were divided into two fractions; one served as a control (open symbols), while the other was irradiated with 400 Gy (closed symbols). Fraction survival is given as the fraction of the unirradiated control. Each value is a mean of three independent experiments, with error bars representing SD. wt, wild-type.
DNA degradation in recB and recD mutants is inhibited by RecA
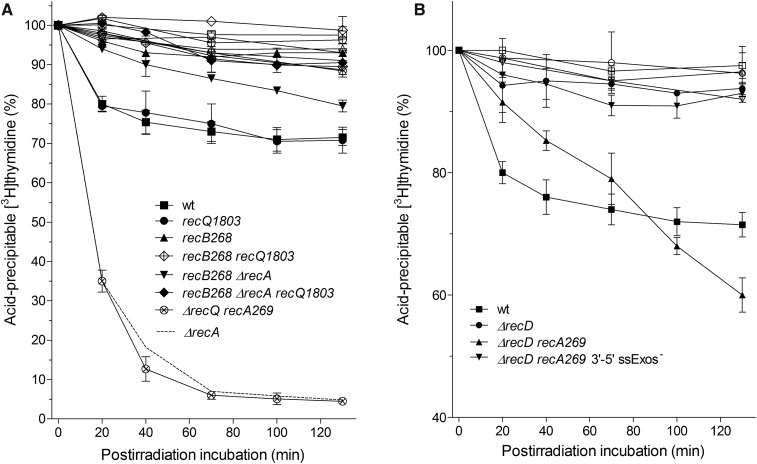
We wanted to determine whether the RecA inhibition of DNA degradation is restricted to RecBCD-catalyzed reactions, or whether it is a more general phenomenon. For this aim, we determined DNA degradation in γ-irradiated recB and recD mutants. No degradation was observed in these mutants (Figure 5, A and B), which is not surprising considering their ExoV− phenotype. However, their recA derivatives did degrade their damaged DNA. A recB recA mutant degraded its DNA continuously during postirradiation incubation, with degradation reaching a level close to that of the wild-type strain after 130 min (Figure 5A). This result indicates that RecA protects DNA from degradation in a recB null mutant. Since in the recB mutant dsDNA ends are processed by the RecQ helicase, we evaluated the role of RecQ in DNA degradation in the recB recA mutant. A triple recB recA recQ mutant degraded ∼10% of its DNA after 130 min incubation, which is about twofold less compared to its parental RecQ+ strain (Figure 5A), indicating that a helicase activity participates in DSB processing reactions that result in degradation of a shattered E. coli chromosome. At the same time, the recQ mutation did not affect DNA degradation in wild-type or recA and recB null mutants (Figure 5A), suggesting that the RecQ helicase role is specifically pronounced in the recB recA mutant.
Figure 5.
DNA degradation in recB (A) and recD (B) mutants is inhibited by RecA protein. Bacterial cultures were divided into two counterparts; one served as a control (open symbols), while the other was irradiated with 400 Gy (closed symbols). The cultures contained [3H]thymidine-labeled chromosome; kinetics of its degradation was monitored during incubation at 37°. (A) AB1157 (□,▪); recQ (○,●); recB (▵,▴); recB recQ (+, ); recB recA (▿,▾); recB recA recQ (◊,♦); recQ recA (×,⊗), and recA (- - -). (B) AB1157 (□,▪); recD (○,●); recD recA (▵,▴); and ExoI− ExoVII− ExoX− SbcCD− RecA− RecD− (▿,▾). Each value is a mean of three independent experiments, with error bars representing SD. wt, wild-type.
); recB recA (▿,▾); recB recA recQ (◊,♦); recQ recA (×,⊗), and recA (- - -). (B) AB1157 (□,▪); recD (○,●); recD recA (▵,▴); and ExoI− ExoVII− ExoX− SbcCD− RecA− RecD− (▿,▾). Each value is a mean of three independent experiments, with error bars representing SD. wt, wild-type.
A recD recA mutant (DE595) also showed constitutive DNA degradation; ∼40% of its DNA was degraded after 2 hr of incubation, thus exceeding the degradation level observed in the wild-type strain (Figure 5B). The RecBC enzyme is a nuclease-free helicase whose unwound products are subject to the activity of ssExos (Đermić 2006; Đermić et al. 2006a; Rinken et al. 1992), which results in limited degradation of the unwound DNA (Rinken et al. 1992). An earlier study (Kuzminov and Stahl 1997) has shown degradation of linearized plasmid DNA in a recD recA but not in a recB recA mutant.
In summary, we have shown that the RecA inhibition of DNA degradation is not only restricted to RecBCD-expressing wild-type bacteria, but also applies to the recB and recD mutants, suggesting that it is a more general phenomenon in E. coli. Also, we have shown that RecA-controlled DNA degradation involves a helicase activity.
DNA degradation in the recB1080 mutant is not inhibited by RecA protein
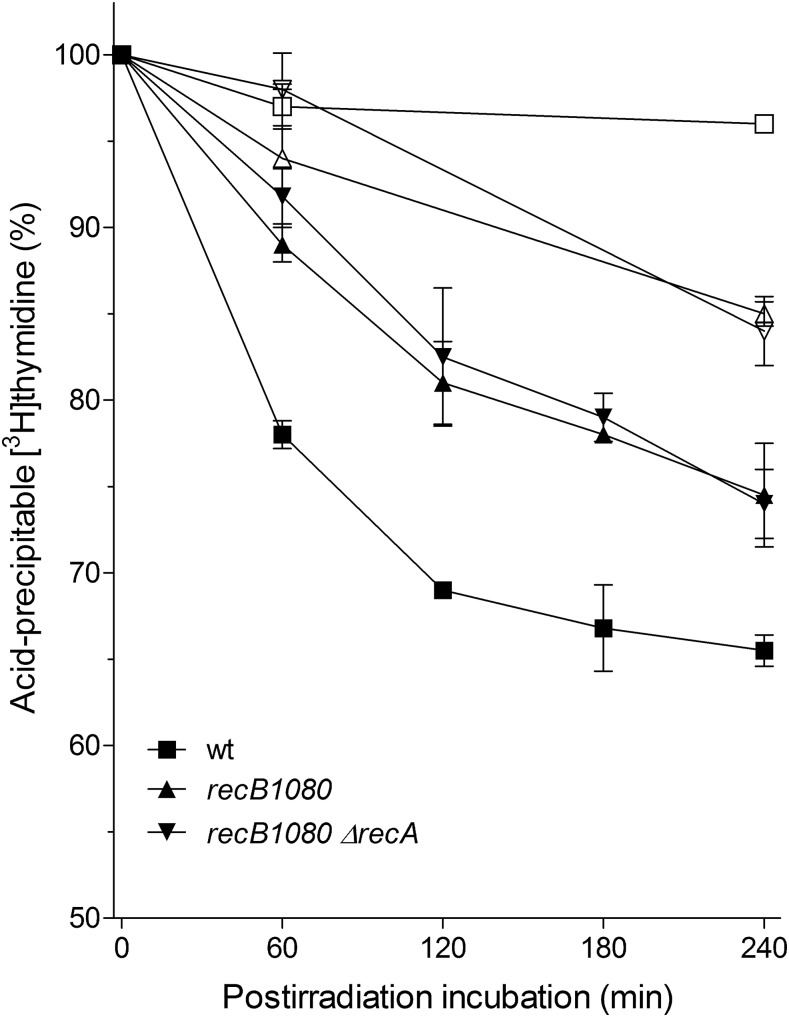
To further characterize the interplay of a helicase activity and RecA polymerization in DNA degradation, we made use of a recB1080 mutant strain RIK174, which produces the RecB1080CD enzyme. This enzyme is a fast and processive helicase, but is also nuclease-free and unable to load RecA onto the unwound 3′ tail (Yu et al. 1998; Anderson et al. 1999). In vitro, the enzyme unwinds the linear DNA duplex, releasing full length, RecA-free ss tails (Yu et al. 1998; Anderson et al. 1999). γ-irradiated RIK174 degraded ∼25% of its DNA after 240 min of incubation, which was ∼70% of DNA degradation in the wild-type bacteria (Figure 6). However, a recB1080 recA mutant showed a similarly low level of DNA degradation as its RecA+ parental strain (Figure 6), suggesting that DNA degradation in the recB1080 mutant is not inhibited by the RecA protein. This result indicates that, during processing of DSBs by a powerful nuclease-free helicase, long unwound tails prevent excessive DNA degradation, in effect relieving the RecA regulation of DNA degradation.
Figure 6.
DNA degradation in the recB1080 mutant is weak and uninhibited by RecA protein. Bacterial cultures were divided into two counterparts; one served as a control (open symbols), while the other was irradiated with 400 Gy (closed symbols). The cultures contained [3H]thymidine-labeled chromosome; kinetics of its degradation was monitored during incubation at 37°. AB1157 (□,▪); recB1080 (▵,▴); and recB1080 recA (▿,▾). Each value is a mean of three independent experiments, with error bars representing SD. wt, wild-type.
Inactivation of 3′–5′ ssExos greatly reduces DNA degradation and makes it independent of RecA
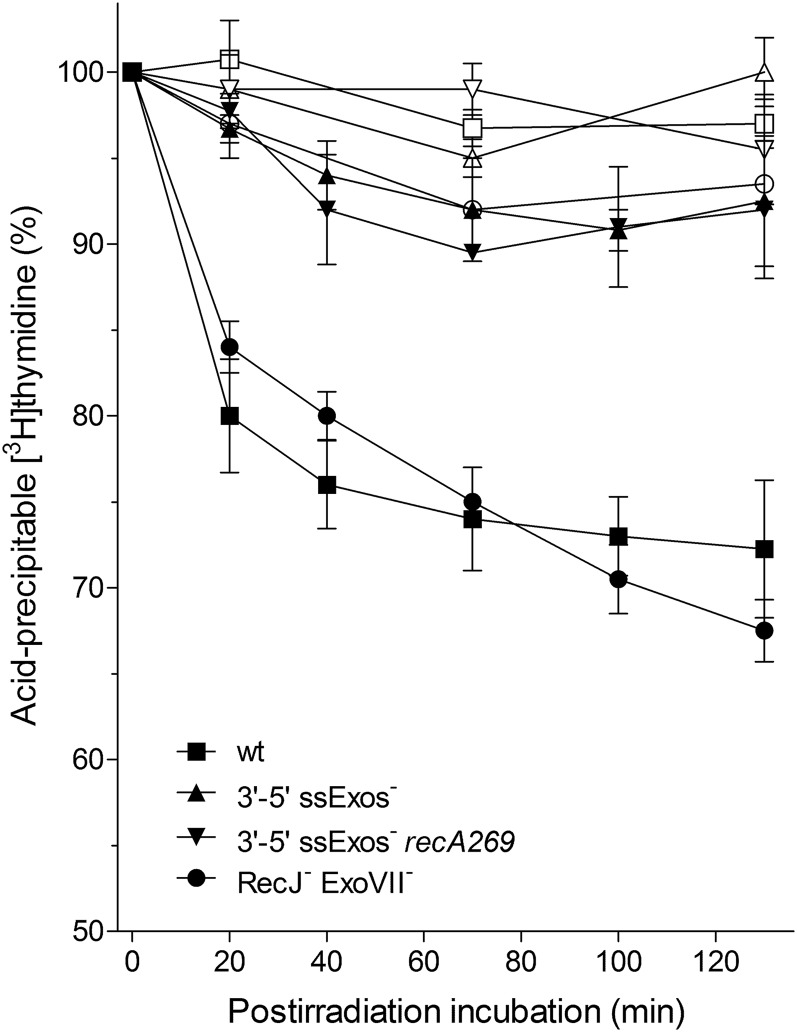
It was shown earlier that inactivation of ExoI, SbcCD, and ExoVII, ssExos that degrade 3′ overhangs, prevents “reckless” DNA degradation in a recA mutant (Zahradka et al. 2009; Repar et al. 2013). We made a quadruple mutant deficient in ExoI, SbcCD, ExoVII, and Exonuclease X (ExoX), ssExos that trim 3′ overhangs (Lovett 2011), and measured its DNA degradation. The mutant degraded ∼10% of its DNA after 130 min of incubation (Figure 7), which is close to the amount of DNA degraded in its unirradiated part (Figure 7), and is greatly reduced compared to degradation in its ssExo+ parental strain. On the other hand, DNA degradation in a strain lacking 5′–3′ ssExos RecJ and ExoVII (Lovett 2011) was similar to that in wild-type bacteria amounting to 31 ± 4% of genomic DNA after 130 min of incubation (Figure 7).
Figure 7.
DNA degradation in a mutant lacking 3′–5′ ssExos is very weak and not inhibited by RecA protein. Bacterial cultures were divided into two fractions; one served as a control (open symbols) while the other was irradiated with 400 Gy (closed symbols). The cultures contained [3H]thymidine-labeled chromosome; kinetics of its degradation were monitored during incubation at 37°. AB1157 (□,▪); ExoI− ExoVII− ExoX− SbcCD− (▵,▴); ExoI− ExoVII− ExoX− SbcCD− RecA− (▿,▾); and RecJ− ExoVII− (○,●). Each value is a mean of three independent experiments, with error bars representing SD. wt, wild-type.
Since the mutant lacking four 3′–5′ ssExos is an ExoV− phenocopy with respect to DNA degradation, although it expresses an intact RecBCD enzyme, we checked the enzyme’s nuclease activity by assessing its ability to inhibit growth of a T4 2 phage. Relative to the titer on the recB mutant, T4 2 phage plating efficiency on the wild-type strain was 0.0067 ± 0.0043, and that on the 3′–5′ ssExos-deficient mutant was 0.019 ± 0.012 (n = 3), which is not significantly different (P = 0.170, two-tailed t-test). This result indicates that RecBCD is proficient in binding to and degrading phage DNA in the mutant lacking 3′–5′ ssExos.
A recA derivative of the quadruple ExoI− SbcCD− ExoVII− ExoX− mutant showed about the same, low-level ∼10% degradation as its RecA+ counterpart (Figure 7), suggesting that DSB processing in bacteria with a preserved 3′ tail is not affected by RecA.
Similarly, DNA degradation in a recD recA mutant was greatly abolished by inactivation of the four 3′–5′ ssExos (Figure 5B). Therefore, we conclude that the constitutive chromosome degradation observed in the irradiated recD recA mutant is dependent on degradation of 3′-ended ss overhangs by the 3′–5′ ssExos.
Hence, our results indicate that 3′ overhangs suppress DSB processing, especially when spared from 3′–5′ ssExos.
Discussion
To gain better insight into the in vivo processing of DSBs, we followed the degradation of radioactively-labeled chromosomal DNA of γ-irradiated E. coli. DSBs in that bacterium are repaired by the RecBCD enzyme, which upon binding to a (nearly blunt) dsDNA end unwinds the DNA duplex and degrades both of the unwound strands. Only after interaction with the χ sequence does the enzyme starts a resection process, meaning that it continues its degradation of the 5′ strand while ceasing trimming of the 3′ strand. Therefore, DNA degradation is an essential and indivisible part of DSB processing in E. coli (there is no situation in which RecBCD activity on DNA is nuclease-free), and by assessing DNA degradation one can indeed get an insight into DSB processing. RecBCD enzyme is the strongest DNase in E. coli (Kuzminov 1999), yet its nuclease activity is augmented in bacteria lacking the RecA protein, wherein it becomes unregulated (“reckless”), leading to complete degradation of a chromosome (Capaldo and Barbour 1975; Skarstad and Boye 1993). “Reckless” degradation in recA null mutants is attributed to either impaired χ regulation of the RecBCD enzyme (Kuzminov et al. 1994; Kuzminov and Stahl 1997) or to a lack of RecA protection of the frayed ends of a processed DNA molecule (Dabert et al. 1992). We have shown here that recruitment of RecA onto ssDNA inhibits degradation at DSBs in wild-type, recB, and recD genetic backgrounds, indicating that the RecA protection of DNA from degradation is a general phenomenon in E. coli. Since there is no χ activity in the recB and recD mutants, and yet degradation of their DNA is still inhibited by RecA, we infer that the physical protection by RecA binding on (3′-terminated) ss overhangs is the main mechanism for RecA inhibition of DNA degradation. A recent study indicated that a RecA-ssDNA complex is resistant to degradation by nucleases (Kohiyama et al. 2013). Analogously, a human RecA ortholog, RAD51 recombinase, prevents excessive DNA nucleolytic degradation in UV-irradiated human cells (Vallerga et al. 2015), indicating conservation of the inhibitory role of recombinase proteins in DNA degradation, thus signifying its importance.
However, our results do not exclude the possibility that RecBCD’s χ activity is indeed impaired in the recA mutant, nor do they determine the possible contribution of each of the two mechanisms to DNA protection in that mutant.
Interestingly, our results show that the ability of the RecA protein to protect DNA from degradation differs from its role in DSB repair. The recA730 lexA3 mutant has greatly suppressed DNA degradation compared with a lexA3 mutant [both contain about the same concentration of RecA(E38K) protein], while having similar γ-survival. This phenotype is not unique as the recB mutant has it too; it also has RecA-inhibited DNA degradation and extremely poor survival. Thus, our results suggest that the RecA role in DNA degradation regulation is less challenging than its role in DSB repair; the former is about preventing 3′–5′ ssExos from degrading the 3′ tail (for which, binding of a couple of RecA molecules would likely suffice), while the latter requires production of a functional RecA nucleofilament. Furthermore, recB and recD null mutants share poor DNA degradation (suppressed by RecA), while the former is deficient for DSB repair and the latter is proficient. Also, recA730 lexA3 and recAo281 lexA3 mutants have about the same amount of DNA degradation, while their γ-survivals greatly differ.
However, we revealed two notable exceptions in the RecA-imposed control of DNA degradation, which enabled further insight into the mechanism of DSB processing regulation in E. coli. Namely, inactivation of four ssExos (ExoI, ExoVII, ExoX, and SbcCD) that degrade a 3′ tail (Lovett 2011) greatly reduced DNA degradation in both RecA+ and RecA− bacteria, making them an ExoV− phenocopy, even though the RecBCD enzyme is active in these cells and able to degrade DNA (as it prevented proliferation of the T4 2 phage, whose genome is blunt-ended). Since RecBCD poorly degrades fragmented chromosomes in cells lacking 3′–5′ ssExos while retaining ExoV activity, we infer that the enzyme is affected in binding to DNA, i.e., creation of blunt DNA ends is suppressed. A previous study showed that a strain lacking three 3′–5′ ssExos (ExoI, ExoVII, and SbcCD) retains nearly wt capacity for DNA repair, whereas its recA derivative had inhibited DNA degradation (ExoV−), thus indicating that these ssExos are not required for blunting the initial irradiation-produced dsDNA ends although being critical for “reckless” DNA degradation (Repar et al. 2013). Because 3′–5′ ssExos inactivation is epistatic to RecA deficiency for DNA degradation, we conclude that RecA inhibits DNA degradation by preventing trimming of 3′-terminated overhangs by 3′–5′ ssExos; hence, when these ssExos are absent, RecA becomes dispensable. Therefore, in cells lacking 3′–5′ ssExos, DSB processing is inhibited by preserved 3′ overhangs, irrespective of RecA protein (Figure 8A). Conversely, RecA deficiency is epistatic to 3′–5′ ssExos deficiency for DNA repair, thus additionally emphasizing differences in roles of the RecA protein in DNA repair and DNA degradation in E. coli.
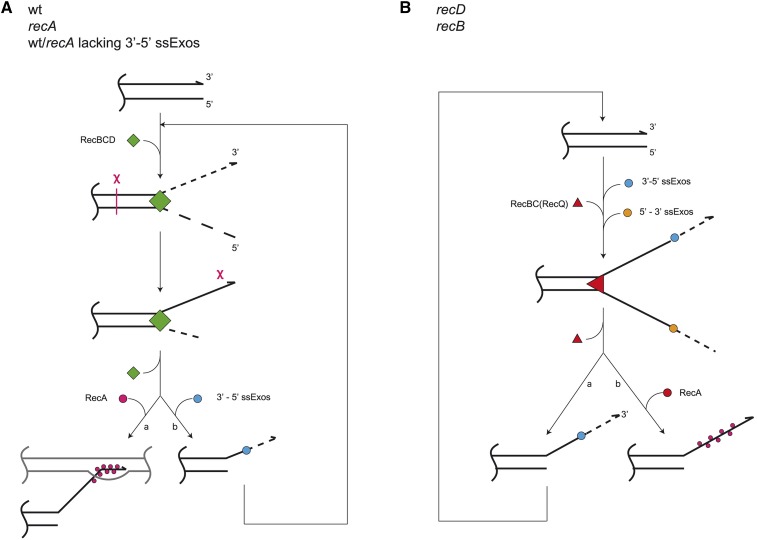
Figure 8.
Processing of a dsDNA end in several E. coli genetic backgrounds. (A) In wt bacteria, dsDNA processing is initiated by binding of RecBCD enzyme onto it, which unwinds the DNA duplex while degrading both unwound strands. Upon interaction with the χ site, the enzyme stops the 3′ strand degradation, while continuing DNA unwinding and degradation of the 5′ strand. χ-modified RecBCD also facilitates RecA loading onto a post-χ 3′ overhang (a). In the recA null mutant, RecBCD degrades both unwound strands before eventually being released from DNA, which might be independent of χ, leaving a protruding 3′ tail. The 3′ tail prevents subsequent reloading of RecBCD onto a processed dsDNA end unless/until 3′–5′ ssExos degrade it (b). Since both RecA+ and RecA− cells have similar, low DNA degradation when 3′–5′ ssExos are inactive [blocked (b) path], these exonucleases apparently act on a post-χ 3′ tail in wt cells [hence path (b) may apply to wt cells too]. (B) In recA derivatives of recB and recD mutants, dsDNA end processing starts with RecQ and RecBC helicase unwinding of the DNA duplex, respectively. The unwound 3′- and 5′-terminated strands are degraded by 3′–5′ and 5′–3′ ssExos, respectively. The degradation of the 5′ strand is apparently faster than that of its complementary 3′ strand (otherwise recD mutant would not be HR proficient) and thus the latter remains after helicase detachment from DNA. The protruding 3′ tail is then degraded by 3′–5′ ssExos (a), therefore enabling creation of a (nearly) blunt DNA end and hence repeated loading of RecQ or RecBC helicase. In RecA+ variants of recB and recD, RecA binds to the 3′ tail (b) and prevents RecQ reloading onto the DNA end in the recB mutant, while blocking its trimming and recreation of the blunt DNA end in the recD mutant and hence inhibits RecBC reloading. dsDNA, double-stranded DNA; ssExos, exonucleases that degrade a 3′ tail; wt, wild-type.
Another situation where DNA degradation is unaffected by RecA concerns the recB1080 mutant, whose recA derivative had about the same, low level of DNA degradation as its parental RecA+ strain. A combination of fast and processive helicase activity and lack of nuclease and RecA loading activities of RecB1080CD enzyme likely results in long, RecA-free overhangs in the recB1080 mutant (as discussed, Ivanković and Đermić 2012; Đermić 2015), analogously to what was observed in vitro (Yu et al. 1998; Anderson et al. 1999). In this mutant, RecA loading is achieved by RecFOR proteins, which perform it from an ss-ds DNA junction (Morimatsu and Kowalczykowski 2003), i.e., from the 5′-end of that tens of kb-long 3′-terminated tail (as discussed, Ivanković and Đermić 2012). Therefore, ssExos of 3′–5′ polarity likely need to digest very long ss stretches in order to reach a growing RecA nucleofilament. This is a heavy task because that ss region is overly reactive; it may get engaged in inter- and intramolecular transactions, resulting in illegitimate recombination intermediates (Ivanković and Đermić 2012) and ds secondary structures, as well as ds products of reannealing with the complementary 5′ tail, respectively. All of them may inhibit the 3′–5′ ssExos, hence increasing the longevity of the 3′-terminated tail. Furthermore, ExoI, the most potent 3′–5′ ssExo (Lovett 2011), degrades the 3′ tail with an ∼275 nt s−1 rate (Brody et al. 1986), which is around four- to eightfold slower than RecBCD helicase (and also less processive) (see below). Therefore, excessive length of the unwound 3′ strands and the mechanism of RecA loading onto them in the recB1080 mutant seemingly protects them from degradation by ssExos, thus making RecA protection dispensable. In fact, nucleolytic degradation of 3′ overhangs by 3′–5′ ssExos is essential for survival of the recB1080 mutant (Ivanković et al. 2017).
Since 3′ tail preservation is common to all the aforementioned situations, we conclude that 3′ overhangs emanating from DSBs are crucial in limiting the extent of DSB processing in E. coli. How do 3′ tails regulate DSB processing? Our results reveal a gradient in intensity of DNA degradation in the recA derivatives of wild-type, recD, and recB strains (∼95, ∼40, and ∼20% degraded DNA, respectively), which unsurprisingly correlates with the activities of their respective DSB processing helicases RecBCD, RecBC, and RecQ. The RecBCD helicase/nuclease processes at least 30 kb of DNA at ∼1000–2000 bp s−1 in vitro (Bianco et al. 2001; Dillingham et al. 2005), the RecBC helicase is about fourfold slower and less processive than RecBCD (Korangy and Julin 1993, 1994), and the RecQ helicase is even slower (∼2 bp s−1, Xu et al. 2003) and less processive than RecBC. However, this proportionality of DNA degradation with the potency of the DSB-processing helicase is disrupted in the recB1080 mutant, whose RecB1080CD enzyme is equally as powerful a helicase as RecBCD (Anderson et al. 1999); and yet, paradoxically, DNA degradation in its recA derivative is very weak, not stronger than that in the parental RecA+ strain, and certainly weaker than those observed in the recA recB and recA recD mutants. Extensive unwound tails produced in recB1080 cells by the powerful RecB1080CD helicase may certainly prevent its reloading onto dsDNA ends because RecBCD and its variants (RecBC and RecB1080CD) bind exclusively onto a (nearly) blunt dsDNA end (Dillingham and Kowalczykowski 2008). On the other hand, continuous DNA degradation for > 2 hr, resulting in a 20–40% degraded genome (1–2 Mbp) in recA derivatives of recB and recD mutants (Figure 8B) reflects perpetual processing of DSBs in those bacteria, which is possible only if their slow and poorly processive helicases (RecQ and RecBC) are repeatedly loaded onto the processed DNA ends. As a consequence, DSB processing catalyzed by repeated reloading of a weak helicase, such as RecQ or RecBC, turns out to be more vigorous than that accomplished by a single-round activity of the powerful helicase RecB1080CD. Consequently, we infer that RecA prevents reloading of RecQ and RecBC helicases in recB and recD mutants, respectively. Inactivation of 3′–5′ ssExos in the recD mutant alleviates the RecA inhibition of DNA degradation, indicating that RecA inhibits DNA degradation by protecting 3′ tails from being degraded by 3′–5′ ssExos. Preserved 3′ tails in turn inhibit RecBC reloading onto the processed DNA end. Lack of DNA degradation in the recB mutant indicates that an optimal substrate for the RecQ binding, a dsDNA end with a 3′ protrusion (Morimatsu and Kowalczykowski 2014), becomes inaccessible to RecQ when the tail is bound by RecA. Furthermore, our results show that 3′–5′ ssExos enable DNA degradation in wild-type cells, suggesting that RecBCD-catalyzed DSB processing is achieved through repeated reloading of the enzyme, which depends on the removal of the 3′ tails. This indicates that, in wild-type E. coli, 3′–5′ ssExos regularly trim RecBCD-produced post-χ 3′ tails (Figure 8A).
However, there is an alternative explanation for the stimulatory effect of 3′–5′ ssExos on DNA degradation. Namely, if RecBCD would release long, acid-precipitable oligonucleotides while processing the DNA duplex (since it has endonucleolytic rather than exonucleolytic activity), 3′–5′ ssExos may be actively trimming those oligonucleotides, thus making them acid-soluble. In this way, low acid-soluble DNA content observed in irradiated 3′–5′ ssExos-deficient cells would mask ongoing DNA degradation in them. However, this hypothesis may be ruled out since both the un- and UV-irradiated ExoI− ExoVII− SbcCD− RecA− mutants have preserved genomic DNA, as assessed by DAPI staining and genome restriction (Repar et al. 2013), suggesting a lack of RecBCD-catalyzed “reckless” DNA degradation.
We show here that DNA degradation in E. coli is inhibited by either DSB-processing helicase inactivation (e.g., by recB mutation in otherwise wild-type cells and by RecQ inactivation in the recB recA mutant), or by preservation of 3′ tails, again leading to the inability of these helicases to reload on the processed dsDNA ends. Our results therefore indicate that repeated helicase loading is the main determinant of the extent of DSB processing, with 3′ overhang metabolism being the crucial factor in helicase reloading. We describe three ways by which the availability of dsDNA ends to a helicase is controlled by 3′ tails: (i) RecA polymerization onto 3′ tails either directly inhibits RecQ loading onto them in the recB null mutant or inhibits their degradation by the 3′–5′ ssExos, thus preventing creation of blunt dsDNA ends and, consequently, reloading of RecBC(D) onto them; (ii) the appearance of blunt dsDNA ends is prevented by inactivation of the 3′–5′ ssExos, hence making shielding RecA binding onto such a protected 3′ overhang dispensable; and (iii) in the recB1080 mutant, lengthy DSB-derived 3′ overhangs are protected from 3′–5′ ssExos in a RecA-independent manner (instead, this is likely achieved by their involvement in transactions that produce ds regions in them), hence preventing recreation of blunt dsDNA ends. Similarly, excessively long 3′ overhangs produced during resection inhibit meiotic DSB repair in eukaryotes (Johnson et al. 2007).
Our collective results indicate that resection of a dsDNA end in E. coli proceeds until a 3′ tail of sufficient length and stability is formed, which then inhibits further end processing by preventing reloading of a DSB-processing helicase. Factors that facilitate 3′ tails’ longevity involve: (i) 3′ tails protection from degradation by 3′–5′ ssExos, by either “insulating” them with the RecA protein or by inactivation of these 3′–5′ ssExos, and (ii) their excessive length.
The 3′ tail regulation of DSB processing that we describe here is reminiscent of DNA end-resection in eukaryotes, with a short 3′ overhang produced during initial DSB resection by the Rad50 and Mre11 nuclease (orthologs of SbcCD) directing a resected end toward HR and microhomology-mediated end joining pathways and away from the nonhomologous end joining repair pathway (Truong et al. 2013). Similarly, the length of (and RecA binding to) the 3′ overhangs created during DNA end resection determines the equilibrium of HR and illegitimate recombination pathways in E. coli (Ivanković and Đermić 2012). Furthermore, SbcCD and its eukaryotic orthologs are analogously required to enable (re)loading of the main DSB processing machines [bacterial RecBCD and eukaryotic EXOI (ExoI)/BLM DNA2 (Sgs1 Dna2)] that perform long-range dsDNA end processing (as discussed recently, Đermić 2015).
Hence, one can note a common regulatory mechanism for DSB processing, with a 3′ tail produced during end resection acting as the main supervisory element by imposing a negative feedback loop on subsequent processing reactions. This mechanism ensures that processing of dsDNA ends, continues until stable and utilizable 3′ tails are produced that enable the efficient repair of DSBs in both bacteria and eukaryotes.
Acknowledgments
We are grateful to Amir Dubravić, Mary Sopta, and Nikola Paić for their help with manuscript preparation. This study was funded by the Croatian Science Foundation, project HRZZ-IP-11-2013-2978.
Footnotes
Communicating editor: R. A. Sclafani
Literature Cited
- Anderson D. G., Kowalczykowski S. C., 1997a The recombination hot spot, Chi, is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 11: 571–581. [DOI] [PubMed] [Google Scholar]
- Anderson D. G., Kowalczykowski S. C., 1997b The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell 90: 77–86. [DOI] [PubMed] [Google Scholar]
- Anderson D. G., Churchill J. J., Kowalczykowski S. C., 1999. A single mutation, RecBD1080A, eliminates RecA protein loading but not Chi recognition by RecBCD enzyme. J. Biol. Chem. 274: 27139–27144. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J., 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36: 525–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P. R., Brewer L. R., Corzett M., Balhorn R., Yeh Y., et al. , 2001. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409: 374–378. [DOI] [PubMed] [Google Scholar]
- Bresler S. E., Noskin L. A., Kuzovleva N. A., Noskina I. G., 1979. Nature of the damage to Escherichia coli DNA induced by gamma radiation. Int. J. Radiat. Biol. 36: 289–300. [DOI] [PubMed] [Google Scholar]
- Brody R. S., Doherty K. G., Zimmerman P. D., 1986. Processivity and kinetics of the reaction of exonuclease I from Escherichia coli with polydeoxyribonucleotides. J. Biol. Chem. 261: 7136–7143. [PubMed] [Google Scholar]
- Capaldo F. N., Barbour S. D., 1975. DNA content, synthesis and integrity in dividing and non-dividing cells of rec- strains of Escherichia coli K12. J. Mol. Biol. 91: 53–66. [DOI] [PubMed] [Google Scholar]
- Churchill J. J., Anderson D. G., Kowalczykowski S. C., 1999. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of Chi, resulting in constitutive recombination activation. Genes Dev. 13: 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. J., 1982. recA operator mutations and their usefulness. Biochimie 64: 669–675. [DOI] [PubMed] [Google Scholar]
- Dabert P., Ehrlich S. D., Gruss A., 1992. χ sequence protects against RecBCD degradation of DNA in vivo. Proc. Natl. Acad. Sci. USA 89: 12073–12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M. J., 2009. A new perspective on radiation resistance based on Deinococcus radiodurans. Nat. Rev. Microbiol. 7: 237–245. [DOI] [PubMed] [Google Scholar]
- Đermić D., 2006. Functions of multiple exonucleases are essential for cell viability, DNA repair and homologous recombination in recD mutants of Escherichia coli. Genetics 172: 2057–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Đermić D., 2015. Double-strand break repair mechanisms in Escherichia coli: recent insights. Adv. Genomics Genet. 5: 35–42. [Google Scholar]
- Đermić D., Halupecki E., Zahradka D., Petranović M., 2005. RecBCD enzyme overproduction impairs DNA repair and homologous recombination in Escherichia coli. Res. Microbiol. 156: 304–311. [DOI] [PubMed] [Google Scholar]
- Đermić D., Zahradka D., Petranović M., 2006a Exonuclease requirements for recombination of λ-phage in recD mutants of Escherichia coli. Genetics 173: 2399–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Đermić D., Đermić E., Zahradka D., Petranović M., Lerš N., 2006b Gamma-irradiated RecD overproducers become permanent recB-/recC- phenocopies for extrachromosomal DNA processing due to prolonged titration of RecBCD enzyme on damaged Escherichia coli chromosome. Biochimie 88: 379–386. [DOI] [PubMed] [Google Scholar]
- Dillingham M. S., Kowalczykowski S. C., 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72: 642–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham M. S., Webb M. R., Kowalczykowski S. C., 2005. Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J. Biol. Chem. 280: 37069–37077. [DOI] [PubMed] [Google Scholar]
- Ivančić-Baće I., Peharec P., Moslavac S., Škrobot N., Salaj-Šmic E., et al. , 2003. RecFOR function is required for DNA repair and recombination in a RecA loading-deficient recB mutant of Escherichia coli. Genetics 163: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanković S., Đermić D., 2012. DNA end resection controls the balance between homologous and illegitimate recombination in Escherichia coli. PLoS One 7: e39030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanković S., Vujaklija D., Đermić D., 2017. Nucleolytic degradation of 3′-ending overhangs is essential for DNA-end resection in RecA-loading deficient recB mutants of Escherichia coli. DNA Repair (Amst.) 57: 56–65. [DOI] [PubMed] [Google Scholar]
- Jockovich M. E., Myers R. S., 2001. Nuclease activity is essential for RecBCD recombination in Escherichia coli. Mol. Microbiol. 41: 949–962. [DOI] [PubMed] [Google Scholar]
- Johnson R., Borde V., Neale M. J., Bishop-Bailey A., North M., et al. , 2007. Excess single-stranded DNA inhibits meiotic double-strand break repair. PLoS Genet. 3: e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohiyama M., Contremoulins V., Baudin X., 2013. Trashing of single-stranded DNA generated during processing of arrested replication fork in E. coli. J. Mol. Biol. 425: 4837–4844. [DOI] [PubMed] [Google Scholar]
- Korangy F., Julin D. A., 1993. Kinetics and processivity of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry 32: 4873–4880. [DOI] [PubMed] [Google Scholar]
- Korangy F., Julin D. A., 1994. Efficiency of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry 33: 9552–9560. [DOI] [PubMed] [Google Scholar]
- Kuzminov A., 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63: 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A., Stahl F. W., 1997. Stability of linear DNA in recA mutant Escherichia coli cells reflects ongoing chromosomal DNA degradation. J. Bacteriol. 179: 880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A., Shabtach E., Stahl F. W., 1994. χ-sites in combination with RecA protein increase survival of linear DNA in Escherichia coli by inactivating exoV activity of RecBCD nuclease. EMBO J. 13: 2764–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery P. E., Kowalczykowski S. C., 1992. Biochemical basis of the constitutive repressor cleavage activity of recA730 protein. A comparison to recA441 and recA803 proteins. J. Biol. Chem. 267: 20648–20658. [PubMed] [Google Scholar]
- Little J. W., 1991. Mechanism of specific LexA cleavage-autodigestion and the role of RecA coprotease. Biochimie. 73: 411–422. [DOI] [PubMed] [Google Scholar]
- Lovett S. T., 2011. The DNA exonucleases of Escherichia coli. EcoSal Plus 4 Available at: .http://www.asmscience.org/content/journal/ecosalplus/10.1128/ecosalplus.4.4.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., 1992. A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Miranda A., Kuzminov A., 2003. Chromosomal lesion suppression and removal in Escherichia coli via linear DNA degradation. Genetics 163: 1255–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu K., Kowalczykowski S. C., 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11: 1337–1347. [DOI] [PubMed] [Google Scholar]
- Morimatsu K., Kowalczykowski S. C., 2014. RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc. Natl. Acad. Sci. USA 111: E5133–E5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persky N. S., Lovett S. T., 2008. Mechanisms of recombination: lessons from E. coli. Crit. Rev. Biochem. Mol. Biol. 43: 347–370. [DOI] [PubMed] [Google Scholar]
- Repar J., Briški N., Buljubašić M., Zahradka K., Zahradka D., 2013. Exonuclease VII is involved in “reckless” DNA degradation in UV-irradiated Escherichia coli. Mutat. Res. 750: 96–104. [DOI] [PubMed] [Google Scholar]
- Rinken R., Thoms B., Wackernagel W., 1992. Evidence that recBC-dependent degradation of duplex DNA in Escherichia coli recD mutants involves DNA unwinding. J. Bacteriol. 174: 5424–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassanfar M., Roberts J. W., 1990. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 212: 79–96. [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C., Cox B. D., Gu F., Luengas E. M., Joshi M. C., et al. , 2013. Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. Elife 2: e01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa H., Iwasaki H., Kato T., Nakata A., 1988. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc. Natl. Acad. Sci. USA 85: 1806–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Boye E., 1993. Degradation of individual chromosomes in recA mutants of Escherichia coli. J. Bacteriol. 175: 5505–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., 2014. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. Perspect. Biol. 6: a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong L. N., Li Y., Shi L. Z., 2013. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA 110: 7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer K. M., Gomez R. F., Sinskey A. J., 1979. Ionizing radiation damage to the folded chromosome of Escherichia coli K-12: sedimentation properties of irradiated nucleoids and chromosomal deoxyribonucleic acid. J. Bacteriol. 138: 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallerga M. B., Mansilla S. F., Federico M. B., Bertolin A. P., Gottifredi V., 2015. Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. Proc. Natl. Acad. Sci. USA 112: E6624–E6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujaklija D., Maček B., 2012. Detecting posttranslational modifications of bacterial SSB proteins, pp. 205–218 in Single-Stranded DNA Binding Proteins in Methods in Molecular Biology, edited by Keck J. Humana Press, Copyright Holder Springer Science+Business Media, LLC, NY. [DOI] [PubMed] [Google Scholar]
- Wallace S. S., 1998. Enzymatic processing of radiation-induced free radical damage in DNA. Radiat. Res. 150: S60–S79. [PubMed] [Google Scholar]
- Wigley D. B., 2013. Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat. Rev. Microbiol. 11: 9–13. [DOI] [PubMed] [Google Scholar]
- Willets N. S., Clark A. J., 1969. Characteristics of some multiply recombination-deficient strains of Escherichia coli. J. Bacteriol. 100: 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. Q., Deprez E., Zhang A. H., Tauc P., Ladjimi M. M., et al. , 2003. The Escherichia coli RecQ helicase functions as a monomer. J. Biol. Chem. 278: 34925–34933. [DOI] [PubMed] [Google Scholar]
- Yu M., Souaya J., Julin D. A., 1998. Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J. Mol. Biol. 283: 797–808. [DOI] [PubMed] [Google Scholar]
- Zahradka K., Buljubašić M., Petranović M., Zahradka D., 2009. Roles of ExoI and SbcCD nucleases in “reckless” DNA degradation in recA mutants of Escherichia coli. J. Bacteriol. 191: 1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelensky A., Kanaar R., Wyman C., 2014. Mediators of homologous DNA pairing. Cold Spring Harb. Perspect. Biol. 6: a016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. All the data necessary for confirming the conclusions presented in this paper are represented in the paper.