Abstract
Here, we present and characterize the spontaneous X-linked recessive mutation casper, which causes oculocutaneous albinism in threespine sticklebacks (Gasterosteus aculeatus). In humans, Hermansky-Pudlak syndrome results in pigmentation defects due to disrupted formation of the melanin-containing lysosomal-related organelle (LRO), the melanosome. casper mutants display not only reduced pigmentation of melanosomes in melanophores, but also reductions in the iridescent silver color from iridophores, while the yellow pigmentation from xanthophores appears unaffected. We mapped casper using high-throughput sequencing of genomic DNA from bulked casper mutants to a region of the stickleback X chromosome (chromosome 19) near the stickleback ortholog of Hermansky-Pudlak syndrome 5 (Hps5). casper mutants have an insertion of a single nucleotide in the sixth exon of Hps5, predicted to generate an early frameshift. Genome editing using CRISPR/Cas9 induced lesions in Hps5 and phenocopied the casper mutation. Injecting single or paired Hps5 guide RNAs revealed higher incidences of genomic deletions from paired guide RNAs compared to single gRNAs. Stickleback Hps5 provides a genetic system where a hemizygous locus in XY males and a diploid locus in XX females can be used to generate an easily scored visible phenotype, facilitating quantitative studies of different genome editing approaches. Lastly, we show the ability to better visualize patterns of fluorescent transgenic reporters in Hps5 mutant fish. Thus, Hps5 mutations present an opportunity to study pigmented LROs in the emerging stickleback model system, as well as a tool to aid in assaying genome editing and visualizing enhancer activity in transgenic fish.
Keywords: genome editing, stickleback, pigmentation, albinism, Hermansky-Pudlak syndrome
The combination of new genome editing methods and next-generation sequencing technologies has ushered in a new era in genetics. High-throughput DNA sequencing enables rapid forward genetic mapping of Mendelian (Schneeberger et al. 2009; Cuperus et al. 2010; Doitsidou et al. 2010; Zuryn et al. 2010; Bowen et al. 2012; Obholzer et al. 2012) and quantitative (Glazer et al. 2014, 2015; Jamann et al. 2015) loci. The remarkably high efficiency of the CRISPR/Cas9 system across diverse species (Friedland et al. 2013; Hwang et al. 2013; Jinek et al. 2013; Guo et al. 2014; Square et al. 2015; Martin et al. 2016) now allows for functional reverse genetic analysis in any species competent for delivery of genome editing reagents. However, the optimization of genome editing protocols is still in its infancy, with the efficiency of DNA double-strand break formation and repair still requiring characterization in many species. Particularly powerful loci for this characterization are those that mutate to cause an obvious viable visible phenotype, such as loci that affect pigmentation (Dahlem et al. 2012; Guo et al. 2014; Irion et al. 2014; Square et al. 2015; Burger et al. 2016; Hoshijima et al. 2016).
The diversity of vertebrate skin pigmentation is due to an interplay between four main groups of pigment-containing cells known as chromatophores (Fujii 2000; Kelsh 2004; Mills and Patterson 2009; Irion et al. 2016). Chromatophores originate from the neural crest, which migrate and then differentiate into pigment cell types during embryonic development (Fujii 2000; Kelsh et al. 2009). Melanophores possess black or dark brown melanin-containing organelles, melanosomes, which are also present in the retinal pigment epithelium (RPE) (Marks and Seabra 2001; Wasmeier et al. 2008). Iridophores appear iridescent and typically silver due to the presence of light-reflective guanine-containing platelets, the size and orientation of which determine the reflective color (Cooper et al. 1990; Oshima 2001). Xanthophores appear yellow-red due to the presence of pteridine within pterinosomes (Ziegler 2003). Erythrophores contain vesicles with red carotenoids obtained through the diet, and are a critical mating cue in sticklebacks (Milinski and Bakker 1990; Wedekind et al. 1998; Fujii 2000). Pigment cells in insects also possess melanin-, ommochrome-, or drosopterin-containing organelles (Shoup 1966).
Pigment-containing organelles belong to a larger class of cellular organelles, the lysosome-related organelles (LROs) (Dell’Angelica et al. 2000; Dell’Angelica 2004). LROs encompass a wide variety of organelles in different cellular contexts, including platelet granules, basophil granules, neutrophil azurophil granules, major histocompatibility complex class II compartments, lamellar bodies, osteoclast granules, and lytic granules (Dell’Angelica et al. 2000; Marks and Seabra 2001; Dell’Angelica 2004). The best studied LRO, the melanosome, has a well characterized biogenesis. Starting at stage I, premelanosomes already have internal vesicles and interluminal fibers that become parallel and organized during stage II, darkening during stage III until they are obscured by melanin in fully formed stage IV melanosomes (Marks and Seabra 2001). Patients with melanosome maturation defects often exhibit defects in other LROs, revealing that different LROs share similar biogenesis pathways (Marks and Seabra 2001).
In a wide range of vertebrates and invertebrates, a highly conserved set of genes including members of biogenesis of LRO complexes (BLOC) 1, 2, and 3 (Helip-Wooley et al. 2007; Wei et al. 2013) regulate the formation and maturation of LROs, including pigment producing LROs, and are required for wild-type pigmentation (Helip-Wooley et al. 2007; Wei et al. 2013). In humans, mutations in the BLOC-2 complex member Hps5 result in Hermansky-Pudlak Syndrome type 5, which is characterized by oculocutaneous albinism, and bleeding diathesis (Huizing et al. 2004). Mutations in orthologs of Hps5 result in oculocutaneous albinism in a wide variety of model organisms, including the ruby-eyed 2 phenotype in mice (Zhang et al. 2003), the snow white phenotype in zebrafish (Daly et al. 2013), the pink phenotype in Drosophila (Falcón-Pérez et al. 2007; Syrzycka et al. 2007), and the translucent phenotype in silkworms (Fujii et al. 2012). In vertebrates, Hps5 is required for the maturation of type I to type II melanosomes (Nguyen et al. 2002), and also binds to and stabilizes other HPS proteins including Hps3 and Hps6 (Daly et al. 2013). In addition to conserved roles in melanosome maturation, Hps5 and Hps6 are required for iridophore development in zebrafish and Xenopus, respectively (Daly et al. 2013; Nakayama et al. 2016).
In zebrafish, the snow white mutant phenotype, oculocutaneous albinism, was shown to result from an I76N point mutation in the WD40 domain of Hps5 (Daly et al. 2013). This mutation results in relatively mild reductions in RPE and melanophore melanization due to fewer and smaller melanosomes, and loss of iridophores. snow white mutants display early larval lethality, representing the only lethal Hps5 allele reported, as well as the only mutation within the N-terminal WD40 domain (Daly et al. 2013).
Here, we report the discovery and characterization of a spontaneous threespine stickleback X-linked recessive albino mutation casper. A mapping-by-sequencing approach revealed casper to be tightly linked to the stickleback ortholog of Hps5. casper mutants have a 1 bp insertion, resulting in a predicted frame-shift mutation in Hps5 that results in an early truncation of the highly conserved protein product. Inducing mutations in Hps5 using the CRISPR/Cas9 system phenocopied the casper mutation. Lastly, we show the usefulness of casper embryos as a tool to visualize fluorescent reporters in adult fish. Together these data provide a new locus in an emergent genetic supermodel (Gibson 2005) to facilitate studies of genome editing, transgene expression, and pigmentation biology.
Materials and Methods
Animal husbandry and imaging
Fish were raised in brackish water (3.5 g/L Instant Ocean salt, 0.217 ml/L 10% sodium bicarbonate) at 18° in 8 hr of light per day in 110 L aquaria. Fry with standard length (SL) less than ∼10 mm were fed a diet of live Artemia, with frozen Daphnia added as fish reached ∼10 mm SL. Adults with SL greater than ∼20 mm were fed a combination of frozen bloodworms and Mysis shrimp. To map the casper mutation, six crosses were generated by crossing four different marine males to heterozygous casper mutant females (Supplemental Material, Table S1). Experiments were approved by the Institutional Animal Care and Use Committees of the University of California-Berkeley (protocol # AUP-2015-01-7117). Embryos were visualized using Montage z-stacks on a Leica M165 FC dissecting microscope, using a GFP2 filter to visualize xanthophores, or on a Keyence VHX-5000 microscope. Adult fish were imaged using a Cannon Powershot S95 digital camera.
DNA purification and sequencing
For sequencing and genotyping, DNA was extracted from caudal fin tissue from the original casper male, the F0 female he was crossed to, as well as 47 individual casper mutant F2 embryos. Caudal fin tissue or embryos were digested for 12 hr at 55° in 600 μl of tail digestion buffer [10 mM Tris pH 8.0, 100 mM NaCl, 10 mM EDTA, 0.05% SDS, 2.5 μl Proteinase K (Ambion AM2546)].
DNA from whole casper mutant F2 embryos was diluted to ∼10 ng/μl for each fish and pooled. Barcoded Nextera libraries (Illumina FC-121-1031) were created from 50 ng of genomic DNA from the pooled embryos, as well as 50 ng of genomic DNA from the original casper male and F0 female mate following the manufacturer’s instructions. Quality was assayed on an Agilent bioanalyzer, and the resulting libraries were sequenced on a single lane of an Illumina HiSeq4000 to generate 100 bp single-end reads.
Bulk segregant analysis
Reads were mapped to a revised assembly of the stickleback genome (Jones et al. 2012; Glazer et al. 2015), using bowtie2 (Langmead and Salzberg 2012) with parameters “-q–sensitive” (Table S2). Resulting SAM files were converted to BAM files, and sorted using Samtools version 0.1.18 (Li et al. 2009). Read groups were added, CIGAR strings fixed, mate pair information was fixed, and PCR duplicates were removed using picard tools v 1.51 (http://broadinstitute.github.io/picard). The Genome Analysis Tool Kit (McKenna et al. 2010; DePristo et al. 2011; Van der Auwera et al. 2013) (GATK)’s (v3.2-2) IndelRealigner (parameter: “-LOD 0.4”), BaseRecalibrator, and PrintReads were used to finalize preprocessing of BAM files. Finally, Unified Genotyper was used to call variants, with parameters “–genotype_likelihoods_model BOTH -stand_call_conf 50.” The resulting VCF file was filtered for variants which had qual score >40, had more than five reads covering the variant, and were not found in the F0 female mated to the original casper male. Analysis was performed using a custom ipython notebook (see Data availability section below for link to custom scripts). Briefly, we computed the proportion of each variant that matched the casper allele, and results were smoothed by plotting the proportion of casper reads within a 50 variant sliding window, advancing five variants at a time. As we found casper to be X-linked, we reasoned that the hemizygous XY casper animals should all be identical in sequence around the causative locus. We computed, as an additional measure, the proportion of variants within the window with reads matching more than one allele, which is expected to be 0 at the causative locus.
PCR, cloning, and Sanger sequencing validation
PCR primers (Table S3) were designed using Primer3 (Rozen and Skaletsky 2000) and ordered from IDT. PCR was performed using Phusion DNA polymerase (Macro laboratory, University of California [UC]-Berkeley) and Phusion Buffer (NEB B0518S). To obtain sequence of single clones, PCR products were purified using a Qiagen PCR purification kit, and digested with XhoI (NEB R0146L) and XbaI (NEB R0145L) in cutsmart buffer for 1 hr at 37°. Digested products were ligated into a pBluescript II SK+ vector cut with XbaI and XhoI, transformed, and plated onto LB agar plates with ampicillin, IPTG, and X-gal. White colonies were picked and used as input to PCR as described above, adding a 5 min incubation at 95° before thermocycling. Resulting reactions were purified using a Qiagen PCR purification kit. Purified PCR product (∼20 ng) was Sanger sequenced by the UC-Berkeley DNA sequencing facility and results visualized using abiview.
Genome editing of Hps5
Genome editing reagents were designed as previously described (Talbot and Amacher 2014). Briefly, pCS2-nCas9n (Addgene 7929) was linearized following digestion with NotI. Linearized plasmid (∼600 ng) was used as input to the mMessage SP6 kit, following the manufacturer’s instructions. mRNA quality was verified by running 0.5 μl of the reaction in 0.1%SDS on a Tris acetic acid EDTA (TAE) gel.
Guide RNAs (gRNAs) were designed using ZiFiT (Sander et al. 2010). DNA oligos were ordered from IDT, and gRNA templates were created with T7 promoters by PCR using Phusion polymerase. Resulting PCR products were gel purified, and 100 ng of the resulting elution was used as input to the MAXIscript T7 kit (Ambion), and guide RNA quality verified by running 0.5 μl of the reaction in a 50% formamide buffer on a TAE gel. Resulting gRNAs were precipitated using lithium chloride, incubated at −80° following addition of 75 μl 100% ethanol, and centrifuged at 15,000 rpm for 1 hr at 4°. Following a wash with 200 μl of 75% ethanol and an additional centrifugation at 15,000 rpm for 10 min at 4°, RNAs were resuspended in 20 μl of DEPC-treated water.
Stickleback embryos at the one cell stage were microinjected as described (Erickson et al. 2016) with some modifications. First, the concentration of gRNA in the 0.2 M KCl injection mixture was increased to 50 ng/μl. The Cas9 mRNA concentration was also doubled to 160 ng/μl, with 0.025% phenol red used as a tracking dye. Embryos were scored for casper-like phenotypes at 4 d postfertilization (dpf). Embryos were scored by eye, with embryos with any sign of mosaic albinism in their RPE classified as “mosaic,” and embryos with ∼75% albino RPE classified as “severe.” DNA from uninjected, wild-type injected, mosaic injected, and casper-like injected embryos was purified at 4–6 dpf, and deletions in Hps5 were validated using Sanger sequencing of PCR products as described above.
Data availability
Scripts used for bulk segregant analysis are available at https://github.com/trahsemaj/CASPER. The original casper male, the F0 mated to the casper male, and bulk segregant sequencing data are available from the SRA (accession number SRP111743).
Results
casper mutants display severely reduced pigmentation early during embryogenesis
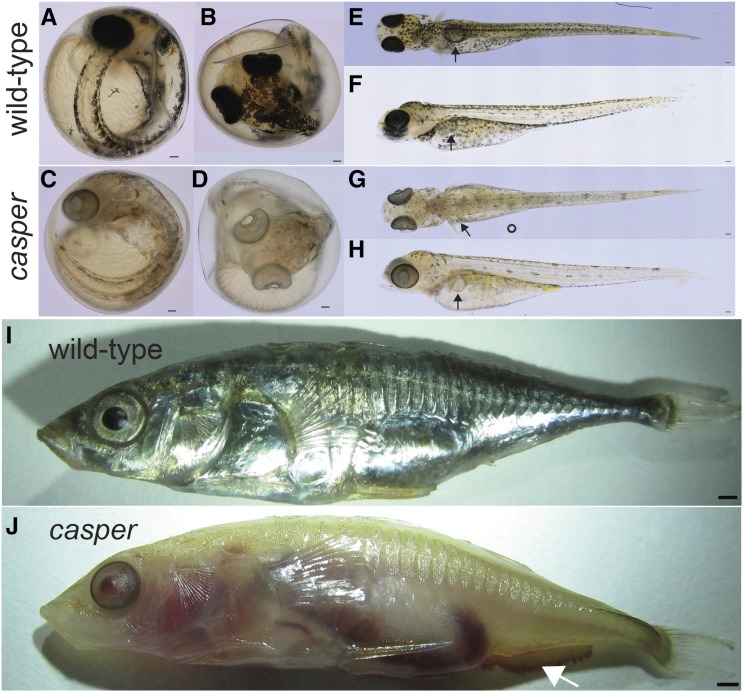
We discovered a single spontaneous mutant male stickleback displaying severe pigmentation defects. We named this mutation casper, and recovered the mutation in subsequent generations (see below). casper mutants display oculocutaneous hypopigmentation in unhatched embryos when pigment first becomes apparent, becoming readily apparent by 7 dpf (Figure 1). Mutants appear fully viable and fertile (see below). casper mutants display severely reduced melanization of their RPE, the most obvious visible phenotype (Figure 1, A–D). Additionally, sexually mature casper males displayed severely reduced pigmentation in their testes relative to their wild-type siblings (Figure S1). Chromatophores are differentially affected in casper mutants. Beginning at the time of their first appearance (4 dpf), melanophores in casper mutants are present, but display severe reductions in melanization relative to their wild-type siblings (Figure 1, E–H). The silver pigmentation from iridophores appears absent from older casper fish (Figure 1, I and J). The red erythrophores, which contain diet-supplied carotenoids (Wedekind et al. 1998), were never observed in the throats of sexually mature casper mutant males. However, the yellow xanthophores (autofluorescent in sticklebacks) appear unaffected by the casper mutation in 10 dpf casper mutants (Figure S2).
Figure 1.
casper mutants display severely reduced eye and body pigmentation. (A–D) casper mutants are hypopigmented at 7 dpf and have severely reduced melanization of RPE: lateral (A, C) and dorsal (B, D) views. (E–H) Hypopigmentation of casper mutants persists at 14 dpf, with severely reduced melanophores but slightly melanized RPE: dorsal (E, G) and lateral (F, H) views. Black arrows indicate inflated swim bladder. (I, J) casper males are translucent at 5 months, with severely reduced iridophores and melanophore pigmentation, and highly reduced melanization of RPE. Mutants also bleed after euthanization (arrow in J). Bars, 100 μm (A–H), 1 mm (I).
casper mutants appear to initially inflate their swim bladder (Figure 1, E–H). Older mutants display variably penetrant minor difficulties in maintaining their position in the water column, suggesting possible swim bladder defects (14/15 1-month-old juvenile casper mutants were found within 1 cm of the bottom of their tank compared to 2/21 wild-type siblings, P < 0.01, Fisher’s exact test). Lastly, casper mutants display a bleeding phenotype, possibly due to a decreased clotting rate, following euthanization in 0.04% tricaine relative to their wild-type siblings (Figure 1, I–J).
casper is a spontaneous, X-linked recessive albino mutation
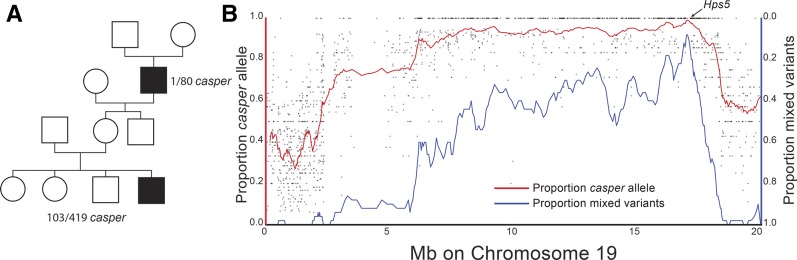
The original casper mutant fish was a single male, and first appeared in a clutch of 80 fish from a cross between a marine male from the Rabbit Slough, Alaska population, and a freshwater creek female from the Cerrito Creek, California population (Figure 2A). To map the locus responsible for the casper phenotype, the original casper male was outcrossed to a female fish from a different marine population (Table S1). All the resulting F1 progeny were phenotypically wild-type, suggesting the casper mutation was either recessive or mosaic, with the germline of the original mutant fish not containing the casper mutation. As sticklebacks have a simple XY sex determination mechanism (Peichel et al. 2004), a spontaneous X-linked recessive mutation would be displayed in the original hemizygous male, but not in F1 progeny. We thus hypothesized that casper was X-linked. To test this hypothesis, we generated six outcrosses of F1 females to four males from two other populations and observed the F2s. Consistent with an X-linked mutation, 103/419 (24.6%) of the F2 offspring were casper mutants (Table S1), and molecular genotyping (Table S3) of 47 of the casper mutants showed that they were all male, confirming the sex-linked nature of casper (P < 1.4 × 10−14, two-tailed binomial test).
Figure 2.
casper is X-linked and maps to a region of chromosome 19 near Hps5. (A) Pedigree of the spontaneous appearance of the original casper mutant and X-linked transmission in subsequent generations. Of the 103 casper mutants, 47 were genotyped, and all 47 were male by molecular genotyping with sex-specific primers. (B) Bulk segregant analysis of 47 casper mutants by high-throughput sequencing. Each point is the proportion of the variant allele with the casper genotype, with the red line showing a sliding window average across 50 variants. The blue line shows the proportion of variants called as heterozygous across a 50-variant sliding window. The peak of both red and blue lines is near the stickleback Hps5 gene. x-axis shows the chromosome 19 revised genome assembly (Glazer et al. 2015). The top of the y-axis for the proportion of mixed variants (blue line) is zero.
We next mapped the casper locus using a bulk segregant approach (Schneeberger et al. 2009; Cuperus et al. 2010; Doitsidou et al. 2010; Zuryn et al. 2010; Bowen et al. 2012; Obholzer et al. 2012). A barcoded Illumina sequencing library was created using genomic DNA pooled from 47 F2 casper males from six different F1 crosses. Additional barcoded libraries were created using DNA from the original casper fish as well as the F0 female crossed to casper, and all libraries were sequenced to moderate (∼10–14×) coverage (Table S2). We mapped casper by examining the proportion of reads at each variant position that matched the casper male’s allele. As an additional measure, we also looked for a loss of variants with mixed mapped reads (reflecting positions where most, or all, mutant male fish have the same X-chromosome genotype), as measured by the proportion of variants which only have reads supporting more than one allele within a 50 variant genomic window (Figure 2B). Both measures had similar peaks along stickleback chromosome 19, the stickleback X chromosome (Peichel et al. 2004), near the stickleback ortholog of a human oculocutaneous albinism gene, Hermansky-Pudlak syndrome 5 (Hps5) (Figure 2B).
casper is the result of the insertion of a single base-pair into the coding sequencing of Hps5
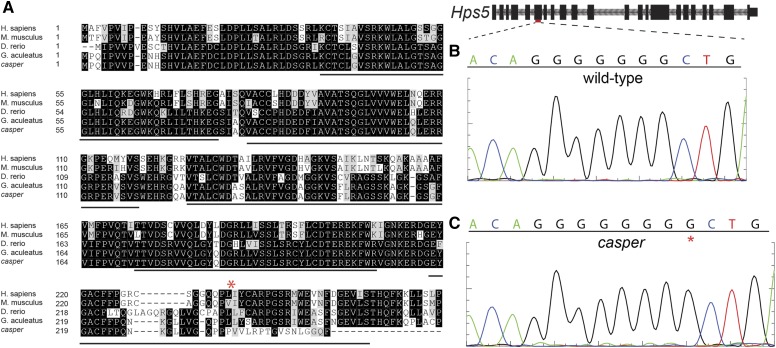
We next sought to determine the mutation responsible for the casper phenotype. Genome-wide variant discovery using high-throughput sequencing data from the original casper male and the F0 wild-type female he was crossed to revealed 14 total variants within the predicted Hps5 coding frame. Of these, 10 were synonymous point mutations, and two were substitutions found in both the affected casper male and unaffected female. The unaffected female had a mutation that results in a substitution from alanine to valine, which appears neutral (score of 0) in the BLOSUM62 matrix (Henikoff and Henikoff 1992), and which is also found in the orthologous mouse Hps5 sequence (Figure 3A). The only remaining, and highest, impact variant was the insertion of a G in the sixth exon of Hps5, resulting in a frameshift and predicted early stop appearing seven codons following the novel insertion (Figure 3A and Figure S3). This variant is present only within the casper male, and not in his unaffected female mate. Sanger sequencing in both the wild-type female (Figure 3B) and casper male (Figure 3C) validated this insertion.
Figure 3.
casper mutants contain a 1 bp insertion in exon 6 of Hps5. (A) A Clustal Omega (Sievers et al. 2011) multiple alignment of predicted amino-terminal HPS5 sequences from vertebrates with a known loss-of-function phenotype, as well as the predicted sequence from the original casper mutant (casper), and his wild-type F0 mate (G. aculeatus). Black lines indicate WD40 repeats predicted in human HPS5, and the red asterisk indicates the position of the casper insertion. See Figure S2 for full amino acid alignment. (B) Sanger sequencing of the red indicated region of Hps5 in wild-type fish. (C) Sanger sequencing of the red indicated region of Hps5 in the original casper mutant. The red “*” indicates the inserted G, which results in a predicted frame-shift and early truncation of the HPS5 protein.
Genome editing of Hps5 phenocopies casper
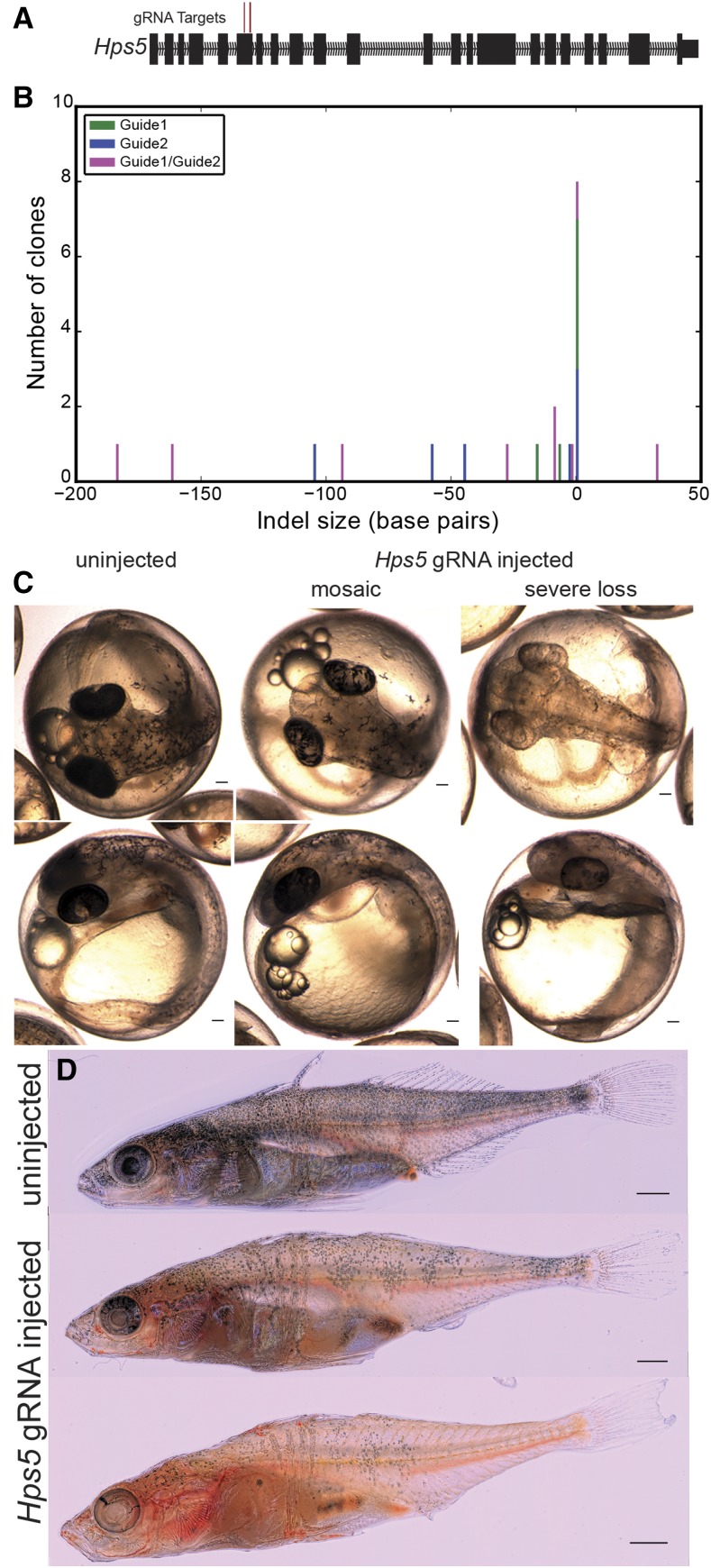
We next tested whether other predicted loss-of-function mutations in stickleback Hps5 could phenocopy casper mutants. The CRISPR/Cas9 system has been shown to be effective in genome editing in a wide range of model organisms, including another teleost, zebrafish (Hwang et al. 2013; Talbot and Amacher 2014). Two guide RNAs (gRNAs) were designed to target the sixth exon of stickleback Hps5 (Figure 4A). Injection of either of the two gRNAs alone as well as Cas9 mRNA at the one-cell stage resulted in a wide range of insertions/deletions (indels) within the Hps5 coding region of representative injected embryos (Figure 4B). Coinjection of the two gRNAs with Cas9 mRNA resulted in local indels around each gRNA target, but also larger deletions between the two, with an overall increase in indel size over single guides (P < 0.05, 1-tailed Mann-Whitney U, Figure 4B).
Figure 4.
CRISPR/Cas9 induced mutations in Hps5 phenocopy the casper mutation. (A) Two guide RNA sequences were targeted to the sixth exon of stickleback Hps5 and were singly or coinjected along with Cas9 mRNA at the one-cell stage. (B) Sanger sequencing of clones derived from single representative Hps5 injected fish reveals the highly efficient and mosaic nature of Cas9 mediated indel formation. (C) Most [29/32 (91%)] embryos injected with Hps5 gRNAs displayed pigmentation reductions, with 14/32 (44%) displaying a mosaic loss of pigment in melanophores and RPE, and 15/32 (47% of F0 injected fish) displayed >75% loss of RPE (severe loss). (D) Adult Hps5 gRNA injected sticklebacks show a mosaic loss of pigmented melanophores and iridophores, and are partially translucent. Bars, 100 μm (C), 1 mm (D).
Hps5 gRNA injected embryos phenocopied casper mutants, with severely reduced melanization in both the RPE and melanophores (Figure 4C). Coinjection of these two gRNAs resulted in highly efficient induction of pigmentation phenotypes. Only 3/32 (9%) of the surviving embryos had fully wild-type pigmentation at 6 dpf, while 15/32 (46.9%) displayed a severe loss of pigment, and 14/32 (44%) appeared mosaic (Figure 4C). Embryos injected with only a single gRNA displayed a similar loss of pigmentation, though with decreased efficiency (P < 0.01, binomial test, Table S4). Overall, we observed severe or mosaic casper-like pigmentation phenotypes in 29/32 (91%) of injected embryos, and 8/9 (90%) of sequenced target regions contained indels near a protospacer adjacent motif (PAM) (Figure 4B). Hps5 injected embryos were viable, and displayed mosaic reduced RPE and melanophore melanization into adulthood, as well as a mosaic loss of iridophore pigmentation (Figure 4D).
Visualizing fluorescent transgenic reporters in casper mutants
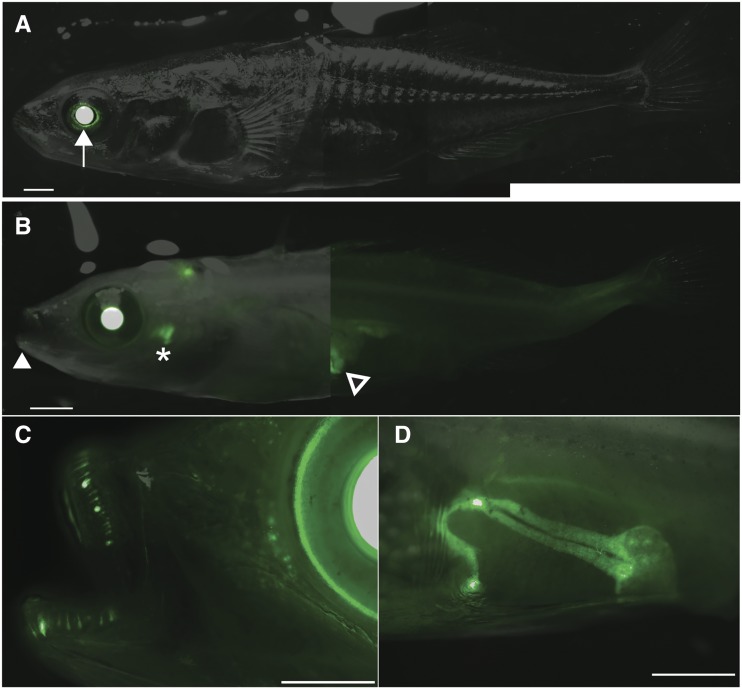
Fish embryos are highly transparent and develop externally, allowing for easy visualization of early embryogenesis. Combined with efficient transgene incorporation using Tol2 transgenesis (Kawakami 2005; Erickson et al. 2016), sticklebacks represent a powerful system for assaying the activity of developmental enhancers using fluorescent reporter constructs (Erickson et al. 2015). However, as fish develop and become more pigmented, imaging becomes extremely difficult, and investigations into late-acting enhancer elements require microdissection. We next sought to test whether the reduced pigmentation of casper embryos would allow for easier imaging of enhancer patterns in juvenile fish.
We crossed a heterozygous casper female to a male carrying a single copy of a fluorescent reporter of a previously described 190 bp Bmp6 enhancer (Erickson et al. 2015), previously described to be active in the fins and teeth. Two months postfertilization, wild-type males showed reported green fluorescent protein (GFP) expression in the lens of the eye—a known internal positive control domain of expression of the zebrafish heat-shock 70-like (Hsp70l) promoter used (Erickson et al. 2015)—with other expression domains obscured by pigmented cells (Figure 5A). casper mutants carrying the transgene showed similar robust lens expression, but also better revealed other visible juvenile expression domains in oral and pharyngeal teeth, as well as revealed a previously unreported major expression pattern in the liver (Figure 5, B–D).
Figure 5.
casper mutants allow improved visualization of fluorescent reporter genes. (A) A fluorescent GFP reporter of a previously described tooth enhancer (Erickson et al. 2015) is only clearly visible in the lenses (white arrow) of wild-type fish. (B) A casper sibling with the same stable integration of the fluorescent reporter reveals more readily apparent oral (white filled arrowhead) and pharyngeal teeth (asterisk) domains, and clearly reveals a major previously unreported liver expression domain (black filled arrowhead). (C) GFP reporter activity during oral tooth development and replacement. (D) The left side of the liver expression domain in casper fish (right visible in B). Bars, 1 mm.
Discussion
Here, we present the first stickleback model of Hermansky-Pudlak syndrome, the spontaneous X-linked recessive casper phenotype, resulting from a frame-shifting insertion in Hps5. Hps5 mutants display oculocutaneous albinism, with severely decreased pigment in melanophores, iridophores, and erythrophores, though, interestingly, not xanthophores. These pigment phenotypes suggest that pterinosomes in xanthophores develop in a Hps5-independent manner, while the melanins, carotenoids, or guanine crystal containing organelles in melanophores, erythrophores, and iridophores develop in a Hps5-dependent manner. Pigment was also drastically reduced in the RPE of Hps5 mutants, suggesting that defects in LRO biogenesis are not restricted to chromatophores, as the RPE does not contain chromatophores (Schraermeyer and Heimann 1999). Additionally, we observed a bleeding phenotype in casper mutants, potentially similar to the bleeding diathesis phenotype seen in human HPS5 mutants (Huizing et al. 2004). Overall, these phenotypes agree with the reported phenotypes of mutations in BLOC-2 complex members in other species (Zhang et al. 2003; Daly et al. 2013; Nakayama et al. 2016).
It is unclear why the stickleback casper mutation in exon 6 of Hps5 causes more severe pigmentation phenotypes than the zebrafish snow white mutation (Daly et al. 2013), which is lethal, unlike the stickleback Hps5 mutations reported here. Whether more N-terminal mutations within or prior to the WD40 domains also cause lethality in sticklebacks, as previously proposed (Daly et al. 2013), could be tested by inducing mutations more N-terminal in Hps5.
Hps5 underlies the casper phenotype
Our mapping-by-sequencing approach using bulk segregant analysis of casper mutants revealed a peak genetic signal near Hps5 on chromosome 19, the stickleback X chromosome. By Sanger sequencing, we found the insertion of a single guanine to a heptaguanine run in the coding frame of Hps5 in casper mutants. This spontaneous insertion might be due to the presence of this homopolymer repeat, as the rate of indel formation is elevated at long homopolymer runs (Montgomery et al. 2013), potentially due to polymerase slippage (Levinson and Gutman 1987). Lastly, we showed injection of Cas9 mRNA and Hps5 guide RNAs resulted in induced mutations in Hps5 and embryos displaying oculocutaneous albinism phenotypes similar to casper mutants, demonstrating Hps5 disruption underlies the casper phenotype. Although our approach using 47 pooled mutants identified a strong candidate gene, future mapping-by-sequencing of other mutations could generate improved genetic resolution by pooling even more mutant DNAs.
Live imaging of fluorescent reporters in casper embryos
casper and Hps5 mutant embryos are both semitransparent even as adults, unlike their wild-type siblings. This transparency allows for better live imaging of fluorescent transgenic reporters in Hps5 mutant fish. Imaging a previously characterized enhancer of Bmp6 (Erickson et al. 2015) in Hps5 mutants, we discovered a major unreported expression domain in the liver. We also found that imaging the previously reported dynamic tooth expression domains (Erickson et al. 2015) to be greatly facilitated by the depigmented phenotype of Hps5 mutants. The viable nature of the casper mutation allows for the creation of stable transgenic lines to more easily visualize reporter gene expression, especially at older postembryonic stages. The X-linked nature of the casper mutation allows for the recovery of casper males from outcrosses to different stable lines within a single generation.
Genome editing with CRISPR/Cas9
We report the first successful generation of loss-of-function mutations using the CRISPR/Cas9 system in sticklebacks. Coinjection of Cas9 mRNA along with either of two guide RNAs (gRNAs) targeted to exon 6 of Hps5 resulted in a high frequency (>90%) of embryos with severe or mosaic pigmentation phenotypes. Furthermore, strong pigment phenotypes were seen in both XY males and XX females, implying that Cas9 is able to induce biallelic hits in stickleback embryos. We also see evidence for high efficiency in our single clone Sanger sequencing, as most (8/9) of our sequenced clones contained induced mutations. This high rate suggests that Hps5 guide RNAs could be used as a marker for other guide RNAs in a co-CRISPR approach (Kim et al. 2014; Kane et al. 2017), with more albino embryos representing embryos with high levels of nuclear Cas9 activity. As stickleback testes are pigmented, screening for albino testes might further enrich for germline mutations from other coinjected gRNAs.
Injection of even a single gRNA is sufficient to induce large (>25 bp) deletions around the genomic target, similar in size to zebrafish reports, but larger than in human cells (Hwang et al. 2013; Paquet et al. 2016). Coinjection of two gRNAs resulted in an increase in efficiently edited embryos, suggesting that coinjection results in a high F0 mutation induction efficiency and allows phenotypic analysis of F0 injected embryos, as we have done here. Furthermore, coinjection of two gRNAs significantly increased the induction of deletions of the intervening sequence between our two Hps5 guide RNAs, showing that Cas9 can efficiently induce genomic deletions in stickleback embryos. These deletions could have a stronger effect on gene function, and allow for easy and inexpensive genotyping of stable mutants. Additionally, these deletions could be targeted to noncoding DNA such as enhancers, which might not be as sensitive to small deletions as a coding frame. Inducing deletions of regulatory elements will allow functional genetic tests of candidate regulatory elements that underlie evolved changes.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.1125/-/DC1.
Acknowledgments
We thank Anthony Lee for first noticing the original casper male. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC-Berkeley, supported by National Institutes of Health (NIH) S10 Instrumentation Grants S10RR029668 and S10RR027303. This work was funded by NIH R01DE021475 (to C.T.M.), and NIH genomics training grant 5T32HG000047-15 (to J.C.H.)
Footnotes
Communicating editor: B. Andrews
Literature Cited
- Bowen M. E., Henke K., Siegfried K. R., Warman M. L., Harris M. P., 2012. Efficient mapping and cloning of mutations in zebrafish by low-coverage whole-genome sequencing. Genetics 190: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A., Lindsay H., Felker A., Hess C., Anders C., et al. , 2016. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 143: 2025–2037. [DOI] [PubMed] [Google Scholar]
- Cooper K. M., Hanlon R. T., Budelmann B. U., 1990. Physiological color change in squid iridophores. II. Ultrastructural mechanisms in Lolliguncula brevis. Cell Tissue Res. 259: 15–24. [DOI] [PubMed] [Google Scholar]
- Cuperus J. T., Montgomery T. A., Fahlgren N., Burke R. T., Townsend T., et al. , 2010. Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc. Natl. Acad. Sci. USA 107: 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem T. J., Hoshijima K., Jurynec M. J., Gunther D., Starker C. G., et al. , 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8: e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly C. M. S., Willer J., Gregg R., Gross J. M., 2013. snow white, a zebrafish model of Hermansky-Pudlak syndrome type 5. Genetics 195: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica E. C., 2004. The building BLOC(k)s of lysosomes and related organelles. Curr. Opin. Cell Biol. 16: 458–464. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica E. C., Mullins C., Caplan S., Bonifacino J. S., 2000. Lysosome-related organelles. FASEB J. 14: 1265–1278. [DOI] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M., Poole R. J., Sarin S., Bigelow H., Hobert O., 2010. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS One 5: e15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson P. A., Cleves P. A., Ellis N. A., Schwalbach K. T., Hart J. C., et al. , 2015. A 190 base pair, TGF-β responsive tooth and fin enhancer is required for stickleback Bmp6 expression. Dev. Biol. 401: 310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson P. A., Ellis N. A., Miller C. T., 2016. Microinjection for transgenesis and genome editing in threespine sticklebacks. J. Vis. Exp. 111 Available at:: https://www.jove.com/video/54055/microinjection-for-transgenesis-genome-editing-threespine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón-Pérez J. M., Romero-Calderón R., Brooks E. S., Krantz D. E., Dell’Angelica E. C., 2007. The Drosophila pigmentation gene pink (p) encodes a homologue of human Hermansky–Pudlak syndrome 5 (HPS5). Traffic 8: 154–168. [DOI] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiácovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R., 2000. The regulation of motile activity in fish chromatophores. Pigment Cell Res. 13: 300–319. [DOI] [PubMed] [Google Scholar]
- Fujii T., Banno Y., Abe H., Katsuma S., Shimada T., 2012. A homolog of the human Hermansky-Pudluck syndrome-5 (HPS5) gene is responsible for the oa larval translucent mutants in the silkworm, Bombyx mori. Genetica 140: 463–468. [DOI] [PubMed] [Google Scholar]
- Gibson G., 2005. The synthesis and evolution of a supermodel. Science 307: 1890–1891. [DOI] [PubMed] [Google Scholar]
- Glazer A. M., Cleves P. A., Erickson P. A., Lam A. Y., Miller C. T., 2014. Parallel developmental genetic features underlie stickleback gill raker evolution. EvoDevo 5: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer A. M., Killingbeck E. E., Mitros T., Rokhsar D. S., Miller C. T., 2015. Genome assembly improvement and mapping convergently evolved skeletal traits in sticklebacks with genotyping-by-sequencing. G3 (Bethesda) 5: 1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Zhang T., Hu Z., Zhang Y., Shi Z., et al. , 2014. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development 141: 707–714. [DOI] [PubMed] [Google Scholar]
- Helip-Wooley A., Westbroek W., Dorward H. M., Koshoffer A., Huizing M., et al. , 2007. Improper trafficking of melanocyte-specific proteins in Hermansky-Pudlak syndrome type-5. J. Invest. Dermatol. 127: 1471–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G., 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima K., Jurynec M. J., Grunwald D. J., 2016. Precise editing of the zebrafish genome made simple and efficient. Dev. Cell 36: 654–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M., Hess R., Dorward H., Claassen D. A., Helip-Wooley A., et al. , 2004. Cellular, molecular and clinical characterization of patients with Hermansky–Pudlak syndrome type 5. Traffic 5: 711–722. [DOI] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U., Krauss J., Nüsslein-Volhard C., 2014. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 141: 4827–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U., Singh A. P., Nüsslein-Volhard C., 2016. The developmental genetics of vertebrate color pattern formation: lessons from zebrafish. Curr. Top. Dev. Biol. 117: 141–169. [DOI] [PubMed] [Google Scholar]
- Jamann T. M., Balint-Kurti P. J., Holland J. B., 2015. QTL mapping using high-throughput sequencing. Methods Mol. Biol. 1284: 257–285. [DOI] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., et al. , 2013. RNA-programmed genome editing in human cells. ELife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E., et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane N. S., Vora M., Varre K. J., Padgett R. W., 2017. Efficient screening of CRISPR/Cas9-induced events in Drosophila using a co-CRISPR strategy. G3 (Bethesda) 7: 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., 2005. Transposon tools and methods in zebrafish. Dev. Dyn. 234: 244–254. [DOI] [PubMed] [Google Scholar]
- Kelsh R. N., 2004. Genetics and evolution of pigment patterns in fish. Pigment Cell Res. 17: 326–336. [DOI] [PubMed] [Google Scholar]
- Kelsh R. N., Harris M. L., Colanesi S., Erickson C. A., 2009. Stripes and belly-spots—a review of pigment cell morphogenesis in vertebrates. Semin. Cell Dev. Biol. 20: 90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D., et al. , 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A., 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4: 203–221. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks M. S., Seabra M. C., 2001. The melanosome: membrane dynamics in black and white. Nat. Rev. Mol. Cell Biol. 2: 738–748. [DOI] [PubMed] [Google Scholar]
- Martin A., Serano J. M., Jarvis E., Bruce H. S., Wang J., et al. , 2016. CRISPR/Cas9 mutagenesis reveals versatile roles of Hox genes in crustacean limb specification and evolution. Curr. Biol. 26: 14–26. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milinski M., Bakker T. C. M., 1990. Female sticklebacks use male coloration in mate choice and hence avoid parasitized males. Nature 344: 330–333. [Google Scholar]
- Mills M. G., Patterson L. B., 2009. Not just black and white: pigment pattern development and evolution in vertebrates. Semin. Cell Dev. Biol. 20: 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery S. B., Goode D. L., Kvikstad E., Albers C. A., Zhang Z. D., et al. , 2013. The origin, evolution, and functional impact of short insertion–deletion variants identified in 179 human genomes. Genome Res. 23: 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Nakajima K., Cox A., Fisher M., Howell M., et al. , 2017. no privacy, a Xenopus tropicalis mutant, is a model of human Hermansky-Pudlak syndrome and allows visualization of internal organogenesis during tadpole development. Dev. Biol. 426: 472–486. [DOI] [PubMed] [Google Scholar]
- Nguyen T., Novak E. K., Kermani M., Fluhr J., Peters L. L., et al. , 2002. Melanosome morphologies in murine models of Hermansky-Pudlak syndrome reflect blocks in organelle development. J. Invest. Dermatol. 119: 1156–1164. [DOI] [PubMed] [Google Scholar]
- Obholzer N., Swinburne I. A., Schwab E., Nechiporuk A. V., Nicolson T., et al. , 2012. Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing. Development 139: 4280–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima N., 2001. Direct reception of light by chromatophores of lower vertebrates. Pigment Cell Res. 14: 312–319. [DOI] [PubMed] [Google Scholar]
- Paquet D., Kwart D., Chen A., Sproul A., Jacob S., et al. , 2016. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533: 125–129. [DOI] [PubMed] [Google Scholar]
- Peichel C. L., Ross J. A., Matson C. K., Dickson M., Grimwood J., et al. , 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. J., 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Sander J. D., Maeder M. L., Reyon D., Voytas D. F., Joung J. K., et al. , 2010. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 38: W462–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A. H., et al. , 2009. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Schraermeyer U., Heimann K., 1999. Current understanding on the role of retinal pigment epithelium and its pigmentation. Pigment Cell Res. 12: 219–236. [DOI] [PubMed] [Google Scholar]
- Shoup J. R., 1966. The development of pigment granules in the eyes of wild type and mutant Drosophila melanogaster. J. Cell Biol. 29: 223–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., et al. , 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Square T., Romášek M., Jandzik D., Cattell M. V., Klymkowsky M., et al. , 2015. CRISPR/Cas9-mediated mutagenesis in the sea lamprey Petromyzon marinus: a powerful tool for understanding ancestral gene functions in vertebrates. Development 142: 4180–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrzycka M., McEachern L. A., Kinneard J., Prabhu K., Fitzpatrick K., et al. , 2007. The pink gene encodes the Drosophila orthologue of the human Hermansky-Pudlak syndrome 5 (HPS5) gene. Genome 50: 548–556. [DOI] [PubMed] [Google Scholar]
- Talbot J. C., Amacher S. L., 2014. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish 11: 583–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G. A., Carneiro M. O., Hartl C., Poplin R., Del Angel G., et al. , 2013. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43: 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmeier C., Hume A. N., Bolasco G., Seabra M. C., 2008. Melanosomes at a glance. J. Cell Sci. 121: 3995–3999. [DOI] [PubMed] [Google Scholar]
- Wedekind C., Meyer P., Frischknecht M., Niggli U. A., Pfander H., 1998. Different carotenoids and potential information content of red coloration of male three-spined stickleback. J. Chem. Ecol. 24: 787–801. [Google Scholar]
- Wei A.-H., He X., Li W., 2013. Hypopigmentation in Hermansky–Pudlak syndrome. J. Dermatol. 40: 325–329. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Zhao B., Li W., Oiso N., Novak E. K., et al. , 2003. Ru2 and Ru encode mouse orthologs of the genes mutated in human Hermansky-Pudlak syndrome types 5 and 6. Nat. Genet. 33: 145–153. [DOI] [PubMed] [Google Scholar]
- Ziegler I., 2003. The pteridine pathway in zebrafish: regulation and specification during the determination of neural crest cell-fate. Pigment Cell Res. 16: 172–182. [DOI] [PubMed] [Google Scholar]
- Zuryn S., Le Gras S., Jamet K., Jarriault S., 2010. A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics 186: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Scripts used for bulk segregant analysis are available at https://github.com/trahsemaj/CASPER. The original casper male, the F0 mated to the casper male, and bulk segregant sequencing data are available from the SRA (accession number SRP111743).