Abstract
Although chromosomal duplications are often deleterious, in some cases they enhance cells’ abilities to tolerate specific genetic or environmental challenges. Identifying the genes that confer these conditionally beneficial effects to particular chromosomal duplications can improve our understanding of the genetic and molecular mechanisms that enable certain aneuploidies to persist in cell populations and contribute to disease and evolution. Here, we perform a screen for spontaneous mutations that improve the tolerance of haploid Saccharomyces cerevisiae to hydrogen peroxide. Chromosome IV duplication is the most frequent mutation, as well as the only change in chromosomal copy number seen in the screen. Using a genetic mapping strategy that involves systematically deleting segments of a duplicated chromosome, we show that the chromosome IV’s duplication effect is largely due to the generation of a second copy of the stress-inducible cytoplasmic thioredoxin peroxidase TSA2. Our findings add to a growing body of literature that shows the conditionally beneficial effects of chromosomal duplication are typically mediated by a small number of genes that enhance tolerance to specific stresses when their copy numbers are increased.
Keywords: aneuploidy chromosomal, duplication natural genetic variation oxidative stress yeast
Abnormalities in chromosomal copy number (or “aneuploidies”) often lead to cancer (Davoli et al. 2013; Potapova et al. 2013; Sheltzer 2013; Durrbaum and Storchova 2015, 2016; Laubert et al. 2015; Mohr et al. 2015; Nicholson and Cimini 2015; Pinto et al. 2015; Santaguida and Amon 2015), developmental defects (Ottesen et al. 2010; Gannon et al. 2011; Siegel and Amon 2012; Akasaka et al. 2013; Bose et al. 2015), premature aging (Andriani et al. 2016; Sunshine et al. 2016), and other health issues in humans. In the budding yeast Saccharomyces cerevisiae, aneuploidies also tend to be deleterious (Torres et al. 2007; Yona et al. 2012; Potapova et al. 2013; Dodgson et al. 2016; Sunshine et al. 2016). However, in some cases, these aneuploidies are conditionally beneficial, as they can enable yeast to tolerate specific loss-of-function mutations or environmental stresses (Selmecki et al. 2009, 2015; Pavelka et al. 2010; Chen et al. 2012a,b; Yona et al. 2012; Tan et al. 2013; Kaya et al. 2015; Liu et al. 2015; Meena et al. 2015; Sirr et al. 2015; Sunshine et al. 2015).
An important question regarding such conditionally beneficial aneuploidies is, do their effects tend to arise due to changes in the copy numbers of one or multiple genes on the aneuploid chromosome(s)? Several studies have attempted to address this question by identifying the specific genes underlying the conditionally beneficial effects of particular chromosomal duplications in budding yeast (Pavelka et al. 2010; Chen et al. 2012a; Kaya et al. 2015; Liu et al. 2015). For example, Kaya et al. found that chromosome XI duplication enabled S. cerevisiae strains lacking all eight thiol peroxidase genes to be nearly as tolerant to oxidative stress as a wild-type strain (Kaya et al. 2015). Two genes mediated the benefit of chromosome XI duplication: CCP1, a hydrogen peroxide scavenger that acts in the mitochondrial intermembrane space, and UTH1, a mitochondrial inner-membrane protein. In another case, Liu et al. showed that chromosome VIII duplication compensates for the absence of essential nuclear pore proteins by causing overexpression of a gene that regulates cell membrane fluidity (Liu et al. 2015). Furthermore, Chen et al. demonstrated that chromosome XV duplication confers resistance to the Hsp90 inhibitor radicicol by increasing the dosage of PDR5 and STI1, which encode a multidrug pump and an Hsp90 cochaperone, respectively (Chen et al. 2012a). Lastly, Pavelka et al. showed that chromosome XIII duplication confers increased resistance to the DNA-damaging agent 4-NQO by increasing the dosage of ATR1, another multidrug pump (Pavelka et al. 2010). These studies suggest that the conditional benefits of aneuploidization are typically mediated by changes in the copy numbers of a small number of genes that allow cells to cope with specific stresses.
Here, we explore how aneuploidies can enable yeast to tolerate environmental stresses to a level beyond that achievable through genetic variation that segregates in natural populations (so-called “natural genetic variation”). We previously found that progeny produced by mating the laboratory strain BY4716 (BY), the vineyard isolate RM11-1a (RM), and the oak isolate YPS163 (YPS) show similar maximal hydrogen peroxide tolerances despite their genetic differences (see Supplemental Material, Figure S1 in File S1), suggesting the extent to which natural genetic variation can increase tolerance to this compound may be constrained (Linder et al. 2016). We attempted to overcome these limits by screening haploid segregants from the BY × RM, BY × YPS, and RM × YPS crosses for spontaneous mutations that increase hydrogen peroxide tolerance beyond the maximal levels seen in our past work. Specifically, we took the single most tolerant F2 segregant that we identified in each of the possible pairwise crosses of the three strains and used these three individuals as the progenitors in a screen for spontaneous mutations that enhance hydrogen peroxide resistance.
We obtained 37 mutants that show increased hydrogen peroxide tolerance relative to their respective progenitors. Using whole genome sequencing, we identified spontaneous mutations that may cause increased hydrogen peroxide tolerance in these mutants. Duplication of chromosome IV was the most frequent mutation, and the only aneuploidy detected in the screen. Consistent with chromosome IV duplication being conditionally beneficial, we found that chromosome IV aneuploids grow worse than their progenitors in the absence of hydrogen peroxide, and that the benefit of chromosome IV disomy occurs on agar plates but not in liquid media. Following on from these discoveries, we determined the genetic basis of this chromosomal duplication’s effect using chromosome- and gene-scale genetic engineering. Employing these techniques, we identified a single gene, the stress-inducible cytoplasmic thioredoxin peroxidase TSA2, which accounts for the majority of the effect of chromosome IV duplication on hydrogen peroxide tolerance. Our findings illustrate how aneuploidies can enable cells to surpass the levels of stress tolerance that are attainable through natural genetic variation and provide further support that the conditionally beneficial effects of aneuploidies tend to have a simple genetic basis.
Materials and Methods
Screen for increased hydrogen peroxide tolerance
Progenitor strains were produced during our past work on the BY × RM, BY × YPS, and RM × YPS crosses (Ehrenreich et al. 2012; Linder et al. 2016) and are described in more detail in Linder et al. (2016). Each progenitor strain was streaked onto yeast extract-peptone-dextrose (YPD) plates and incubated for 2 d at 30°. To maximize biological independence among mutations obtained from the screen, 24 different colonies per progenitor were individually inoculated into 800 μl of YPD broth. These cultures were outgrown for 2 d at 30° with shaking at 200 rpm. A total of 20 μl from each culture were then diluted using 80 μl of sterile water and spread onto YPD plates containing different doses of hydrogen peroxide. These plates were incubated at 30° for 4–6 d, so that slow-growing mutants would have enough time to form visible colonies. Glycerol freezer stocks were then made for mutants that formed visible colonies on doses at least 1 mM higher than the minimum inhibitory concentration (MIC) of their progenitor and stored at −80°. After this initial screen, all mutants were then phenotyped side-by-side with their respective progenitors across a broad range of hydrogen peroxide doses to confirm their increased tolerance. Mutants that grew on doses at least 0.5 mM higher than their progenitor were saved for downstream analysis. Fourteen BY × RM, nine RM × YPS, and 14 BY × YPS derived mutants met this requirement.
Whole genome sequencing of progenitors and mutants
Archived mutants, as well as their progenitors, were inoculated into YPD liquid and outgrown for 2 d at 30°. For each mutant, DNA was extracted using the Qiagen DNeasy kit and a whole genome library was prepared using the Illumina Nextera kit. Each library was constructed using limited cycle PCR to add adapters to Nextera-treated genomic DNA according to manufacturer recommendations. Two unique 8-bp barcodes were incorporated into each library, so that dual indexed sequencing could be performed. Sequencing was conducted on an Illumina NextSeq 500 instrument at the USC Epigenome Center. Reads were then demultiplexed using custom Python scripts. The average per site coverages of the progenitors and mutants were 116× and 130×, respectively (see Note S1 in File S1).
Analysis of sequencing data
To facilitate high confidence identification of mutations using the short-read sequencing data, progenitor-specific reference genomes were generated. Burrows–Wheeler Aligner (BWA-MEM) software (Li and Durbin 2009) was used to map reads from the progenitors to the BY genome. The parameters used for alignment were as follows: “bwa –mem –t 6 ref.fsa read1.f1 read2.fq > output.sam.” To remove duplicate reads, the rmdup command was implemented in SAMtools (Li et al. 2009). To create Mpileup files, SAMtools was then run with the command “samtools mpileup –f ref.fsa read.rmdp.srt.bam > output.mp.” We then identified genetic differences between a given progenitor and BY, integrated these differences into the BY genome, and then remapped reads for that progenitor to the modified genome sequence. This process was repeated for up to 10 cycles, and the output genome was then used as the reference for mapping reads from mutants derived from that progenitor. Reads from mutant strains were aligned to the progenitor reference genomes using BWA-MEM and the same parameters described above, followed by the generation of Mpileup files with SAMtools and the same parameters reported above. Point mutations and small indels were identified as differences from the corresponding progenitor that were seen in at least 90% of the reads at a particular site, whereas aneuploidies were detected based on changes in coverage across all or part of a chromosome. Custom Python scripts were used to identify these mutations, as well as to calculate per site or per genomic window sequencing coverage.
Gene ontology enrichment analysis
Gene ontology (GO) analysis was carried out on the Saccharomyces Genome Database website (http://www.yeastgenome.org/cgi-bin/GO/goTermFinder.pl), using GO Term Finder version 0.83 with the molecular function category selected. All genes listed in Table S1 and Table S2 in File S1 were included in the analysis.
PCR-mediated chromosomal deletion
Similar to Sugiyama et al. (2008), PCR-mediated chromosomal deletion (PCD) was implemented using constructs with three segments, in the following order: 300–600 bp of sequence homologous to the desired integration site on chromosome IV, a kanMX cassette, and a synthetic telomere seed sequence consisting of six repeats of the motif 5′-CCCCAA-3′ (Figure S2 in File S1). To generate this construct, the region containing the integration site was PCR amplified from genomic DNA using a reverse primer that was tailed with 30 bases of sequence identity to the kanMX construct. At the same time, kanMX was PCR amplified from a plasmid using a forward primer with 30 bases of sequence identity to the integration site and a reverse primer containing the synthetic telomere seed sequence. Integration site and kanMX/synthetic telomere seed sequences were then joined using overlap fusion PCR (Sugiyama et al. 2005). This was done by mixing the two products in equimolar fractions in combination with the forward primer for the integration site and the reverse primer for the kanMX/synthetic telomere seed sequence, and conducting PCR. The cycling parameters for overlap fusion PCR were as follows: initial denaturation at 98° for 3 min, followed by 30 cycles of 98° for 30 sec, 63° for 30 sec, and 72° for 1.5 min. The final step was a 5 min extension at 72°. Throughout this process, all PCR steps were implemented using NEB High-Fidelity Phusion polymerase and all PCR products were purified using either Qiagen QIAquick Gel Extraction or QIAquick PCR Purification kits. A standard lithium acetate–based technique was used to transform PCD constructs into cells, with ∼5 μg of construct employed (Gietz and Schiestl 2007). Transformants were recovered using selection for G418 resistance on YPD plates, verified by colony PCR or genome sequencing, and archived at −80° in glycerol solution. Primers used to construct PCD products are listed in Table S3 in File S1.
Deletion of individual genes
Targeted gene deletions were performed using the kanMX cassette. Constructs for deleting ADA2, ECM11, GUK1, NHX1, and UTP6 were generated by amplifying the kanMX cassette from a plasmid using tailed primers. Each tail contained 30–60 bases of sequence homology to the target gene’s flanking regions. Specifically, the forward and reverse primers were designed to have tails identical to the region immediately upstream of the translation start site and downstream of the stop codon, respectively. Regarding deletions of PPN1, TOM1, TSA2, RPS17B, RPS18A, YDR455C, and YHP1 transformations using knockout cassettes tailed with only 60–120 bp of total homology to the targeted region were relatively inefficient. To increase the efficiency of these knockouts, gene deletion products were generated in a manner analogous to making the PCD constructs described above. Then, 300–600 bp of sequence targeting a region just upstream of the translation start site was fused to the kanMX cassette, with the caveat that the reverse primer for kanMX amplification was tailed with 30–60 bases of homology to a region just downstream of the gene being deleted. Additionally, several 100 bp of sequence up- and downstream of TSA2 were deleted by the same method, using a modified targeting sequence and downstream homology tail. The same lithium acetate–based transformation methods described for PCD were used for all individual gene knockouts. Transformants were verified by colony PCR. Primers used for individual gene knockouts are listed in Table S4 in File S1.
Plasmid-based increase of TSA2 copy number
Low-copy plasmids were constructed by cloning the complete TSA2 locus (promoter, coding region, and 3′UTR) from the BY × RM haploid progenitor into the multiple cloning site of pRS410, a gift from Fred Cross purchased through Addgene (Addgene plasmid #11258). This plasmid contains CEN6/ARS4, which results in maintenance at low-copy number in S. cerevisiae, as well as AmpR and KanR2, which enable selection of transformed bacteria on ampicillin and transformed yeast on G418, respectively. Restriction sites for NotI and XhoI were added as 5′ tails to the primers used to amplify the TSA2 locus. The pRS410 plasmid and modified TSA2 PCR product were separately and sequentially digested with each enzyme at 37°, after which, the reactions were quenched by a 20-min incubation at 65°. After the second digestion step, the doubly digested plasmid and insert were ligated overnight. A small volume of the ligation reaction was then used to transform competent Escherichia coli obtained from Invitrogen. Transformants were selected on LB plates supplemented with ampicillin. Successful integration of the TSA2 locus was confirmed by PCR amplifying DNA extracted from transformants using M13 primers, which flank the multiple cloning site. Plasmids were purified from transformed strains using the Qiagen midi plasmid purification kit and transformed into euploid progenitor strains. Based on Sanger sequencing, the cloned TSA2 locus was found to have the same sequence as the BY × RM progenitor. Primers used for construction of plasmids are listed in Table S4 in File S1.
qPCR analysis of TSA2 copy number
TSA2 copy number was checked in overexpression transformants, as well as the BY × RM F2 progenitor and a BY × RM mutant disomic for chromosome IV, using qPCR. DNA was extracted from the transformants using the Qiagen QIAamp kit. Transformants were grown on selection plates containing G418. After 3 d at 30°, three colonies from each overexpression transformant and four colonies from the BY × RM progenitor and disomic mutant strains were transferred to 96-deep-well plates, with 800 μl of YPD, and incubated at 30° for 2 d with shaking, after which strains were either pinned onto YPD plates supplemented with hydrogen peroxide or transferred to 96-deep-well plates containing YPD broth supplemented with hydrogen peroxide. The remaining cells from each well were transferred to Eppendorf tubes, spun down at >13,000 rpm for 10 sec, after which the supernatant was removed and cells were resuspended in spheroplasting solution containing zymolyase. An RNase treatment was then performed to eliminate any transcripts that could potentially be amplified along with the genomic copies of TSA2 and our reference gene ACT1. The KAPA SYBR Fast qPCR kit was used for qPCR analysis on the Agilent Mx3005p. 96-well semiskirted PCR plates were employed and Bio-Rad Microseal “C” Film was used to cover them. Cycling parameters were as follows: 95° for 2 min, followed by 40 cycles of 95° for 3 sec and 60° for 30 sec. At the end of each cycle, fluorescence measurements were then taken for each well. After the final cycle, a 10-min extension step was run at 72°, followed by a melting curve analysis from 55 to 95°, by 0.4° increments. After qPCR, products were checked on 1.5% agarose gels to ensure primer specificity. TSA2 abundance in each sample was measured relative to ACT1. All experiments were conducted in at least biological triplicate. Primers used for qPCR are listed in Table S4 in File S1.
Phenotyping of transformants
Transformants were outgrown for 2 d in 800 μl of YPD broth incubated at 30° with shaking. As controls for batch effects, each time one or more transformants were phenotyped, their euploid and aneuploid progenitors were also examined. After the liquid outgrowth step, strains were pinned onto YPD plates supplemented with different hydrogen peroxide doses. Alternatively, in some experiments, strains were then transferred to liquid media supplemented with a range of doses of hydrogen peroxide for 3 d, after which the OD630 of 50 μl of culture from each well diluted in 150 μl of water was measured using a plate reader. These strains were incubated at 30° with shaking. Each experiment was done in at least biological triplicate. Agar plates were incubated for 3 d at 30° and then imaged on a GelDoc imaging device using a 0.5 sec exposure time. MIC was determined as the lowest hydrogen peroxide dose at which a given strain could not grow.
Data availability
All sequence data are available through the Sequence Read Archive under Bioproject number PRJNA338809, study accession number SRP081591, and sample accession numbers SRX2018688–SRX2018727 and SRX2037407–SRX2037410.
Results and Discussion
Screen for spontaneous mutations that increase hydrogen peroxide tolerance
A total of 24 independent cultures of each of the three haploid progenitor strains were examined after 2 d of outgrowth using selection on agar plates supplemented with hydrogen peroxide (see Materials and Methods). All mutants (37 total) that exhibited an increase in MIC at least 0.5 mM higher than their corresponding progenitor were analyzed further (see Figure S3, A–C in File S1; Materials and Methods). BY × RM, RM × YPS, and BY × YPS derived mutants were, on average, 2.3 mM (32%), 2.1 mM (25%), and 0.6 mM (7%) more tolerant than their progenitor, respectively (see Figure S3, A–C in File S1).
The most frequently identified mutation is a chromosomal duplication
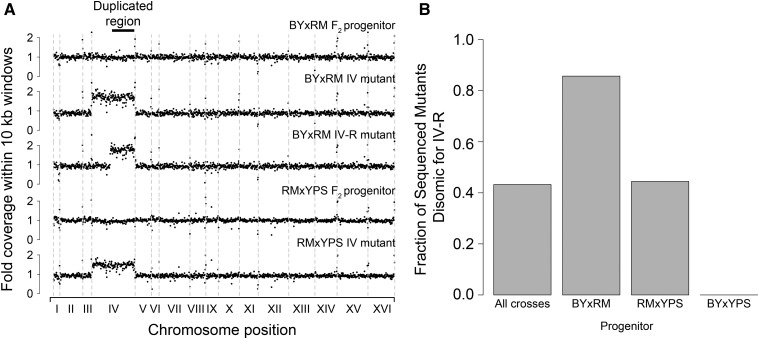
Analysis of genome-wide sequencing coverage indicated that 43% of the mutants carried two complete copies of chromosome IV (Figure 1). No other aneuploidies were detected. The disomy was common among the BY × RM (79%) and RM × YPS derived (45%) mutants, but was absent from the BY × YPS derived mutants (Figure 1B). Given that the BY × RM and RM × YPS derived mutants also showed higher average gains in tolerance, this finding is consistent with duplication of chromosome IV conferring a sizable increase in tolerance (see Figure S3, A–C in File S1).
Figure 1.
Chromosome IV duplication is the most frequent mutational event in a screen for spontaneous hydrogen peroxide resistance mutations. (A) Genome-wide coverage plots are provided for the BY × RM and RM × YPS F2 progenitors, as well as representative aneuploid or segmental duplication mutants derived from them. Site-specific coverages are scaled to the average per site coverage seen across the entire genome for a given strain. (B) Bar plots show the fraction of sequenced mutants that were disomic for the right arm of chromosome IV both across the entire screen and by individual progenitor.
A single segmental duplication was also detected; this was found in a BY × RM derived mutant that possessed two copies of only the right arm of chromosome IV (chromosome IV-R; Figure 1A). The segmental duplication spanned ∼890 kb (58% of chromosome IV). Including this mutant, 86% of the BY × RM derived mutants were disomic for chromosome IV-R.
Additionally, 39 unique point mutations were identified among the mutants (see Table S1 and Table S2 in File S1). Twenty-two of these point mutations were nonsynonymous, eight were synonymous, and nine occurred in intergenic regions. Some point mutations were located in genes that affect hydrogen peroxide tolerance, including a SUMO E3 ligase involved in DNA repair (MMS21), an activator of cytochrome oxidase 1 translation (PET309), a subunit of cytochrome c oxidase (COX1), and a negative regulator of Ras-cAMP-PKA signaling (GPB2; see Table S1 and Table S2 in File S1). At false discovery rates of 0.17 or lower, no specific GO enrichments were seen among the genes harboring point mutations (see Materials and Methods). These results are consistent with our past finding that genetic perturbation of many different cellular processes can influence hydrogen peroxide tolerance (Linder et al. 2016), but could also have been driven by passenger mutations that do not play causal roles in hydrogen peroxide tolerance.
Duplication of chromosome IV-R was the most frequently detected mutation in our screen. However, strains carrying point mutations were found among the BY × RM and RM × YPS derived mutants that were as tolerant to hydrogen peroxide as strains containing the chromosome IV-R duplication (see Figure S3, A and B in File S1). In these genetic backgrounds, duplication of chromosome IV-R may occur more frequently than spontaneous point mutations that increase hydrogen peroxide tolerance (Kaya et al. 2015; Liu et al. 2015).
Chromosome IV-R duplication is conditionally beneficial
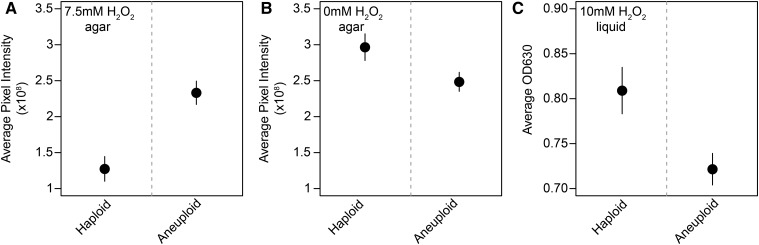
Chromosome IV-R duplication only conferred a growth benefit when hydrogen peroxide exposure occurred on agar plates (Figure 2A and Figure S4, A and B in File S1). In other conditions, chromosome IV-R duplication appeared to have a fitness cost. Similar to past reports that chromosomal duplications are usually deleterious (Pavelka et al. 2010; Sunshine et al. 2016), chromosome IV-R duplication reduced growth both on agar plates without hydrogen peroxide and in liquid medium containing hydrogen peroxide (Figure 2, B and C and Figure S4, A and B in File S1).
Figure 2.
Duplication of chromosome IV is conditionally beneficial. (A) Replicates of a strain fully disomic for chromosome IV show significantly improved growth compared to euploid replicates when grown on agar plates supplemented with 7.5 mM hydrogen peroxide. However, when these strains are grown on agar plates with rich medium (B), euploid individuals show superior overall growth as compared to individuals disomic for chromosome IV. When both strains are exposed to 10 mM of hydrogen peroxide in liquid culture (C), euploid individuals also show higher growth than individuals disomic for chromosome IV. In (A) and (B), end-point colony pixel intensity measurements were generated using plate growth assays and image analysis in ImageJ. Mean and 95% confidence intervals are plotted. These measurements are based on four biological replicates per strain, which were each grown in three technical replicates (see Note S5 in File S1). In (C), individuals were exposed to hydrogen peroxide in liquid media for 3 d, after which the OD630 was measured for each culture. Measurements were again based on four biological and three technical replicates per strain (see Materials and Methods).
Identification of a single region responsible for most of the effect of chromosome IV-R duplication
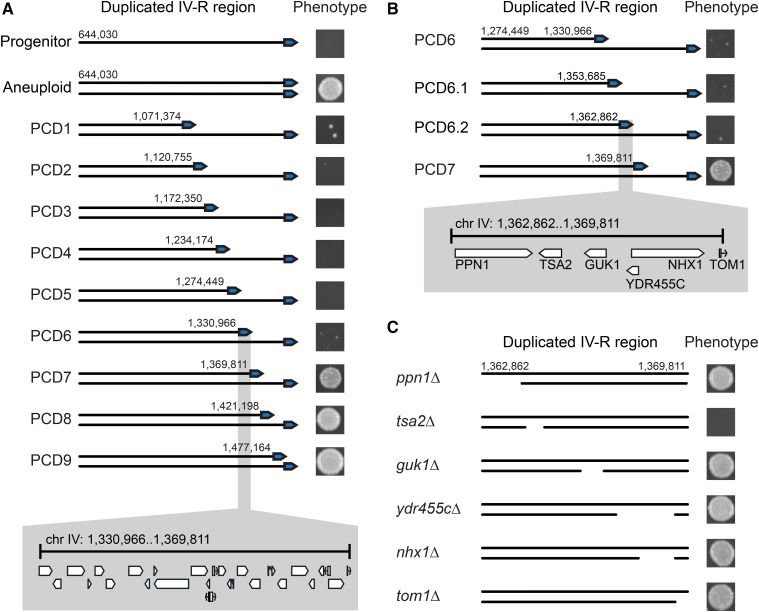
To map the conditional growth benefit of chromosome IV-R duplication to specific genes, we adapted a technique known as PCD, which involves eliminating segments of a chromosome that are distal to a centromere by inserting a drug resistance cassette linked to a synthetic telomere seed sequence (Figure S2 in File S1; Materials and Methods) (Sugiyama et al. 2005, 2008; Kaboli et al. 2016). Colony PCR was used both to confirm correct placement of the deletion cassette as well as to verify that a single copy of the deleted region remained (see Materials and Methods).
We first used PCD to delete the right half of chromosome IV-R from a BY × RM derived aneuploid (Figure 3A; Materials and Methods). This genetic change, which was confirmed by whole genome sequencing (see Figure S5 in File S1; Materials and Methods), caused a reversion to the hydrogen peroxide tolerance exhibited by the euploid BY × RM progenitor prior to the screen (see Figure S6A in File S1). This result indicates that the deleted chromosomal segment is required for the aneuploidy’s effect.
Figure 3.
Genetic dissection of the chromosome IV duplication’s effect on hydrogen peroxide tolerance. (A) PCDs were staggered nearly every 50 kb along chromosome IV-R in a BY × RM derived aneuploid. Phenotyping of partially aneuploid strains generated by PCD identified a single genomic interval with a large effect on hydrogen peroxide tolerance when strains were examined at a dose of 7.5 mM. This ∼40 kb region contains 23 genes and three dubious ORFs. (B) Two additional PCD strains were generated within the previously mentioned window and examined at 7.5 mM. This narrowed the interval to 7 kb that contained five genes and a dubious ORF. (C) Individual gene deletions revealed that TSA2 is largely responsible for the increase in tolerance conferred by duplication of chromosome IV-R. In (A and B), representative images of colonies grown at 7.5 mM of hydrogen peroxide are shown next to their associated PCD strains; numbers adjacent to the telomere indicate the starting position of each PCD on chromosome IV. In (C), representative images of colonies grown at 7.5 mM of hydrogen peroxide are again shown next to their associated individual gene deletion strains, with deleted regions indicated by gaps on the duplicated chromosome.
We next generated a panel of PCD strains, with large-scale deletions staggered, on average, every 50.7 kb along the distal half of chromosome IV-R. This led to the identification of a single 40 kb region that recapitulates most of the effect of the chromosome IV-R disomy (Figure 3A and Figure S6A in File S1). It is important to note that, although chromosome-scale deletions of this 40 kb region appeared to phenocopy the progenitor at certain hydrogen peroxide doses (Figure 3, A and B), the average MICs of these strains were higher than that of their progenitor (Figure S6, A and B in File S1). Possible explanations for this result include the presence of one or more additional point mutations that contribute to hydrogen peroxide tolerance in the mutant, an additional gene or regulatory element on chromosome IV-R whose dosage contributes to tolerance, and nonlinear relationships between DNA content and hydrogen peroxide tolerance, as the chromosome-scale deletion strains remain aneuploid at a large portion of chromosome IV (see Note S2 in File S1). We note that batch effects resulted in slight differences in the growth of the euploid progenitor at 7.5 mM hydrogen peroxide across separate experiments, but that growth differences between genotypes remained qualitatively consistent (Figure 3A, Figure S4A in File S1, and Figure S10 in File S1).
Duplication of TSA2 mediates the conditionally beneficial effect of the aneuploidy
To further resolve this window, we performed two additional PCD transformations, which fine-mapped the causal interval to roughly 7 kb, spanning positions 1,362,862 to 1,369,812 bp (Figure 3B and Figure S6B in File S1). This interval contains five genes—the polyphosphatase PPN1, the cytoplasmic thioredoxin peroxidase TSA2, the guanylate kinase GUK1, the ion exchanger NHX1, and the E3 ubiquitin ligase TOM1—as well as a dubious ORF (YDR455C). We used standard techniques to individually delete each of these genes from a BY × RM derived aneuploid, again using colony PCR to verify both correct placement of the deletion cassette and that a single copy of the gene remained (see Materials and Methods). Also, because telomeres can influence the transcription of genes >20 kb away (Gottschling et al. 1990; Aparicio and Gottschling 1994), we deleted the six genes upstream of PPN1 (Figure 3B and Figure S7 in File S1).
The only gene deletion that showed a phenotypic effect was TSA2, which encodes a cytoplasmic thioredoxin peroxidase (Figure 3, B and C and Figure S7 in File S1) (Gasch et al. 2000; Park et al. 2000; Wong et al. 2002; Munhoz and Netto 2004; Ogusucu et al. 2007; Nielsen et al. 2016). Previous work has shown that deleting TSA2 leads to a decrease in hydrogen peroxide tolerance (Wong et al. 2002). Loss of the TSA2 coding region eliminated the majority of the aneuploidy’s effect (Figure 3C and Figure S7 in File S1), proving a causal role for TSA2 in the conditional benefit conferred by chromosome IV-R duplication. Knockout of TSA2 in other mutants, including a fully disomic BY × RM mutant, a partially disomic BY × RM mutant, and a fully disomic RM × YPS mutant, confirmed that the effect of TSA2 was reproducible across different aneuploid individuals recovered from our screen (Figure S8 in File S1; see Note S3 in File S1).
Plasmid-based overexpression of TSA2 increases tolerance of hydrogen peroxide
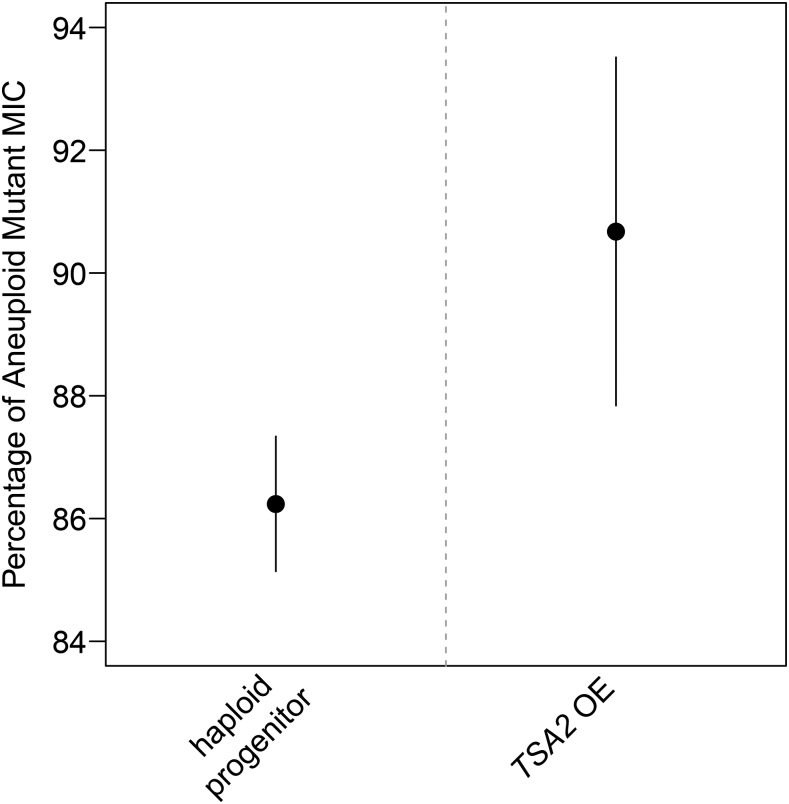
To further confirm that an increase in copy number of TSA2 has a beneficial effect on hydrogen peroxide tolerance, the complete TSA2 locus was cloned into a low-copy CEN plasmid, which was subsequently transformed into the BY × RM euploid progenitor (see Materials and Methods). Increased copy number of TSA2 in the euploid progenitor was confirmed through qPCR (Figure S9 in File S1; Materials and Methods). Plasmid-based overexpression of TSA2 in the euploid progenitor resulted in a significant increase in tolerance (Figure 4 and Figure S10 in File S1). However, the low-copy number plasmid did not fully recapitulate the effect of having a second chromosomal copy of the gene, further suggesting that TSA2 does not fully explain the effects of chromosome IV disomy on hydrogen peroxide tolerance (Figure 4, Figure S10 in File S1, and Figure S11 in File S1). In line with this possibility, TSA2 overexpression strains did not show the increased susceptibility to hydrogen peroxide that was exhibited by aneuploid mutants in liquid media (Figure S12 in File S1).
Figure 4.
Plasmid-based overexpression of TSA2 partially recapitulates the effect of having two chromosomal copies of this gene. The entirety of the TSA2 locus was cloned into a CEN plasmid that was then transformed into the haploid progenitor (see Materials and Methods). Two independently generated TSA2 overexpressing strains were screened for hydrogen peroxide tolerance alongside the haploid progenitor and disomic strain. Shown are 95% confidence intervals for the MIC of the overexpressing strains as a percentage of the MIC of the aneuploid strain. These measurements are based on three biological replicates, which were each grown in three technical replicates.
Conclusion
Previously, we found that the maximal hydrogen peroxide tolerances of segregants from the BY × RM, BY × YPS, and RM × YPS crosses are comparable (see Figure S1 in File S1) (Linder et al. 2016), suggesting that the level of resistance achievable through natural genetic variation may be constrained to some degree. To surpass the levels of tolerance seen in our prior study, we conducted a screen for spontaneous mutations that confer higher hydrogen peroxide resistance than we previously observed. We employed segregants that had maximal tolerances as the progenitors in our mutagenesis screen because these individuals carry combinations of natural genetic variants that lead to resistance and we wanted to find mutations that provide even greater tolerance than these allele combinations.
Duplication of chromosome IV-R was the most common mutation in our screen. As with other chromosomal duplications (Pavelka et al. 2010; Sunshine et al. 2015), the beneficial effect of the chromosome IV-R disomy is conditional: it depends on both the presence of hydrogen peroxide and exposure to hydrogen peroxide on agar plates, and may be dependent on genetic background. Regarding this latter point, the present data cannot differentiate whether the progenitors in our screen varied in their genome stabilities or in their abilities to beneficially utilize the chromosome IV-R disomy. Assessment of genotype at TSA2 indicates that preexisting variation at this locus probably does not explain why we observed chromosome IV aneuploids in the BY × RM and RM × YPS crosses, but not the BY × YPS cross (see Note S4 in File S1).
Using chromosome- and gene-scale deletions, we determined that increased copy number of a single detoxifying gene, TSA2, explains the majority of the benefit conferred by duplication of chromosome IV-R. TSA2 is unique among budding yeast’s cytoplasmic thioredoxin peroxidases, as it is the only one that shows markedly increased activation in response to hydrogen peroxide (Gasch et al. 2000; Park et al. 2000; Wong et al. 2002; Munhoz and Netto 2004; Ogusucu et al. 2007; Nielsen et al. 2016). Our results are consistent with recent studies from other groups showing that typically, a small number of genes, usually one or two, mediate the conditionally beneficial effects of aneuploidies (Pavelka et al. 2010; Chen et al. 2012a; Kaya et al. 2015; Liu et al. 2015).
In summary, our work speaks to challenges in enhancing particular traits using natural genetic variation, spontaneous mutations, or a combination of the two. Indeed, the maximal trait values achievable through natural genetic variation may be limited because of both the specific alleles present in a population and features of a system that prevent extreme phenotypic levels from occurring. Trying to overcome these limits may be achievable using spontaneous mutations, which have not experienced the same selection pressures as natural variants and may be more likely to have large effects. However, our work suggests that the most likely mutational event to underlie such phenotypic increases is chromosomal duplication. Although these duplications have a limitation in that they can be easily lost (Berman 2016), they may represent a transient state that can facilitate the acquisition of other mutations that provide a more permanent solution to stressful conditions (Yona et al. 2012).
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.300069/-/DC1.
Acknowledgments
We thank Jonathan Lee, Martin Mullis, and Rachel Schell for reviewing a draft of this manuscript. This work was supported by grants from the National Institutes of Health (R01GM110255), National Science Foundation (MCB1330874), and Alfred P. Sloan Foundation to I.M.E.
Footnotes
Communicating editor: D. Gresham
Literature Cited
- Akasaka N., Tohyama J., Ogawa A., Takachi T., Watanabe A., et al. , 2013. Refractory infantile spasms associated with mosaic variegated aneuploidy syndrome. Pediatr. Neurol. 49: 364–367. [DOI] [PubMed] [Google Scholar]
- Andriani G. A., Faggioli F., Baker D., Dolle M. E., Sellers R. S., et al. , 2016. Whole chromosome aneuploidy in the brain of Bub1bH/H and Ercc1-/Delta7 mice. Hum. Mol. Genet. 25: 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O. M., Gottschling D. E., 1994. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 8: 1133–1146. [DOI] [PubMed] [Google Scholar]
- Berman J., 2016. Ploidy plasticity: a rapid and reversible strategy for adaptation to stress. FEMS Yeast Res. 16: fow020. [DOI] [PubMed] [Google Scholar]
- Bose D., Krishnamurthy V., Venkatesh K. S., Aiyaz M., Shetty M., et al. , 2015. Molecular delineation of partial trisomy 14q and partial trisomy 12p in a patient with dysmorphic features, heart defect and developmental delay. Cytogenet. Genome Res. 145: 14–18. [DOI] [PubMed] [Google Scholar]
- Chen G., Bradford W. D., Seidel C. W., Li R., 2012a Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Rubinstein B., Li R., 2012b Whole chromosome aneuploidy: big mutations drive adaptation by phenotypic leap. Bioessays 34: 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T., Xu A. W., Mengwasser K. E., Sack L. M., Yoon J. C., et al. , 2013. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155: 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson S. E., Kim S., Costanzo M., Baryshnikova A., Morse D. L., et al. , 2016. Chromosome-specific and global effects of aneuploidy in Saccharomyces cerevisiae. Genetics 202: 1395–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrbaum M., Storchova Z., 2015. Consequences of aneuploidy in cancer: transcriptome and beyond. Recent Results Cancer Res. 200: 195–224. [DOI] [PubMed] [Google Scholar]
- Durrbaum M., Storchova Z., 2016. Effects of aneuploidy on gene expression: implications for cancer. FEBS J. 283: 791–802. [DOI] [PubMed] [Google Scholar]
- Ehrenreich I. M., Bloom J., Torabi N., Wang X., Jia Y., et al. , 2012. Genetic architecture of highly complex chemical resistance traits across four yeast strains. PLoS Genet. 8: e1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon W. T., Martinez J. E., Anderson S. J., Swingle H. M., 2011. Craniofacial dysmorphism and developmental disorders among children with chromosomal microdeletions and duplications of unknown significance. J. Dev. Behav. Pediatr. 32: 600–604. [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., et al. , 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 31–34. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A., 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Kaboli S., Miyamoto T., Sunada K., Sasano Y., Sugiyama M., et al. , 2016. Improved stress resistance and ethanol production by segmental haploidization of the diploid genome in Saccharomyces cerevisiae. J. Biosci. Bioeng. 121: 638–644. [DOI] [PubMed] [Google Scholar]
- Kaya A., Gerashchenko M. V., Seim I., Labarre J., Toledano M. B., et al. , 2015. Adaptive aneuploidy protects against thiol peroxidase deficiency by increasing respiration via key mitochondrial proteins. Proc. Natl. Acad. Sci. USA 112: 10685–10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubert T., Freitag-Wolf S., Linnebacher M., Konig A., Vollmar B., et al. , 2015. Stage-specific frequency and prognostic significance of aneuploidy in patients with sporadic colorectal cancer–a meta-analysis and current overview. Int. J. Colorectal Dis. 30: 1015–1028. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder R. A., Seidl F., Ha K., Ehrenreich I. M., 2016. The complex genetic and molecular basis of a model quantitative trait. Mol. Biol. Cell 27: 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Yong M. Y., Yurieva M., Srinivasan K. G., Liu J., et al. , 2015. Gene essentiality is a quantitative property linked to cellular evolvability. Cell 163: 1388–1399. [DOI] [PubMed] [Google Scholar]
- Meena J. K., Cerutti A., Beichler C., Morita Y., Bruhn C., et al. , 2015. Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion. EMBO J. 34: 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr M., Zaenker K. S., Dittmar T., 2015. Fusion in cancer: an explanatory model for aneuploidy, metastasis formation, and drug resistance. Methods Mol. Biol. 1313: 21–40. [DOI] [PubMed] [Google Scholar]
- Munhoz D. C., Netto L. E., 2004. Cytosolic thioredoxin peroxidase I and II are important defenses of yeast against organic hydroperoxide insult: catalases and peroxiredoxins cooperate in the decomposition of H2O2 by yeast. J. Biol. Chem. 279: 35219–35227. [DOI] [PubMed] [Google Scholar]
- Nicholson J. M., Cimini D., 2015. Link between aneuploidy and chromosome instability. Int. Rev. Cell Mol. Biol. 315: 299–317. [DOI] [PubMed] [Google Scholar]
- Nielsen M. H., Kidmose R. T., Jenner L. B., 2016. Structure of TSA2 reveals novel features of the active-site loop of peroxiredoxins. Acta Crystallogr D Struct. Biol. 72: 158–167. [DOI] [PubMed] [Google Scholar]
- Ogusucu R., Rettori D., Munhoz D. C., Netto L. E., Augusto O., 2007. Reactions of yeast thioredoxin peroxidases I and II with hydrogen peroxide and peroxynitrite: rate constants by competitive kinetics. Free Radic. Biol. Med. 42: 326–334. [DOI] [PubMed] [Google Scholar]
- Ottesen A. M., Aksglaede L., Garn I., Tartaglia N., Tassone F., et al. , 2010. Increased number of sex chromosomes affects height in a nonlinear fashion: a study of 305 patients with sex chromosome aneuploidy. Am. J. Med. Genet. A. 152A: 1206–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. G., Cha M. K., Jeong W., Kim I. H., 2000. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 275: 5723–5732. [DOI] [PubMed] [Google Scholar]
- Pavelka N., Rancati G., Zhu J., Bradford W. D., Saraf A., et al. , 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A. E., Pereira T., Silva G. L., Andre S., 2015. Aneuploidy identifies subsets of patients with poor clinical outcome in grade 1 and grade 2 breast cancer. Breast 24: 449–455. [DOI] [PubMed] [Google Scholar]
- Potapova T. A., Zhu J., Li R., 2013. Aneuploidy and chromosomal instability: a vicious cycle driving cellular evolution and cancer genome chaos. Cancer Metastasis Rev. 32: 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Amon A., 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 16: 473–485. [DOI] [PubMed] [Google Scholar]
- Selmecki A. M., Dulmage K., Cowen L. E., Anderson J. B., Berman J., 2009. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 5: e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A. M., Maruvka Y. E., Richmond P. A., Guillet M., Shoresh N., et al. , 2015. Polyploidy can drive rapid adaptation in yeast. Nature 519: 349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer J. M., 2013. A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res. 73: 6401–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel J. J., Amon A., 2012. New insights into the troubles of aneuploidy. Annu. Rev. Cell Dev. Biol. 28: 189–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirr A., Cromie G. A., Jeffery E. W., Gilbert T. L., Ludlow C. L., et al. , 2015. Allelic variation, aneuploidy, and nongenetic mechanisms suppress a monogenic trait in yeast. Genetics 199: 247–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M., Ikushima S., Nakazawa T., Kaneko Y., Harashima S., 2005. PCR-mediated repeated chromosome splitting in Saccharomyces cerevisiae. Biotechniques 38: 909–914. [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Nakazawa T., Murakami K., Sumiya T., Nakamura A., et al. , 2008. PCR-mediated one-step deletion of targeted chromosomal regions in haploid Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 80: 545–553. [DOI] [PubMed] [Google Scholar]
- Sunshine A. B., Payen C., Ong G. T., Liachko I., Tan K. M., et al. , 2015. The fitness consequences of aneuploidy are driven by condition-dependent gene effects. PLoS Biol. 13: e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine A. B., Ong G. T., Nickerson D. P., Carr D., Murakami C. J., et al. , 2016. Aneuploidy shortens replicative lifespan in Saccharomyces cerevisiae. Aging Cell 15: 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z., Hays M., Cromie G. A., Jeffery E. W., Scott A. C., et al. , 2013. Aneuploidy underlies a multicellular phenotypic switch. Proc. Natl. Acad. Sci. USA 110: 12367–12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E. M., Sokolsky T., Tucker C. M., Chan L. Y., Boselli M., et al. , 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. [DOI] [PubMed] [Google Scholar]
- Wong C. M., Zhou Y., Ng R. W., Kung Hf H. F., Jin D. Y., 2002. Cooperation of yeast peroxiredoxins Tsa1p and Tsa2p in the cellular defense against oxidative and nitrosative stress. J. Biol. Chem. 277: 5385–5394. [DOI] [PubMed] [Google Scholar]
- Yona A. H., Manor Y. S., Herbst R. H., Romano G. H., Mitchell A., et al. , 2012. Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl. Acad. Sci. USA 109: 21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data are available through the Sequence Read Archive under Bioproject number PRJNA338809, study accession number SRP081591, and sample accession numbers SRX2018688–SRX2018727 and SRX2037407–SRX2037410.