Abstract
Day after day, the importance of relying on nature in many fields such as food, medical, pharmaceutical industries, and others is increasing. Essential oils (EOs) are considered as one of the most significant natural products for use as antimicrobials, antioxidants, antitumorals, and anti-inflammatories. Optimizing the usage of EOs is a big challenge faced by the scientific researchers because of the complexity of chemical composition of every EO, in addition to the difficulties to determine the best in inhibiting the bacterial activity. The goal of this article is to present a new computational tool based on two methodologies: reduction by using rough sets and optimization with particle swarm optimization. The developed tool dubbed as Essential Oil Reduction and Optimization Tool is applied on 24 types of EOs that have been tested toward 17 different species of bacteria.
Keywords: essential oils, reduction, optimization, rough sets, particle swarm optimization
Introduction
Nature is rich in antibacterial, antifungal, and antioxidant products. Plants are considered as one of the important anti-organism producers. Plants produce various types of secondary metabolites that are characterized by their biological properties, for use against predators and microbial pathogens. One of these secondary metabolites, which have an effective antimicrobial activity, is the “essential oil” (EO).1 EOs, also known as essences, volatile oils, etheric oils, or aetheroleum, are natural products that can be produced from different parts of the plants (e.g., flowers, leaves, stems, seeds, fruit, or barks of aromatic plants). The EOs consist of several types of compounds, which are usually liquid, volatile, limpid, with lower density than water, and soluble in organic solvents. Extracting the EOs from different parts of the plants can be done by various chemical techniques such as “steam distillation”, “solvent extraction”, “expression under pressure”, “supercritical fluid”, and “subcritical water extraction”.2
Thus, due to the efficiency of EOs as antimicrobials, antioxidants, antitumoral, antifungal and anti-inflammatories, they are widely used in different fields such as the cosmetic industry (e.g., ingredients of fragrances, decorative cosmetic, fine fragrances, and flavoring), the food industry (e.g., aromas and flavors), the pharmaceutical industry (e.g., active components of medicines and as antibacterials/antimicrobials), and aromatherapy.3
Although the EOs are characterized by their efficiency as antibacterial organisms, their effective degree is varied according to their chemical composition, which may vary due to their agriculture, geographic location, seasonal variability, and climatic factors and according to the part of the plant (fruit, leaves, etc.).4 Nowadays, the determination of the bioactivity of the EOs from their chemical composition is a well-established idea among the scientific community. However, practically, the traditional experimental methods usually have many limitations. Characterizing the complexity of the chemical interactions among the components of EOs is a significant limitation. In addition, defining the relations between these components and the attacked microbes is another limitation.3 Thus, the importance of using the computational models instead has increased. Developing a computational tool to select the EOs that have antimicrobial activities depending on knowing their effective compounds only and without complex laboratory analysis can save money and time and enhance consistency of final products. Generally, the computational tools (e.g., artificial neural networks [ANNs], genetic algorithm [GA], and particle swarm optimization [PSO]), especially those that are based on mathematical or statistical models (e.g., rough sets [RS] and fuzzy logic), can enhance the ability of determining the relationships among the EOs’ compounds such as the chemical interactions, synergisms, and antagonisms. Accordingly, these tools may be very useful in classifying, optimizing, and predicting the antibacterial activity of a certain EO depending on the historical analysis of other EOs.5
One of the main challenges faced by scientific researchers in determining the antimicrobial activity of the EOs is the huge number of compounds that constitute the oil and which compounds have the significant antimicrobial effects. In this article, a new computational tool is presented based on RS and PSO algorithms. The proposed tool uses the RS algorithm as a reduction mathematical model to solve the problem of the huge number of EO compounds, which accordingly improves the optimization process that is based on PSO to determine the most proper EO for every type of bacteria.
Related work
In spite of the importance of applying artificial computing in biological systems, it is still not widespread in usage. Some researchers have tended to use the artificial computing tools in predicting the antimicrobial effects of EOs, while others have tried to use these tools in classifying the EOs according to their antibacterial activities.
Daynac et al have developed a computational model to evaluate the use of the ANNs for predicting the antimicrobial activity of EOs. The suggested model was fast artificial neural network (FANN) software that used 49 EOs with definition of their chemical compositions as the network inputs; the output data reflected the antimicrobial activity of these EOs against four common pathogens: “Staphylococcus aureus”, “Escherichia coli”, “Candida albicans”, and “Clostridium perfringens”. The ANN has proved its efficiency in predicting >70% of the antimicrobial activities within a 10 mm maximum error range by disk diffusion assays.5
Neves et al have presented a decision system for predicting the biological activity of the Schinus characterized by a variability in its chemical composition depending on its geographical and seasonal growing. The proposed study was conducted with the objective to characterize the founding of a computational framework that uses the knowledge representation and reasoning techniques to set the structure of the information and the associated inference mechanisms by using a logic programming-based approach to the knowledge representation and reasoning complemented with a computational framework based on ANNs. This model has achieved an accuracy of ~85% in predicting the pharmacological activity of the Schinus EOs.2
Another direction of using the artificial computing is the optimization process. Rajkovic et al have developed two optimization models to predict the antifungal effects for mixing the EOs of “Thymus vulgaris L.” and “Cinnamomum cassia L.”. The two proposed models are the response surface methodology (RSM) and the ANN combined with the GA. Both of the two models have tended to optimize the variables of the mixture process (i.e., time of action, concentration of individual or mixture EOs, and their mass ratio). Based on the mean relative percent deviation (MRPD), both of the two models have provided a good quality of prediction for the antifungal effect in terms of the three predefined independent variables. Nevertheless, due to the low value of the MRPD for the ANN – GA model, it has achieved more accuracy in predicting the antifungal effect of the EO mixture than the RSM model (±10.7% for RSM and ±0.1% ANN – GA).6
Problem formulation
The EO is a complex mixture of a huge number of compounds. Some of these compounds are characterized by their high antimicrobial activity such as aldehydes, phenols, and terpene alcohols (e.g., cinnamaldehyde, citral, carvacrol, eugenol, and thymol), while other compounds such as ketones or esters (e.g., β-myrcene, α-thujone, and geranyl acetate) have a limited activity against the microbes. On the other hand, EOs that contain terpene hydrocarbons were usually inactive in facing the microbial infection.7 The traditional chemical methods used for extracting and analyzing the EOs to determine their chemical composition are usually expensive and complicated due to the variation in the structure and concentration percentage of the EO compositions.3 Developing a computational tool may have a significant role in decreasing the time and cost of chemical analysis and in determining the most proper EO that has the optimal antibacterial activity, especially for the scientific industries that are based on using the EOs as antibacterials or antioxidants.
This article presents a new optimization tool based on EO reduction (Essential Oil Reduction and Optimization Tool [EOROT]) for enhancing the classification process of the EOs according to their antibacterial influence rate. The developed EOROT tool is based on three main phases. Determining the EOs’ chemical composition is the first phase. Second, reducing the huge number of these chemical compounds by specifying the main antibacterial compounds and establishing some logic rules to finding the relation between these compounds. Finally, finding the EOs that have high antibacterial activities and searching for the most-suited EO for every type of the bacteria. Figure 1 shows a brief illustration for the methodology of EOROT.
Figure 1.
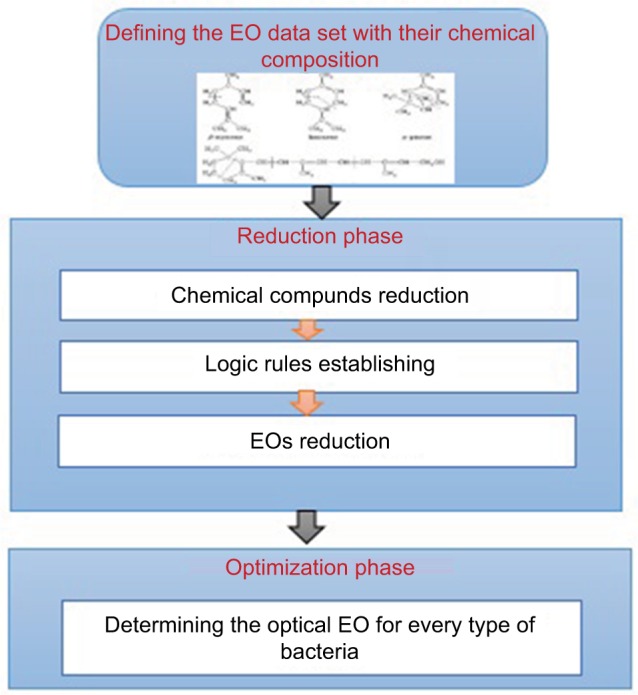
Methodology of the proposed EOROT.
Abbreviations: EOROT, Essential Oil Reduction and Optimization Tool; EO, essential oil.
Data set of EOs
Initially, analyzing the EOs is the first stage in the “EOROT” algorithm. The target of this stage is to determine all the compounds and their concentration percentages in the composition of EOs. This stage is the base of the reduction process, which will specify the significant compounds and remove the noninfluential ones.
The experimental study in this research is based on 24 types of plant EOs as a sample for antibacterial EOs, which are common in inhibiting the same types of bacteria. These EOs have >850 compounds; Table 1 lists the number of the identified compounds in every EO and their concentration percentage (the complete data is available in the Supplementary material section).8–26
Table 1.
Number of identified compounds in the EOs
| No. | EO | Number of identified compounds | Percentage total of identified compounds |
|---|---|---|---|
| 1 | Cupressus arizonica Green (leaves)8 | 87 | 99.4 |
| 2 | Ocimum basilicum9 | 29 | 98 |
| 3 | Pimpinella anisetum (Turkey)10 | 22 | 99.5 |
| 4 | Pimpinella flabellifolia (Turkey)10 | 19 | 99.7 |
| 5 | Salvia aucheri var. aucheri11 | 14 | 97.2 |
| 6 | Salvia aramiensis (Turkish flora)11 | 15 | 98.5 |
| 7 | Salvia pilifera (Turkish flora)11 | 30 | 98.2 |
| 8 | Dictamnus dasycarpus12 | 47 | 88.9 |
| 9 | Satureja subspicata Vis.13 | 24 | 97.47 |
| 10 | Achillea pachycephala Rech.f.14 | 53 | 98.2 |
| 11 | Achillea santolina L.14 | 52 | 96.1 |
| 12 | Achillea biebersteini15 | 23 | 84.2 |
| 13 | Cinnamomum zeylanicum16 | 39 | 98.6 |
| 14 | Laurus nobilis L.17 | 28 | 82.54 |
| 15 | Cymbopogon citratus18 | 42 | 97.2 |
| 16 | Origanum vulgare subsp. vulgare19 | 43 | 95.84 |
| 17 | Rosmarinus officinalis20 | 25 | 95.6 |
| 18 | Satureja montana L.21 | 49 | 98.8 |
| 19 | Lippia grandis Schauer (Verbenaceae)22 | 29 | 95.16 |
| 20 | Thymus serpyllum23 | 46 | 99.67 |
| 21 | T. serpyllum (Aureus)23 | 43 | 99.49 |
| 22 | Achillea millefolium subsp. millefolium24 | 36 | 89.8 |
| 23 | Salvia tomentosa25 | 44 | 97.7 |
| 24 | Mentha spicata26 | 18 | 99.89 |
Abbreviation: EO, essential oil.
Reduction and rules of extraction methodology
Recently, the emergence of the huge real-time data sets has increased the need for using the data mining and machine learning, which have proved their efficiency in dealing with these types of data. However, many data sets contain unnecessary features that may cause undesirable increase in the processing time. Therefore, the need for reducing the dimension size of the data set becomes a necessary stage in the data mining process.27 The reduction process refers to simplifying a data set by reducing its dimensionality through removing a subset of attributes from the original data set while retaining the same predictive accuracy. Generally, the reduction has a number of important benefits for the machine learning and data mining algorithms. It reduces the computation time of the induction algorithm, saves the computational cost, and also improves the accuracy of the induced rules.28 RS approach was originally proposed by Pawlak.29 It is defined as a mathematical approach that can obtain high-quality classifiers called reduct from the total data set attributes. This “reduct” can be used to predict the decision outputs just like using the original set.28 In general, in the RS approach, the information is represented in the form of condition attributes C and decision attributes D.30 The relation between the condition and the decision attributes in RS is defined by the indiscernibility degree of the data points handled using the concept of sets’ approximation. There are two main types of the approximation in RS, upper and lower, which are denoted as and , respectively; they are defined as follows in Equations 1 and 2:
| (1) |
| (2) |
According to the determined upper and lower approximation of the sets, the accuracy of the classification results can be quantified as follows:
| (3) |
These approximations are the base of finding the reduction for data set. They can also be used to deduce the rules between the condition and decision attributes. The syntax of the decision rules usually has the form of logic expression or if … then rule.30,31
Optimization methodology
The aim of the optimization is to determine the best-suited solution to a problem under a given set of constraints. The optimization problem is represented as an intelligent search problem where one or more agents are used to determine the optimal solution on the search landscape for the desired problem population.32 There are several kinds of numerical optimization methods such as neural network, gradient-based search, GA, ant colony, and PSO.32,33 PSO is one of the most famous optimization methods that has proved its efficiency in searching for the optima. Generally, it is a population-based algorithm that can be easily implemented and applied to solve various optimization problems by searching for the optimal solution within the available solutions based on social behavior patterns of organisms that live and interact within large groups. PSO incorporates swarming behaviors observed in flocks of birds, schools of fish, or swarms of bees.34
Proposed EOROT
The suggested EOs presented in Table 1 are commonly characterized by their antibacterial effects, especially for certain types of bacteria listed in Table 2. The influence of the EO on inhibiting the bacteria activity can be measured by several methods such as the minimum bactericidal concentration (MBC) and minimum inhibitory concentration (MIC).8 The selected 24 EOs were tested on 17 types of bacteria, and the activities of the EOs against these bacteria were studied via the presence or the absence of inhibition zones according to MIC measure, as displayed in Table 2.8–26 The MIC can be defined as the lowest concentration of the EO at which the bacteria do not show visible growth after overnight incubation.8
Table 2.
EOs and their MIC percentage rate on the selected bacteria
| No. | EO/MIC | Bacteria
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli | Pseudomonas aeruginosa | Enterococcus faecalis | Staphylococcus aureus | Salmonella typhimurium | Klebsiella pneumonia | Streptococcus paeumoniae | Bacillus cereus | Acinetobacter lwoffii | Clostridium perfringens | Candida albicans | Candida krusei | Bacillus subtilis | Listeria monocytogenes | Proteus vulgaris | Listeria plantarum | Yersinia enterocolitica | ||
| 1 | Cupressus arizonica Green (leaves) | 0.38 | 23.6 | 1.53 | 0.98 | 0.38 | 0.49 | 6.12 | X | X | X | X | X | X | X | X | X | X |
| 2 | Ocimum basilicum | 2.7 | X | X | 1.3 | X | X | X | X | X | X | X | 1.4 | 3.4 | X | X | X | X |
| 3 | Pimpinella anisetum (Turkey) | 72 | X | X | X | X | 72 | 8 | 72 | 18 | 4.5 | 36 | 36 | X | X | X | X | X |
| 4 | Pimpinella Flabellifolia (Turkey) | X | X | X | X | X | 72 | 9 | 72 | 36 | 2.25 | 36 | 36 | X | X | X | X | X |
| 5 | Salvia aucheri var. aucheri (Turkish flora) | 36 | X | X | X | X | 72 | 36 | 72 | 9 | 36 | 18 | 18 | X | X | X | X | X |
| 6 | Salvia aramiensis (Turkish flora) | 36 | X | X | X | X | 36 | 18 | 36 | 4.5 | 18 | 18 | 9 | X | X | X | X | X |
| 7 | Salvia pilifera (Turkish flora) | >72 | X | X | X | X | 72 | 72 | >72 | 18 | 72 | 36 | 18 | X | X | X | X | X |
| 8 | Dictamnus dasycarpus | 6.2 | 25 | 25 | 3.13 | 25 | X | X | X | X | X | 6.25 | X | X | X | X | X | X |
| 9 | Satureja subspicata Vis. | 0.78 | 12.5 | 0.78 | 0.09 | 6.25 | 6.25 | X | 3.12 | X | 0.3 | 0.09 | X | 3.12 | X | X | X | X |
| 10 | Achillea pachycephala Rech.f. | 6.5 | X | X | 6.25 | 1.56 | 3.12 | X | 12.5 | X | X | X | X | 6.25 | 1.56 | X | X | X |
| 11 | Achillea santolina L. | 6.25 | X | X | 6.25 | 1.56 | 6.25 | X | 12.5 | X | X | X | X | 12.5 | X | 0.78 | X | X |
| 12 | Achillea biebersteini Afan | 18 | >72 | X | 36 | X | X | X | 9 | X | X | X | 0.15 | X | X | X | X | X |
| 13 | Cinnamomum zeylanicum | 5 | X | X | 5 | 5 | X | X | 2.5 | X | X | X | X | X | X | X | 2.5 | 0.07 |
| 14 | Laurus nobilis L. | 2.5 | X | X | 10 | 5 | X | X | 5 | X | X | X | X | X | X | X | 2.5 | 0.62 |
| 15 | Cymbopogon citratus | 0.62 | X | X | 0.31 | 1.25 | X | X | 0.15 | X | X | X | X | X | X | X | 0.62 | 0.08 |
| 16 | Origanum vulgare subsp. vulgare | 5 | X | X | 5 | 5 | X | X | 2.5 | X | X | X | X | X | X | X | 2.5 | 0.075 |
| 17 | Rosmarinus officinalis | 2.5 | X | X | 10 | 10 | X | X | 5 | X | X | X | X | X | X | X | 2.5 | 0.07 |
| 18 | Satureja montana L. | 1.56 | 0.2 | X | 0.78 | X | X | X | 1.56 | X | 0.39 | X | X | X | 0.78 | X | X | X |
| 19 | Lippia grandis Schauer (Verbenaceae) | 1.15 | 0.57 | X | 1.15 | X | 1.15 | X | X | X | X | X | X | X | X | X | X | X |
| 20 | Thymus serpyllum | 0.25 | 0.13 | X | 0.1 | X | X | X | 0.1 | X | X | X | X | X | X | 0.18 | X | X |
| 21 | T. serpyllum (Aureus) | 0.05 | 0.13 | X | 0.1 | X | X | X | 0.31 | X | X | 0.16 | X | X | X | 0.39 | X | X |
| 22 | Achillea millefolium subsp. millefolium | X | X | 12 | X | X | 72 | 4.5 | 72 | 18 | 9 | X | 18 | X | X | X | X | X |
| 23 | Salvia tomentosa | X | X | X | 18 | X | 72 | 2.25 | 9 | 18 | 0.45 | 18 | 36 | X | X | X | X | X |
| 24 | Mentha spicata | X | X | X | 10 | 10 | X | X | 2.5 | X | X | X | X | 2.5 | 2.5 | X | X | X |
Abbreviations: EO, essential oil; MIC, minimum inhibitory concentration.
It is commonly known that the activity of the bacteria may be repressed by more than one type of anti-organisms, as shown in Table 2. The EO, which is labeled as one of the most famous anti-organisms (anti-bacteria), can inhibit the bacteria growing with variant ratios according to the MIC rate. Testing the EOs against the bacteria and measuring the MIC can be done via many types of antimicrobial screening methods such as well-diffusion method and disk diffusion method (DDM). This research is based on the second method.24 Some EOs have limited impact on specific types of bacteria, while others have wide anti-bacterial impact. Thus, it is a difficult issue to determine the EOs that contain the suitable antibacterial compounds that can achieve the desired inhibition for the bacterial growth due to the huge number of compounds in their chemical composition as shown in Table 1. Herein, the importance of the reduction process appears in an attempt to abstract the chemical compounds and retain only those that have a high antibacterial impact.
The reduction process based on the RS theory is the first step in the proposed EOROT algorithm. It is divided into three phases. 1) It begins with reducing the number of the compounds for every EO by keeping only the compounds that affect the EO concentration. This process depends on defining the threshold value for the compound’s concentration percentage by 2% and ignoring the compounds that do not exceed this threshold. 2) After the reduction of compounds, the RS starts establishing some logic rules; these rules can help in determining the EOs that have a high antibacterial effect generally without testing them on certain types of bacteria as defined in the following equation:
| (4) |
where the symbols A, B, C, D, E, and F are the concentration percentage rates of the compound in the EO and the symbols V.H, H, N, and L are the antibacterial activity degrees for these compounds. Table 3 displays these symbols and their significances. 3) The last phase in the reduction process is applying the extracted rules to constringe the number of EOs by finding the significant antibacterial EOs only. The pseudo code of the EOROT algorithm is presented in Box 1.
Table 3.
Significance of compound concentration percentage and antibacterial degree
| Symbol | Significance of concentration percentage | Symbol | Significance of antibacterial activity degree |
|---|---|---|---|
| A | ≥70 | V.H | Very high |
| B | <70 and ≥50 | H | High |
| C | <50 and ≥30 | N | Normal |
| D | <30 and ≥10 | L | Low |
| E | <10 and ≥2 |
After completing the reduction phase, the optimization process begins its role in the EOROT algorithm. Generally, the optimization process is a popular computational technique that tends to find the optimal solution to the current problem. The main purpose of the optimization phase in EOROT is to find the proper EO that can inhibit the bacteria with the best MIC. PSO is the optimization algorithm that is used in EOROT; it is based on defining the population of the current problem that consists of some particles (i.e., bacteria and EOs) represented by a matrix P = [Ni * Mj], where N is the bacteria and M is the EO. For every bacteria, the target is to find the inhibitory EO with minimum MIC. Thus, the searching process begins by initializing the bacteria and the EO’s MIC degree and changing the velocity of the search until the optimal solution is reached. This method reduces time and cost substantially, where the algorithm takes into consideration how to optimize the number of EOs that affects the bacteria to achieve a high utilization of one EO in inhibiting multiple types of bacteria. Figure 2 displays how the PSO works in order to find the optimal EO for the selected bacteria.
Figure 2.
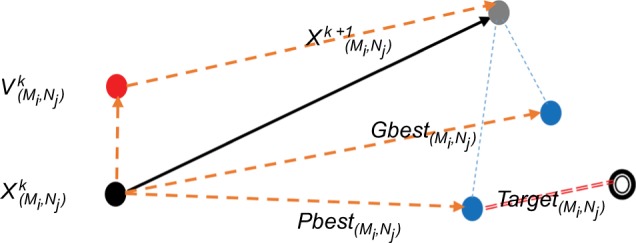
Scenario of PSO processing on the selected population of bacteria and EOs.
Abbreviations: PSO, particle swarm optimization; EO, essential oil.
In Figure 2, represents the initial value of the particles (i.e., the initial bacteria and EO), and are the best suggested positions for the current particle, and the is the optimal solution (i.e., the EO with the minimum MIC that can inhibit this bacteria). In the search process, every particle moves with a certain velocity that determines its new position, and the search will stop when the new position becomes close to the target.
Implementation and results analysis of EOROT
In this section, the EOROT will be practically applied on the 24 EOs, which are represented in Table 1. The suggested EOs have >850 compounds; many of them are not vital compounds in the chemical compositions. As discussed in the previous section, the first phase in EOROT is the reduction process that is based on the reduction of compounds and establishing of logic rules. Table 4 displays the results after implementing the process of compounds’ reduction.
Table 4.
Compounds of the EOs after the EOROT reduction process
| No. | EO | Vital compounds in the EO | Compound percentage in the EO | Antibacterial degree of the compound | Number of compounds after reduction | Percentage of compounds after reduction |
|---|---|---|---|---|---|---|
| 1 | Cupressus arizonica Green (leaves) | α-Pinene | 20.0 | High | 9 | 68.3 |
| Umbellulone | 18.4 | High | ||||
| cis-Muurola-4(14),5-diene | 9.4 | Low | ||||
| Limonene | 5.8 | High | ||||
| cis-Muurola-5-en-4b-ol | 3.5 | Low | ||||
| cis-Muurola-3,5-diene | 3.3 | None | ||||
| Sabinene | 2.9 | High | ||||
| Epizonarene | 2.8 | Normal | ||||
| trans-Calamenene | 2.2 | Low | ||||
| 2 | Ocimum basilicum | Linalool | 60.6 | High | 5 | 83.2 |
| Cadinol, epi-α | 8.6 | High | ||||
| α-Bergamotene | 7.8 | High | ||||
| c-Cadinene | 3.2 | High | ||||
| Camphor | 3 | High | ||||
| 3 | Pimpinella Anisetum (Turkey) | (E)-anethole | 82.8 | High | 2 | 97.3 |
| Methyl chavicol | 14.5 | Low | ||||
| 4 | Pimpinella Flabellifolia (Turkey) | Limonene | 47 | High | 5 | 97.4 |
| (E)-anethole | 37.9 | High | ||||
| α-Pinene | 6 | High | ||||
| c-Terpinene | 3.5 | None | ||||
| β-Pinene | 3 | High | ||||
| 5 | Salvia aucheri var. aucheri | 1,8-Cineole | 30.5 | High | 8 | 86.6 |
| Camphor | 21.3 | High | ||||
| Borneol | 8.5 | High | ||||
| Camphene | 7.8 | Low | ||||
| α-Pinene | 7.6 | High | ||||
| β-Pinene | 5.7 | High | ||||
| α-Terpinyl acetate | 2.6 | High | ||||
| Spathulenol | 2.6 | Normal | ||||
| 6 | Salvia aramiensis (Turkish Flora) | 1,8-Cineole | 46 | High | 8 | 82.3 |
| β-Pinene | 10.3 | High | ||||
| Camphor | 8.7 | High | ||||
| α-Pinene | 4.9 | High | ||||
| Camphene | 4.2 | Low | ||||
| Borneol | 3.6 | High | ||||
| Tricyclene | 2.5 | Low | ||||
| Myrcene | 2 | Low | ||||
| 7 | Salvia pilifera (Turkish flora) | α-Thujene | 36.1 | Low | 9 | 77.7 |
| α-Pinene | 13.8 | High | ||||
| 1,8-Cineole | 9.2 | High | ||||
| Piperitenone | 4.7 | High | ||||
| trans-Thujone | 3.6 | High | ||||
| Terpinen-4-ol | 3.2 | None | ||||
| c-Muurolene | 2.6 | None | ||||
| α-Terpineol | 2.3 | None | ||||
| Myrcene | 2.2 | Low | ||||
| 8 | Dictamnus dasycarpus | syn-7-Hydroxy-7-anisylnorbornene | 29.4 | High | 8 | 70.3 |
| Pregeijerene | 15.5 | High | ||||
| Geijerene | 11.4 | High | ||||
| Dictamnol | 3.5 | None | ||||
| β-Phellandrene | 3.3 | High | ||||
| 8S,13-Cedranediol | 2.7 | None | ||||
| Elemol | 2.4 | None | ||||
| 8-epi-Dictamnol | 2.1 | None | ||||
| 9 | Satureja subspicata Vis. | Carvacrol | 16.76 | Very high | 13 | 84.61 |
| α-Pinene | 13.58 | High | ||||
| p-Cymene | 10.76 | High | ||||
| c-Terpinene | 9.54 | None | ||||
| Thymol methyl ether | 8.83 | Very high | ||||
| Myrcene | 4.82 | Low | ||||
| Linalool | 3.94 | High | ||||
| β-Caryophyllene | 3.76 | High | ||||
| Limonene | 3.45 | High | ||||
| Geranyl acetate | 2.81 | Low | ||||
| Nerol | 2.13 | Very high | ||||
| Thymol | 2.12 | Very high | ||||
| Borneol | 2.11 | High | ||||
| 10 | Achillea pachycephala Rech.f. | 1,8-Cineole | 16.4 | High | 11 | 61.8 |
| Camphor | 11.2 | Camphor | ||||
| Camphene | 7.2 | Low | ||||
| Borneol | 5.2 | High | ||||
| Sabinene | 4.6 | None | ||||
| Terpinen-4-ol | 4.3 | None | ||||
| E-Pinocarveol | 3.2 | High | ||||
| a-Pinene | 3.1 | High | ||||
| Linalool | 2.4 | High | ||||
| p-Cymene | 2.1 | High | ||||
| Octanoic acid | 2.1 | Low | ||||
| 11 | Achillea santolina L. | Fragranyl acetate | 28.4 | High | 10 | 64.3 |
| Fragranol | 8.1 | High | ||||
| Terpinen-4-ol | 6.4 | None | ||||
| 1,8-Cineole | 5 | High | ||||
| Isoborneol | 4.2 | High | ||||
| Z-Sabinene hydrate | 3.5 | None | ||||
| β-Pinene | 2 | High | ||||
| Thymol | 2.5 | Very high | ||||
| Artemisia alcohol | 2.1 | High | ||||
| Lavandulyl | 2.1 | High | ||||
| 2-methylbutanoate | ||||||
| 12 | Achillea biebersteini | Piperitone | 34.9 | High | 7 | 74.8 |
| Eucalyptol | 13 | High | ||||
| Camphor | 8.8 | Camphor | ||||
| Chrysanthenone | 8.2 | High | ||||
| Borneol | 4.4 | High | ||||
| a-Pinene | 3.1 | High | ||||
| a-Terpineol | 2.4 | High | ||||
| 13 | Cinnamomum zeylanicum | (E)-cinnamaldehyde | 37.6 | Very high | 6 | 85 |
| Cinnamyl acetate | 23.7 | High | ||||
| Cinnamyl benzoate | 16.4 | High | ||||
| a-Pinene | 2.7 | High | ||||
| Benzyl benzoate | 2.4 | High | ||||
| Linalool | 2.2 | High | ||||
| 14 | Laurus nobilis L. | 1,8-Cineole | 46.61 | High | 6 | 85.04 |
| Sabinene | 14.05 | High | ||||
| α-Terpinyl acetate | 11.94 | High | ||||
| α-Terpineol | 6.83 | High | ||||
| α-Pinene | 3.66 | High | ||||
| Methyl eugenol | 2 | High | ||||
| 15 | Cymbopogon citratus | Geranial | 43.95 | Low | 4 | 81.32 |
| Neral | 31.05 | High | ||||
| β-Myrcene | 3.18 | Low | ||||
| Nerol | 3.14 | Very high | ||||
| 16 | Origanum vulgare subsp. vulgare | Thymol | 37.129 | Very high | 9 | 81.502 |
| Gamma-terpinene | 9.668 | High | ||||
| Carvacrol | 9.573 | Very high | ||||
| Carvacrol, methyl ether | 6.88 | None | ||||
| cis-alpha-bisabolene | 6.805 | None | ||||
| Eucalyptol (1,8-cineole) | 3.823 | High | ||||
| p-Cymene | 3.583 | High | ||||
| Elemol | 2.041 | None | ||||
| trans-Caryophyllene | 2 | Low | ||||
| 17 | Rosmarinus officinalis | 1,8-Cineole | 40.7 | High | 6 | 83.6 |
| Camphor | 17.9 | High | ||||
| α-Pinene | 10.3 | High | ||||
| Camphene | 6.3 | Low | ||||
| Borneol | 5.2 | High | ||||
| α-Terpineol | 3.2 | High | ||||
| 18 | Satureja montana L. | Carvacrol | 24.46 | Very high | 10 | 88.46 |
| Linalool | 17.94 | High | ||||
| cis-Sabinene hydrate | 14.61 | High | ||||
| Terpinen-4-ol | 10.6 | High | ||||
| p-Cymene | 5.38 | High | ||||
| Borneol | 3.62 | High | ||||
| b-Caryophyllene | 3.37 | High | ||||
| Caryophyllene oxide | 3.3 | High | ||||
| Elemol | 2.72 | None | ||||
| Thymol | 2.46 | Very high | ||||
| 19 | Lippia grandis Schauer (Verbenaceae) | Carvacrol | 37.12 | Very high | 9 | 78.56 |
| p-Cymene | 11.64 | High | ||||
| Thymol | 7.83 | Very high | ||||
| 3-tert-Butyl-4-methoxyphenol | 6.91 | Very high | ||||
| β-Caryophyllene | 3.93 | High | ||||
| β-Bisabolene | 3.72 | Low | ||||
| Linalool | 3.32 | High | ||||
| γ-Terpinene | 2.07 | High | ||||
| 20 | Thymus serpyllum | Carvacrol | 37.49 | Very high | 8 | 78.1 |
| γ-Terpinene | 10.79 | High | ||||
| β-Caryophyllene | 6.51 | High | ||||
| p-Cymene | 6.06 | High | ||||
| (E)-β-Ocimene | 4.63 | High | ||||
| β-Bisabolene | 4.51 | None | ||||
| Carvacrol methyl ether | 4.4 | None | ||||
| Eucalyptol | 3.71 | High | ||||
| 21 | T. serpyllum (Aureus) | Carvacrol | 44.93 | Very high | 8 | 82.19 |
| γ-Terpinene | 10.08 | High | ||||
| p-Cymene | 7.39 | High | ||||
| β-Caryophyllene | 6.77 | High | ||||
| 3-Octanone | 6.19 | None | ||||
| Carvacrol methyl ether | 2.58 | None | ||||
| α-Terpinene | 2.15 | None | ||||
| Borneol | 2.1 | High | ||||
| 22 | Achillea millefolium subsp. millefolium | Eucalyptol (1,8-cineole) | 24.6 | High | 12 | 79.3 |
| Camphor | 16.7 | High | ||||
| α-Terpineol | 10.2 | High | ||||
| β-Pinene | 4.2 | High | ||||
| Borneol | 4 | High | ||||
| Bisabolol oxide II | 3.8 | Low | ||||
| Bisabolone oxide | 3.3 | Low | ||||
| Sabinene | 2.8 | High | ||||
| Terpinen-4-ol | 2.8 | None | ||||
| α-Pinene | 2.4 | High | ||||
| Camphene | 2.4 | High | ||||
| α-Bisabolol | 2.1 | Low | ||||
| 23 | Salvia tomentosa | β-Pinene | 39.7 | High | 9 | 75.8 |
| α-Pinene | 10.9 | High | ||||
| Camphor | 9.7 | High | ||||
| Borneol | 4.3 | High | ||||
| Camphene | 2.4 | None | ||||
| Caryophyllene | 2.3 | High | ||||
| Viridiflorol | 2.3 | None | ||||
| Limonene | 2.2 | High | ||||
| α-Caryophyllene | 2 | High | ||||
| 24 | Mentha spicata | Carvone | 78.76 | High | 2 | 90.26 |
| Limonene | 11.5 | High |
Abbreviation: EO, essential oil.
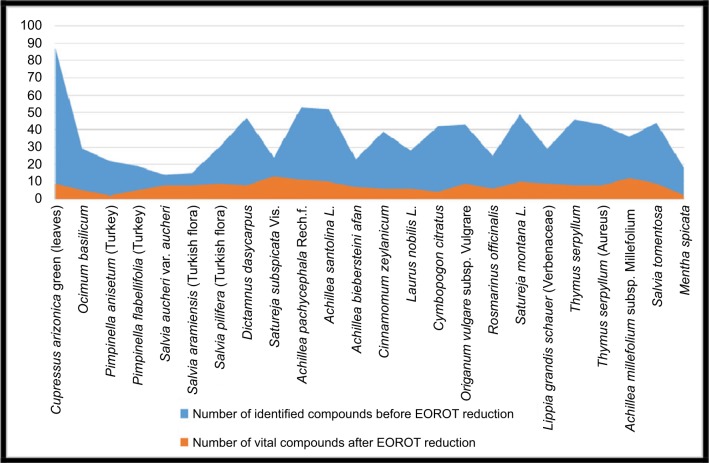
By completing the compound’s reduction process, the noninfluential compounds in the EO’s composition will be neglected according to the indiscernibility relation in the RS algorithm that preserves only the core of these compounds that have a vital impact in the EO’s activity. Figure 3 displays the difference between the number of EOs’ compounds before and after applying the reduction process using EOROT.
Figure 3.
Comparison between the EOs’ numbers before and after the reduction process.
Abbreviations: EO, essential oil; EOROT, Essential Oil Reduction and Optimization Tool.
After reducing the EOs’ compounds, the EOROT continues its job by filtering the 24 EOs to find the ones that have a high antibacterial activity. The logic rules that are defined in Equation 4 will be applied to calculate the accuracy rate a for every EO, which is considered as the measurement parameter in determining the antibacterial activity degree for the EO. Accuracy rate a is predefined in our algorithm with the value 0.6; thus, the EOs that have an accuracy rate <0.6 will be neglected as they do not contain effective antibacterial compounds. Table 5 presents the final results of the EOROT reduction process. As it is displayed, the number of EOs is reduced to 19 and thus achieves a high accuracy rate in antibacterial activity.
Table 5.
EOs after completing the EOROT reduction process
| No. | EO | Accuracy rate |
|---|---|---|
| 1 | Ocimum basilicum | 0.9 |
| 2 | Pimpinella anisetum (Turkey) | 0.84 |
| 3 | Salvia aucheri var. aucheri | 0.74 |
| 4 | Salvia aramiensis (Turkish flora) | 0.71 |
| 5 | Dictamnus dasycarpus | 0.63 |
| 6 | Satureja subspicata Vis. | 0.69 |
| 7 | Achillea pachycephala Rech.f. | 0.68 |
| 8 | Achillea santolina L. | 0.70 |
| 9 | Achillea biebersteini Afan | 0.89 |
| 10 | Cinnamomum zeylanicum | 0.85 |
| 11 | Laurus nobilis L. | 0.89 |
| 12 | Cymbopogon citratus | 0.77 |
| 13 | Rosmarinus officinalis | 0.85 |
| 14 | Satureja montana L. | 0.79 |
| 15 | Lippia grandis Schauer (Verbenaceae) | 0.79 |
| 16 | Thymus serpyllum | 0.657 |
| 17 | T. serpyllum (Aureus) | 0.695 |
| 18 | Salvia tomentosa | 0.71 |
| 19 | Mentha spicata | 0.9 |
Abbreviations: EO, essential oil; EOROT, Essential Oil Reduction and Optimization Tool.
Box 1. Pseudo code of EOROT.
| Reduction stage |
| 1. Set the information system of antibacterial plants A=(U,A) |
| 2. Define the indiscernibility matrix M(A)=(cij) |
| 3. Build the discernibility function FA for the information system A as in Equation (1). |
| 4. Reduce M attributes using laws of Rough sets (Upper, and Lower Laws). |
| 5. Define d as number of non-empty rows of reduced M. |
| 6. Build families sets of R0, R1, R2,………… Rd in the as follows: |
| 7. Begin: |
| 8. R0 is empty |
| 9. For i = 1 to d |
| 10. Ri=Si ∪ Ti, where Si={R∈Ri−1:R ∩ Ci ≠∅}, and |
| 11. Calculate the accuracy α for each Ri |
| 12. End |
| 13. If αi < 0.6 |
| 14. Remove dispensable attribute form each element of Rd |
| 15. REDA (A)= Rd |
| Optimization Stage |
| 16. Set the Population P as a matrix P = [Ni*Mj] where N is the bacteria type and M is the plant |
| 17. Set the particle is Pij which is the bacteria i on plant j |
| 18. For each particle |
| 19. Initialize position and velocity |
| 20. End For |
| 21. Do |
| 22. For each particle |
| 23. Find in the particle neighborhood, the particles with the best fitness as Pbest and Gbest. |
| 24. Calculate Pi velocity according to the velocity equation |
| 25. Vij(k+1)=wvij+c1r1[pbest-xij(k)]+c2r2[gbest-xij(k)] |
| 26. Update Pi position according to the position equation |
| 27. Xij(k+1)=xij(k)+vij(k+1) |
| 28. If the new position for Pi is less than its current position then |
| 29. Modify the velocity and position for Pi and Pbest and Gbest |
| 30. Else |
| 31. Modify the velocity of Pi and keep its old position |
| 32. End For |
| 33. While maximum iterations or minimum error criteria is not attained |
Abbreviation: EOROT, Essential Oil Reduction and Optimization Tool.
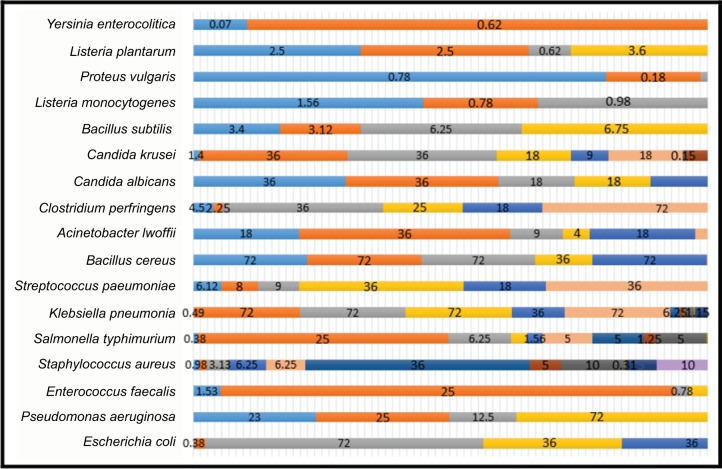
The extracted EOs from the reduction process (i.e., 19 EOs) are tested toward 17 types of bacteria, and as it is displayed in Table 2, each type of bacteria can be discouraged by more than an EO with different ratios of MIC. Figure 4 gives a brief overview of the MIC values for the EOs on every type of bacteria.
Figure 4.
MIC values for the EOs on every type of bacteria.
Abbreviations: MIC, minimum inhibitory concentration; EO, essential oil.
Therefore, the optimization process, which is considered as the last and substantial phase in EOROT tool, begins its work. The target of the optimization is to determine the optimal solution to inhibit the growth of the existing bacteria. Thus, deciding the optimal solution is related to two issues: EOROT will search for the EO that achieves the bacterial activity’s inhibition with the less MIC rate, and at the same time, it will try to maximize the utilization of the selected EO in inhibiting multiple types of the existing bacteria as displayed in Table 6.
Table 6.
Optimal MIC for inhibiting the bacteria activity
| Bacteria | EO | MIC |
|---|---|---|
| Escherichia coli | Thymus serpyllum (Aureus) | 0.05 |
| Pseudomonas aeruginosa | T. serpyllum (Aureus) | 0.13 |
| Bacillus cereus | T. serpyllum (Aureus) | 0.31 |
| Proteus vulgaris | T. serpyllum (Aureus) | 0.39 |
| Enterococcus faecalis | Satureja subspicata Vis. | 0.78 |
| Staphylococcus aureus | S. subspicata Vis. | 0.09 |
| Clostridium perfringens | S. subspicata Vis. | 0.3 |
| Candida albicans | S. subspicata Vis. | 0.09 |
| Salmonella typhimurium | Cupressus arizonica Green (leaves) | 0.38 |
| Klebsiella pneumonia | C. arizonica Green (leaves) | 0.49 |
| Streptococcus paeumoniae | C. arizonica Green (leaves) | 6.12 |
| Listeria plantarum | Cymbopogon citratus | 0.62 |
| Yersinia enterocolitica | C. citratus | 0.08 |
| Acinetobacter lwoffii | Salvia aramiensis (Turkish flora) | 4.5 |
| Candida krusei | Achillea biebersteini Afan | 0.15 |
| Bacillus subtilis | Mentha spicata | 2.5 |
| Listeria monocytogenes | Satureja montana L. | 0.78 |
Abbreviations: MIC, minimum inhibitory concentration; EO, essential oil.
Thus, it could be concluded that the proposed computational tool “EOROT” enhances the process of finding the optimal EO that can inhibit the growth activity of the bacteria with the best MIC rate. In addition, EOROT takes into consideration how to boost the utilization of the available EOs, for instance Thymus serpyllum can be used to inhibit the activity of E. coli, Pseudomonas aeruginosa, Bacillus cereus, and Proteus vulgaris as evidenced in Table 6.
Therefore, according to the extracted information from the EOROT tool, it can also be used to predict the efficiency of the EOs in inhibiting other types of bacterial activities.
Conclusion
Reliance on natural products has become an important trend in many fields as the alternative medicine, which depends on using the natural anti-organisms in fighting bacteria, fungi and tumoral infections. One of the most efficient anti-organisms extracted from the plants is the EO that proves its efficiency as antibacterial agents. This article presents a new computational tool known as EOROT in an attempt to determine the best and optimal EO in inhibiting the activity of the bacteria growth. Furthermore, EOROT has the ability to increase the EO’s utilization by using it in inhibiting multiple kinds of bacteria, which, in turn, will save the cost of materials. On the other hand, the extracted information from the EOROT tool could be used to predict the efficiency of the EOs in inhibiting the other types of bacterial activities that may decrease the time also. The weak point in this research is the limited number of EOs and types of bacteria used in the case study, with the research based on only 24 EOs tested toward 17 kinds of bacteria. Thus, in the future work, the EOROT will be developed to handle a larger number of data and also it will be enhanced to predict the activity of the EOs.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zuzarte M, Salgueiro L. Bioactive Essential Oils and Cancer. Springer; 2015. Chapter 2: Essential oils chemistry; pp. 19–28. [Google Scholar]
- 2.Neves J, Martins M, Candeias F, et al. Logic programming and artificial neural networks in pharmacological screening of Schinus essential oils. Int J Biol Biomol Agric Food Biotechnol Eng. 2015;9(7):728–733. [Google Scholar]
- 3.Chamorro E, Zambón S, Morales W. Gas Chromatography in Plant Science, Wine Technology, Toxicology and Some Specific Applications. InTech; 2012. Study of the chemical composition of essential oils by gas chromatography; pp. 307–324. [Google Scholar]
- 4.Maida I, Nostro A, Pesavento G, et al. Exploring the anti-Burkholderia cepacia complex activity of essential oils: a preliminary analysis. Evid Based Complement Alternat Med. 2014;2014:1–10. doi: 10.1155/2014/573518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daynac M, Cortes-Cabrera A, Prieto J. Application of artificial intelligence to the prediction of the antimicrobial activity of essential oils. Evid Based Complement Alternat Med. 2015;2015:1–10. doi: 10.1155/2015/561024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajkovic K, Pekmezovic M, Barac A, Nikodinovic-Runic J. Inhibitory effect of thyme and cinnamon essential oils on Aspergillus flavus: optimization and activity prediction model development. Ind Crops Prod. 2015;65:7–13. [Google Scholar]
- 7.Dhifi W, Bellili S, Jazi S, Bahloul N, Mnif W. Essential oils’ chemical characterization and investigation of some biological activities: a critical review. MDPI Med J. 2015;3(25):1–16. doi: 10.3390/medicines3040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chéraif I, Jannet H, Hammami M, Khouja M, Mighri Z. Chemical composition and antimicrobial activity of essential oils of Cupressus arizonica Greene. Biochemical Systematics and Ecology. 2007;35:813–820. [Google Scholar]
- 9.Hussain A, Anwar F, Sherazi S, Przybylski R. Chemical composition, antioxidant and antimicrobial activities of basil (Ocimum basilicum) essential oils depends on seasonal variations. Food Chem. 2008;108(3):986–995. doi: 10.1016/j.foodchem.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Tepe B, Akpulat H, Sokmen M, et al. Screening of the antioxidative and antimicrobial properties of the essential oils of Pimpinella anisetum and Pimpinella flabellifolia from Turkey. Food Chem. 2006;97:719–724. [Google Scholar]
- 11.Kelen M, Tepe B. Chemical composition, antioxidant and antimicrobial properties of the essential oils of three Salvia species from Turkish flora. Bioresour Technol. 2008;99(10):4096–4104. doi: 10.1016/j.biortech.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Lei J, Yu J, Yu H, Liao Z. Composition, cytotoxicity and antimicrobial activity of essential oil from Dictamnus dasycarpus. Food Chem. 2008;107:1205–1209. [Google Scholar]
- 13.Skocibusic M, Bezic N, Dunkic V. Phytochemical composition and antimicrobial activities of the essential oils from Satureja subspicata Vis. growing in Croatia. Food Chem. 2006;96:20–28. [Google Scholar]
- 14.Motavalizadehkakhky A, Shafaghat A, Zamani H, et al. Compositions and the in vitro antimicrobial activities of the essential oils and extracts of two Achillea species from Iran. J Med Plants Res. 2013;7(19):1280–1292. [Google Scholar]
- 15.Sokmen A, Sokmen M, Daferera D, et al. The in vitro antioxidant and antimicrobial activities of the essential oil and methanol extracts of Achillea biebersteini Afan (Asteraceae) Phytother Res. 2004;18(6):451–456. doi: 10.1002/ptr.1438. [DOI] [PubMed] [Google Scholar]
- 16.Boniface Y, Philippe S, Rose de Lima H, et al. Chemical composition and antimicrobial activities of Cinnamomum zeylanicum Blume dry leaves essential oil against food-borne pathogens and adulterated microorganisms. Int Res J Biol Sci. 2012;1(6):18–25. [Google Scholar]
- 17.Cherrat L, Espina L, Bakkali M, Garcia-Gonzalo D, Pagan R, Laglaoui A. Chemical composition and antioxidant properties of Laurus nobilis L. and Myrtus communis L. essential oils from Morocco and evaluation of their antimicrobial activity acting alone or in combined processes for food preservation. Soc Chem Ind. 2013;94:1197–1204. doi: 10.1002/jsfa.6397. [DOI] [PubMed] [Google Scholar]
- 18.Tajidin E, Ahmad S, Rosenani A, Azimah H, Munirah M. Chemical composition and citral content in lemongrass (Cymbopogon citratus) essential oil at three maturity stages. Afr J Biotechnol. 2012;11(11):2685–2693. [Google Scholar]
- 19.Vazirian M, Mohammadi M, Farzaei M, Amin G, Amanzadeh Y. Chemical composition and antioxidant activity of Origanum vulgare subsp. vulgare essential oil from Iran. Res J Pharmacogn. 2015;2(1):41–46. [Google Scholar]
- 20.Zaouali Y, Bouzaine T, Boussaid M. Essential oils composition in two Rosmarinus officinalis L. varieties and incidence for antimicrobial and antioxidant activities. Food Chem Toxicol. 2010;48(11):3, 144–3152. doi: 10.1016/j.fct.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Mihajilov-Krstev T, Radnovic D, Kitic D, et al. Chemical composition, antimicrobial, antioxidative and anticholinesterase activity of Satureja montana L. ssp montana essential oil. Cent Eur J Biol. 2014;9(7):668–677. [Google Scholar]
- 22.Sarrazin S, Oliveira R, Barata L, Mourão R. Chemical composition and antimicrobial activity of the essential oil of Lippia grandis Schauer (Verbenaceae) from the western Amazon. Food Chem. 2012;134(3):1474–1478. doi: 10.1016/j.foodchem.2012.03.058. [DOI] [PubMed] [Google Scholar]
- 23.Wesołowska A, Grzeszczuk M, Jadczak D, Nawrotek P, Struk M. Comparison of the chemical composition and antimicrobial activity of Thymus serpyllum essential oils. Not Bot Horti Agrobo. 2015;43(2):432–438. [Google Scholar]
- 24.Candan F, Unlu M, Tepe B, et al. Antioxidant and antimicrobial activity of the essential oil and methanol extracts of Achillea mille-folium subsp. millefolium Afan. (Asteraceae) J Ethnopharmacol. 2003;87(2–3):215–220. doi: 10.1016/s0378-8741(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 25.Tepe B, Daferera D, Sokmen A, Sokmen M, Polissiou M. Antimicrobial and antioxidant activities of the essential oil and various extracts of Salvia tomentosa Miller (Lamiaceae) Food Chem. 2005;90:333–340. [Google Scholar]
- 26.Shahbazi Y. Chemical composition and in vitro antibacterial activity of Mentha spicata essential oil against common food-borne pathogenic bacteria. J Pathog. 2015;2015:916305. doi: 10.1155/2015/916305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedar A, Omar M, Sewisy A. Rough sets attribute reduction using an accelerated genetic algorithm. 16th IEEE/ACIS International Conference on Software Engineering, Artificial Intelligence, Networking and Parallel/Distributed Computing (SNPD); 2015. [Google Scholar]
- 28.Huang K, Chang T, Chang S. Rough set-based dataset reduction method using swarm algorithm and cluster validation function; 48th Hawaii International Conference on System Sciences; 2015. pp. 1483–1492. [Google Scholar]
- 29.Pawlak Z. Rough sets. Int J Inform Comput Sci. 1982;11:341–356. [Google Scholar]
- 30.Pałkowski Ł, Błaszczyński J, Krysiński J, et al. Application of rough set theory to prediction of antimicrobial activity of bis-quaternary ammonium chlorides. Lect Notes Comput Sci. 2012;7414:107–116. [Google Scholar]
- 31.Bing Z, Fu J. Algorithm on rule extraction based on rough set and neural network theory. The 6th International Forum on Strategic Technology; 2011. pp. 1137–1140. [Google Scholar]
- 32.Das S, Abraham A, Konar A. Particle swarm optimization and differential evolution algorithms: technical analysis, applications and hybridization perspectives. Adv Comput Intell Ind Syst Stud Comput Intell. 2008;116:1–38. [Google Scholar]
- 33.Crisan M. Convergence and Hybrid Information Technologies. Vukovar: Intech; 2010. [Google Scholar]
- 34.El-Attar N. Prediction Resources Scheduling in Cloud Computing Systems. Germany: Lambert Academic Publishing; 2016. [Google Scholar]