Abstract
After fertilization, the genomes derived from an oocyte and spermatozoon are in a transcriptionally silent state before becoming activated at a species-specific time. In mice, the initiation of transcription occurs at the mid-one-cell stage, which represents the start of the gene expression program. A recent RNA sequencing analysis revealed that the gene expression pattern of one-cell embryos is unique and changes dramatically at the two-cell stage. However, the mechanism regulating this alteration has not yet been elucidated. It has been shown that chromatin structure and epigenetic factors change dynamically between the one- and two-cell stages. In this article, we review the characteristics of transcription, chromatin structure, and epigenetic factors in one- and two-cell mouse embryos and discuss the involvement of chromatin structure and epigenetic factors in the alteration of transcription that occurs between these stages.
Keywords: Chromatin structure, Epigenetic factors, Preimplantation development, Zygotic gene activation
Upon fertilization, an oocyte and a spermatozoon fuse to form a one-cell embryo. The maternal genome, derived from the oocyte, undergoes chromosomal segregation to extrude half of the chromosomes as a second polar body. In the paternal genome, derived from the spermatozoon, protamine, which constitutes the compact chromatin structure, is removed and replaced with maternally supplied histones to establish nucleosomes. The paternal and maternal genomes independently form pronuclei in one-cell embryos. Syngamy occurs during the M phase, where a nucleus containing both the maternal and paternal genomes is formed after cleavage into a two-cell embryo. During the one- and two-cell stages after fertilization, dynamic alterations in transcription, chromatin structure, and epigenetic factors occur, which are referred to as gene expression reprogramming and genome remodeling. In this review, we outline these alterations and discuss the involvement of chromatin structure and epigenetic factors in the regulation of transcription. We focus on mouse embryos because the mouse is currently the most studied species in terms of gene expression and epigenetic factor during the early preimplantation stage.
Characteristics of Transcription in One- and Two-cell Embryos
Dramatic changes in gene expression occur after fertilization. In oocytes, genes are actively transcribed during the growth phase, but transcription ceases once the oocytes are fully grown. After fertilization, transcription restarts during the mid S-phase of the one-cell stage, but the transcriptional activity is low. A drastic activation of gene expression is seen during the mid-to-late two-cell stage. Therefore, the relatively low levels of gene activation at the one-cell stage and the high levels at the two-cell stage are referred to as minor and major zygotic gene activation (ZGA), respectively (Table 1) [1, 2]. Gene expression patterns are very different between these two phases [3]. During minor ZGA, low levels of transcription occur for more than 90% of all genes [4, 5], as well as from intergenic regions including retrotransposons [3]. However, during major ZGA, the percentage of transcribed genes decreases to less than 80%, but the levels of transcription of some genes increase greatly [4, 5]; furthermore, transcription from the intergenic regions decreases [3]. The regulation of transcription is different between the one- and two-cell stages (Table 1). Reporter gene analysis has revealed that transcription does not require an enhancer but only a core promoter at the one-cell stage; however, a proximal promoter and enhancers are required for transcription at the two-cell stage [3, 6, 7]. This type of transcriptional regulation, being dependent on only a core promoter, seems to induce cryptic initiation of transcription and was observed in embryos at the one-cell, but not two-cell, stage [3]. Thus, the transition from promiscuous transcription across the genome to more regulated transcription occurs between the one- and two-cell stages.
Table 1. Summary of the differences in transcription, chromatin structure, and epigenetic factors between one- and two-cell embryos.
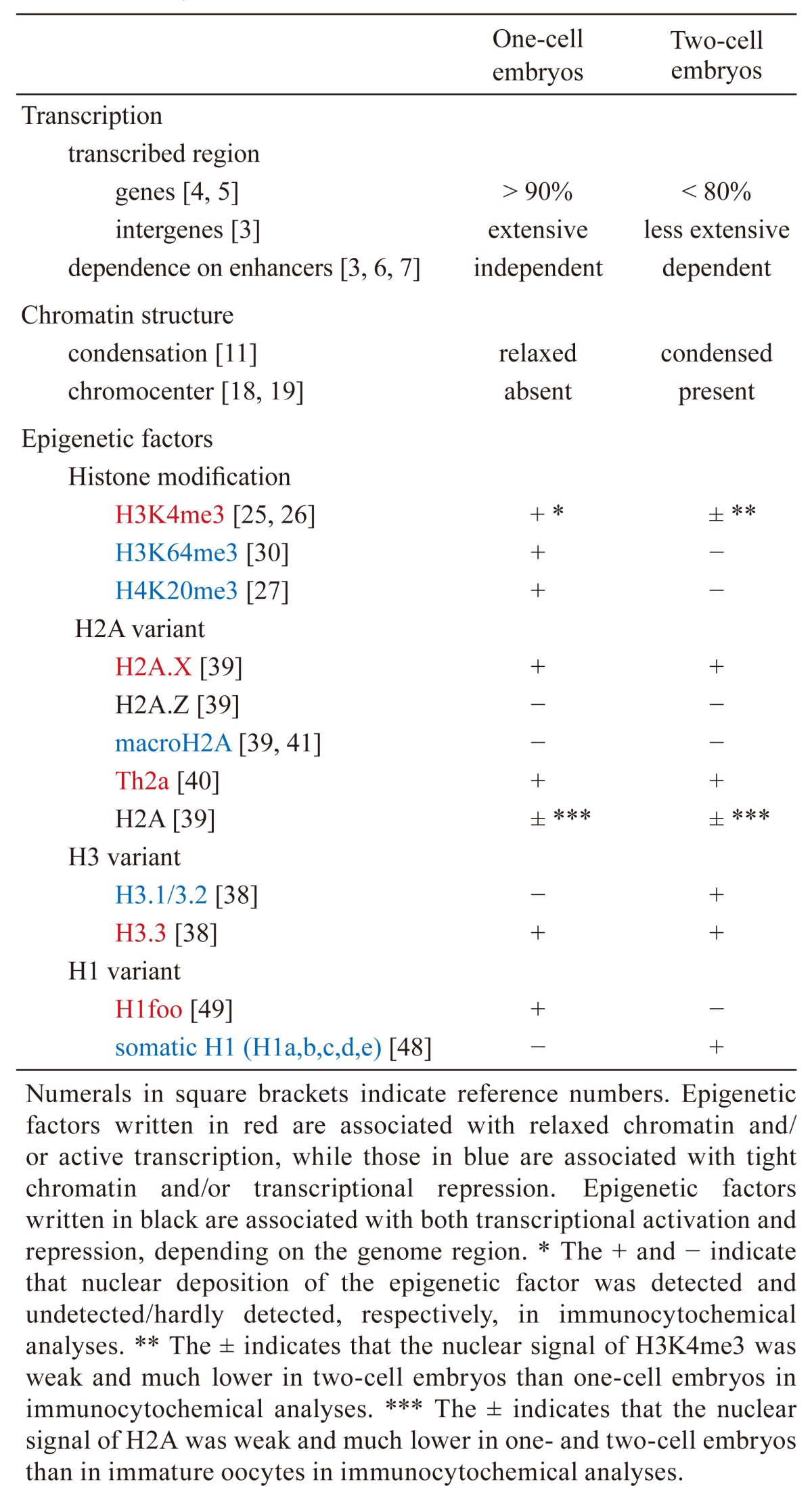
In one-cell embryos, the pattern and regulation of transcription are asymmetric between the male and female pronuclei (Table 2). It has been demonstrated through an in vitro transcription assay that the male pronucleus has higher transcriptional activity than the female pronucleus [1]. In a reporter gene analysis, the male pronucleus showed transcriptional activity in the absence of an enhancer [3, 6] and the addition of an enhancer did not affect its activity [7]. On the other hand, the presence of an enhancer increased the transcriptional activity of the female pronucleus, but the degree of this increase was much smaller than that seen in two-cell embryos [7, 8]. Therefore, the degree of dependence on enhancers in the female pronucleus seems to be intermediate between the male pronucleus and the two-cell embryo. RNA sequencing analysis has revealed many genes with differential expression between parthenogenetic embryos and normal embryos [9], suggesting that the gene expression pattern is different between male and female pronuclei. It has also been reported that major satellites are more transcribed in the male pronucleus than the female one [10].
Table 2. Summary of the differences in transcription, chromatin structure, and epigenetic factors between the male and female pronuclei.
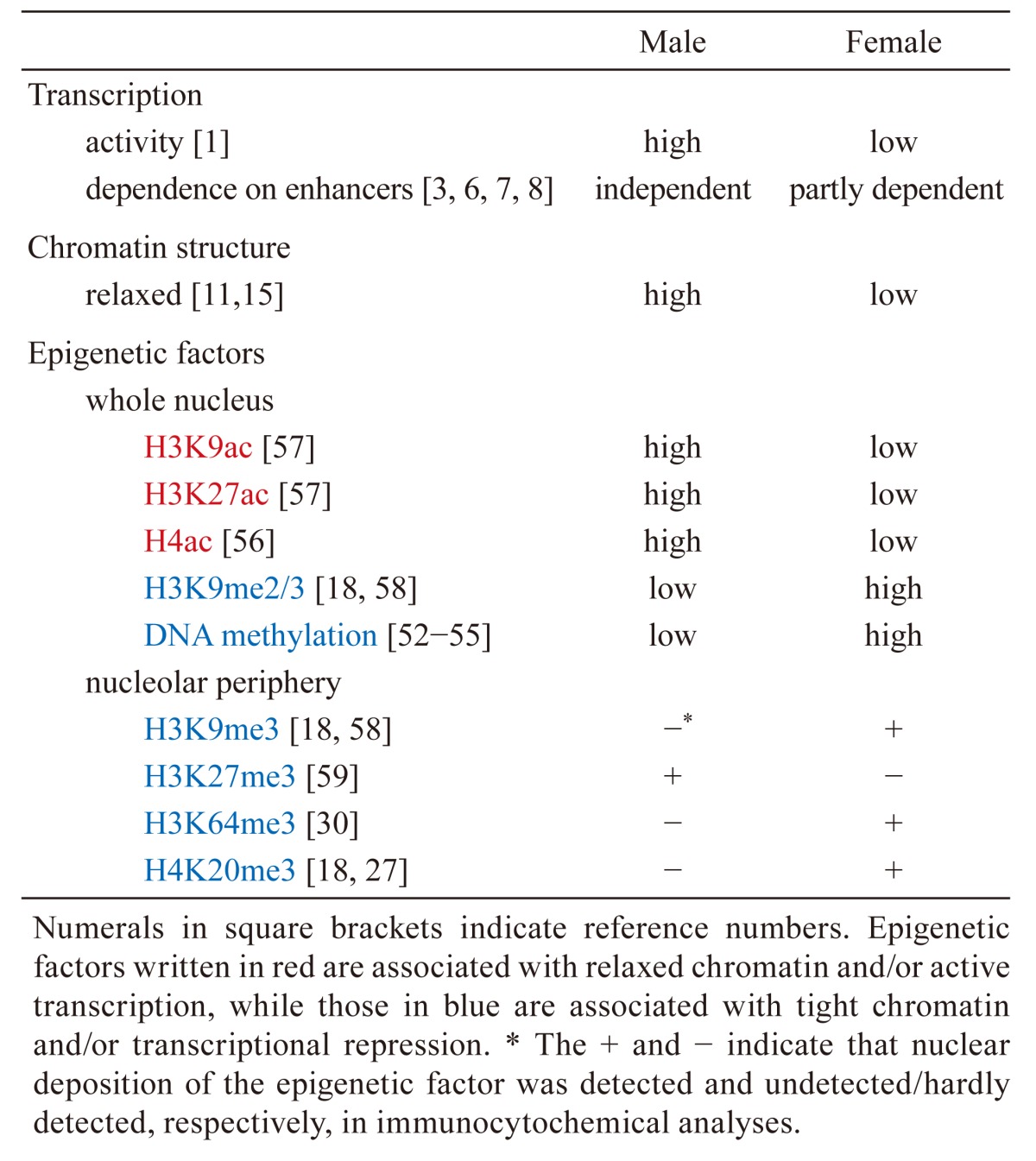
Chromatin Structure in One- and Two-cell Embryos
Several studies have shown that the chromatin structure changes drastically from the one- to two-cell stage (Table 1). Fluorescence recovery after photobleaching (FRAP) has demonstrated that the mobility of histones is extremely high in one-cell embryos and decreases in two-cell embryos [11]. Since the mobility of histones and the degree of chromatin looseness are positively correlated [12, 13], it is likely that the chromatin structure is extremely relaxed in one-cell embryos and then becomes more condensed in two-cell embryos. Electron spectroscopic imaging has also shown that the chromatin is highly dispersed in one-cell stage embryos but becomes more condensed in two-cell stage embryos [14]. Interestingly, FRAP has also indicated that the level of relaxation is different between the parental pronuclei (Table 2). Compared with the male pronucleus, histone mobility was lower in the female pronucleus but still higher than in two-cell embryos [11]. Thus, the degree of chromatin relaxation in the female pronucleus seems to be intermediate to those of the male pronucleus and two-cell embryo nucleus. Consistent with these results, chromatin was more sensitive to DNase I in the male pronucleus than in the female pronucleus [15], suggesting again that the chromatin structure is more relaxed in the male pronucleus than in the female pronucleus. A recent analysis of three-dimensional genome architecture using high-resolution chromosome conformation capture also revealed that the pattern of genome compartmentalization is different between the male and female pronuclei [16]. A genome-wide analysis of chromatin accessibility, using an assay for transposase-accessible chromatin in combination with high throughput sequencing, showed that chromatin obtained a wide open configuration within the transcribed region during minor ZGA [17]. Finally, several chromocenters, where pericentromeric heterochromatin congregates, can be observed as densely stained foci by DNA staining using 4′,6-diamidino-2-phenylindole and DNA-fluorescent in situ hybridization analysis of major satellites in somatic cells. No focus of chromocenter has been observed in the pronuclei of one-cell embryos [18, 19], but foci do appear at the late two-cell stage. Taken together, these results suggest that the chromatin structure is globally relaxed at the one-cell stage and becomes relatively condensed at the two-cell stage.
In general, relaxed chromatin is believed to facilitate the access of transcription factors to their target DNA sequences [20, 21]. This is supported by the finding that most upstream regions of active genes have a relaxed chromatin configuration [22], which may induce a low level of promiscuous transcription across the genome in one-cell embryos [5]. This hypothesis explains why enhancers are not required for transcription in one-cell embryos, since one of the functions of enhancers is to relax chromatin [23, 24] and the chromatin of one-cell embryos is already relaxed. At the two-cell stage however, the chromatin condenses and enhancers are required for transcription. Although major satellites are actively transcribed in one- and early two-cell stage embryos, chromocenters are organized in late two-cell stage embryos and the level of transcription from major satellite is rapidly decreased after the two-cell stage [19]. In addition, the male pronucleus of one-cell embryos has a looser chromatin structure and a lower degree of dependence on enhancers compared with the female pronucleus [7, 8, 11]. This more relaxed chromatin state seems to explain the difference in transcriptional regulation between the male and female pronuclei.
Epigenetic Factors in One- and Two-cell Embryos
In addition to transcription levels and chromatin structure, various epigenetic factors have been reported to change dramatically between the one- and two-cell stages (Table 1). Immunocytochemistry using an antibody against H3K4me3, which is involved in active transcription, revealed that the nuclear level of this modification decreases during the one- and two cell stages [25, 26]. H4K20me3 signals were observed in the pericentromeric regions of only the female pronucleus of one-cell embryos and disappeared at the two-cell stage [18, 27]. Ectopic expression of Suv4-20h2, which is a methyltransferase acting on H4K20me3, maintained the level of H4K20me3 during the one- and two-cell stages and induced developmental arrest, suggesting that the loss of H4K20me3 in one- and two-cell embryos is necessary for preimplantation development [28]. Nuclear localization of H3K64me3, which is involved in pericentromeric heterochromatin formation [29], developed a pattern similar to that of H4K20me3 in one- and two-cell embryos [30]. Finally, a genome-wide analysis of the distribution of H3K4me3 using chromatin immunoprecipitation (ChIP) has shown that H3K4me3 peaks in the vicinity of the transcription start sites (TSSs) of active genes in two-cell embryos and somatic cells. H3K4me3 peaks were absent in the TSSs of one-cell embryos but were instead present as broad signals covering vast regions [26, 31].
In addition to changes in histone modifications, histone variants change dramatically between the one- and two-cell stages (Table 1). There are three major non-centromeric histone H3 variants: H3.1, H3.2, and H3.3. H3.3 is deposited abundantly in the upstream regions of active genes [32,33,34] and is preferentially modified with H3K9ac, H3K14ac, and H3K79me2, which promote transcription [35, 36], suggesting that H3.3 is involved in the formation of relaxed chromatin and active transcription. On the other hand, H3.1/3.2 possess modifications involved in transcriptional repression, such as H3K9me2 and H3K27me2/3 [35, 37]. We previously analyzed the nuclear incorporation of flag-tagged H3 variants by microinjecting their encoding cRNAs into embryos and found that H3.2 and H3.3, but not H3.1, were efficiently incorporated into the pronucleus of one-cell stage embryos [38]. However, we have recently found that H3.3 is much more efficiently incorporated than H3.2 when the concentration of microinjected cRNA is reduced (unpublished data). We have also found by immunocytochemistry using an antibody against H3.1/3.2 that the nuclear level of H3.1/3.2 is very low at the one-cell stage and increases at the two-cell stage (unpublished data). In addition to H3 variants, H2A variants have also shown an uneven pattern of nuclear deposition in one- and two-cell embryos. In these embryos, only H2A.X and TH2A were detected clearly, whereas H2A.Z and macroH2A were not [39,40,41]. A low level of canonical H2A was detected when compared with oocytes [39]. In general, macroH2A is involved in the repression of transcription, whereas H2A.Z is abundantly localized in the promoter regions, but not the gene bodies, of active genes and is involved in transcriptional regulation [34, 42,43,44,45]. H2A.X is involved in DNA repair [46]. Although the function of H2A.X in the regulation of gene expression is not clear, it has been reported that H2A.X is abundantly localized in actively transcribed genes [34]. TH2A is involved in the formation of loose chromatin [40]. Therefore, the composition of H3.3/H2A.X and/or H3.3/TH2A in the nucleosome may be responsible for the formation of extremely relaxed chromatin in one-cell embryos.
Linker histone H1 plays an important role in the construction of higher-order chromatin structures. The linker histone H1 family consists of seven somatic H1 variants which are ubiquitously expressed in somatic cells and four germ cell-specific variants [47]. In immunocytochemical analyses for these variants in one-cell embryos, the somatic H1 variants H1a, b, c, d, and e were not detected in the pronuclei [48], while H1foo, an oocyte-specific H1 variant, was detected clearly [49]. However, the nuclear level of H1foo decreased drastically at the two-cell stage [49], whereas the nuclear level of somatic H1 variants increased [48]. Interestingly, it has been suggested that the somatic H1 variants and H1foo are involved in the formation of condensed and relaxed chromatin, respectively [50, 51].
Epigenetic factors show different patterns of localization between male and female pronuclei in one-cell embryos (Table 2). Previously, it was reported that the male, but not the female, pronucleus undergoes active DNA demethylation [52]; however, recent studies have revealed that a conversion of methyl to hydroxymethyl groups, rather than just a removal of methyl groups, occurs in the male pronuclei [53,54,55]. It has also been reported that various histone modifications are asymmetric between the male and female pronucleus. The levels of histone H4, H3K9, and K27 acetylation associated with active transcription are higher in the male pronucleus than in the female pronucleus [36, 56, 57]. On the other hand, the levels of histone methylation (H3K9me2/3) associated with transcriptional repression are lower in the male pronucleus compared with the female pronucleus [18, 37, 58]. The pericentromeric regions of the genome are localized in the periphery of the nucleolus in one-cell embryos. This region is enriched in H3K9me3, H3K64me3, and H4K20me3 in the female pronucleus but not in the male pronucleus [18, 27, 30, 58]. On the other hand, H3K27me3 is abundantly localized in this region of the male pronucleus only [59]. Regarding histone variants, H3.3 was found predominantly in the male pronucleus soon after fertilization [38].
Distinctive patterns of nuclear localization of various epigenetic factors seem to be involved in the establishment of unique characteristics of chromatin structure and transcription in one- and two-cell embryos. In one-cell embryos, the epigenetic factors that are associated with relaxed chromatin and active transcription, i.e. H3K4me3, H3.3, H2A.X, and H1foo, are abundantly deposited in the pronucleus, whereas macroH2A, H3.1, and H3.2, which are associated with condensed chromatin and gene repression, are absent or present at low levels. At the two-cell stage, the levels of H3.1 and H3.2 increase while those of H3K4me3 and H1foo decrease, which may condense the chromatin structure. In one-cell embryos, the absence of H3K4me3 peaks in the vicinity of the TSSs of active genes but instead having a broad signal presence covering vast regions may contribute to enhancer-independent and cryptic transcription. MacroH2A, which is associated with tight chromatin, is absent from the nucleus at the two-cell stage as well as from the pronucleus at the one-cell stage and then appears at the late preimplantation stage [39, 41]. Although macroH2A does not seem to be involved in the change in chromatin structure between the one- and two-cell stages, it may be involved in the formation of a tight chromatin structure in the late preimplantation stage. Notably, FRAP has shown that the chromatin structure in two-cell embryos is still much more relaxed than that seen in late-preimplantation-stage embryos [11], even though it is more condensed than that of one-cell embryos.
In one-cell embryos, the differences in chromatin structure and transcriptional activity between male and female pronuclei may be caused by asymmetric localization of certain epigenetic factors. DNA methylation and H3K9me2/3, which are associated with tight chromatin and a transcriptionally repressive state, were detected in the female pronucleus only. This may induce more condensed chromatin and lower levels of transcriptional activity in the female pronucleus than in the male pronucleus. Furthermore, asymmetric localization of H3K9me3, H3K64me3, and H4K20me3 in the periphery of the nucleolus may be associated with the different levels of transcription from major satellites between the parental pronuclei.
Future Prospects
As described above, epigenetic factors are altered dramatically from the one- to two-cell stage. However, little is known about the precise roles of these epigenetic factors in the regulation of transcription, chromatin structure, and development in one- and two-cell embryos, since most studies have examined the changes in these factors but not their functions. An analysis of the transcriptional activity and/or chromatin structure of embryos with either a knockdown or knockout of epigenetic factors of interest is needed. The recent development of the CRISPR-Cas9 system will facilitate the generation of knockout animals for various epigenetic factors. Furthermore, recent technical advances have enabled ChIP-seq experiments using a smaller number of cells. Using these technologies, knowledge gained regarding the genome-wide distribution of various epigenetic factors will help elucidate the whole picture of chromatin structure and transcriptional regulation during the one- and two-cell stages.
Acknowledgments
This work was supported in part by Grants-in-Aid (to FA) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (#16H01215, #25252054 and #20175160).
References
- 1.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 181: 296–307. [DOI] [PubMed] [Google Scholar]
- 2.Schultz RM. Regulation of zygotic gene activation in the mouse. BioEssays 1993; 15: 531–538. [DOI] [PubMed] [Google Scholar]
- 3.Abe K, Yamamoto R, Franke V, Cao M, Suzuki Y, Suzuki MG, Vlahovicek K, Svoboda P, Schultz RM, Aoki F. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3 processing. EMBO J 2015; 34: 1523–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto R, Abe K, Suzuki Y, Suzuki MG, Aoki F. Characterization of gene expression in mouse embryos at the 1-cell stage. J Reprod Dev 2016; 62: 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto R, Aoki F. A unique mechanism regulating gene expression in 1-cell embryos. J Reprod Dev 2017; 63: 9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamamoto G, Suzuki T, Suzuki MG, Aoki F. Regulation of transketolase like 1 gene expression in the murine one-cell stage embryos. PLoS ONE 2014; 9: e82087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiekowski M, Miranda M, DePamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Dev Biol 1993; 159: 366–378. [DOI] [PubMed] [Google Scholar]
- 8.Majumder S, Zhao Z, Kaneko K, DePamphilis ML. Developmental acquisition of enhancer function requires a unique coactivator activity. EMBO J 1997; 16: 1721–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park SJ, Komata M, Inoue F, Yamada K, Nakai K, Ohsugi M, Shirahige K. Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev 2013; 27: 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, van Lohuizen M, Peters AH. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet 2008; 40: 411–420. [DOI] [PubMed] [Google Scholar]
- 11.Ooga M, Fulka H, Hashimoto S, Suzuki MG, Aoki F. Analysis of chromatin structure in mouse preimplantation embryos by fluorescent recovery after photobleaching. Epigenetics 2016; 11: 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin RM, Cardoso MC. Chromatin condensation modulates access and binding of nuclear proteins. FASEB J 2010; 24: 1066–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chalut KJ, Höpfler M, Lautenschläger F, Boyde L, Chan CJ, Ekpenyong A, Martinez-Arias A, Guck J. Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells. Biophys J 2012; 103: 2060–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS ONE 2010; 5: e10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho T, Sakai S, Nagata M, Aoki F. Involvement of chromatin structure in the regulation of mouse zygotic gene activation. Anim Sci J 2002; 73: 113–122. [Google Scholar]
- 16.Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 2017; 544: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, Li W, Li Y, Ma J, Peng X, Zheng H, Ming J, Zhang W, Zhang J, Tian G, Xu F, Chang Z, Na J, Yang X, Xie W. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 2016; 534: 652–657. [DOI] [PubMed] [Google Scholar]
- 18.Probst AV, Santos F, Reik W, Almouzni G, Dean W. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma 2007; 116: 403–415. [DOI] [PubMed] [Google Scholar]
- 19.Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell 2010; 19: 625–638. [DOI] [PubMed] [Google Scholar]
- 20.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 1993; 72: 73–84. [DOI] [PubMed] [Google Scholar]
- 21.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol 2011; 12: 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell 2008; 132: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majumder S, Miranda M, DePamphilis ML. Analysis of gene expression in mouse preimplantation embryos demonstrates that the primary role of enhancers is to relieve repression of promoters. EMBO J 1993; 12: 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem 2003; 72: 449–479. [DOI] [PubMed] [Google Scholar]
- 25.Bošković A, Bender A, Gall L, Ziegler-Birling C, Beaujean N, Torres-Padilla ME. Analysis of active chromatin modifications in early mammalian embryos reveals uncoupling of H2A.Z acetylation and H3K36 trimethylation from embryonic genome activation. Epigenetics 2012; 7: 747–757. [DOI] [PubMed] [Google Scholar]
- 26.Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, Preissl S, Jermstad I, Haugen MH, Suganthan R, Bjørås M, Hansen K, Dalen KT, Fedorcsak P, Ren B, Klungland A. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016; 537: 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kourmouli N, Jeppesen P, Mahadevhaiah S, Burgoyne P, Wu R, Gilbert DM, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, Shi W, Fundele R, Singh PB. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J Cell Sci 2004; 117: 2491–2501. [DOI] [PubMed] [Google Scholar]
- 28.Eid A, Rodriguez-Terrones D, Burton A, Torres-Padilla ME. SUV420 activity in the preimplantation mouse embryo controls timely replication. Genes Dev 2016; 30: 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange UC, Siebert S, Wossidlo M, Weiss T, Ziegler-Birling C, Walter J, Torres-Padilla ME, Daujat S, Schneider R. Dissecting the role of H3K64me3 in mouse pericentromeric heterochromatin. Nat Commun 2013; 4: 2233. [DOI] [PubMed] [Google Scholar]
- 30.Daujat S, Weiss T, Mohn F, Lange UC, Ziegler-Birling C, Zeissler U, Lappe M, Schübeler D, Torres-Padilla ME, Schneider R. H3K64 trimethylation marks heterochromatin and is dynamically remodeled during developmental reprogramming. Nat Struct Mol Biol 2009; 16: 777–781. [DOI] [PubMed] [Google Scholar]
- 31.Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, Liu W, Kou X, Zhao Y, He W, Li C, Chen B, Li Y, Wang Q, Ma J, Yin Q, Kee K, Meng A, Gao S, Xu F, Na J, Xie W. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 2016; 537: 553–557. [DOI] [PubMed] [Google Scholar]
- 32.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark nucleosome-free regions of active promoters and other regulatory regions. Nat Genet 2009; 41: 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010; 140: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yukawa M, Akiyama T, Franke V, Mise N, Isagawa T, Suzuki Y, Suzuki MG, Vlahovicek K, Abe K, Aburatani H, Aoki F. Genome-wide analysis of the chromatin composition of histone H2A and H3 variants in mouse embryonic stem cells. PLoS ONE 2014; 9: e92689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem 2006; 281: 559–568. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence M, Daujat S, Schneider R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet 2016; 32: 42–56. [DOI] [PubMed] [Google Scholar]
- 37.Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the H3 barcode hypothesis. Proc Natl Acad Sci USA 2006; 103: 6428–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akiyama T, Suzuki O, Matsuda J, Aoki F. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet 2011; 7: e1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development 2010; 137: 3785–3794. [DOI] [PubMed] [Google Scholar]
- 40.Shinagawa T, Takagi T, Tsukamoto D, Tomaru C, Huynh LM, Sivaraman P, Kumarevel T, Inoue K, Nakato R, Katou Y, Sado T, Takahashi S, Ogura A, Shirahige K, Ishii S. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell 2014; 14: 217–227. [DOI] [PubMed] [Google Scholar]
- 41.Chang CC, Ma Y, Jacobs S, Tian XC, Yang X, Rasmussen TP. A maternal store of macroH2A is removed from pronuclei prior to onset of somatic macroH2A expression in preimplantation embryos. Dev Biol 2005; 278: 367–380. [DOI] [PubMed] [Google Scholar]
- 42.Angelov D, Molla A, Perche PY, Hans F, Côté J, Khochbin S, Bouvet P, Dimitrov S. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell 2003; 11: 1033–1041. [DOI] [PubMed] [Google Scholar]
- 43.Perche PY, Vourch C, Konecny L, Souchier C, Robert-Nicoud M, Dimitrov S, Khochbin S. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr Biol 2000; 10: 1531–1534. [DOI] [PubMed] [Google Scholar]
- 44.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129: 823–837. [DOI] [PubMed] [Google Scholar]
- 45.Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 2012; 151: 1608–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004; 3: 959–967. [DOI] [PubMed] [Google Scholar]
- 47.Pan C, Fan Y. Role of H1 linker histones in mammalian development and stem cell differentiation. Biochim Biophys Acta 2016; 1859: 496–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu G, Ghadam P, Sirotkin A, Khochbin S, Skoultchi AI, Clarke HJ. Mouse oocytes and early embryos express multiple histone H1 subtypes. Biol Reprod 2003; 68: 1569–1576. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka M, Hennebold JD, Macfarlane J, Adashi EY. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development 2001; 128: 655–664. [DOI] [PubMed] [Google Scholar]
- 50.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 2005; 123: 1199–1212. [DOI] [PubMed] [Google Scholar]
- 51.Kunitomi A, Yuasa S, Sugiyama F, Saito Y, Seki T, Kusumoto D, Kashimura S, Takei M, Tohyama S, Hashimoto H, Egashira T, Tanimoto Y, Mizuno S, Tanaka S, Okuno H, Yamazawa K, Watanabe H, Oda M, Kaneda R, Matsuzaki Y, Nagai T, Okano H, Yagami K, Tanaka M, Fukuda K. H1foo has a pivotal role in qualifying induced pluripotent stem cells. Stem Cell Rep 2016; 6: 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature 2000; 403: 501–502. [DOI] [PubMed] [Google Scholar]
- 53.Iqbal K, Jin SG, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA 2011; 108: 3642–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun 2011; 2: 241. [DOI] [PubMed] [Google Scholar]
- 55.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 2011; 334: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 1997; 124: 4615–4625. [DOI] [PubMed] [Google Scholar]
- 57.Hayashi-Takanaka Y, Yamagata K, Wakayama T, Stasevich TJ, Kainuma T, Tsurimoto T, Tachibana M, Shinkai Y, Kurumizaka H, Nozaki N, Kimura H. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res 2011; 39: 6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol 2005; 280: 225–236. [DOI] [PubMed] [Google Scholar]
- 59.Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol 2010; 12: 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]


