Abstract
Although more than 100 imprinted genes have already been identified in the mouse and human genomes, little is known about genomic imprinting in cattle. For a better understanding of these genes in cattle, parthenogenetically activated bovine blastocysts were transferred to recipient cows to obtain parthenotes, and fibroblasts derived from a Day 40 (Day 0 being the day of parthenogenetic activation) parthenogenetic embryo (BpEFs) were successfully obtained. Bovine embryonic fibroblasts (BEFs) were also isolated from a normal fertilized embryo obtained from an artificially inseminated cow. The expression of imprinted genes was analyzed by RT-PCR. Paternally expressed genes (PEGs) in mouse (viz., IGF2, PEG3, ZAC1, NDN, DLK1, SGCE, and PEG10) were expressed in BEFs, but not in BpEFs, suggesting that these genes are also imprinted in cattle. However, other PEGs in mouse (viz., IMPACT, MAGEL2, SNRPN, and PEG1/MEST) were expressed in both BEFs and BpEFs. These genes may not be imprinted in BEFs. The expression of seven maternally expressed genes in mouse was also analyzed, and only CDKN1C was not expressed in BpEFs. The DNA methylation patterns of repetitive elements (Satellite I, Satellite II, alpha-satellite, and Art2) were not different between the BEFs and BpEFs; however, the differentially methylated region (DMR) of paternally methylated H19 was hypomethylated, whereas those of maternally methylated PEG3 and PEG10 were hypermethylated in BpEFs, as expected. The methylation of the SNRPN DMR was not different between the BEFs and BpEFs, in accordance with the SNRPN expression levels in both cell types. The XIST gene, which is essential for X chromosome inactivation in females, was expressed in BpEFs, whereas its DMR was half-methylated, suggesting that X chromosome inactivation is normal in these cells. Microarray analysis was also applied to identify novel PEGs that should be expressed only in BEFs but not in BpEFs. More than 300 PEG candidate genes, including IGF2, PEG3, and PEG10, were obtained. These results illustrate the epigenetic characteristic of bovine parthenogenetic embryos and contribute to the identification of novel imprinted genes in cattle.
Keywords: Bovine, DNA methylation, Embryonic fibroblast, Imprinted genes, Parthenotes
It is known that parental genomes are not functionally equivalent in mammalian species, and both paternal and maternal genomes are necessary for proper embryonic development to term [1, 2]. This phenomenon is called genomic imprinting, which is a mammalian-specific gene expression regulatory system causing paternal- or maternal-specific monoallelic gene expression [3]. More than 100 imprinted genes have been found and confirmed in both the mouse and the human genome to date; however, only 20 imprinted genes have been identified in cattle (http://www.geneimprint.com/site/genes-by-species). Imprinted genes are important for embryonic/postnatal development, placentation, metabolism, animal behavior, and numerous physiological functions [4]. The molecular mechanism controlling imprinted monoallelic gene expression is mainly DNA methylation and histone modifications. It is known that the paternal genome is methylated in spermatogenesis, and the maternal genome is methylated in oogenesis. After fertilization, the allele-specific DNA methylation mark is maintained during the developmental stages and erased in primordial germ cells to establish another methylation imprint mark (depending on the sex) and then passed on to the next generation [5]. Because of this cycle, some imprinted genes are methylated only in sperm, and others are methylated only in oocytes. As a parthenogenetic embryo is derived from the oocyte without fertilization, the methylation patterns in the parthenogenetic embryo are supposed to be similar to those of oocytes. Consequently, only imprinted genes of the mother are expressed, whereas those of the father are not expressed in parthenogenetic embryos. For that reason, parthenogenetic and androgenetic embryos were used for identifying novel imprinted genes in the mouse genome, and several paternally expressed genes (PEGs) and maternally expressed genes (MEGs) were discovered [6, 7]. In cattle, Hansmann et al. [8] used parthenogenetic embryos for characterizing the bovine intergenic IGF2-H19 imprinting control region. However, most of the studies on imprinted gene expression using bovine parthenogenetic embryos focused on preimplantation development up to the blastocyst stage in vitro [9, 10]. Only a few studies have analyzed the developmental potential of bovine parthenogenetic embryos after implantation [11, 12], and the molecular characteristics are still unanswered.
It has been reported that the murine parthenogenetic embryo develops to around embryonic Day 9.5, with poor placentation and heart beating, but dies soon after that stage, and no murine parthenogenetic embryo has developed to term [1, 2, 13, 14]. Kono et al. [15] succeeded in producing the first parthenogenetic mouse, called “Kaguya,” by manipulating the genetically modified maternal genome. In cattle, Fukui et al. [11] reported that in vitro-matured parthenogenetic embryos could develop to 48 days after their transfer to heifers. In pigs, Kure-Bayashi et al. [16] obtained heart-beating fetuses at 29 days after activation. Wang et al. [17] reported the expression of imprinted genes and DNA methylation patterns in porcine parthenogenetic fetuses and placentas. However, no parthenogenetic embryonic fibroblast has been isolated and analyzed in cattle.
In this study, we isolated bovine embryonic fibroblasts from an embryonic Day 40 (D40) parthenogenetic embryo (BpEFs) and a normal embryo (BEFs), and analyzed their imprinted gene expression patterns and DNA methylation levels. We also applied a DNA microarray for identifying novel bovine PEG candidates, which are theoretically not expressed in parthenogenetic embryos.
Materials and Methods
Culture of bovine embryonic fibroblasts
The embryonic D40 parthenogenetic embryo was produced and isolated as described previously [18, 19]. Oocytes were collected from slaughterhouse-derived ovaries (Holstein or Japanese Black cows). The normal D40 embryo was obtained from the slaughterhouse. For both types of embryos, extraembryonic tissues were not collected. The embryos were minced aseptically with a pair of sterile ophthalmic scissors and then trypsinized to single cells with 0.05% trypsin/ethylenediaminetetraacetic acid (EDTA) solution (Thermo Fisher Scientific, San Jose, CA, USA) for 15 min at 37°C. The cells were incubated in Dulbecco’s modified Eagle’s medium (Wako Pure Chemical Industries, Osaka, Japan) supplemented with 10% fetal bovine serum and a penicillin/streptomycin solution (Thermo Fisher Scientific) at 37°C in a humidified atmosphere with 5% CO2. The total cell number and dead/live cell number were calculated using the Tali™ Image-Based Cytometer (Thermo Fisher Scientific) after application of the Tali® Viability Kit - Dead Cell Red (Thermo Fisher Scientific). The cells were passaged using 0.05% trypsin/EDTA solution before reaching confluence. All animal experiments were approved by the Committee for the Care and Use of Experimental Animals at the Institute of Livestock and Grassland Science, Japan.
DNA methylation analysis
Genomic DNA was extracted by using the NucleoSpin® Tissue Kit (TaKaRa Bio, Shiga, Japan) according to the manufacturer’s instructions. Bisulfite conversion was carried out by using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s instructions. The PCR amplifications were performed in a 20 µl volume containing 50 pmol of each primer (Table 1), using the EmeraldAmp® PCR Master Mix (TaKaRa Bio), at the following conditions: 94°C for 2 min, followed by 30 cycles of 94°C for 30 sec, 45–55°C (depending on the primer sets; see Table 1) for 30 sec, and 72°C for 30 sec. Differentially methylated regions (DMRs) of PEG3, SNRPN, PEG10, XIST, NANOG, and OCT4 were amplified by nested PCR assay (2 µl of the first PCR solution was used as a template for the second round of PCR). For bisulfite sequencing analysis, the amplified PCR products were cloned into the pGEM-T-easy vector (Promega, Madison, WI, USA) and then sent for sequencing (Greiner Bio-One, Frickenhausen, Germany). At least 12 clones were sequenced from each sample. Sequenced clones were analyzed with the QUMA (QUantification for Methylation Analysis) program [20]. The Mann-Whitney U-test was used for statistical analysis, and P < 0.05 denoted a statistically significant difference.
Table 1. Primer sequences and annealing temperature used in this study.
| Gene | Forward and reverse primer sequences | Product (bp) | Annealing (ºC) |
| Primers for RT-PCR | |||
| IGF2 | 5'ACCCTCCAGTTTGTCTGTGG3' | 349 | 60 |
| 5'GGTGACTCTTGGCCTCTCTG3' | |||
| PEG3 | 5'CTTCGCGGTCATTTCTGAGT3' | 282 | 60 |
| 5'TTGTCCTTGCCGTACATCTTC3' | |||
| ZAC1 | 5'GGGAAGAAGTACAACACCATGC3' | 249 | 63 |
| 5'CTGTGTGGACCACCAGGT3' | |||
| NDN | 5'GTGAARGATGTCATCGGCAG3' | 590 | 60 |
| 5'GTCCTCWGAGACACTGYTGC3' | |||
| DLK1 | 5'GTGACCAGTGCGTGACCTTT3' | 454 | 54 |
| 5'GCAGGTCTTGTCCATGAAGC3' | |||
| SGCE | 5'CCCGTTACCCTATCAAGCAG3' | 557 | 56 |
| 5'GGCAGCACATGATATAAGCG 3' | |||
| PEG10 | 5'TCAACCTTTTGGGGGCTGTT3' | 284 | 60 |
| 5'GGGGTAAGAAGAAGGGCCAC3' | |||
| IMPACT | 5'TGGCGAGGAGTGGTGTGTCA3' | 594 | 68 |
| 5'GGCATAGATGTTGTGGGTGG3' | |||
| MAGEL2 | 5'CTGATGGTGGTTCTGAGCCT3' | 257 | 60 |
| 5'CAGGACAATCATCTTGCTGG3' | |||
| SNRPN | 5'TGGGAAGGAGCAGCAAGGTG3' | 532 | 62 |
| 5'TGGTCAACTGATGGTGGCGG3' | |||
| PEG1/MEST | 5'CGCCGAGATCGTCTCCGCAG3' | 377 | 58 |
| 5'CTCCACGATGCTGGCCTGCTC3' | |||
| IGF2R | 5'CTTGGCGGACCGGCACTTCAACTACACCTCACTGA3' | 282 | 60 |
| 5'CCTGCGGCTGCGGTGCACACCCCCACACTGTAG3' | |||
| H19 | 5'GATGGTGCTACCCAGCTCAT3' | 231 | 60 |
| 5'CCTTCCAGAGCTGATTCCTG3' | |||
| GNAS | 5'GAAGGACAAGCAGGTCTACC3' | 675 | 60 |
| 5'GACCATGTTGTAGCTGCTG 3' | |||
| UBE3A | 5'GGAGTTGATGAGGGAGGTGTT3' | 635 | 58 |
| 5'TCTGTAGTTTCTTCTAGTGCTTGGA3' | |||
| GRB10 | 5'GAAGATGGGACAAGCAAAGT3' | 290 | 58 |
| 5'CTGGCACCAAGTAACCATCTG3' | |||
| GTL2 | 5'CCCACCAGCAAACAAAGCAAC3' | 174 | 60 |
| 5'CATCAAGGCAAAAAGCACATCG3' | |||
| CDKN1C | 5'AGAGCCTCGTGCTCAAAGAG3' | 137 | 60 |
| 5'TTTAAACACGAAACCGAACG3' | |||
| beta-actin | 5'CAGAAGGACTCGTACGTGGG3' | 200 | 60 |
| 5'TTGGCCTTAGGGTTCAGGG3' | |||
| XIST | 5'AGCATTGCTTAGCATGGCTC' | 365 | 60 |
| 5'TGGCTGTGACCGATTCTACC3' | |||
| VIMENTIN | 5'AAATCCAGGAGCTTCAGGCC3' | 501 | 60 |
| 5'CCTCTCCTTCCAGCAGCTTC3' | |||
| CYTOKERATIN18 | 5'CCGCATCGTTCTGCAGATTG3' | 551 | 60 |
| 5'CATATCGGGCCTCCACTTCC3' | |||
| OCT4 | 5'GGCAAACGATCAAGCAGTG3' | 317 | 60 |
| 5'TAATCCCAAAGGCCTGGTAC3' | |||
| NANOG | 5'GTGTTTGGTGAACTCTCCTG3' | 307 | 60 |
| 5'GGGAATTGAAATACTTGACAG3' | |||
| SOX2 | 5'GGTTGACATCGTTGGTAATTTATAATAGC3' | 88 | 60 |
| 5'CACAGTAATTTCATGTTGGTTTTTCA3' | |||
| Primers for Bisulfite PCR | |||
| Satellite I | 5'AATACCTCTAATTTCAAACT3' | 211 | 46 |
| 5'TTTGTGAATGTAGTTAATA3' | |||
| Satellite II | 5'CAACCCATAATCAATAAACTC3' | 297 | 46 |
| 5'GTTGAGGTAGTAGTTAGGTA3' | |||
| apha-satellite | 5'AATAATTCCACATTCCRTAAAACCC3' | 153 | 55 |
| 5'GATGTTTTYGGGGAGAGAGG3' | |||
| Art2 | 5'TTAAATTCAATTCAATCACTCAATCA3' | 224 | 55 |
| 5'TTTATGTGAAGAGTTGATTTATTGGA3' | |||
| H19 DMR | 5'TTTTGTGGATTATTGTGGTATT3' | 198 | 55 |
| 5'ATCTTAAACTAATCTCCCAACCC3' | |||
| PEG3 DMR (1st) | 5'GGAGGAAGAAGTTGGAGTAGA3' | – | 51 |
| 5'CCCCTACCCAAAATAATCAAC3' | |||
| PEG3 DMR (2nd) | 5'GATATGTTTATTTTTGGTTGTTGG3' | 280 | 51 |
| 5'ACCCTAATCCCAAACTCCAACT3' | |||
| PEG10 DMR (1st) | 5'GGATATGAGTTATAGATTAATTT3' | – | 45 |
| 5'CACTCATCAATCTATAATTCATA3' | |||
| PEG10 DMR (2nd) | 5'GTAGATGTGTTGTAAAGTGATATT3' | 267 | 47 |
| 5'ACAATTCATACTAATTTCAATAC3' | |||
| SNRPN DMR (1st) | 5'GGAAAGTTTGAGGAAATTTGAATAAGG3' | – | 55 |
| 5'CAAATACCCCCAAAACCTAACAAAA3' | |||
| SNRPN DMR (2nd) | 5'TTGGGAGGTATTATTTTGGGTTGAAG3' | 548 | 55 |
| 5'AAAAAATCAATCCAACCCCAAACCTC3' | |||
| XIST DMR (1st) | 5'GGGTGTTTTTGTTTTAGTGTGTAGTA3' | – | 51 |
| 5'CTTTAATACCACCCACTAAAATTAATAC3' | |||
| XIST DMR (2nd) | 5'TTGTTATATAGTAAAAGATGGT3' | 405 | 46 |
| 5'ACCAATCCTAACTAACTAAATA3' | |||
| NANOG DMR (1st) | UAGAGTGAATTAAAGAGGAAAATGG3' | – | 51 |
| 5'TATAAAAATAAAAACCATCCAATCCA3' | |||
| NANOG DMR (2nd) | 5'GTAGTTTTTGTTATAAATTAGTTTGA3' | 361 | 51 |
| 5'AAATAAAACTCAACCATACTTAACC3' | |||
| OCT4 DMR (1st) | 5'GGGTGGAGAGTAATTTTGAGGG3' | – | 51 |
| 5'TAATACTAACTAATAATAAATAACC3' | |||
| OCT4 DMR (2nd) | 5'GAAGTTGGATAAGGAGAAGTTGGAG3' | 316 | 51 |
| 5'AATAAAAAAACCTACTTAACAAAAACC3' | |||
Gene expression analysis
Total RNA was isolated by using the NucleoSpin® RNA Kit (TaKaRa Bio) according to the manufacturer’s instructions. A 500 ng sample of the RNA was used for cDNA synthesis, carried out with the PrimeScript™ RT Reagent Kit (TaKaRa Bio). The cDNA (2 µl) was mixed with EmeraldAmp® PCR Master Mix (TaKaRa Bio) containing 50 pmol of each primer (Table 1), and RT-PCR was performed under the following conditions: 94°C for 2 min, followed by 30 cycles of 94°C for 30 sec, 54–68°C (depending on the primer sets; see Table 1) for 30 sec, and 72°C for 30 sec, with a final extension at 72°C for 5 min. The expression patterns were visualized by 2% agarose gel electrophoresis.
For the microarray analysis, total RNA was isolated by using the miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNase treatment was carried out with the TURBO DNA-free™ Kit (Thermo Fisher Scientific). The RNA samples were then sent to the Chemicals Evaluation and Research Institute (Tokyo, Japan), and Agilent Bovine DNA Microarray 44K (G2519F#23647) v2.0 (Agilent Technologies, Santa Clara, CA, USA) was used for the microarray analysis. GeneSpring GX 14.5 software (Agilent Technologies) was used for the normalization and statistical analysis.
Results
Cell growth analysis
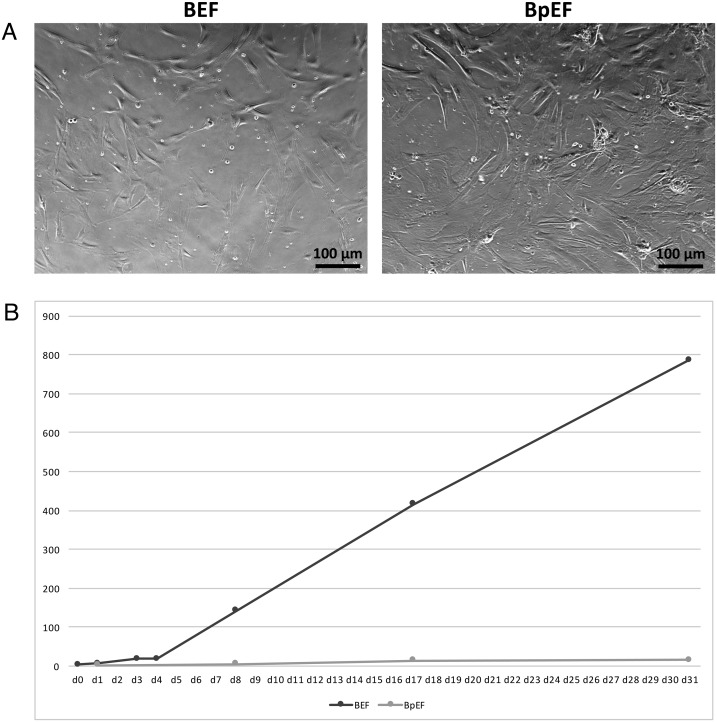
A bovine parthenogenetic embryo of approximately 2.5 cm in length was obtained at 40 days after parthenogenetic activation (Fig. 1), from which BpEFs were isolated. These cells and normal BEFs were separately cultured and passaged before reaching confluence. The morphology of both cell types was very similar, being bipolar/multipolar and elongated in shape and attached to the bottom of the culture dish (Fig. 2A). The number of live cells was counted and is visualized in Fig. 2B. Obviously, the growth rate of the BpEFs was much slower than that of the BEFs. After 6–7 passages, the BpEFs began to die.
Fig. 1.
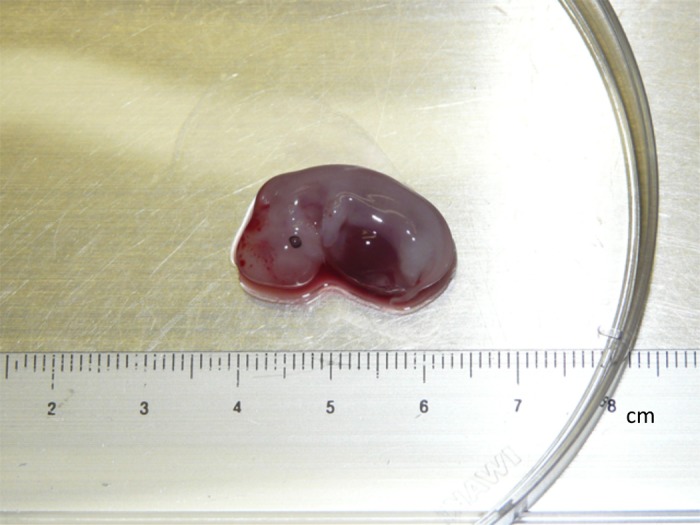
Image of a Day 40 (D40) parthenogenetic embryo.
Fig. 2.
Culture of bovine embryonic fibroblasts (BEFs) and bovine parthenogenetic embryonic fibroblasts (BpEFs) in vitro. (A) Bright field microscopic images of BEFs (left) and BpEFs (right). (B) Growth of BEFs and BpEFs in maintenance culture. BpEFs grew much slower than BEFs. X-axis: culture days; Y-axis: × 106 cells.
Gene expression analysis by RT-PCR
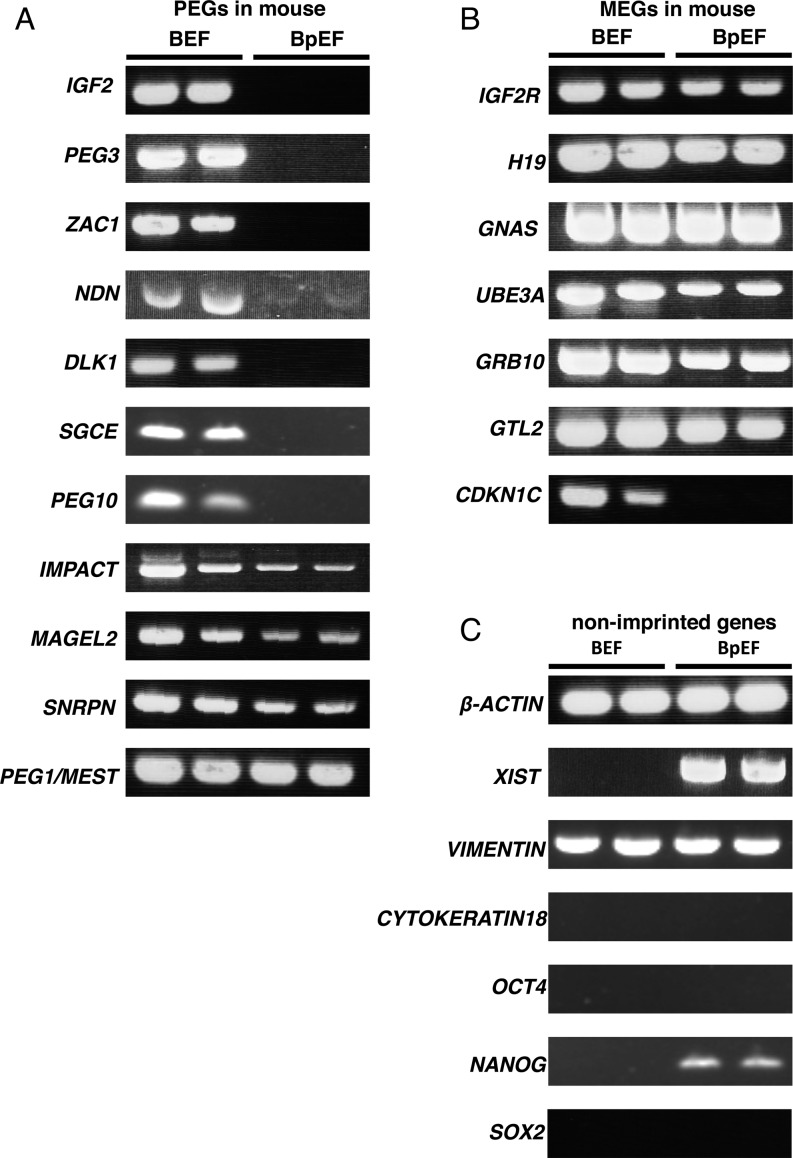
We analyzed the bovine expression patterns of 11 paternally expressed and seven maternally expressed homologous genes in mouse. We also analyzed the expression of the beta-actin gene as the housekeeping gene, XIST as the X chromosome inactivation marker, and OCT4, NANOG, and SOX2 as the pluripotent-related genes. Furthermore, we analyzed the expression of the epithelial marker gene encoding cytokeratin 18, which is not expressed in fibroblasts, and the mesenchymal marker gene encoding vimentin, which is expressed in fibroblasts. As shown in Fig. 3, PEGs were not expressed in BpEFs as expected, but MEGs showed biallelic expression in these cells, although two-fold expression changes cannot be determined by the RT-PCR method. For mouse PEGs, we found that IGF2, PEG3, ZAC1, NDN, DLK1, SGCE, and PEG10 were expressed only in BEFs; however, IMPACT, MAGEL2, SNRPN, and PEG1/MEST were expressed in both BpEFs and BEFs (Fig. 4A). For mouse MEGs, IGF2R, H19, GNAS, UBE3A, GRB10, and GTL2 were expressed in both BpEFs and BEFs; however, CDKN1C was expressed only in BEFs, like the PEGs (Fig. 4B). The housekeeping gene beta-actin was expressed in both BEFs and BpEFs, and XIST was expressed only in BpEFs as expected, because the BEFs used in this study were male (Fig. 4C). The expression patterns of the genes encoding cytokeratin 18 and vimentin confirmed that the cell type derived from both embryos were fibroblasts (Fig. 4C). Pluripotent-related genes OCT4 and SOX2 were not expressed in both types of cells, whereas NANOG was expressed only in BpEFs (Fig. 4C).
Fig. 3.
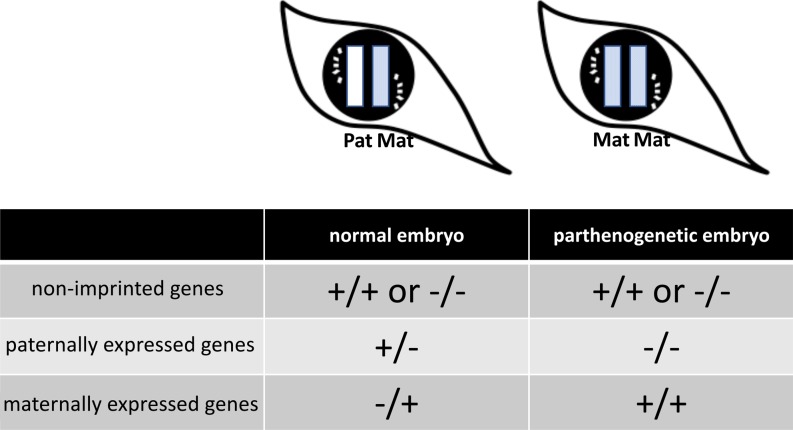
Schematic diagram of imprinted gene expression patterns in a normal embryo and a parthenogenetic embryo. +, expressed; –, not expressed. Pat, paternal allele; Mat, maternal allele. Theoretically, in the parthenogenetic embryo, paternally expressed genes are not expressed and maternally expressed genes are expressed two-fold.
Fig. 4.
Gene expression analysis of imprinted and non-imprinted genes in bovine embryonic fibroblasts (BEFs) and bovine parthenogenetic embryonic fibroblasts (BpEFs) by RT-PCR. The left two bands are BEFs, and the right two bands are BpEFs. (A) Paternally expressed genes (PEGs) in mouse. (B) Maternally expressed genes (MEGs) in mouse. (C) Non-imprinted genes.
DNA methylation analysis by bisulfite sequencing
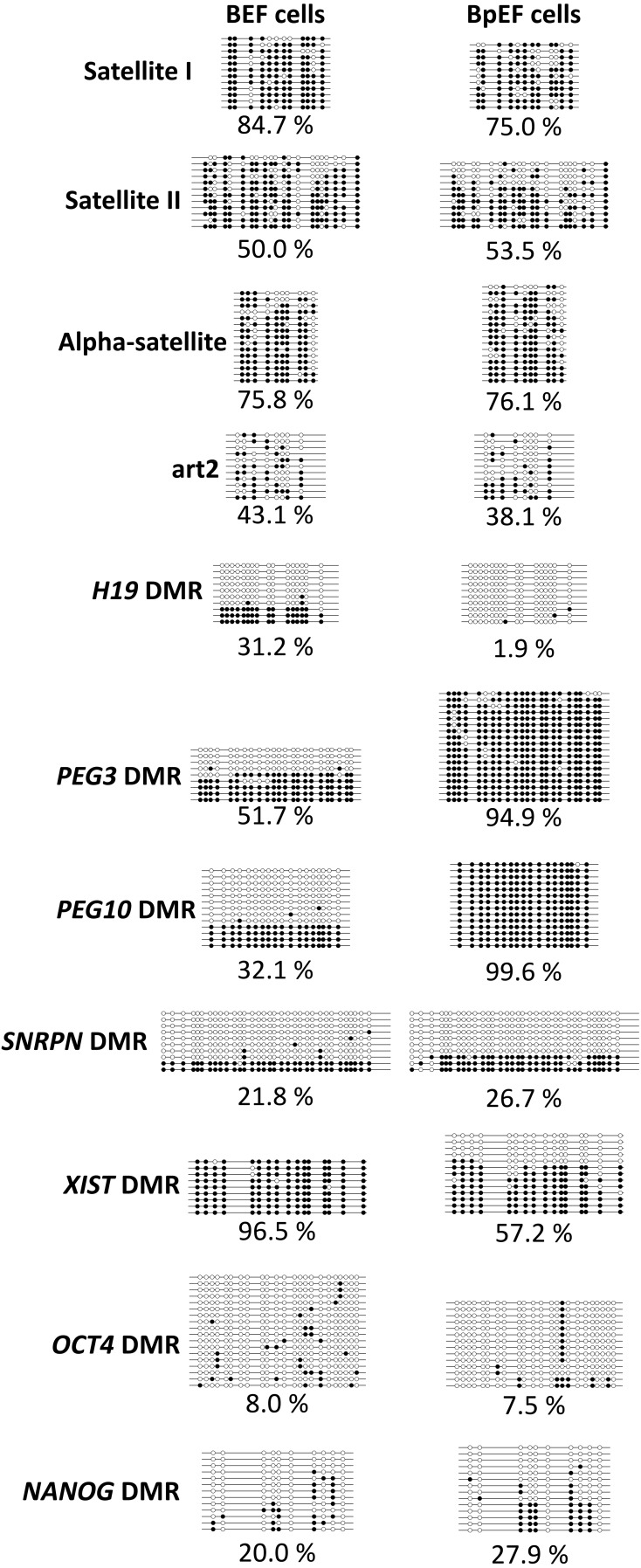
We next analyzed the DNA methylation patterns of repetitive elements and imprinted and non-imprinted genes in BEFs and BpEFs. Satellite I, Satellite II, and alpha-satellite sequences are centromeric repeat elements, and art2 sequences are interspersed repetitive sequences. There were no differences in DNA methylation levels between BEFs and BpEFs (Fig. 5). On the other hand, the imprinted gene methylation patterns were significantly different. As shown in Fig. 5, the H19 DMR was 30% methylated in BEFs, with completely unmethylated clones (Fig. 5; horizontal line with white circles only) and methylated clones (Fig. 5; horizontal line with black circles only), and was almost unmethylated in BpEFs. Completely methylated clones represent paternal alleles, and completely unmethylated clones represent paternal alleles; however, paternal and maternal alleles cannot be distinguished unless there is a single nucleotide polymorphism (SNP) found between the parental alleles. In BEFs, we could not detect any SNPs between the paternal and maternal alleles in all genes examined. PEG3 and PEG10 DMRs were also 30–50% methylated in BEFs (again, completely unmethylated clones and completely methylated clones) but were almost fully methylated in BpEFs. SNRPN DMR methylation was not different between the BEFs and BpEFs, whereas the XIST DMR was almost fully methylated in BEFs but half methylated in BpEFs. The methylation levels of OCT4 and NANOG DMRs were not different between the two types of cells.
Fig. 5.
DNA methylation patterns analyzed by bisulfite sequencing in bovine embryonic fibroblasts (BEFs) and bovine parthenogenetic embryonic fibroblasts (BpEFs). Each circle shows CpG dinucleotides. White circles represent unmethylated cytosines, and black circles represent methylated cytosines. Each line indicates an individual clone (allele) that was sequenced. The percentage of methylation is indicated below.
Microarray analysis
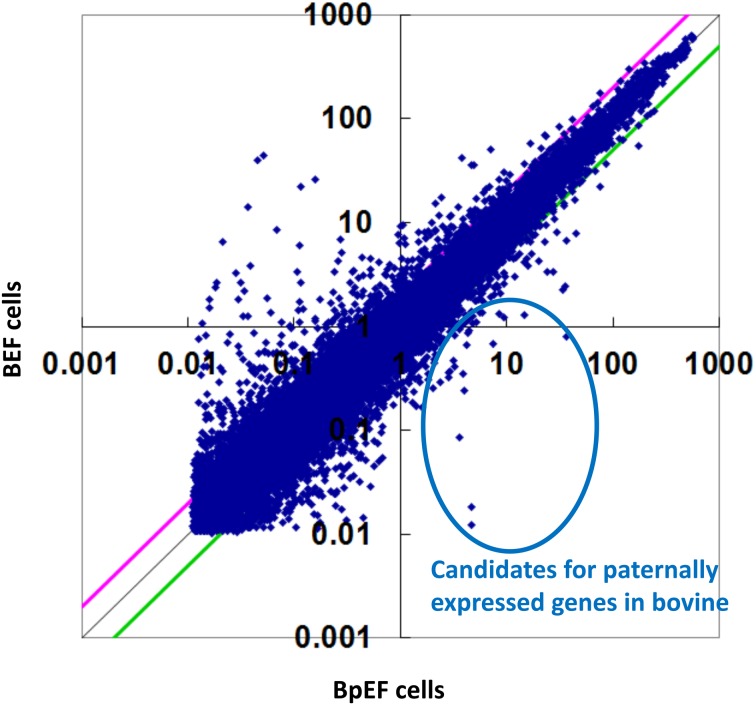
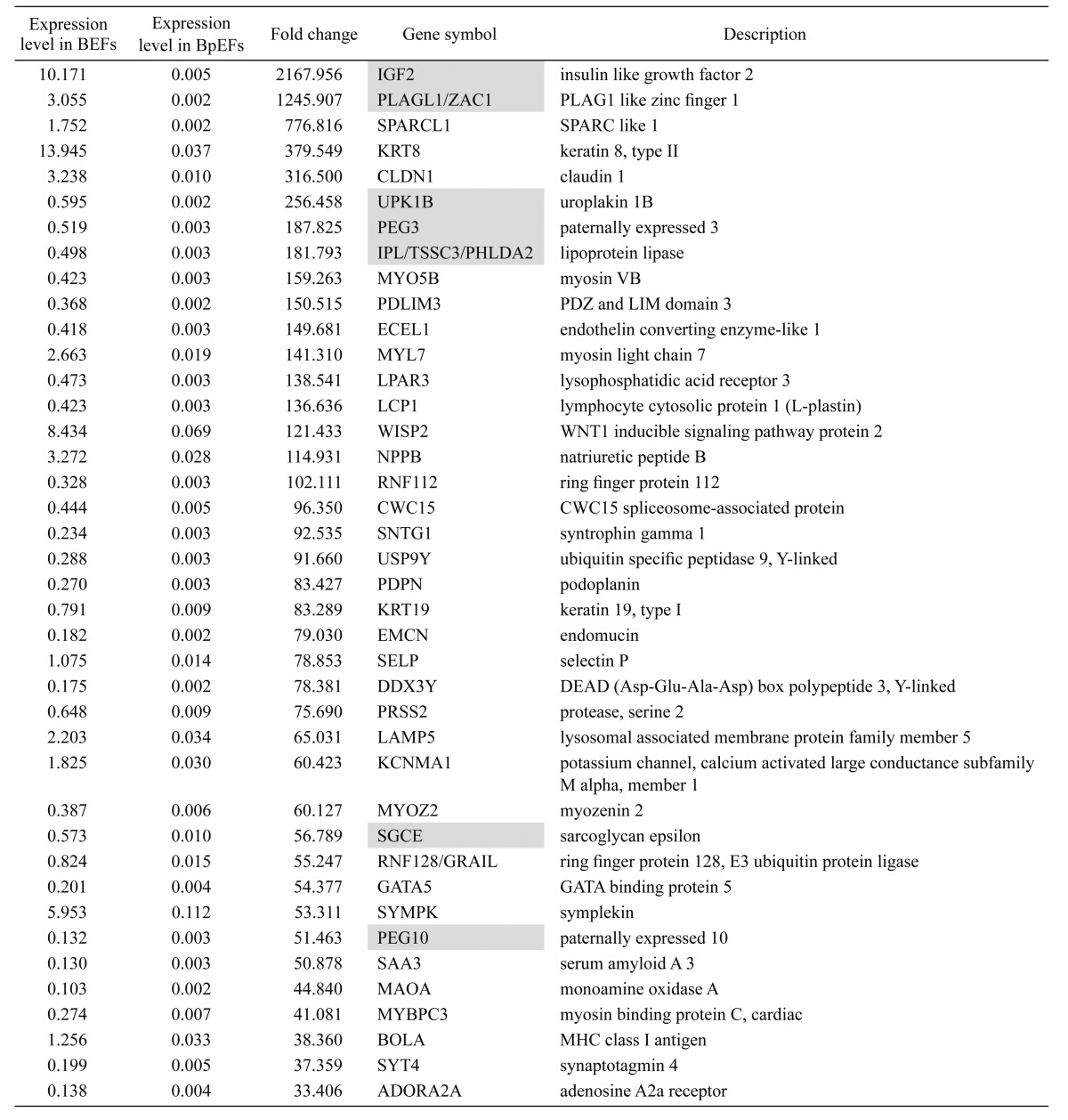
The Agilent bovine DNA microarray was used for genome-wide screening of bovine PEG candidates (expressed only in the normal embryo, but not in the parthenogenetic embryo). More than 30,000 probes were detected, of which 388 were detected only in BEFs (Fig. 6). These probes contained the known imprinted genes IGF2, PLAGL1/ZAC1, PEG3, IPL/TSSC3/PHLDA2, SGCE, and PEG10. The top 40 genes with their relative expression levels, fold changes, gene symbols, and descriptions are listed in Table 2.
Fig. 6.
Scatterplot of the microarray data. Genes expressed only in bovine embryonic fibroblasts (BEFs) but not in bovine parthenogenetic embryonic fibroblasts (BpEFs) are candidates for bovine paternally expressed genes (PEGs).
Table 2. Bovine paternally expressed gene (PEG) candidates (reported imprinted genes are highlighted).
Discussion
The length and developmental features of the D40 parthenogenetic embryo were not different to those of the normal embryo, and the isolated fibroblasts were morphologically indistinguishable from normal embryonic fibroblasts. However, the speed of BpEF growth in vitro was much slower. This was also observed for murine parthenogenetic/androgenetic embryonic fibroblasts [21]. However, murine parthenogenetic embryonic stem (ES) cells can be maintained in a similar way to normal ES cells [22], suggesting the existence of different cell senescence mechanisms in pluripotent cells.
The expression patterns of imprinted genes in BpEFs were almost the same as those in mouse; however, some PEGs in mouse (IMPACT, MAGEL2, SNRPN, and PEG1/MEST) were unexpectedly expressed in the BpEFs. The methylation patterns of H19, PEG3, and PEG10 DMRs in BpEFs were very similar to those of oocytes, whereas that of the SNRPN DMR was different. Interestingly, SNRPN DMR methylation was the same in both BpEFs and BEFs, suggesting that the expression of this gene is controlled by DMR methylation. The reason why the SNRPN DMR is not fully methylated in BpEFs is not known, although this DMR is reported to be fully methylated in bovine oocytes [23]. Hypomethylation at the H19 DMR and repressed expression of the IGF2 gene in BpEFs are concordant with previous studies in which IGF2 and H19 expression levels were regulated by DNA methylation at the H19 DMR in mouse [24, 25]. The variable DNA methylation patterns observed at H19, PEG3, and SNRPN DMRs in BEFs might be explained by a PCR bias or the heterogeneity of cell types and DNA methylation levels. PCR bias can be determined by analyzing SNPs between paternal and maternal alleles; however, we did not detect any SNPs in this study. The PEG3 and PEG10 gene expression patterns and DNA methylation patterns were strongly correlated, as in the study of mouse [26, 27]. CDKN1C, also known as p57KIP2, is a MEG in mouse and reported to be a MEG in cows as well [28]; however, we unexpectedly observed that this gene is not expressed in BpEFs, like a PEG. As monoallelic expression of imprinted genes is stage- and tissue-dependent, it is possible that CDKN1C is not imprinted in BpEFs and/or affected by other gene expression changes. We also analyzed the expression patterns and DNA methylation levels of non-imprinted genes OCT4, NANOG, and SOX2, which are well-known pluripotency-related transcription factors. OCT4 and NANOG are expressed only in early embryogenesis and germ cell lineages, and are methylated and repressed in somatic cell lineages [29, 30]. For that reason, demethylation of OCT4 and NANOG is an important feature of ES cells and induced pluripotent stem cells [31]. Interestingly, although the OCT4 DMR was almost unmethylated, the gene was not expressed in both cell types. The NANOG DMR was 20% methylated in BEFs and 28% methylated in BpEFs, but its expression was observed in BpEFs only. This correlation cannot be explained by DNA methylation patterns only, as OCT4 and NANOG expression levels are also regulated by a transcription factor network other than DNA methylation [32]. Given the reports that NANOG expression is regulated by OCT4 and SOX2 in mouse and human ES cells [33], the reason why NANOG was expressed only in BpEFs with no expression of OCT4 and SOX2 is not known. However, NANOG expression in the absence of OCT4 and SOX2 expression in human mesenchymal stem cells has been reported [34]. It is possible that contamination of tissue stem cells and/or in vitro growth conditions is involved in the expression of NANOG in BpEFs. Future studies will help in elucidating the regulation of NANOG in cattle.
DNA methylation levels in repetitive elements are considered to reflect genome-wide DNA methylation levels. Herein, we studied DNA methylation levels of three centromeric repeat elements (Satellite I, Satellite II, and alpha-satellite) and the interspersed repeat element art2 in BEFs and BpEFs. There were no significant differences between them, suggesting that genome-wide DNA methylation changes did not occur in BpEFs. However, the DNA methylation levels of these four repetitive elements in mature oocytes [35] were slightly (10–15%) lower than those in BpEFs and BEFs in this study, suggesting the gain of methylation during embryogenesis [36].
The XIST gene is a non-coding RNA and essential for X-chromosome inactivation processes in females to compensate for the gene dosages on the X chromosome in males [37]. XIST is expressed from the inactivated X chromosome in female cells and is repressed in male cells. DNA methylation at the XIST DMR is essential to repress XIST gene expression [38], and we found almost 100% methylation at the XIST DMR and repressed XIST gene expression in the male BEFs. In BpEFs, the XIST DMR was half methylated, with completely unmethylated alleles (putative inactive X chromosomes) and completely fully methylated alleles (putative active X chromosomes), with expression of the XIST gene, suggesting that the X chromosome inactivation mechanisms are normal. This finding is contrary to those found for murine and porcine parthenogenetic embryos [17, 39, 40]. Wang et al. [17] reported that in porcine parthenogenetic fetuses and placentas at Day 28 of gestation, MEGs were overexpressed and PEGs were underexpressed. They also reported that genome-wide DNA methylation levels were not different between parthenogenetic and normal embryos. These results are similar to our observations; however, they reported demethylation of XIST DMRs in the parthenogenetic fetuses and placentas in contrast to the normal ones. As we analyzed fibroblasts in only one male bovine normal embryo, analysis of female cells is needed to compare XIST DMR patterns in cattle. More detailed analysis of X chromosome inactivation in BpEFs is also needed to elucidate species-specific differences in mammalian X chromosome inactivation mechanisms [41].
By DNA microarray analysis, we could list PEG candidates in cattle, which were expressed in BEFs but not in BpEFs. However, most of them are considered to be secondarily affected genes rather than PEGs. For example, of the top 40 PEG candidates listed in Table 2, only seven genes were reported to be imprinted in cattle. For example, KRT8 and KRT19 were expressed only in BEFs, and Kim et al. [19] reported that disruption of imprinting regulator Rex1/Zfp42 results in upregulation of KRT8, KRT18, and KRT19 genes in Rex1-null mouse blastocysts. Even if these keratin genes were not reported to be imprinted in mice and cattle, their expression might be regulated by imprinted gene(s). IPL/TSSC3/PHLDA2 is reported to be maternally expressed in bovine embryo and extraembryonic tissue of the 14-week-old fetus; however, the expression pattern is skewed in different bovine fetal tissues at different developmental stages [42]. In adult bovine tissues, PHLDA2 is reported to be biallelically expressed, suggesting development-stage-specific expression [43]. Detailed analysis of PEG candidates is needed to identify novel ones in cattle.
In summary, we have reported the first epigenetic analysis of bovine parthenogenetic embryonic fibroblasts. We determined the expression of 18 imprinted genes and six non-imprinted genes, and the DNA methylation patterns of four repetitive elements, four imprinted genes, and four non-imprinted genes. The data demonstrate that some imprinted gene expression patterns and DNA methylation patterns were different between mice and cattle; however, X chromosome inactivation mechanisms seemed to be normal in bovine parthenogenetic embryos. Our results have shown that the bovine parthenogenetic embryo is a useful material for analyzing the evolutionary aspect of genomic imprinting in mammals. However, in this study, we analyzed cultured fibroblast cells derived from only one parthenogenetic embryo and one normal embryo, and therefore further study using more embryos for direct analysis is needed to confirm the results.
Acknowledgments
This work was supported by a grant from the National Agriculture and Food Research Organization (NARO) Japan and JSPS KAKENHI (grant no. JP15K07764). The authors are grateful to Drs. Masaya Geshi and Hitomi Takanashi for the pregnancy appraisal, and the members of the Animal Breeding and Reproduction Research Division, Institute of Livestock and Grassland Science, NARO.
References
- 1.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 1984; 308: 548–550. [DOI] [PubMed] [Google Scholar]
- 2.McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 1984; 37: 179–183. [DOI] [PubMed] [Google Scholar]
- 3.Miyoshi N, Barton SC, Kaneda M, Hajkova P, Surani MA. The continuing quest to comprehend genomic imprinting. Cytogenet Genome Res 2006; 113: 6–11. [DOI] [PubMed] [Google Scholar]
- 4.Kaneda M. Genomic imprinting in mammals-epigenetic parental memories. Differentiation 2011; 82: 51–56. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 2008; 9: 129–140. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, Viville S, Barton SC, Ishino F, Surani MA. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat Genet 1995; 11: 52–59. [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 2000; 5: 211–220. [DOI] [PubMed] [Google Scholar]
- 8.Hansmann T, Heinzmann J, Wrenzycki C, Zechner U, Niemann H, Haaf T. Characterization of differentially methylated regions in 3 bovine imprinted genes: a model for studying human germ-cell and embryo development. Cytogenet Genome Res 2011; 132: 239–247. [DOI] [PubMed] [Google Scholar]
- 9.Cruz NT, Wilson KJ, Cooney MA, Tecirlioglu RT, Lagutina I, Galli C, Holland MK, French AJ. Putative imprinted gene expression in uniparental bovine embryo models. Reprod Fertil Dev 2008; 20: 589–597. [DOI] [PubMed] [Google Scholar]
- 10.Ruddock NT, Wilson KJ, Cooney MA, Korfiatis NA, Tecirlioglu RT, French AJ. Analysis of imprinted messenger RNA expression during bovine preimplantation development. Biol Reprod 2004; 70: 1131–1135. [DOI] [PubMed] [Google Scholar]
- 11.Fukui Y, Sawai K, Furudate M, Sato N, Iwazumi Y, Ohsaki K. Parthenogenetic development of bovine oocytes treated with ethanol and cytochalasin B after in vitro maturation. Mol Reprod Dev 1992; 33: 357–362. [DOI] [PubMed] [Google Scholar]
- 12.Tveden-Nyborg PY, Alexopoulos NI, Cooney MA, French AJ, Tecirlioglu RT, Holland MK, Thomsen PD, D'Cruz NT. Analysis of the expression of putatively imprinted genes in bovine peri-implantation embryos. Theriogenology 2008; 70: 1119–1128. [DOI] [PubMed] [Google Scholar]
- 13.Surani MA, Barton SC. Development of gynogenetic eggs in the mouse: implications for parthenogenetic embryos. Science 1983; 222: 1034–1036. [DOI] [PubMed] [Google Scholar]
- 14.Surani MA, Kothary R, Allen ND, Singh PB, Fundele R, Ferguson-Smith AC, Barton SC. Genome imprinting and development in the mouse. Dev Suppl 1990; 89–98. [PubMed] [Google Scholar]
- 15.Kono T, Obata Y, Wu Q, Niwa K, Ono Y, Yamamoto Y, Park ES, Seo JS, Ogawa H. Birth of parthenogenetic mice that can develop to adulthood. Nature 2004; 428: 860–864. [DOI] [PubMed] [Google Scholar]
- 16.Kure-bayashi S, Miyake M, Okada K, Kato S. Successful implantation of in vitro-matured, electro-activated oocytes in the pig. Theriogenology 2000; 53: 1105–1119. [DOI] [PubMed] [Google Scholar]
- 17.Wang D, Chen X, Song Y, Lv Q, Lai L, Li Z. Disruption of imprinted gene expression and DNA methylation status in porcine parthenogenetic fetuses and placentas. Gene 2014; 547: 351–358. [DOI] [PubMed] [Google Scholar]
- 18.Nagai K, Sata R, Takahashi H, Okano A, Kawashima C, Miyamoto A, Geshi M. Production of trophoblastic vesicles derived from Day 7 and 8 blastocysts of in vitro origin and the effect of intrauterine transfer on the interestrous intervals in Japanese black heifers. J Reprod Dev 2009; 55: 454–459. [DOI] [PubMed] [Google Scholar]
- 19.Saito K, Mori M, Sumio Y.Prolongation of interestrus intervals following intra-uterine transfer of parthenogenetic blastocysts in Japanese Brown cows. Research bulletin of the Kumamoto Prefectural Agricultural Research Center 2009; 16: 48-51. [Google Scholar]
- 20.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res 2008; 36: W170−175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez L, Kozlov S, Piras G, Stewart CL. Paternal and maternal genomes confer opposite effects on proliferation, cell-cycle length, senescence, and tumor formation. Proc Natl Acad Sci USA 2003; 100: 13344–13349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufman MH, Robertson EJ, Handyside AH, Evans MJ. Establishment of pluripotential cell lines from haploid mouse embryos. J Embryol Exp Morphol 1983; 73: 249–261. [PubMed] [Google Scholar]
- 23.Lucifero D, Suzuki J, Bordignon V, Martel J, Vigneault C, Therrien J, Filion F, Smith LC, Trasler JM. Bovine SNRPN methylation imprint in oocytes and day 17 in vitro-produced and somatic cell nuclear transfer embryos. Biol Reprod 2006; 75: 531–538. [DOI] [PubMed] [Google Scholar]
- 24.Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet 1995; 9: 407–413. [DOI] [PubMed] [Google Scholar]
- 25.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 1998; 12: 3693–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li LL, Szeto IY, Cattanach BM, Ishino F, Surani MA. Organization and parent-of-origin-specific methylation of imprinted Peg3 gene on mouse proximal chromosome 7. Genomics 2000; 63: 333–340. [DOI] [PubMed] [Google Scholar]
- 27.Ono R, Shiura H, Aburatani H, Kohda T, Kaneko-Ishino T, Ishino F. Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6. Genome Res 2003; 13: 1696–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robbins KM, Chen Z, Wells KD, Rivera RM. Expression of KCNQ1OT1, CDKN1C, H19, and PLAGL1 and the methylation patterns at the KvDMR1 and H19/IGF2 imprinting control regions is conserved between human and bovine. J Biomed Sci 2012; 19: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuji-Takayama K, Inoue T, Ijiri Y, Otani T, Motoda R, Nakamura S, Orita K. Demethylating agent, 5-azacytidine, reverses differentiation of embryonic stem cells. Biochem Biophys Res Commun 2004; 323: 86–90. [DOI] [PubMed] [Google Scholar]
- 30.Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, Zeng R, Jing NH, Chen T, Li E, Sasaki H, Xu GL. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol 2007; 27: 8748–8759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676. [DOI] [PubMed] [Google Scholar]
- 32.Niwa H. The pluripotency transcription factor network at work in reprogramming. Curr Opin Genet Dev 2014; 28: 25–31. [DOI] [PubMed] [Google Scholar]
- 33.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem 2005; 280: 24731–24737. [DOI] [PubMed] [Google Scholar]
- 34.Pierantozzi E, Gava B, Manini I, Roviello F, Marotta G, Chiavarelli M, Sorrentino V. Pluripotency regulators in human mesenchymal stem cells: expression of NANOG but not of OCT-4 and SOX-2. Stem Cells Dev 2011; 20: 915–923. [DOI] [PubMed] [Google Scholar]
- 35.Kaneda M, Watanabe S, Akagi S, Inaba Y, Geshi M, Nagai T. Proper reprogramming of imprinted and non-imprinted genes in cloned cattle gametogenesis. Anim Sci J 2017. doi: 10.1111/asj.12846 (in press). [DOI] [PubMed] [Google Scholar]
- 36.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 2002; 241: 172–182. [DOI] [PubMed] [Google Scholar]
- 37.Sado T, Brockdorff N. Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philos Trans R Soc Lond B Biol Sci 2013; 368: 20110325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norris DP, Patel D, Kay GF, Penny GD, Brockdorff N, Sheardown SA, Rastan S. Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell 1994; 77: 41–51. [DOI] [PubMed] [Google Scholar]
- 39.Nesterova TB, Barton SC, Surani MA, Brockdorff N. Loss of Xist imprinting in diploid parthenogenetic preimplantation embryos. Dev Biol 2001; 235: 343–350. [DOI] [PubMed] [Google Scholar]
- 40.Tada T, Takagi N. Early development and X-chromosome inactivation in mouse parthenogenetic embryos. Mol Reprod Dev 1992; 31: 20–27. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V, Heard E. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 2011; 474: 239–240. [DOI] [PubMed] [Google Scholar]
- 42.Sikora KM, Magee DA, Berkowicz EW, Lonergan P, Evans AC, Carter F, Comte A, Waters SM, MacHugh DE, Spillane C. PHLDA2 is an imprinted gene in cattle. Anim Genet 2012; 43: 587–590. [DOI] [PubMed] [Google Scholar]
- 43.Wang M, Li D, Zhang M, Yang W, Wu G, Cui Y, Li S. Biallelic expression of Tssc4, Nap1l4, Phlda2 and Osbpl5 in adult cattle. J Genet 2015; 94: 391–395. [DOI] [PubMed] [Google Scholar]